Abstract
Aebp2 encodes an evolutionarily conserved zinc finger protein that has not been well studied so far, yet recent studies indicated that this gene is closely associated with the Polycomb Repressive Complex 2 (PRC2). Thus, the current study characterized the basic aspects of this gene, including alternative promoters and protein isoforms. According to the results, Aebp2 is controlled through three alternative promoters, deriving three different transcripts encoding the embryonic (32 kDa) and somatic (52 kDa) forms. Chromatin Immuno-Precipitation (ChIP) experiments revealed that AEBP2 binds to its own promoter as well as the promoters of Jarid2 and Snai2. While the embryonic form acts as a transcriptional repressor for Snai2, the somatic form functions as a transcriptional activator for Jarid2, Aebp2 and Snai2. Cell migration assays also demonstrated that the Aebp2 somatic form has an enhancing activity in cell migration. This is consistent with the functional association of Aebp2 with migratory neural crest cells. These results suggest that the two protein isoforms of AEBP2 may have opposite functions for the PcG target genes, and may play significant roles in cell migration during development.
Keywords: Aebp2, Polycomb Repressive Complex 2, cell migration, neural crest cells
Introduction
The Polycomb Repressive Complex 2 (PRC2) is an epigenetic modifier involved in defining and maintaining cell fate during the development of multicellular organisms. Ezh2 (Enhancer of zeste homolog 2), a histone methyltransferase, interacts with Eed (embryonic ectoderm development) and Suz12 (suppressor of zeste 12) to form PRC2, and adds di- and tri- methylation marks on the lysine 27 of histone H3 (H3K27me2/3) [1]. The modification mark, H3K27me3, is associated with global transcriptional repression of many developmental genes, such as homeotic genes [1-6]. Other co-factors also interact with the PRC2, including RbAp48 (Retinoblastoma-associated protein 48), Aebp2 (Adipocyte Enhancer Binding Protein 2) and Jarid2 [7]. Among these factors, Jarid2 and Aebp2 are known to bind DNA [8,9], and thus can serve as components responsible for the targeting of PRC2 [8]. However, whether Aebp2 or Jarid2 targets PRC2 has been controversial [10]. Recently, a 3D electron-microscopic model of PRC2 demonstrated that AEBP2 is a major allosteric modulator stabilizing the overall conformation of PRC2 [11].
AEBP2 was initially discovered as a DNA-binding repressor for the aP2 gene encoding a fatty-acid binding protein in adipocytes [12]. AEBP2 is composed of four major domains: acidic, neutral, zinc finger and basic domains [8]. In mammals, the zinc finger and basic domains of AEBP2 interact with the other proteins of PRC2 [9,11,13]. Consistent with mammalian PRC2, the fly homologue of Aebp2, Jing [14], also interacts with Jarid2 and the other factors of the PRC2 [15]. According to the results from the knock-in mouse model of Aebp2, this PcG gene is highly expressed within the neural crest cells (NCCs) of developing embryos [16]. Consistent with this, the mutant heterozygotes for Aebp2 tend to exhibit a set of phenotypes that are similar to neural crest cell defects observed in humans, such as megacolon, hearing defects and hypopigmentation [16]. This suggests a major role played by Aebp2 and thus by PRC2 in the development and migration processes of NCCs during mammalian development [16].
In this study, we performed several sets of experiments characterizing the fundamental aspects of Aebp2 in order to better understand its functional roles as a transcription factor in PRC2 and in neural crest cells. According to the results, the transcription of Aebp2 is regulated through three alternative promoters, subsequently producing two protein isoforms (52 and 32 kDa). The results also indicate that the embryonic form (32 kDa) is a transcriptional repressor, as seen in previous studies [12], whereas the somatic form (52 kDa) is a potent transcriptional activator. Furthermore, in vitro assays demonstrated that the somatic form has an ability to enhance cell migration, suggesting significant roles played by Aebp2 in neural crest cell migration.
