Abstract
H/ACA ribonucleoproteins (RNPs) are known as one of two major classes of small nucleolar RNPs. They predominantly guide the site-directed pseudouridylation of target RNAs, such as ribosomal and spliceosomal small nuclear RNAs. In addition, they process ribosomal RNA and stabilize vertebrate telomerase RNA. Taken together, the function of H/ACA RNPs is essential for ribosome biogenesis, pre-mRNA splicing, and telomere maintenance. Every cell contains one to two hundred different species of H/ACA RNPs, each consisting of the same four core proteins and one function-specifying H/ACA RNA. Most of these RNPs reside in nucleoli and Cajal bodies and mediate the isomerization of specific uridines to pseudouridines. Catalysis of the reaction is mediated by the putative pseudouridylase NAP57 (dyskerin, Cbf5p). Unexpectedly, mutations in this housekeeping enzyme are the major determinants of the inherited bone marrow failure syndrome dyskeratosis congenita. This review details the many diverse functions of H/ACA RNPs, some yet to be uncovered, with an emphasis on the role of the RNP proteins. The multiple functions of H/ACA RNPs appear to be reflected in the complex phenotype of dyskeratosis congenita.
Introduction
H/ACA ribonucleoproteins (RNPs) were identified some eight years ago as small nucleolar (sno)RNPs (Balakin et al., 1996; Ganot et al., 1997b). The defining molecules of these RNPs are their small nucleolar RNAs (snoRNAs). SnoRNAs can be divided into the two major classes, H/ACA and C/D, and function predominantly in the modification of ribosomal RNA (rRNA). The over 200 H/ACA and C/D snoRNAs specify (by site-directed base pairing) a similar number of nucleotides in mammalian rRNA for pseudouridylation and 2’-O-methylation, respectively. Most snoRNAs are short, 60 - 150 nucleotides in length, and share a conserved secondary structure and short sequence elements termed H and ACA in the case of H/ACA and C and D in the case of C/D snoRNAs. All H/ACA snoRNAs assemble with the same four core proteins to form H/ACA snoRNPs and all C/D snoRNAs assemble with another set of four core proteins to form C/D snoRNPs. Although these snoRNPs are the founding members of H/ACA and C/D RNPs, other members have been identified outside of the nucleolus and/or with additional functions. Several reviews have covered the many intriguing aspects of snoRNPs, such as the location of snoRNA genes in introns of mammalian genes, the evolution of snoRNPs, the importance of nucleotide modifications, etc. (Bachellerie et al., 2002; Decatur and Fournier, 2003; Filipowicz and Pogacic, 2002; Henras et al., 2004b; Kiss, 2002; Kiss, 2004; Lafontaine and Tollervey, 1998; Ofengand, 2002; Smith and Steitz, 1997; Tran et al., 2004). Our understanding of C/D snoRNPs has always been a step ahead of that of H/ACA snoRNPs. However, the identification of vertebrate telomerase as an H/ACA RNP and the association of H/ACA RNPs with the bone marrow failure syndrome dyskeratosis congenita (DC) have guided H/ACA RNPs into the limelight. This review focuses on the H/ACA class of RNPs, our expanding knowledge of their functions, the contribution of their protein components, and how H/ACA RNPs may be affected in DC.
H/ACA RNAs
H/ACA RNAs allow us to distinguish different H/ACA RNPs. At present there are four classes of H/ACA RNAs of known function but additional H/ACA RNAs of unknown function(s) have been identified and are referred to as orphan H/ACA RNAs (Fig. 1). All H/ACA RNAs share a consensus 5’-hairpin-hinge-hairpin-tail-3’ secondary structure with the conserved nucleotides ANANNA situated in the hinge region and ACA exactly three positions from the 3’ end (Fig. 2A) (Balakin et al., 1996; Ganot et al., 1997b). They can be divided into two groups, guide and non-guide RNAs.
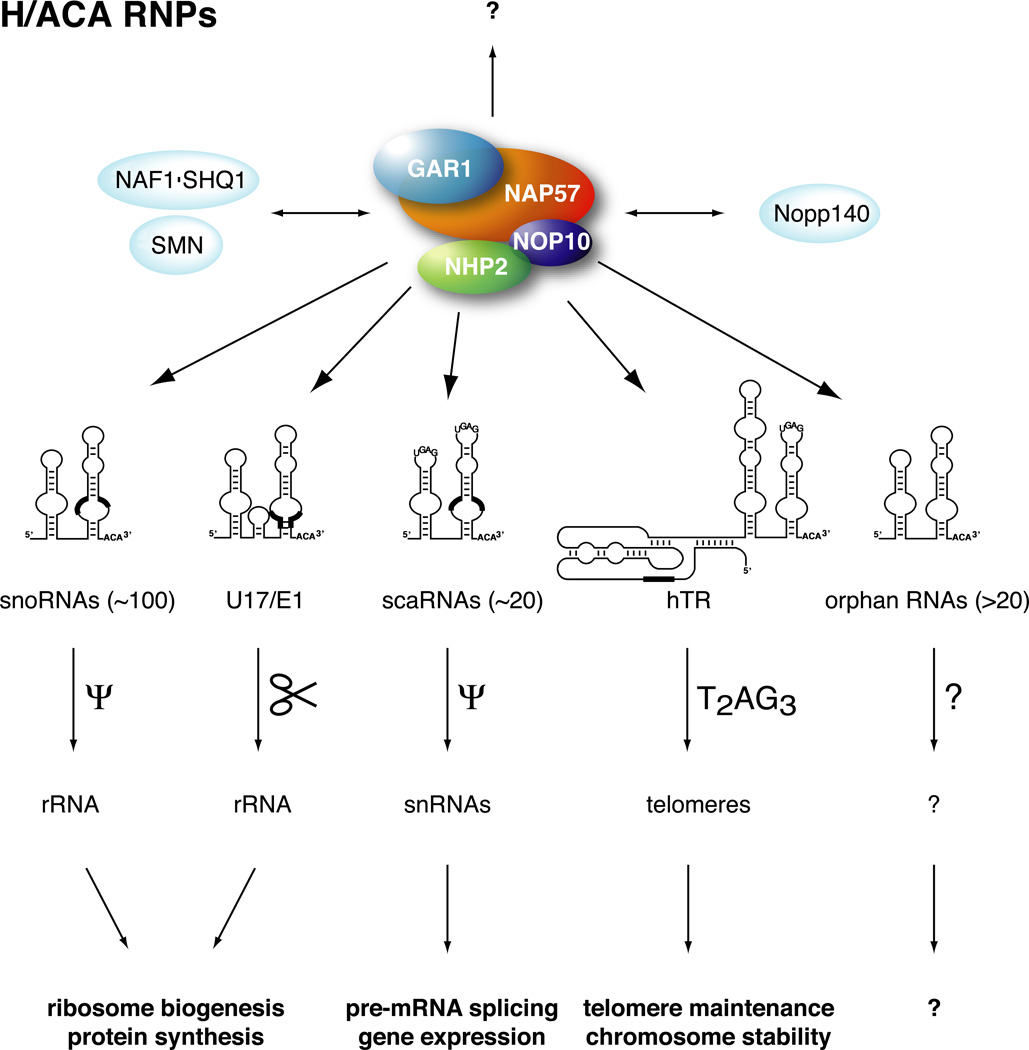
Figure 1. Schematic overview of mammalian H/ACA RNP proteins, RNAs, targets, and functions.
four H/ACA core proteins forming a protein only complex (solid colors) and interacting proteins (light blue) are depicted. Interactions between the core proteins are based on experimental evidence, i.e., GAR1 and NOP10 associate independently with NAP57 while NHP2 requires the prior association of NAP57 and NOP10. At least one set of four core proteins associates with each individual H/ACA RNA to form an RNP (arrows). RNP-independent functions of the proteins are also possible (question mark). The basic secondary structures of the five types of H/ACA RNAs and their names and numbers are shown underneath the proteins. Note hairpin sizes vary and insertions may also be present in individual RNAs. The location of antisense elements, highly conserved sequences, and the template region of hTR are indicated by thick black lines. Cajal body localization consensus elements are drawn in the stemloops (UGAG). Below the RNAs, the targets of the various RNPs, ribosomal RNA (rRNA), small nuclear RNAs (snRNAs), and telomeres, which are pseudouridylated (Ψ), processed (scissors), and extended by T2AG3 repeats, respectively, are shown. Finally, the cellular processes affected by the function of the individual RNPs are indicated at the bottom. Question marks refer to unknown targets and functions. In the case of one orphan H/ACA RNA these seem to be brain-specific (see text).
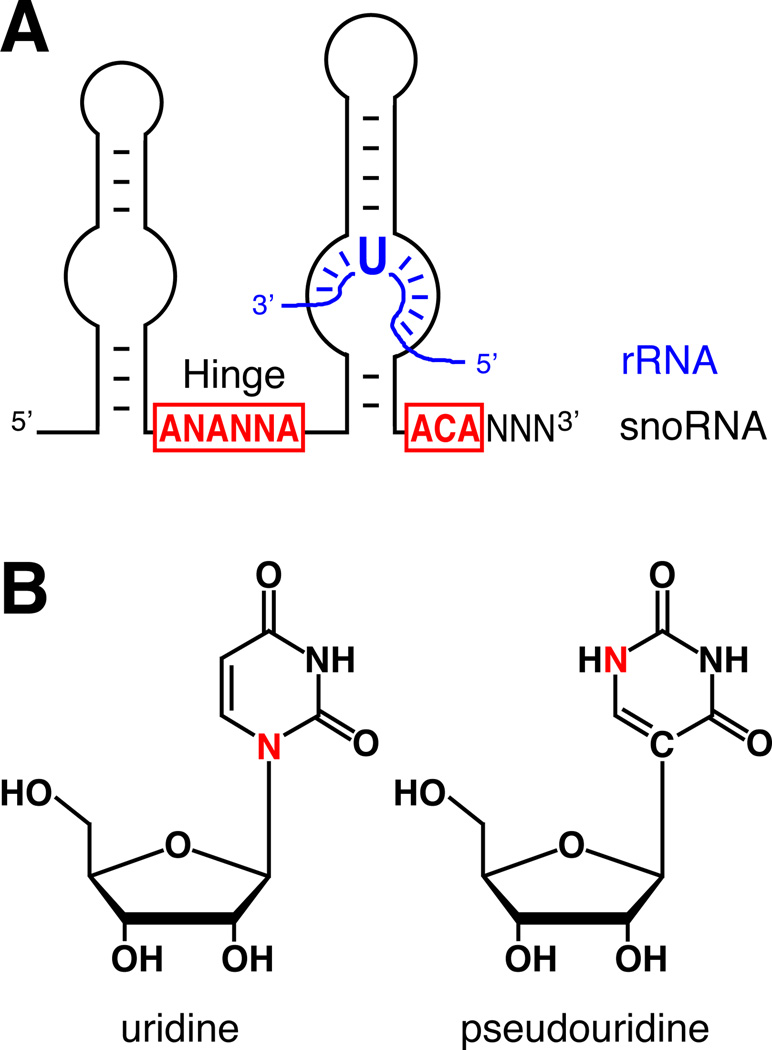
Figure 2. Structures of H/ACA RNA-rRNA hybrid, uridine, and pseudouridine.
(A) Schematic representation of the basic two-hairpin structure of H/ACA RNAs containing the conserved sequence elements ANANNA in the hinge region (Hinge) and ACA three nucleotides from the 3’ end in red. The name of the RNAs is based on the hinge region and the ACA triplet. An rRNA hybridizing to a bulge in the 3’ hairpin with the unpaired uridine (U) to be targeted for pseudouridylation is shown (blue). Note for simplification, a second unpaired nucleotide 3’ to the target uridine is not depicted. Also, H/ACA RNAs guide pseudouridylation by the bulge in the 3’ and/or 5’ hairpin. (B) Structural formulas of uridine (left) and pseudouridine (right). Note the different location of one of the base nitrogens (red) and the nitrogen-carbon versus carbon-carbon glycosidic bond between the two isomers.
Guide RNAs
Guide H/ACA RNAs specify uridines in target RNAs for conversion to pseudouridines by site-directed base pairing using two 3 to 10 nucleotide long antisense elements (Ganot et al., 1997a; Ni et al., 1997). These elements are complementary to the sequences flanking the target uridine and are situated in a bulge of one and/or the other hairpin of an H/ACA RNA, at either side of the base of a stem forming a pseudouridylation pocket (Fig. 2A). In this manner, guide RNAs hybridize to target RNAs over a stretch of 6 to 20 nucleotides bracing the target uridine. To date, the target RNAs identified are rRNAs and spliceosomal small nuclear RNAs (snRNAs) (Ganot et al., 1997a; Hüttenhofer et al., 2001; Jady and Kiss, 2001; Ni et al., 1997). In mammals, the isomerization to pseudouridines of approximately 100 uridines in rRNAs and 27 in snRNAs is guided by a similar number of H/ACA RNAs. While the rRNA guides are referred to as snoRNAs based on their accumulation in nucleoli, snRNA guides are also known as small Cajal body specific RNAs (scaRNAs) due to their concentration in Cajal bodies (small nuclear organelles, ~0.5–1µm in diameter, that function, at least in part, as maturation sites for snRNPs, particularly, in snRNA modification) (Darzacq et al., 2002). ScaRNAs contain a short Cajal body targeting sequence (CAB box) in the terminal loops (of one) of their hairpins (Richard et al., 2003). The CAB box conforms to a 5’-UGAG-3’ consensus with the third adenine and fourth guanine nucleotides being most highly conserved. Remarkably, in some cases, the conserved H/ACA two-hairpin secondary structure of scaRNAs expands by tandem duplication or integration into a C/D RNA (Darzacq et al., 2002; Jady and Kiss, 2001; Kiss et al., 2004; Kiss et al., 2002). Finally, a notable scaRNA (U100) was identified that bears all the hallmarks of a guide RNA for the pseudouridylation of uridine in position 9 of U6 snRNA, which, however, is not modified (Vitali et al., 2003). Therefore, U100 may guide a function other than pseudouridylation, such as chaperoning U6 for RNP assembly.
Non-guide RNAs
Non-guide H/ACA RNAs comprise snoRNA U17/E1 and mammalian telomerase RNA (Kiss and Filipowicz, 1993; Mitchell et al., 1999a; Ruff et al., 1993). Rather than modifying rRNA, U17/E1 (snR30 in yeast) is required for processing of pre-rRNA to produce 18S rRNA (Fig. 1) (Morrissey and Tollervey, 1993). This makes snR30 (U17/E1) essential for ribosome production and consequently viability (Bally et al., 1988). Recently, two highly conserved sequence elements were discovered in the bulge of the 3’ hairpin of U17/E1/snR30 that, despite lack of complementarity to pre-rRNA, proved essential for the early cleavages of 35S pre-rRNA (Atzorn et al., 2004). These 7 and 9 nucleotide long elements are conserved in all U17/E1 homologs that can be identified in budding and fission yeast, in all vertebrates, and even in the unicellular ciliated protozoan Tetrahymena thermophila (Atzorn et al., 2004; Cervelli et al., 2003).
Human telomerase RNA (hTR) provides the template for the replication of chromosome ends. It extends for 451 nucleotides and the last 240 form a consensus H/ACA two-hairpin structure while the 5’ half folds into a pseudoknot containing the template for the reverse transcriptase (Fig. 1) (Mitchell et al., 1999a). The H/ACA domain of hTR is required in vivo for hTR accumulation and stability. As in the case of U17/E1, there is no indication that hTR serves as a pseudouridylation guide. In fact, in vitro hTR fails to modify a synthetic substrate that conforms to the standard rules, i.e., complementarity to the upper halves of the bulge in either hairpin (C. Wang and U.T. Meier, unpublished results). Moreover, hTR is a scaRNA since it features a CAB box and localizes to Cajal bodies in a cell cycle dependent manner (Jady et al., 2004; Zhu et al., 2004).
Orphan RNAs
Finally, screens of cDNA libraries (generated from size selected RNAs and RNAs precipitated with H/ACA core proteins) for novel H/ACA RNAs in mouse and human uncovered 12 and 14 H/ACA RNAs, respectively, that lack complementarity to any of the stable non-coding RNAs, e.g., rRNAs, snRNAs, and snoRNAs (Hüttenhofer et al., 2001; Kiss et al., 2004; Vitali et al., 2003). These H/ACA RNAs belong to a growing family of orphan H/ACA RNAs. They could guide the pseudouridylation of mRNAs and/or yet to be identified stable RNAs or they could function in a pseudouridylation guide-independent fashion, like U17/E1 and hTR. The possibility that some of these functions/pseudouridylation targets could be tissue-specific was highlighted by the identification of the orphan H/ACA RNA HBI-36 (Cavaille et al., 2000). HBI-36 is expressed from the second intron of the serotonin C2 receptor gene. Like this brain-specific gene, HBI-36 is predominantly expressed in the choroid plexus. Although no target RNA has been identified for HBI-36, these findings illustrate how one tissue could be specifically affected by a general impairment of H/ACA RNPs, e.g., by mutation of one of the core proteins. Taken together, while the full functional realm of H/ACA RNAs remains to be elucidated, they consist of at least four distinct functional classes; those that pseudouridylate rRNA, those that process rRNA, those that pseudouridylate snRNAs, and hTR required for telomere maintenance (Fig. 1).
H/ACA ribonucleoproteins
H/ACA RNAs, like most cellular RNAs, associate with proteins to form RNPs. Specifically, each of these one to two hundred different H/ACA RNAs associates with the same four core proteins, NAP57 (dyskerin, Cbf5p), GAR1, NHP2, and NOP10 to form an H/ACA RNP (Fig. 1) (Balakin et al., 1996; Ganot et al., 1997b; Henras et al., 1998; Lafontaine et al., 1998; Watkins et al., 1998). These proteins are evolutionarily highly conserved with orthologs in yeast and archaea (Henras et al., 1998; Meier and Blobel, 1994; Rozhdestvensky et al., 2003; Watanabe and Gray, 2000; Watkins et al., 1998). As judged from genetic depletion studies in yeast, all four core proteins are essential for viability and, with the exception of GAR1, required for the stability of H/ACA RNAs (Bousquet-Antonelli et al., 1997; Dez et al., 2001; Henras et al., 1998; Lafontaine et al., 1998; Watkins et al., 1998). Similarly, mammalian NAP57, NOP10, and NHP2 form a core trimer in vitro that specifically associates with H/ACA RNAs in the absence of GAR1 (Wang and Meier, 2004). Once assembled, pseudouridylation competent H/ACA RNPs are stable and do not exchange their RNAs suggesting that new particle formation requires de novo synthesis (Wang and Meier, 2004). When studying H/ACA core proteins, it is important to keep in mind that they are part of a structurally similar and stable but functionally heterogeneous group of RNPs as defined by their RNAs. For example, all core proteins concentrate in both nucleoli and Cajal bodies of every cell (Girard et al., 1992; Henras et al., 1998; Meier and Blobel, 1994; Pogacic et al., 2000), but their associated H/ACA RNAs differ between the two organelles, e.g., snoRNAs in nucleoli and scaRNAs in Cajal bodies. Nevertheless, the stable core of H/ACA RNPs consists of five components, four proteins and one RNA (Fig. 1). Although the stoichiometry of the five components needs to be established, estimates from electron micrographs of purified yeast H/ACA RNPs are consistent with a particle size that accommodates one RNA and two of each of the four core proteins (Watkins et al., 1998). In particular, based on the bipartite structure of these particles and the structural and functional duplicity of the H/ACA RNA hairpins, one complement of four proteins could associate with each hairpin (Lübben et al., 1995; Watkins et al., 1998).