Results
Alternative promoters and protein isoforms of Aebp2
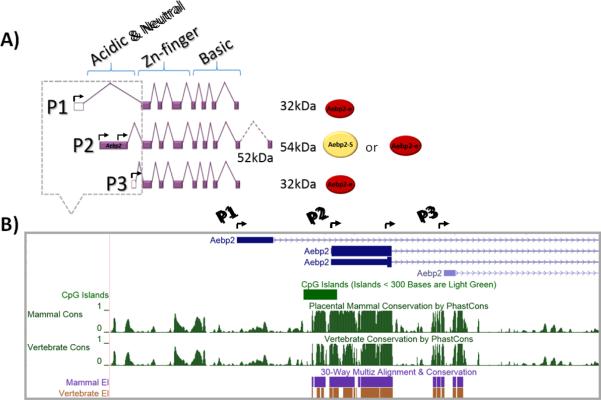
The 5’ genomic region of mouse Aebp2 was carefully examined using the UCSC genome browser to identify all the promoters responsible for its transcription. First, transcription start sites (TSSs) were identified through aligning all the cDNA sequences against the genomic sequence of the Aebp2 locus. Three different TSSs are found in the 5’-side of mouse Aebp2 (Fig. 1A). The genomic regions surrounding these TSSs are termed P1, P2 and P3 promoters of Aebp2. Second, these promoters (P1-3) were also examined in terms of their sequence composition and evolutionary conservation levels (Fig. 1B and Supplemental Material 7). The P1 promoter contains a 400-bp region displaying sequence similarity to rodent SINEs (Short Interspersed DNA Elements). The SINE-derived P1 sequence is also found in the homologous region of rat Aebp2. However, this sequence is limited to the rodent lineage only; it is not found in other mammals, such as humans and cows. The P2 promoter region, about 1.2 kb in length, shows typical features of CpG islands, such as high ratios of CG/AT and high frequencies of CpG dinucleotide ranging from 73.5-82.9% observed CpG site over expected CpG site per 200 nucleotides (Supplemental Material 7). The P2 promoter displays high levels of sequence conservation throughout all the vertebrates. Similar to the P2 promoter, the P3 promoter region also show some CpG richness (61-68.9% observed CpG site over expected per 200 nucleotides) but with a much shorter length, approximately 500 bp. The P3 promoter is conserved among all the placental mammals. According to the results from previous studies [8,16], two protein isoforms of AEBP2 exist in mammals: somatic (52 kDa) and embryonic (32 kDa) forms. As predicted, the transcripts driven by these three promoters have two ORFs (Open Reading Frames). The transcripts driven by both P1 and P3 promoters have one ORF, which corresponds to the embryonic form (32 kDa). On the other hand, the transcript from the P2 promoter harbors a longer ORF corresponding to the somatic form (52 kDa). In sum, the transcription of mouse Aebp2 appears to be driven by three alternative promoters showing different sequence compositions and evolutionary conservation levels.
Fig. 1. Alternative promoters and protein isoforms of mouse Aebp2.
A) The alternative transcripts driven by the three promoters (P1-3) of mouse Aebp2 are predicted to produce two protein isoforms. The untranslated and translated regions are marked with open and filled boxes, respectively. The 32 kDa protein (Aebp2-e) includes a zinc finger domain and a basic domain, while the 52 kDa protein (Aebp2-S) includes an additional acidic and neutral domain along with the zinc finger and basic domains. The transcription start sties (TSSs) are marked with arrows. B) The genomic structure of the three promoters of mouse Aebp2. The relative positions of the three promoters are presented along with their sequence conservation levels among different vertebrates. The P1 promoter is rodent-specific whereas the P2 promoter is conserved throughout all vertebrates. The P3 promoter is also conserved among placental mammals.
Expression levels of Aebp2 transcripts in adult tissues
To further characterize the identified promoters of Aebp2, the expression levels and patterns of the transcripts driven by these promoters were analyzed using qRT-PCR. First, total RNA isolated from the heart, kidney, lung, and thymus of one-month-old mice were used for measuring the expression levels of the three alternative transcripts (Fig. 2). The relative expression levels were derived after normalization with β-actin as an internal control. Then, the average ΔΔCt values of P1, P2, and P3 transcripts were calculated from each tissue, and then compared among the individual organs. As shown in Figure 2, the expression levels of the P2 transcript were similar between the different organs. On the other hand, the expression levels of P1 and P3 transcripts were much higher in the lung than those observed in the heart, kidney and thymus (Fig. 2). This indicates ubiquitous expression of the P2 transcript whereas somewhat higher expression levels of P1 and P3 transcripts in the lung. In a given tissue, the expression level of one transcript was also compared with those from the two remaining transcripts, which is evident through different values on the Y axis in Figure 2. Within each tested organ, the relative expression levels of the P2 transcript were the highest, followed by the P1 and P3 transcripts. The expression levels of the P1 and P3 transcripts are approximately 10-100 fold lower than those of the P2 transcript in most tissues. This indicates that the majority of the Aebp2 transcripts in adult organs are derived from the P2 promoter.