Functional importance
Ribosome biogenesis
Proper activity of H/ACA RNPs impacts three basic cellular functions, ribosome biogenesis, pre-mRNA splicing, and telomere maintenance (Fig. 1). In ribosome biogenesis, H/ACA RNPs function in pseudouridylation and processing of rRNA. The majority of H/ACA RNPs direct the site-specific pseudouridylation of ~100 uridines in vertebrate pre-rRNA. Pseudouridine is an isomer of uridine with its uracil attached via a carbon-carbon instead of a nitrogen-carbon glycosidic bond (Fig. 2B). Although pseudouridines represent the most abundant modified nucleosides in cellular RNAs, their precise functions remain elusive. On a molecular level, the additional imino group (Fig. 2B, red) affects the local environment by increasing the rigidity of the nucleotide and by stabilizing local base stacking (Arnez and Steitz, 1994; Charette and Gray, 2000; Davis, 1995; Meroueh et al., 2000).
The following points indicate the importance of these local effects of pseudouridines for ribosome function. First, rRNAs of all organisms contain pseudouridines and their number increases over evolution, e.g., 11 in E. coli rRNA, 44 in S. cerevisiae, and ~91 in humans (Maden, 1990; Ofengand, 2002). Second, even though rRNA is pseudouridylated at the level of its precursor, only uridines in regions that form part of mature rRNA are isomerized suggesting a function in the ribosome (Brand et al., 1979; Jeanteur et al., 1968). Third, most pseudouridines (and 2’-O-methyl groups) cluster in functionally important regions of rRNA, e.g., the peptidyl transferase center and contact regions between the large and small ribosomal subunits (Bakin et al., 1994; Decatur and Fournier, 2002). Fourth, omission of some of the pseudouridines in the peptidyl transferase center of bacterial and yeast ribosomes (by deletion of the site-specific pseudouridine synthases and the specific H/ACA snoRNAs, respectively) impairs growth (King et al., 2003; Raychaudhuri et al., 1998). In one case, this growth defect could be attributed to functionally impaired ribosomes (King et al., 2003). Similarly, global inhibition of rRNA pseudouridylation below detection limits by site-directed mutagenesis of the yeast pseudouridylase Cbf5p essentially renders cells inviable above and below 25°C (Zebarjadian et al., 1999). However, lack of pseudouridines in other positions of rRNA, even if highly conserved, had minor or no effects on growth, detectable only in competitive growth experiments in which the wild type cells outcompeted those lacking certain pseudouridines in rRNA (Badis et al., 2003; King et al., 2003; Ofengand, 2002; Parker et al., 1988; Samarsky et al., 1995). Overall, these data point to a requirement for pseudouridines in certain positions of rRNA while their impact in other positions may be minute and only detectable over a long period of time and with sensitive assays.
Perhaps the most important impact of H/ACA RNPs on ribosome biogenesis is that on pre-rRNA processing. Although only one mammalian H/ACA RNP (U17/E1) mediates this process, it stands out because its activity is essential for ribosome biogenesis and consequently cell viability, as demonstrated with its yeast ortholog snR30 (Bally et al., 1988; Morrissey and Tollervey, 1993). H/ACA RNP U17/E1, therefore, warrants particular attention when globally impacting H/ACA RNPs, i.e., via mutations in one of their core proteins.
Pre-mRNA splicing
Pre-mRNA splicing is affected by the pseudouridylation of spliceosomal snRNAs by vertebrate H/ACA RNPs (Fig. 1). With the exception of U11 and U6atac, all snRNAs of major and minor spliceosomes contain at least one pseudouridine (Massenet et al., 1998). As in the case of rRNA, pseudouridines cluster in functionally important regions of snRNAs, e.g., in nucleotides that base-pair with other snRNAs or with intronic consensus sites of pre-mRNAs. Best studied are the functions of the 13 pseudouridines of U2 snRNA. Yi-Tao Yu and coworkers documented in an elegant set of experiments the requirement of U2 modifications near its central branch site recognition region for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes (Yu et al., 1998; Zhao and Yu, 2004). In contrast, in HeLa nuclear extracts, modifications at the 5’ end of U2 may be essential for splicing (Donmez et al., 2004). A functional mechanism for the highly conserved pseudouridine of U2 that lies opposite the branch site adenosine of introns of pre-mRNA has been proposed. This pseudouridine dramatically alters the local structure of a model RNA duplex, which may help make the nucleophile accessible for the first step of splicing (Newby and Greenbaum, 2001; Newby and Greenbaum, 2002). Surprisingly, yeast can survive without this particular pseudouridine in its U2 snRNA (Behm-Ansmant et al., 2003; Ma et al., 2003). Although we are far from understanding the specific roles of the hundreds of pseudouridines, these findings exemplify how a pseudouridine can impact the structure and function of the RNA it resides in. Since in vertebrates pseudouridylation of snRNAs is mediated by H/ACA RNPs (Darzacq et al., 2002; Hüttenhofer et al., 2001; Jady and Kiss, 2001; Kiss et al., 2004; Kiss et al., 2002), the preceding observations illustrate how an effect on all H/ACA RNPs could be particularly deleterious to cells by inhibition of splicing.
In addition to spliceosomal snRNAs, some snoRNAs (U3 and U8) involved in pre-rRNA processing also contain pseudouridines (Kato and Harada, 1984; Reddy et al., 1979). However, there is no insight as to the function of these modifications. Given that most snoRNAs or, for that matter, other RNAs have not been analyzed for their state of modification, could there be other stable cellular RNAs and possibly even mRNAs that are pseudouridylated? For example, a C/D RNP mediates 2’-O-methylation of archaeal pre-tRNATrp (Bortolin et al., 2003; Singh et al., 2004). Analogously, the formation of some pseudouridines in tRNAs could be catalyzed by H/ACA RNPs.
Telomere maintenance
Telomere maintenance depends on one H/ACA RNP, the vertebrate telomerase RNP. In vivo, the H/ACA domain of hTR is essential for hTR accumulation, hTR 3’ end processing, and telomerase activity (Mitchell et al., 1999a). For example, stable expression in yeast of the mature form of heterologous hTR, like that of all H/ACA RNAs, depends on its association with the three H/ACA core proteins Cbf5p, Nhp2p, and Nop10p (Dez et al., 2001). Therefore, proper assembly and function of H/ACA RNPs will also affect telomere function and consequently play crucial roles in cellular senescence and cancer (de Lange, 2002).
In summary, H/ACA RNPs are multifunctional particles with new functions possibly to be identified. Of all their presently known functions, the role of H/ACA RNPs in ribosome biogenesis and pre-mRNA splicing (through rRNA processing and snRNA pseudouridylation), due to the essential nature of these processes, is likely to have the most immediate impact on cell growth and viability.
Dyskeratosis congenita
As a target of the inherited disorder dyskeratosis congenita (DC), the functional importance of H/ACA RNPs is further highlighted. DC (aka Zinsser-Engmann-Cole syndrome) manifests itself first by the three cutaneous features of nail dystrophy, abnormal skin pigmentation, and mucosal leucoplakia, hence the name (Cole et al., 1930; Engmann, 1926; Zinsser, 1910). The majority of patients (85.5%) develop bone marrow failure, usually before the age of 30, as the leading cause of premature mortality. DC also is characterized by a predisposition to malignancy in rapidly dividing tissues (for recent reviews see Dokal and Vulliamy, 2003; Marrone and Mason, 2003). In addition, many other somatic abnormalities have been reported, such as epiphora, mental retardation, pulmonary disease, hair loss, and short stature marking DC as a multi-system disorder and contributing to the general heterogeneity of its phenotype.
DC is inherited in three patterns, X-linked, autosomal dominant, and autosomal recessive. X-linked DC (X-DC) is the most frequent and severe form followed by autosomal recessive (AR) and autosomal dominant (AD) forms (Dokal, 2000). AD-DC is the mildest form and can go unnoticed until the 5th decade of life. X-DC is caused by mutations in the gene DKC1 encoding NAP57 (dyskerin) and AD-DC by mutations in the gene encoding hTR (Heiss et al., 1998; Vulliamy et al., 2001a). The gene affected in AR-DC has yet to be mapped. Therefore two components of H/ACA RNPs are implicated in the molecular mechanism of DC; a core protein that is part of all H/ACA RNPs (X-DC) and hTR, the RNA component of one specific H/ACA RNP, telomerase (AD-DC). The discovery of NAP57 as part of the telomerase RNP along with the documentation of shortened telomeres in cells from patients with X-DC and AD-DC led to the theory that DC in general was caused by telomere dysfunction (Mitchell et al., 1999a; Mitchell et al., 1999b; Vulliamy et al., 2001a; Vulliamy et al., 2001b). However, given the broad range of functions of H/ACA RNPs as outlined in the previous section, it could not be excluded that other activities of H/ACA RNPs might be impaired in X-DC and contribute to its complex phenotype. Indeed, two recent studies in mice now support this suspicion.