Fig. 2. Expression levels of the three alternative transcripts of Aebp2 in adult mouse organs.
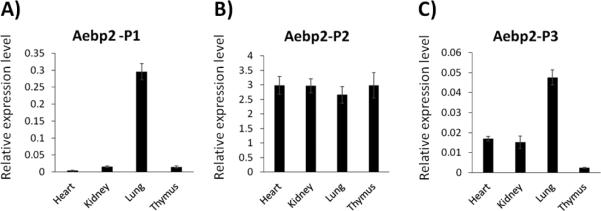
A series of qRT-PCR analyses were performed to measure the relative expression levels of the alternative transcripts of Aebp2 in the four major organs of adult mice. A) The P1 promoter-driven transcript of Aebp2 („P1 transcript’ hereafter) is expressed at the highest levels in the lung. B) The P2 transcript is expressed at the similar levels between the heart, kidney, lung and thymus. C) The P3 transcript is expressed also at the highest in the lung.
AEBP2 binding to active and repressive promoter regions
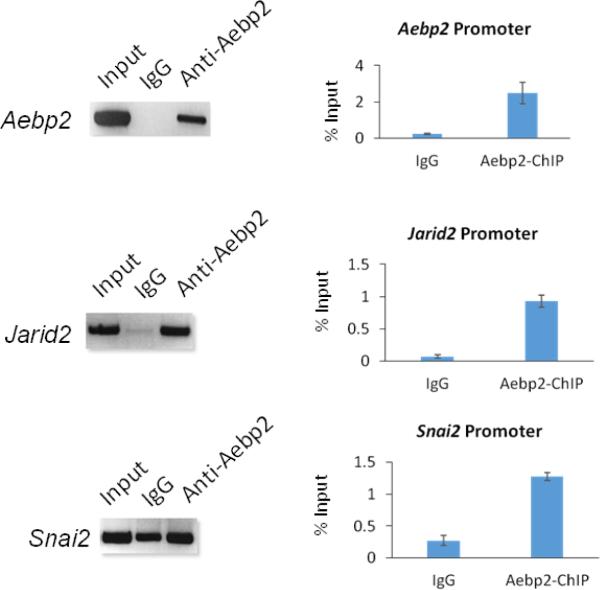
Chromatin Immuno-Precipitation (ChIP) experiments were performed to identify the downstream genes of Aebp2 using a similar scheme as previously described [8]. In brief, this modified scheme includes a restriction enzyme digestion step with 4-bp cutters, such as Tsp509I and AluI, to shorten the length of ChIP DNA before the following elution step. Eluted DNA was subsequently sequenced using a NGS platform. The antibody used for ChIP assay was targeted to recognize the somatic form of AEBP2, but due to the homologous zinc finger and basic domains in the two protein isoforms, both embryonic and somatic forms were detected with this antibody (Supplemental Material 2). This series of ChIP-Seq experiments using the thymus of one-month-old mice derived a relatively small set of potential target loci of AEBP2 (134 loci neighboring 218 genes, Supplemental Material 1). Most AEBP2 bound regions from the ChIP data are promoter regions that are enriched with transcriptionally active histone marks in the thymus (Supplemental Material 3 & 6). The DNA binding motif “CTT” was enriched in the DNA fragments derived from these ChIP peaks (Supplemental Material 4). Among these candidate AEBP2 binding regions, two genomic regions immediately stood out due to their functional connection to PRC2: the promoter regions of Jarid2 and Aebp2 itself (Supplemental Material 5). In addition, the promoter region of Snai2, a well-known target gene of the PRC2 in NCCs, also showed some levels of AEBP2 binding (data not shown). The AEBP2 binding to the promoters of Jarid2, Aebp2 and Snai2 were further verified by independent ChIP assays and quantified by qPCR. The results indeed confirmed that AEBP2 binds to these promoter regions (Fig. 3). Overall, this series of ChIP experiments identified the promoter regions of Jarid2, Aebp2 and Snai2 as the in vivo target loci of AEBP2. It is interesting to note that AEBP2 binds to its own promoter regions, suggesting that Aebp2 may be auto-regulated.
Fig. 3. AEBP2 binding to the promoter regions of Aebp2, Jarid2 and Snai2.
The potential binding of AEBP2 to the three promoter regions was confirmed through performing individual ChIP experiments using the chromatin prepared from the thymus of one-month-old adult mice. Each locus was analyzed first by PCR at a fixed number of 40 cycles using a set of templates derived from Input, IgG, anti-Aebp2 antibody (left panel). This was further analyzed with qPCR showing the enrichment levels relative to that of Input with error bars (right panel). The enrichment levels showed 10 fold (Aebp2), 13 fold (Jarid2), and 4 fold (Snai2) higher than those of IgG. This further confirmed the binding of AEBP2 to the promoter regions of Aebp2, Jarid2 and Snai2.
Transcriptional regulatory roles of the somatic and embryonic isoforms of AEBP2
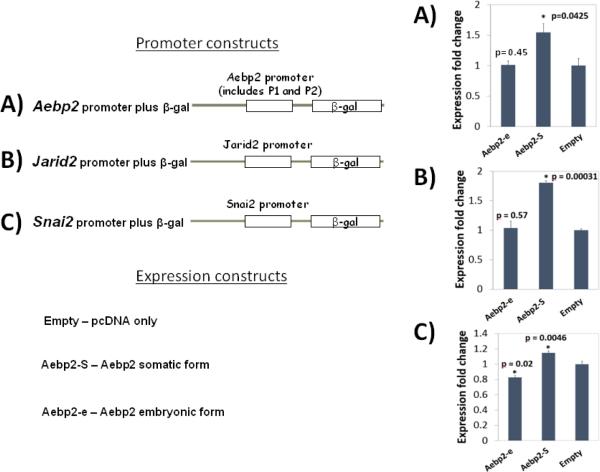
The functions of the somatic and embryonic forms of AEBP2 were tested through cotransfection experiments using the following two sets of expression and promoter constructs. Expression constructs include pcDNA-empty, AEBP2-somatic and AEBP2-embryonic. Promoter constructs include a set of promoterless β-Gal reporters containing the promoters of Jarid2, Aebp2 and Snai2. The co-transfection of these two sets to HEK293T cells was also accompanied with an internal control (pGL3-Luc) to monitor transfection efficiency. After transfection, proper expression of the somatic and embryonic forms was also confirmed through a series of western blotting (Supplemental Material 2). The three promoter constructs displayed relatively high levels of promoter strength in HEK293T cells based on the readily detectable β-Gal activity. Aebp2's promoter showed the highest reporter activity, followed by the Jarid2's and Snai2's promoters (Fig. 4). Overall, this series of co-transfection experiments derived the following conclusions. Co-transfection of the somatic form of AEBP2 resulted in up-regulated β-Gal expression in the promoter constructs of Aebp2 (1.53 fold; p=0.0425), Jarid2 (1.8 fold; p=0.00031), and Snai2 (1.1 fold; p=0.0046), suggesting that the somatic form functions as a transcriptional activator for all three promoters. In contrast, co-transfection of the embryonic form resulted in down-regulation of β-Gal expression in the promoter construct of Snai2 (0.83 fold; p=0.0046) but no change in the promoter constructs of Aebp2 and Jarid2 (1.06; p=0.45 and 0.98; p=0.57, respectively). This suggests that the embryonic form may be a transcriptional repressor for Snai2 in this cell line. Since the changes in the reporter gene expression is very marginal, it is possible that other factors may be needed to enhance the transcriptional regulatory activity. Nevertheless, each promoter assay was repeated three independent times, yet the overall pattern was consistent. Thus, the results from one of the three independent trials were presented in Figure 4 with the error bar representing variable promoter strength values in triplicates per assay. In sum, the somatic form of AEBP2 appears to be a transcriptional activator for Jarid2, Aebp2 and Snai2, whereas the embryonic form is likely a transcriptional repressor for Snai2.
Fig. 4. AEBP2 involvement in the transcriptional activity of Jarid2, Snai2 and Aebp2.
The expression vectors producing the somatic (Aebp2-S) and embryonic (Aebp2-e) form of AEBP2 were co-transfected to HEK293T cells along with the β-Gal reporter constructs containing the promoters of Jarid2 (A), Aebp2 (B) and Snai2 (C). The somatic form (Aebp2-S) increased the transcriptional activity of all three promoters, indicating that the somatic form functions as an activator. In contrast, the embryonic form (Aebp2-e) decreased the transcriptional activity of the promoters of Snai2, but not Aebp2 and Jarid2. This indicates that the embryonic form may be a locus-specific repressor.