Mice deficient for telomerase RNA demonstrated that telomere attrition can lead to chromosome instability, cancer and early onset of aging phenotypes, all features observed in human DC (Blasco et al., 1997). However, due to a peculiarity of laboratory mice (their telomeres are three to four times longer than those of humans) these phenotypes manifested themselves only after four to six generations. It was remarkable, therefore, when a hypomorphic NAP57 mouse model closely replicated the phenotype of DC in the first generation in the absence of any telomere effects (Ruggero et al., 2003). These mice showed reduced pseudouridylation of rRNA, impaired pre-rRNA processing, and impaired translation suggesting that impairment of non-telomerase targets also resulted in a DC phenotype (Ruggero et al., 2003). In a second approach, Mochizuki et al. (2004) introduced two NAP57 mutations that are observed in patients presenting with typical X-DC into murine embryonic stem (ES) cells. Both ES cell lines showed reduced levels of certain H/ACA snoRNAs (though not the same) and a decrease in rRNA processing and overall levels of pseudouridine in rRNA. Only one of the two ES cell lines, however, exhibited reduced amounts of hTR, telomerase activity, and shortened telomeres. Again, these data documented that functions of H/ACA RNPs other than those associated with telomerase are affected in X-DC (Mochizuki et al., 2004). Interestingly, an effect on ribosome biogenesis may be a general hallmark of rare bone marrow failure disorders, as about 25% of cases of Diamond-Blackfan anemia are caused by mutations in the RPS19 gene encoding a protein of the small ribosomal subunit (Cmejla et al., 2000; Draptchinskaia et al., 1999; Ramenghi et al., 2000; Willig et al., 1999). How this translates into the complex phenotypes of these disorders remains to be elucidated. Nevertheless, these studies teach us that it is important to understand the molecular impact of X-DC mutations on each individual H/ACA RNP to determine if and how it is affected.
H/ACA proteins
Although a plethora of work and reviews on structure, function, and biogenesis of H/ACA RNAs has been published (see introduction), much less is known about the H/ACA core proteins and how they form H/ACA RNPs. This is mostly due to the fact that while individual H/ACA RNAs can readily be studied, the core proteins are always part of a mixed population of over one hundred different but related complexes that complicate the readout of any experiment. This section reviews the individual H/ACA RNP core proteins.
NAP57
NAP57 is perhaps the best-known H/ACA protein because it is the target of X-DC and the putative pseudouridylase of H/ACA RNPs. Originally NAP57 was recognized in yeast as the centromere binding factor Cbf5p (Jiang et al., 1993). Mammalian NAP57 was identified as a Nopp140 associated protein of 57kD (Meier and Blobel, 1994) and later also named dyskerin due to its link to X-DC (Heiss et al., 1998). Nopp140 appears to be a chaperone of both H/ACA and C/D RNPs with which it interacts without being an integral part (see below; Yang et al., 2000). NAP57, like all snoRNP-associated proteins, is phylogenetically highly conserved. Over most of its sequence it is 70% identical to the yeast Cbf5p and 34% identical over a considerable stretch to the bacterial pseudouridine synthase TruB (Fig. 3A; Meier and Blobel, 1994; Nurse et al., 1995). Its function as a pseudouridylase is supported by the negative effect on rRNA pseudouridylation of genetic depletion of Cbf5p and its fly ortholog minifly and of point mutations in its catalytic domain (Giordano et al., 1999; Lafontaine et al., 1998; Zebarjadian et al., 1999). NAP57 and its orthologs are an integral part of all H/ACA snoRNPs tested, including vertebrate telomerase (Lafontaine et al., 1998; Mitchell et al., 1999b; Yang et al., 2000). In its amino terminal half, NAP57 contains two short motifs that are conserved in most pseudouridine synthases (Fig. 3A, Ψ) and in the carboxy terminal half, it has an RNA binding domain conserved in pseudouridylases and archaeosine transglycosylases (Fig. 3A, PUA) (Aravind and Koonin, 1999; Gustafsson et al., 1996; Koonin, 1996). The second Ψ motif contains a universally conserved aspartate (D125 in human NAP57) that apparently forms a covalent enzyme-substrate intermediate during the isomerization of uridine to pseudouridine (Fig. 3A, asterisk) (Gu et al., 1999; Hoang and Ferre-D'Amare, 2001; Huang et al., 1998; Spedaliere et al., 2004). The carboxy terminus of NAP57 is highly charged with three lysine clusters separated by acidic stretches (Fig. 3A, K). A fourth lysine cluster is present in its amino terminus and most clusters can function as nuclear localization sequences (Heiss et al., 1999; Youssoufian et al., 1999). NAP57 is concentrated in the dense fibrillar component of nucleoli and in Cajal bodies together with the other H/ACA core components (Fig. 4) (Meier and Blobel, 1994).
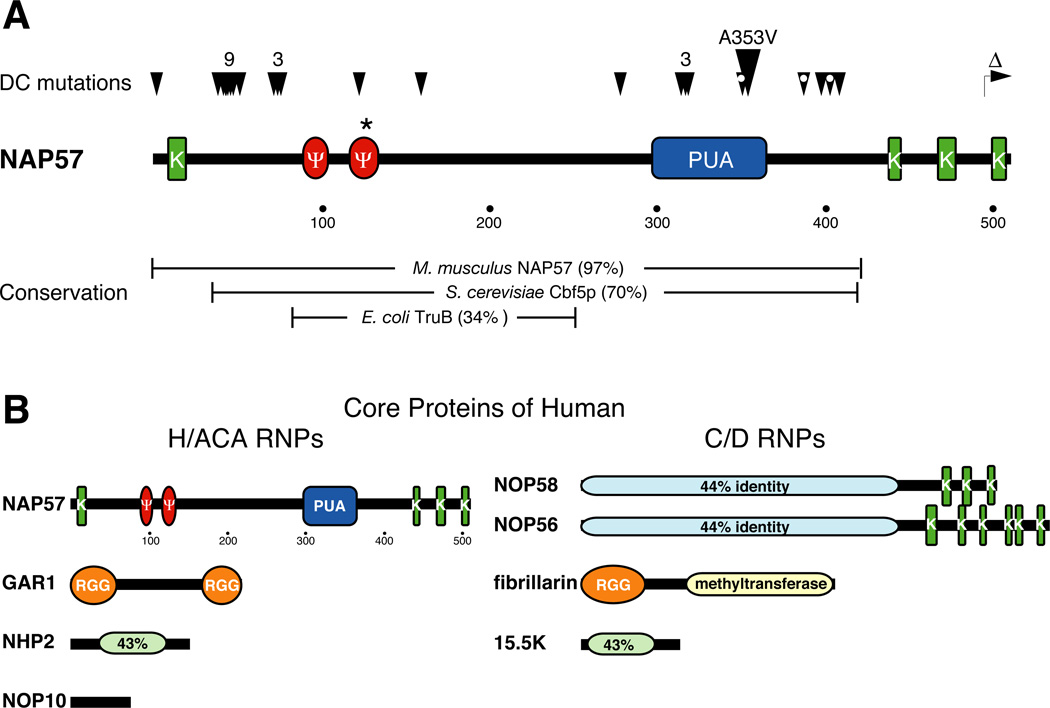
Figure 3. Domains of H/ACA and C/D core proteins.
(A) Schematic of human NAP57 drawn to scale (amino acid positions marked under sequence) indicating various sequence elements, lysine-rich stretches (green, K) often separated by acidic serine clusters, two motifs conserved in most pseudouridylases (red, Ψ), and a domain conserved in pseudouridylases and archaeosine transglycosylases (blue, PUA). The catalytic aspartate at amino acid 125 is highlighted (asterisk). The positions of mutations identified in patients with DC are indicated above the sequence (arrowheads). The total number of tightly clustered mutations is printed over the arrowheads. The most frequent mutation, A353V, observed in ~40% of X-DC cases is enlarged. White dots in the arrowheads indicate that this residue has been mutated two different amino acids. Additionally, a carboxyterminal truncation is specified (Δ). The percent identity of human NAP57 to its mouse (M. musculus), yeast (S. cerevisiae), and bacterial (E. coli) homologs over a certain range (brackets) is given below. (B) Comparison of the domains of core proteins of H/ACA (left) and C/D RNPs (right). The sequences are drawn to scale and the domains are as in (A). In addition, glycine-arginine-rich (RGG) and conserved methyltransferase domains are indicated. Sequence identity between two proteins is given in light green or blue. Note 15.5K is also known as NHP2L1/NHPX and Snu13p in yeast.
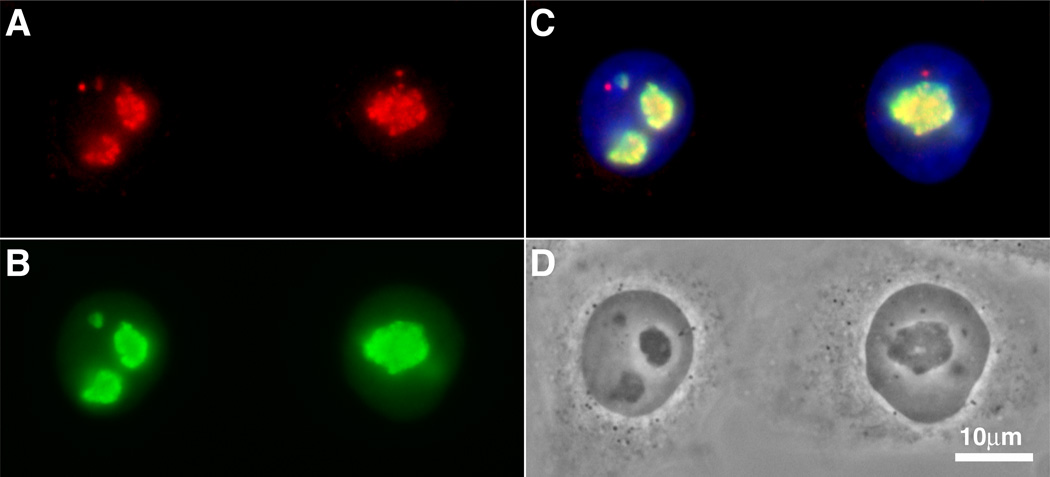
Figure 4. Double immunofluorescence of NAP57 and nucleolin in HeLa cells.
Indirect immunolocalization of NAP57 (A) and nucleolin (B) in fixed and permeabilized HeLa cells. (C) Merged immunofluorescence of (A) and (B) combined with DNA stain (blue). (D) Phase contrast image of the same two cells shown in the other panels. Note the granular staining of NAP57 reflecting its localization to the dense fibrillar component of nucleoli as compared to the more uniform staining of nucleolin, which is present in all parts of nucleoli. Note how the green of the nucleolin label extends beyond that of the partially colocalizing NAP57 (yellow in the merged image C). In addition and unlike nucleolin, NAP57 also concentrates in Cajal bodies (extra nucleolar red dots, particularly well visible in the merged image C).
Additional NAP57 functions
NAP57 exerts at least two functions, one as enzyme and one as crucial building block of H/ACA RNPs. Given the essential functions of some of the H/ACA RNAs (see above), it is not surprising that NAP57 is encoded by an essential gene in yeast, fly, and mice, where a knockout is embryonically lethal (Giordano et al., 1999; He et al., 2002; Jiang et al., 1993; Phillips et al., 1998). However, additional functions may contribute to the importance of NAP57 and the complex phenotype of X-DC caused by its mutation. These functions are mostly inferred from genetic evidence from the yeast ortholog Cbf5p. A centromeric function is implied by the suppression of a temperature-sensitive mutation in one of the subunits of the yeast centromere DNA binding complex CBF3 by the overexpression of Cbf5p (Jiang et al., 1993). Cbf5p involvement with the nuclear cap-binding complex of pre-mRNAs and snRNAs is suggested by its synthetic lethal phenotype with a deletion of both subunits together (Fortes et al., 1999). An association of Cbf5p with nuclear organization is supported by a mutation in CBF5 that (likely due to reduced expression) disrupts nucleolar localization of pre-tRNAs and suppresses a transcriptional silencing effect of tRNA genes on nearby RNA polymerase II promoters (Kendall et al., 2000). Finally, a link of Cbf5p to rRNA transcription is established by the suppression of a temperature-sensitive CBF5 allele, cbf5-1, by the overexpression of the RNA polymerase I transcription factor Rrn3p (Cadwell et al., 1997). Importantly, because rRNA pseudouridylation is normal in a cbf5-1 strain and the lethal phenotype of cbf5-1 at the nonpermissive temperature is partially rescued by transcription of rRNA from an RNA polymerase II-driven promoter, Cbf5p must also play an essential role in polymerase I-driven rRNA transcription (Cadwell et al., 1997; Zebarjadian et al., 1999). Similarly, a second essential function for Cbf5p is implied from complementation studies with rat NAP57. Specifically, heterologous expression of NAP57 rescues the growth arrest and loss of H/ACA RNAs caused by genetic depletion of Cbf5p, but not the lethal phenotype of cbf5-1 at nonpermissive temperature or that of a cbf5 deletion (Yang et al., 2000). Presumably, genetic depletion leaves behind an imperceptible but essential fraction of Cbf5p whose function NAP57 fails to rescue. Although there are clear functional differences between yeast Cbf5p and vertebrate NAP57, these data suggest functions for the two orthologs in addition to those associated with H/ACA RNAs. In fact, it is not clear if any Cbf5p/NAP57 exists free in the cell independent from H/ACA RNPs, although glycerol gradient analysis of nuclear extracts suggests that it would have to be a minor amount (Wang et al., 2002). Interestingly, all additional functions of Cbf5p described above appear to be gene dosage dependent. This is also reflected in the severe effects caused by decreased expression of NAP57 in the X-DC mouse model, in flies, and in humans with a DC mutation in the NAP57 promoter resulting in its reduced expression (Giordano et al., 1999; Kauffman et al., 2003; Knight et al., 2001; Ruggero et al., 2003; Salowsky et al., 2002). Therefore, NAP57 is a multifunctional protein whose dosage is extremely important to the well being of cells and organisms. In the case of AD-DC, where hTR is mutated, slight changes in NAP57 levels could explain effects on NAP57 related functions. For example, AD-DC mutations in hTR could cause its degradation and free up an incremental but critical amount of NAP57 thereby leading to phenotypes other than those attributed to lack of telomerase activity. Because hTR represents a minor fraction of cellular H/ACA RNAs, this hypothesis may be far-fetched, but it is testable. For instance, mice carrying AD-DC mutations in telomerase RNA should manifest a DC phenotype in the first generations before telomere effects are noticeable.
X-DC mutations
Most of the over 30 DC mutations identified in NAP57 are missense mutations, often leading to conservative amino acid exchanges. In addition, a short carboxyterminal truncation, two intronic base changes and a mutation in the promoter have been identified (Knight et al., 1999; Knight et al., 2001; Salowsky et al., 2002; Vulliamy et al., 1999). Although distributed throughout the open reading frame, DC mutations tend to cluster in two places, an amino terminal region outside the conserved Ψ motifs and between amino acids 300 and 400 comprising the conserved PUA domain (Fig. 3A). Many of these mutations are quite subtle and their impact on the structure and function of NAP57 are not immediately obvious. This is perhaps best illustrated by the alanine 353 to valine mutation, which accounts for ~40% of X-DC cases (Dokal and Vulliamy, 2003). The most surprising aspect of this mutation is that in the yeast ortholog Cbf5p, the corresponding residue is a valine. Therefore, yeast Cbf5p contains compensatory mutations or this X-DC mutation affects a mammalian-specific NAP57 function, such as its association with hTR and scaRNAs or a putative extra-RNP activity. Indeed, impact of DC mutations on NAP57-specific and H/ACA RNP-independent functions is a reasonable possibility because no DC mutations have been identified in any of the genes encoding the other three core proteins (Marrone and Mason, 2003). In contrast, Fanconi Anemia, another inherited bone marrow failure syndrome, is associated with mutations in every gene of a multi-subunit complex (reviewed in Tischkowitz and Dokal, 2004).
Additional core proteins
In addition to NAP57, three small basic proteins associate with all H/ACA RNAs, GAR1 (22.3kD), NHP2 (17.2kD), and NOP10 (7.7kD) (Fig. 3B; Balakin et al., 1996; Ganot et al., 1997b; Henras et al., 1998; Watkins et al., 1998). GAR1 consists of a central core domain flanked by two glycine-arginine-rich (GAR) domains (Girard et al., 1992). The core domain of yeast Gar1p is sufficient for viability and binds H/ACA snoRNAs in vitro (Bagni and Lapeyre, 1998; Girard et al., 1994). NHP2 was originally identified as a nonhistone chromatin protein (Kolodrubetz et al., 1988). It is homologous to the ribosomal protein L30 and to 15.5K/NHP2L1/NHPX (Snu13p in yeast) shared between C/D snoRNPs and U4 spliceosomal snRNPs (Henras et al., 1998; Leung and Lamond, 2002; Nottrott et al., 1999; Watkins et al., 1998; Watkins et al., 2000). Unlike its homologs, NHP2 alone does not bind to a specific RNA motif but associates non-specifically with RNA secondary structures (Henras et al., 2001; Wang and Meier, 2004). NOP10, although conserved like the other core proteins, contains no known motifs and measures only 64 amino acids (Henras et al., 1998). Like NAP57, all these proteins are concentrated in nucleoli and Cajal bodies of mammalian cells reflecting the location of H/ACA RNPs (Fig. 4) (Pogacic et al., 2000).
Comparison to C/D core proteins
Although H/ACA and C/D RNPs both function in site-directed RNA modification, they consist of different classes of RNAs and proteins. However, both contain four core proteins that are homologous or share domains with each other (Fig. 3B). Interestingly, the putative enzymes of the two types of RNPs, the pseudouridylase NAP57 and the methylase fibrillarin, do not share their domains with each other but with NOP56 and NOP58 and with GAR1, respectively. Specifically, NAP57 shares its lysine-rich carboxyl terminus with NOP56 and NOP58, while both GAR1 and fibrillarin contain glycine-arginine-rich domains. NHP2 shares its RNA binding capacity, but not specificity, and overall structure with the 15.5K/NHP2L1/NHPX C/D core protein. Indeed, both proteins have a common relative in archaea, L7Ae, which is part of archaeal H/ACA and C/D RNPs and ribosomes (Rozhdestvensky et al., 2003; Tran et al., 2004). NOP10 apparently lacks a homologous counterpart in C/D snoRNPs. It is not clear if the core proteins of these two diverse RNPs acquired homologous domains because of their related working environment or if they evolved from distant common ancestors, as in the case of NHP2 and 15.5K/NHP2L1/NHPX (Fig. 3B).
H/ACA RNP assembly
The association of the four core proteins with H/ACA RNAs was originally demonstrated by coprecipitation and copurification in yeast (Balakin et al., 1996; Ganot et al., 1997b; Henras et al., 1998; Lafontaine et al., 1998; Lübben et al., 1995; Watkins et al., 1998) and subsequently confirmed in mammalian cells (Dragon et al., 2000; Mitchell et al., 1999b; Pogacic et al., 2000; Yang et al., 2000). Only recently was the assembly pathway of H/ACA RNPs elucidated using in vitro translated proteins followed by immunoprecipitation (Wang and Meier, 2004). In contrast to the assembly of other RNPs, which is frequently initiated by protein-RNA interactions, e.g., in the case of 15.5K and C/D snoRNAs (Watkins et al., 2002), H/ACA core proteins form a protein-only complex (Fig. 1). Specifically, GAR1 and NOP10 associate with NAP57 independently while binding of NHP2 requires prior interaction between NOP10 and NAP57 (Wang and Meier, 2004). The specificity for H/ACA RNA recognition is provided by the core trimer of NAP57-NOP10-NHP2 and is GAR1 independent. Step-wise disassembly of affinity purified H/ACA RNPs confirms these interactions in yeast suggesting that they, like the individual proteins and RNAs, are phylogenetically conserved (Henras et al., 2004a). Although the existence of additional H/ACA core proteins cannot be excluded, the combination of all these studies strongly indicates that NAP57, NOP10, NHP2, and GAR1 constitute the complete set of H/ACA core proteins.