Aebp2 functional involvement in cell migration
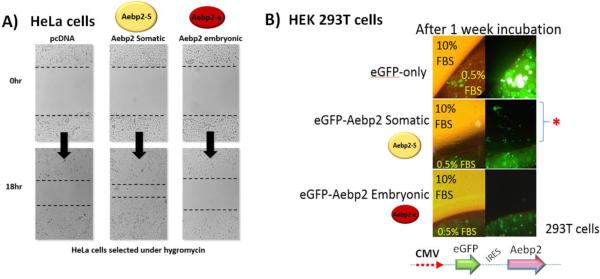
The known functions of both Aebp2 and Jing, a homologue in fly, are closely associated with migratory cells, suggesting that Aebp2 might be involved in cell migration. This is further supported by potential control of Aebp2 over Snai2, a master gene for EMT (Epithelial-to-Mesenchymal Transition) and cell migration [17]. Thus, we performed two series of cell migration assays to test this possibility. First, scratch assays were performed using HeLa cells that had been stably transfected with a set of the expression constructs containing: Aebp2-somatic, Aebp2-embryonic and pcDNA-empty constructs (Fig. 5A). The cells were plated to 100% confluence under hygromycin selection, and were treated with serum starvation to be synchronized to G1 phase. After a scratch was made with a pipet tip, the rate of cell migration was measured as a wound-healing process. The cells with the Aebp2-somatic construct showed 2-fold accelerated wound closure as compared to those with the Aebp2-embryonic or pcDNA-empty constructs. This demonstrated that the somatic form of AEBP2 has an enhancing activity in cell migration. Second, this observation was further tested through another independent method, agarose droplet assay with HEK293T cells (Fig. 5B). To visualize cell migration, each of the somatic and embryonic forms of AEBP2 was co-expressed along with an eGFP cassette as a bicistronic transcript with IRES (internal ribosomal entry site). Proper expression of protein products was also confirmed through western blotting (Supplemental Material 2). Inspection of the cells that had been incubated for one week derived the following observations. The HEK293T cells expressing the somatic form migrated out of the agarose droplets whereas the other cells with the pcDNA-empty or the Aebp2 embryonic form remained trapped in the agarose droplets. Thus, this set of migration assays again confirmed a similar conclusion that the somatic form has an enhancing effect on cell migration. Taken together, the results from both migration assays demonstrated that the somatic form of AEBP2 has a capability to enhance cell migration, further supporting potential roles of Aebp2 in cell migration.
Fig. 5. AEBP2 involvement in cell migration.
A) Scratch assay. HeLa cells were individually transfected with the following constructs: pcDNA empty, Aebp2 somatic form (Aebp2-S), and Aebp2 embryonic form (Aebp2-e). After a scratch was formed on a layer of confluent cells using p10 pipet tips, the migration of the cells was monitored. The HeLa cells expressing the somatic form showed a two-fold increase in cell migration compared to the cells with the pcDNA empty vector 18 hours after the scratch formation. However, the cells with the embryonic form did not show any difference compared to the cells with the pcDNA empty vector. B) Agarose droplet assay. The HEK 293T cells transfected individually with pcDNA-eGFP, Aebp2 somatic-eGFP and Aebp2 embryonic-eGFP were trapped in the 0.25% agarose droplets containing 0.5% FBS. After one-week incubation, the cells with Aebp2 somatic-eGFP migrated out of the agarose droplets, confirming the enhancing effect of the somatic form on cell migration. However, the cells with both pcDNA-eGFP and Aebp2 embryonic-eGFP did not migrate out of the droplets.
Discussion
In this study, we characterized in detail the basic aspects of mouse Aebp2 with various experimental approaches. According to the results, the Aebp2 locus is controlled through three different promoters, and subsequently derives three alternative transcripts. These transcripts are responsible for producing the somatic and embryonic forms of the AEBP2 protein. ChIP experiments revealed that AEBP2 binds to its own promoter as well as the promoters of Jarid2 and Snai2. A series of co-transfection experiments further demonstrated that the somatic form of AEBP2 is a transcriptional activator for Jarid2, Aebp2 and Snai2 while the embryonic form is a repressor for Snai2. Two sets of cell migration assays also confirmed potential roles of the somatic form of AEBP2 in cell migration, consistent with the close association of Aebp2 with migratory neural crest cells.