H/ACA RNP interacting proteins
Several proteins have been identified that interact with H/ACA RNPs without being an integral part. Nopp140, SMN, NAF1, and SHQ1 have been associated with H/ACA RNPs based on their ability to interact with one or several of the core components (Fig. 1; Dez et al., 2002; Fatica et al., 2002; Meier and Blobel, 1994; Pellizzoni et al., 2001; Whitehead et al., 2002; Yang et al., 2002). Additionally, Rnt1p, the yeast RNase responsible for the earliest cleavage event at the 3’ end of pre-rRNA, physically interacts with Gar1p and seems involved in the nuclear import of H/ACA RNP core proteins (Tremblay et al., 2002).
NAF1-SHQ1 complex
The nuclear assembly factor, Naf1p, and the factor required for snoRNA of the H/ACA class quantitative accumulation, Shq1p, are two interacting proteins in yeast nuclei whose genetic depletion causes specific loss of all H/ACA snoRNAs (Dez et al., 2002; Fatica et al., 2002; Yang et al., 2002). While both proteins associate with Nhp2p, Naf1p additionally binds the three remaining H/ACA core proteins. However, neither protein is part of mature H/ACA RNPs. Naf1p shares an RNA binding domain with GAR1 and binds non-specifically to secondary structure elements in RNA, analogous to Nhp2p (Fatica et al., 2002; Henras et al., 2001). Based on these and other qualities, Naf1p and Shq1p have been implicated in the assembly of H/ACA RNPs (Dez et al., 2002; Fatica et al., 2002; Yang et al., 2002). Because both yeast proteins are phylogenetically conserved, they likely harbor a similar function in mammalian cells.
SMN
The survival of motor neurons protein, SMN, is affected in autosomal recessive spinal muscular atrophy (the leading genetic cause for infant death) and forms the core of a multiprotein complex that assembles Sm proteins onto spliceosomal snRNAs (Gubitz et al., 2004; Meister et al., 2002; Terns and Terns, 2001). SMN interacts with Sm proteins via their glycine-arginine-rich domains. Similarly, SMN binds to the related domains of GAR1 and fibrillarin and, therefore, may also be involved in assembly of snoRNPs (Jones et al., 2001; Pellizzoni et al., 2001; Whitehead et al., 2002). However, such an activity would be restricted to higher eukaryotes as yeast lacks a recognizable SMN homolog.
Nopp140
The nucleolar and Cajal body phosphoprotein Nopp140 is currently the only protein that associates with mature H/ACA RNPs. It is not a stable component of H/ACA RNPs because a concentration of 500mM salt dissociates it from the RNPs without affecting their integrity (Meier and Blobel, 1992; Wang et al., 2002; Yang et al., 2000). In fact, NAP57 was identified as a Nopp140 associated protein (Meier and Blobel, 1994). However, the interaction with H/ACA RNPs may occur through several components because individual core proteins or RNAs fail to interact in two- or three-hybrid assays, respectively (Wang et al., 2002). Loss of Nopp140-H/ACA RNP interaction by dephosphorylation of Nopp140 suggests that the association is governed by phosphorylation (Wang et al., 2002). Nopp140 also interacts with C/D snoRNPs and may function as a general chaperone of snoRNPs because its depletion from nucleoli and Cajal bodies co-depletes H/ACA and C/D RNPs (Isaac et al., 1998; Yang et al., 2000). Recent data suggests an involvement of Nopp140 in the assembly of C/D RNPs (Watkins et al., 2004). Additionally, Nopp140 has been implicated in rRNA transcription (Chen et al., 1999), as a transcriptional coactivator and protein kinase A dependent mediator of the acute phase response in liver (Chiu et al., 2002; Miau et al., 1997), and in the induction of intranuclear membrane cisternae in endometrial cells that may be important for human blastocyst attachment (Isaac et al., 2001). Through Nopp140, therefore, H/ACA RNPs could impact a whole range of cellular and tissue-specific processes. Like most other proteins affiliated with H/ACA RNPs, Nopp140 is evolutionary conserved and has a counterpart in yeast, Srp40p (Meier, 1996).
Proteins specific to a H/ACA RNP
Given the large number and variety of H/ACA RNAs, it is likely that certain H/ACA RNPs harbor specific proteins that are an integral part of the particle. Currently, the only such H/ACA RNP-specific protein known is the catalytic subunit of the telomerase RNP, the reverse transcriptase TERT (Lingner et al., 1997; Weinrich et al., 1997). Interestingly however, although hTR is present in most somatic cells, presumably stabilized by the four H/ACA core proteins (see above), TERT is not (reviewed in Cong et al., 2002).
Evolution
The diverse mammalian H/ACA RNPs highlighted in this review originate from single protein enzymes that evolved to accommodate multiple functions (reviewed in Lafontaine and Tollervey, 1998). In bacteria, distinct enzymes both recognize the uridine for isomerization and catalyze the pseudouridylation of rRNA and tRNA. Mammalian NAP57 and yeast Cbf5p are most closely related to the E. coli enzyme TruB responsible for formation of the almost universally conserved pseudouridine at position 55 in tRNA (Fig. 3A). In eukaryotes, a distinct enzyme also catalyzes this modification, while NAP57 and Cbf5p are integral parts of H/ACA RNPs. The origin of H/ACA RNPs appears to predate the split between archaea and eukaryotes, as they have also been identified in archae-, but not eubacteria (Rozhdestvensky et al., 2003; Watanabe and Gray, 2000). H/ACA RNPs seem to have evolved with increasing numbers of pseudouridines in target RNAs as illustrated by striking differences between yeast and mammals. In particular, from yeast to mammals, the numbers of pseudouridines in rRNAs and snRNAs increases from 44 to ~91 and from 6 to 27, respectively (Massenet et al., 1998; Ofengand, 2002). In both organisms pseudouridylation of rRNA is mediated by H/ACA RNPs while that of snRNAs, in yeast, unlike mammals, is catalyzed by distinct enzymes (Behm-Ansmant et al., 2003; Ma et al., 2003; Massenet et al., 1999; Zhao et al., 2002). Moreover and unlike vertebrate H/ACA telomerase RNA, yeast telomerase RNA forms a snRNP-like particle with Sm proteins (Mitchell et al., 1999a; Seto et al., 1999). Evidently, H/ACA RNPs have adopted a whole range of functions across evolution. This is perhaps best illustrated by the discovery of a brain-specific H/ACA RNA in mammals (Cavaille et al., 2000) despite the fact that all previous activities of H/ACA RNPs are housekeeping functions important for the viability of every single cell.
Conclusions and perspectives
H/ACA RNPs perform multiple basic functions in all cells from archaea to mammals. Their functions are defined by one of over one hundred H/ACA RNAs, each associated with the same four core proteins. Although simple five-component particles, they are functionally diverse and complex. Therefore, how will mutations in one component affect the different but similar RNPs? This question of course is at the heart of X-DC where NAP57 is mutated. Genetic depletion studies in yeast teach us that NAP57 is essential for the stability of all H/ACA RNPs. Analysis of X-DC patient samples and transgenic mouse ES cells harboring X-DC mutations suggests that mutations in NAP57 affect different H/ACA RNPs to various degrees. However, while peripheral lymphocytes of all DC patients, X-DC and AD-DC, show telomere shortening (Vulliamy et al., 2001a; Vulliamy et al., 2001b), some disagreement remains as to the impact on other H/ACA RNP functions (Meier, 2003). Some studies report no effect on snoRNAs and scaRNAs and consequently rRNA and snRNA pseudouridylation (Mitchell et al., 1999b; Wong et al., 2004). However, studies with a X-DC mouse model and transgenic ES cells find impaired rRNA pseudouridylation and reduced amounts of certain H/ACA snoRNAs (Mochizuki et al., 2004; Ruggero et al., 2003). Similarly, extracts from X-DC patient cell lines compared to those from the carrier mother, exhibit reduced in vitro pseudouridylation of certain rRNA target sites that correlates with a reduced amount of the corresponding H/ACA snoRNA detected on northern blots (S. Roy, C. Wang, and U.T. Meier, unpublished results). Obviously, further experimentation is indicated with a larger sampling of patient cells and analysis of all potential functions affected by H/ACA RNPs. In particular and to get at the molecular mechanism of DC and H/ACA RNPs in general, it will be important to study the impact of NAP57 mutations on the structure and assembly of individual H/ACA RNPs.
Finally, X-DC is a fascinating example of an inherited disease with a complex phenotype that is caused by mutations in a protein involved in some of the most basic functions of every cell. This is similar to laminopathies characterized by mutations in lamin A, a structural protein of the nuclear envelope, that cause various muscular and lipodystrophies and even progeria, a severe form of accelerated aging (reviewed in Burke and Stewart, 2002; Hutchison and Worman, 2004). The dissection of the molecular mechanisms of these diseases is teaching us about expected and unexpected functions of these housekeeping proteins in mammalian organisms. Since the study of snoRNAs and snoRNPs has already yielded many surprises and even revision of dogmas, we can only look forward to what other targets and functions of H/ACA RNPs will be revealed.
Acknowledgements
This review is dedicated to the memory of Jim Ofengand. I thank Charles Query, Susan Smith, Jon Warner, and the members of my laboratory for critical comments on the manuscript and Nupur Kittur for performing the immunofluorescence experiments in Fig. 4. The work in the author’s laboratory is supported by grants from the American Cancer Society and the National Heart Lung and Blood Institute.
References
- Aravind L, Koonin EV. Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol. 1999;48:291–302. doi: 10.1007/pl00006472. [DOI] [PubMed] [Google Scholar]
- Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- Atzorn V, Fragapane P, Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell. Biol. 2004;24:1769–1778. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Badis G, Fromont-Racine M, Jacquier A. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA. 2003;9:771–779. doi: 10.1261/rna.5240503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Lapeyre B. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. J. Biol. Chem. 1998;273:10868–10873. doi: 10.1074/jbc.273.18.10868. [DOI] [PubMed] [Google Scholar]
- Bakin A, Lane BG, Ofengand J. Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry. 1994;33:13475–13483. doi: 10.1021/bi00249a036. [DOI] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- Bally M, Hughes J, Cesareni G. SnR30: a new, essential small nuclear RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:5291–5303. doi: 10.1093/nar/16.12.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Urban A, Ma X, Yu YT, Motorin Y, Branlant C. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA. 2003;9:1371–1382. doi: 10.1261/rna.5520403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Bortolin ML, Bachellerie JP, Clouet-d'Orval B. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp) Nucleic Acids Res. 2003;31:6524–6535. doi: 10.1093/nar/gkg860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Henry Y, Gélugne J-P, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997;16:4770–4776. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand RC, Klootwijk J, Sibum CP, Planta RJ. Pseudouridylation of yeast ribosomal precursor RNA. Nucleic Acids Res. 1979;7:121–34. doi: 10.1093/nar/7.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Stewart CL. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell. Biol. 2002;3:575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- Cadwell C, Yoon H-J, Zebarjadian Y, Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 1997;17:6175–6183. doi: 10.1128/mcb.17.10.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervelli M, Oliverio M, Bellini A, Bologna M, Cecconi F, Mariottini P. Structural and sequence evolution of U17 small nucleolar RNA (snoRNA) and its phylogenetic congruence in chelonians. J. Mol. Evol. 2003;57:73–84. doi: 10.1007/s00239-003-2453-2. [DOI] [PubMed] [Google Scholar]
- Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- Chen HK, Pai CY, Huang JY, Yeh NH. Human Nopp140, which interacts with RNA polymerase I: implications for rRNA gene transcription and nucleolar structural organization. Mol. Cell. Biol. 1999;19:8536–8546. doi: 10.1128/mcb.19.12.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CM, Tsay YG, Chang CJ, Lee SC. Nopp140 is a mediator of the protein kinase A signaling pathway that activates the acute phase response alpha1-acid glycoprotein gene. J. Biol. Chem. 2002;277:39102–39111. doi: 10.1074/jbc.M205915200. [DOI] [PubMed] [Google Scholar]
- Cmejla R, Blafkova J, Stopka T, Zavadil J, Pospisilova D, Mihal V, Petrtylova K, Jelinek J. Ribosomal protein S19 gene mutations in patients with diamond-blackfan anemia and identification of ribosomal protein S19 pseudogenes. Blood Cells Mol. Dis. 2000;26:124–132. doi: 10.1006/bcmd.2000.0286. [DOI] [PubMed] [Google Scholar]
- Cole HN, Rauschkolb JE, Toomey J. Dyskeratosis congenita with pigmentation, dystrophy unguis and leukokeratosis oris. Arch. Dermatol. Syph. 1930;21:71–95. doi: 10.1001/archderm.1955.01540280027005. [DOI] [PubMed] [Google Scholar]
- Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2'-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- Dez C, Henras A, Faucon B, Lafontaine D, Caizergues-Ferrer M, Henry Y. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 2001;29:598–603. doi: 10.1093/nar/29.3.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Noaillac-Depeyre J, Caizergues-Ferrer M, Henry Y. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 2002;22:7053–7065. doi: 10.1128/MCB.22.20.7053-7065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokal I. Dyskeratosis congenita in all its forms. Br. J. Haematol. 2000;110:768–779. doi: 10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- Dokal I, Vulliamy T. Dyskeratosis congenita: its link to telomerase and aplastic anaemia. Blood Rev. 2003;17:217–225. doi: 10.1016/s0268-960x(03)00020-1. [DOI] [PubMed] [Google Scholar]
- Donmez G, Hartmuth K, Luhrmann R. Modified nucleotides at the 5' end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F, Pogacic V, Filipowicz W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 2000;20:3037–3048. doi: 10.1128/mcb.20.9.3037-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Engmann MF. A unique case of reticular pigmentation of the skin with atrophy. Arch. Dermatol. Syph. 1926;13:685–687. [Google Scholar]
- Fatica A, Dlakic M, Tollervey D. Naf1 p is a box H/ACA snoRNP assembly factor. RNA. 2002;8:1502–1514. [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj IW. Genetic and physical interactions involving the yeast nuclear cap- binding complex. Mol. Cell. Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997a;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997b;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- Giordano E, Peluso I, Senger S, Furia M. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell Biol. 1999;144:1123–1133. doi: 10.1083/jcb.144.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J-P, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard JP, Bagni C, Caizergues-Ferrer M, Amalric F, Lapeyre B. Identification of a segment of the small nucleolar ribonucleoprotein-associated protein GAR1 that is sufficient for nucleolar accumulation. J. Biol. Chem. 1994;269:18499–18506. [PubMed] [Google Scholar]
- Gu X, Liu Y, Santi DV. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc. Natl. Acad. Sci. U S A. 1999;96:14270–14275. doi: 10.1073/pnas.96.25.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp. Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Gustafsson C, Reid R, Greene PJ, Santi DV. Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucleic Acids Res. 1996;24:3756–3762. doi: 10.1093/nar/24.19.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Navarrete S, Jasinski M, Vulliamy T, Dokal I, Bessler M, Mason PJ. Targeted disruption of Dkc1, the gene mutated in X-linked dyskeratosis congenita, causes embryonic lethality in mice. Oncogene. 2002;21:7740–7744. doi: 10.1038/sj.onc.1205969. [DOI] [PubMed] [Google Scholar]
- Heiss NS, Girod A, Salowsky R, Wiemann S, Pepperkok R, Poustka A. Dyskerin localizes to the nucleolus and its mislocalization is unlikely to play a role in the pathogenesis of dyskeratosis congenita. Hum. Mol. Genet. 1999;8:2515–2524. doi: 10.1093/hmg/8.13.2515. [DOI] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- Henras A, Dez C, Noaillac-Depeyre J, Henry Y, Caizergues-Ferrer M. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res. 2001;29:2733–2746. doi: 10.1093/nar/29.13.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Capeyrou R, Henry Y, Caizergues-Ferrer M. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs. RNA. 2004a;10:1704–1712. doi: 10.1261/rna.7770604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Dez C, Henry Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 2004b;14:335–343. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Hoang C, Ferre-D'Amare AR. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- Huang L, Pookanjanatavip M, Gu X, Santi DV. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37:344–351. doi: 10.1021/bi971874+. [DOI] [PubMed] [Google Scholar]
- Hutchison CJ, Worman HJ. A-type lamins: guardians of the soma? Nat. Cell Biol. 2004;6:1062–1067. doi: 10.1038/ncb1104-1062. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C, Pollard JW, Meier UT. Intranuclear endoplasmic reticulum induced by Nopp140 mimics the nucleolar channel system of human endometrium. J. Cell Sci. 2001;114:4253–4264. doi: 10.1242/jcs.114.23.4253. [DOI] [PubMed] [Google Scholar]
- Isaac C, Yang Y, Meier UT. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Kiss T. A small nucleolar guide RNA functions both in 2'-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 2001;20:541–551. doi: 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanteur P, Amaldi F, Attardi G. Partial sequence analysis of ribosomal RNA from HeLa cells. II. Evidence for sequences of non-ribosmal type in 45 and 32 s ribosomal RNA precursors. J. Mol. Biol. 1968;33:757–775. doi: 10.1016/0022-2836(68)90318-5. [DOI] [PubMed] [Google Scholar]
- Jiang W, Middleton K, Yoon H-J, Fouquet C, Carbon J. An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Mol. Cell. Biol. 1993;13:4884–4893. doi: 10.1128/mcb.13.8.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KW, Gorzynski K, Hales CM, Fischer U, Badbanchi F, Terns RM, Terns MP. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- Kato N, Harada F. Nucleotide sequence of nuclear 5.4 S RNA of mouse cells. Biochim. Biophys. Acta. 1984;782:127–131. doi: 10.1016/0167-4781(84)90015-0. [DOI] [PubMed] [Google Scholar]
- Kauffman T, Tran J, DiNardo S. Mutations in Nop60B, the Drosophila homolog of human dyskeratosis congenita 1, affect the maintenance of the germ-line stem cell lineage during spermatogenesis. Dev. Biol. 2003;253:189–199. doi: 10.1016/s0012-1606(02)00013-1. [DOI] [PubMed] [Google Scholar]
- Kendall A, Hull MW, Bertrand E, Good PD, Singer RH, Engelke DR. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc. Natl. Acad. Sci. USA. 2000;97:13108–13113. doi: 10.1073/pnas.240454997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 2004;24:5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss AM, Jady BE, Darzacq X, Verheggen C, Bertrand E, Kiss T. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 2002;30:4643–4649. doi: 10.1093/nar/gkf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Kiss T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- Kiss T, Filipowicz W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993;12:2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Heiss NS, Vulliamy TJ, Greschner S, Stavrides G, Pai GS, Lestringant G, Varma N, Mason PJ, Dokal I, Poustka A. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am. J. Hum. Genet. 1999;65:50–58. doi: 10.1086/302446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Vulliamy TJ, Morgan B, Devriendt K, Mason PJ, Dokal I. Identification of novel DKC1 mutations in patients with dyskeratosis congenita: implications for pathophysiology and diagnosis. Hum. Genet. 2001;108:299–303. doi: 10.1007/s004390100494. [DOI] [PubMed] [Google Scholar]