The mouse Aebp2 locus is regulated through three alternative promoters according to the current study (Fig. 1). The P2 promoter is predicted to be the main one responsible for the two protein isoforms, the 52 kDa somatic and 32 kDa embryonic forms, while the P1 and P3 promoters may contribute to generation of the embryonic form. The CpG composition of P1, P2, and P3 are 57%, 80%, and 65% respectively (Supplemental Material 7). Based on our data (Fig. 2) and the EST database, the P2 promoter with the highest CpG content most likely functions as a housekeeping gene, while P1 and P3 may function tissue/developmental specifically. The DNA methylation profile of P2 and P3 promoters are unmethylated in most tissues, while P1 shows variable DNA methylation levels ranging from 0-80 % in germ cells and somatic cells (Kim et al., unpublished). P3 promoter is more evolutionarily conserved than the P1 promoter, thus is more likely to be involved in embryonic development. This evolutionary conservation agrees well with the fundamental roles played by Aebp2 in the various aspects of vertebrate biology. In contrast to P2 and P3 promoters, the P1 promoter appears to be very lineage-specific without any evolutionary conservation. In mice, the 5’- and 3’-side sequences of the P1 promoter are similar to B1 and B2 SINE elements, respectively, suggesting that these opportunistic retrotransposons might have adapted as a promoter for the Aebp2 locus during rodent evolution. Interestingly, a similar situation has also occurred independently in the primate lineage. In humans, another retrotransposon, L1, is located upstream of the P2 and P3 promoters, and derives the transcription of AEBP2 as a separate promoter [18]. The relatively recent origin of these elements during mammalian evolution suggests that the functional roles of the P1 promoter are likely related to some unknown lineage-specific aspects of Aebp2. Also, its relative functional contribution to the Aebp2 locus might be minor compared to those of the P2 and P3 promoters. The recent evolutionary origin of the P1 promoter is overall intriguing but enigmatic at the same time, and may require further investigation in the future. In summary, the Aebp2 locus is controlled through three alternative promoters, and the two promoters, P2 and P3, are likely responsible for the production of the somatic and embryonic forms of AEBP2 protein, respectively.
According to the results, the Aebp2 locus produces two protein isoforms with opposing functions: the somatic form as an activator and the embryonic form as a repressor (Fig. 4). First, the amino acid sequences of the additional acidic domain in the Aebp2 somatic form contain consecutive glutamate (EEE) and aspartate (DD) residues. These types of domains have been described in other proteins as “acid blobs” or “negative noodles” [19] and were characterized to have activation functions in GCN4 and GAL4 proteins [20-21]. The repressor function of AEBP2 has been previously demonstrated multiple times through in vitro biochemical experiments and also through its genetic association with the PRC2 [6,9,13,15]. In contrast, the activator function by the somatic form has not been noticed previously, although there have been several hints for this function for Aebp2. Second, the heterozygous mutants for mouse Aebp2 have been shown to further repress its downstream genes, suggesting an activator role for Aebp2 [16]. Similar observations have also been made for Jarid2. In the case of Jarid2, mutations have been shown to derive both de-repression and further repression of PcG target genes. Thus, it has been proposed that Jarid2 may function as a „molecular rheostat’ in which the protein levels of JARID2 can inhibit or enhance the histone-modifying activity of the PRC2 [9,15].