- Kolodrubetz D, Haggren W, Burgum A. Amino-terminal sequence of a Saccharomyces cerevisiae nuclear protein, NHP6, shows significant identity to bovine HMG1. FEBS Lett. 1988;238:175–179. doi: 10.1016/0014-5793(88)80251-5. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Pseudouridine synthases: four families of enzumes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem. Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Lamond AI. In vivo analysis of NHPX reveals a novel nucleolar localization pathway involving a transient accumulation in splicing speckles. J. Cell Biol. 2002;157:615–629. doi: 10.1083/jcb.200201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lübben B, Fabrizio P, Kastner B, Lührmann R. Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:11549–11554. doi: 10.1074/jbc.270.19.11549. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhao X, Yu YT. Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 2003;22:1889–1897. doi: 10.1093/emboj/cdg191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden BH. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acids Res. Mol. Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- Marrone A, Mason PJ. Dyskeratosis congenita. Cell. Mol. Life Sci. 2003;60:507–517. doi: 10.1007/s000180300042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol. 1999;19:2142–2154. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S, Mougin A, Branlant C. Posttranscriptional modifications in the U small nuclear RNAs. In: Grosjean H, Benne R, editors. The modification and editing of RNA. Washington, DC: ASM Press; 1998. pp. 201–228. [Google Scholar]
- Meier UT. Comparison of the rat nucleolar protein Nopp140 to its yeast homolog SRP40: Differential phosphorylation in vertebrates and yeast. J. Biol. Chem. 1996;271:19376–19384. [PubMed] [Google Scholar]
- Meier UT. Dissecting dyskeratosis. Nat. Genet. 2003;33:116–117. doi: 10.1038/ng0203-116. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. [correction appeared in 140: 447] 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- Meroueh M, Grohar PJ, Qiu J, SantaLucia J, Jr, Scaringe SA, Chow CS. Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucleic Acids Res. 2000;28:2075–2083. doi: 10.1093/nar/28.10.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miau L-H, Chang C-J, Tsai W-H, Lee S-C. Identification and characterization of a nucleolar phosphoprotein Nopp140, as a transcription factor. Mol. Cell. Biol. 1997;17:230–239. doi: 10.1128/mcb.17.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3' end. Mol. Cell. Biol. 1999a;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999b;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Mochizuki Y, He J, Kulkarni S, Bessler M, Mason PJ. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc. Natl. Acad. Sci. USA. 2004;101:10756–10761. doi: 10.1073/pnas.0402560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JP, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA. 2001;7:833–845. doi: 10.1017/s1355838201002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat. Struct. Biol. 2002;9:958–965. doi: 10.1038/nsb873. [DOI] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Luhrmann R. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5' stem-loop of U4 snRNA. EMBO J. 1999;18:6119–6133. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse K, Wrzesinski J, Bakin A, Lane BG, Ofengand J. Purification, cloning, and properties of the tRNA Y55 synthase from. Escherichia coli. RNA. 1995;1:102–112. [PMC free article] [PubMed] [Google Scholar]
- Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 2002;514:17–25. doi: 10.1016/s0014-5793(02)02305-0. [DOI] [PubMed] [Google Scholar]
- Parker R, Simmons T, Shuster EO, Siliciano PG, Guthrie C. Genetic analysis of small nuclear RNAs in Saccharomyces cerevisiae: viable sextuple mutant. Mol. Cell. Biol. 1988;8:3150–3159. doi: 10.1128/mcb.8.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Baccon J, Charroux B, Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 2001;11:1079–1088. doi: 10.1016/s0960-9822(01)00316-5. [DOI] [PubMed] [Google Scholar]
- Phillips B, Billin AN, Cadwell C, Buchholz R, Erickson C, Merriam JR, Carbon J, Poole SJ. The Nop60B gene of Drosophila encodes an essential nucleolar protein that functions in yeast. Mol. Gen. Genet. 1998;260:20–29. doi: 10.1007/s004380050866. [DOI] [PubMed] [Google Scholar]
- Pogacic V, Dragon F, Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramenghi U, Campagnoli MF, Garelli E, Carando A, Brusco A, Bagnara GP, Strippoli P, Izzi GC, Brandalise S, Riccardi R, Dianzani I. Diamond-Blackfan anemia: report of seven further mutations in the RPS19 gene and evidence of mutation heterogeneity in the Italian population. Blood Cells Mol. Dis. 2000;26:417–422. doi: 10.1006/bcmd.2000.0324. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Conrad J, Hall BG, Ofengand J. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA. 1998;4:1407–1417. doi: 10.1017/s1355838298981146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R, Henning D, Busch H. Nucleotide sequence of nucleolar U3B RNA. J. Biol. Chem. 1979;254:11097–11105. [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–4293. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff EA, Rimoldi OJ, Raghu B, Eliceiri GL. Three small nucleolar RNAs of unique nucleotide sequences. Proc. Natl. Acad. Sci. USA. 1993;90:635–638. doi: 10.1073/pnas.90.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- Salowsky R, Heiss NS, Benner A, Wittig R, Poustka A. Basal transcription activity of the dyskeratosis congenita gene is mediated by Sp1 and Sp3 and a patient mutation in a Sp1 binding site is associated with decreased promoter activity. Gene. 2002;293:9–19. doi: 10.1016/s0378-1119(02)00725-4. [DOI] [PubMed] [Google Scholar]
- Samarsky DA, Balakin AG, Fournier MJ. Characterization of three new snRNAs from Saccharomyces cerevisiae: snR34, snR35 and snR36. Nucleic Acids Res. 1995;23:2548–2554. doi: 10.1093/nar/23.13.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Singh SK, Gurha P, Tran EJ, Maxwell ES, Gupta R. Sequential 2'-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J. Biol. Chem. 2004;279:47661–47671. doi: 10.1074/jbc.M408868200. [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Spedaliere CJ, Ginter JM, Johnston MV, Mueller EG. The pseudouridine synthases: revisiting a mechanism that seemed settled. J. Am. Chem. Soc. 2004;126:12758–12759. doi: 10.1021/ja046375s. [DOI] [PubMed] [Google Scholar]
- Terns MP, Terns RM. Macromolecular complexes: SMN--the master assembler. Curr. Biol. 2001;11:R862–R864. doi: 10.1016/s0960-9822(01)00517-6. [DOI] [PubMed] [Google Scholar]
- Tischkowitz M, Dokal I. Fanconi anaemia and leukaemia - clinical and molecular aspects. Br. J. Haematol. 2004;126:176–191. doi: 10.1111/j.1365-2141.2004.05023.x. [DOI] [PubMed] [Google Scholar]
- Tran E, Brown J, Maxwell ES. Evolutionary origins of the RNA-guided nucleotide-modification complexes: from the primitive translation apparatus? Trends Biochem. Sci. 2004;29:343–350. doi: 10.1016/j.tibs.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Tremblay A, Lamontagne B, Catala M, Yam Y, Larose S, Good L, Elela SA. A physical interaction between Gar1p and Rnt1pi is required for the nuclear import of H/ACA small nucleolar RNA-associated proteins. Mol. Cell. Biol. 2002;22:4792–4802. doi: 10.1128/MCB.22.13.4792-4802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P, Royo H, Seitz H, Bachellerie JP, Huttenhofer A, Cavaille J. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 2003;31:6543–6551. doi: 10.1093/nar/gkg849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001a;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- Vulliamy TJ, Knight SW, Heiss NS, Smith OP, Poustka A, Dokal I, Mason PJ. Dyskeratosis congenita caused by a 3' deletion: germline and somatic mosaicism in a female carrier. Blood. 1999;94:1254–1260. [PubMed] [Google Scholar]
- Vulliamy TJ, Knight SW, Mason PJ, Dokal I. Very short telomeres in the peripheral blood of patients with X-linked and autosomal dyskeratosis congenita. Blood Cells Mol. Dis. 2001b;27:353–357. doi: 10.1006/bcmd.2001.0389. [DOI] [PubMed] [Google Scholar]
- Wang C, Meier UT. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Query CC, Meier UT. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol. Cell. Biol. 2002;22:8457–8466. doi: 10.1128/MCB.22.24.8457-8466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Gray MW. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res. 2000;28:2342–2352. doi: 10.1093/nar/28.12.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA- binding protein, are present together with Gar1p in all box H/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- Whitehead SE, Jones KW, Zhang X, Cheng X, Terns RM, Terns MP. Determinants of the interaction of the spinal muscular atrophy disease protein SMN with the dimethylarginine-modified box H/ACA small nucleolar ribonucleoprotein GAR1. J. Biol. Chem. 2002;277:48087–48093. doi: 10.1074/jbc.M204551200. [DOI] [PubMed] [Google Scholar]
- Willig TN, Draptchinskaia N, Dianzani I, Ball S, Niemeyer C, Ramenghi U, Orfali K, Gustavsson P, Garelli E, Brusco A, Tiemann C, Perignon JL, Bouchier C, Cicchiello L, Dahl N, Mohandas N, Tchernia G. Mutations in ribosomal protein S19 gene and diamond blackfan anemia: wide variations in phenotypic expression. Blood. 1999;94:4294–4306. [PubMed] [Google Scholar]
- Wong JM, Kyasa MJ, Hutchins L, Collins K. Telomerase RNA deficiency in peripheral blood mononuclear cells in X-linked dyskeratosis congenita. Hum. Genet. 2004;115:448–455. doi: 10.1007/s00439-004-1178-7. [DOI] [PubMed] [Google Scholar]
- Yang PK, Rotondo G, Porras T, Legrain P, Chanfreau G. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 2002;277:45235–45242. doi: 10.1074/jbc.M207669200. [DOI] [PubMed] [Google Scholar]
- Yang Y, Isaac C, Wang C, Dragon F, Pogacic V, Meier UT. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Biol. Cell. 2000;11:567–577. doi: 10.1091/mbc.11.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H, Gharibyan V, Qatanani M. Analysis of epitope-tagged forms of the dyskeratosis congenital protein (dyskerin): identification of a nuclear localization signal. Blood Cells Mol. Dis. 1999;25:305–309. doi: 10.1006/bcmd.1999.0258. [DOI] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li ZH, Terns RM, Terns MP, Yu YT. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. RNA. 2002;8:1515–1525. [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu YT. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA. 2004;10:681–690. doi: 10.1261/rna.5159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsser F. Atrophia cutis reticularis com pigmentatione, dystrophia et leukoplakia oris. Ikonogr. Dermatol. 1910;5:219–223. [Google Scholar]