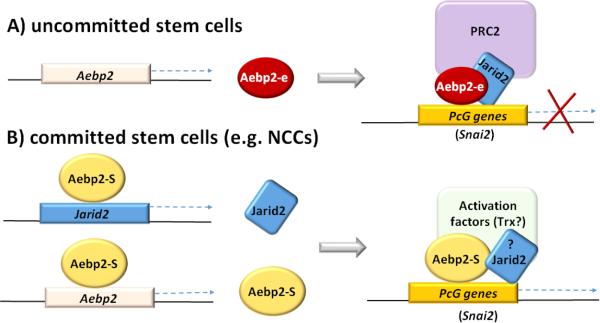
However, it is currently unknown what controls the protein levels of JARID2. According to the results from the ChIP assay (Fig. 3), the promoters of Jarid2 and Aebp2 itself appear to be the targets of the AEBP2 protein. Furthermore, the somatic form of AEBP2 functions as an activator for the transcription of both loci (Fig. 4). Given these lines of evidence, the conflicting observations associated with JARID2 and the PRC2 may be explained through the two protein isoforms of AEBP2 (Fig. 6). In uncommitted ES cells, the embryonic form of AEBP2 together with JARID2 recruit the PRC2 to PcG target genes, resulting in global repression of developmental regulators, such as Snai2. In committed lineage-specific stem cells, the somatic form increases the amount of itself as well as JARID2 to form a potential activation complex, resulting in the activation of some of the PcG target genes. One good example would be the activated Snai2 in neural crest cells. In this model, the somatic and embryonic form of AEBP2 share the same set of target genes, and thus compete for binding to these target genes (Fig. 6). In this case, the relative protein levels of the embryonic to somatic forms would be a key factor deciding the functional consequence of AEBP2 binding to a given gene. This might be particularly true during the transition period from the uncommitted to committed state of stem cells. Overall, we believe that this model provides a plausible explanation for some of the conflicting observations associated with Aebp2 and Jarid2. Nevertheless, this model also needs to be further refined with additional data. In particular, one of the key data would be the identity and composition of a predicted activation complex associated with the somatic form of AEBP2.
Fig. 6. A model for the repressor and activator roles of AEBP2.
A) A repressor role through the PRC2 in uncommitted stem cells. The embryonic from of AEBP2 and JARID2 are responsible for recruiting the PCR2 to PcG genes for their temporary repression in uncommitted stem cells. B) An activator role through a hypothetical activation complex in committed stem cells. The somatic form activates the transcription of itself and Jarid2 to form an activator complex, which competes and eventually replaces the PRC2 to turn on some of the temporary repressed PcG genes in committed stem cell lineages.
Aebp2 appears to play a very unique role at the cellular level, enhancing the migration capability of cells (Fig. 5). This unique role of Aebp2 is consistent with the other observations described below. First, the fly homologue of Aebp2, Jing, was initially discovered due to its involvement in cell migration. A loss-of-function mutation on Jing caused a deficiency in movement in the border cells in eggs, which was the basis for the name „Jing’ (meaning stillness in Chinese). The border cell migration also prerequisites the transition process from epithelial to mesenchymal cells, thus indicating Jing's involvement in this EMT process [14]. One of key genes in this process is Snai2 in both flies and vertebrates, yet this gene turns out to be a major target of AEBP2 according to the results from ChIP experiments (Fig. 3). In fact, the somatic form of AEBP2, the one involved in cell migration, was shown to activate the transcription of Snai2, hinting at a potential regulatory network connecting Aebp2 to Snai2 for the EMT process. Also, a two fold up-regulation of endogenous Snai2 gene expression was seen in AEBP2 somatic form overexpressing cell lines (Supplemental Material 8). Second, Aebp2 displays very unique expression patterns during embryogenesis: high levels of expression in neural crest cells (NCCs) [16]. Vertebrates’ NCCs are another cell population that goes through a similar set of processes as shown in the border cells in flies, such as EMT and migration processes [22]. The functional involvement of Aebp2 in NCCs has been indeed demonstrated through the phenotypes of its mutant mouse model, displaying various cell migration defects [16]. The cell migration defects observed in vivo at the organismal level are overall in agreement with the fact that the somatic form of AEBP2 has an enhancing activity in cell migration. According to the results from that mutant model, both the somatic and embryonic forms are disrupted, yet many NCC genes were further down-regulated. This suggests defects in the activator function of AEBP2. Therefore, the somatic form is most likely the one regulating the transcription of NCC genes in that migratory cell population, which is again consistent with the observed migratory role played by the somatic from. Conditionally deleting the AEBP2 somatic form in epithelial cells undergoing mesenchymal transition, would further demonstrate the function of this protein isoform in EMT during neural crest cell development. In summary, this series of independent observations provide a testable model for the mechanisms by which Aebp2 regulates the migration and development process of NCCs. In that regard, dissecting the exact functions of the somatic and embryonic forms of AEBP2 through conditional mutagenesis experiments would be a very exciting research direction in the near future.
Materials and Methods
cDNA synthesis and qRT-PCR
Heart, kidney, lung, and thymus were harvested from one-month-old mice, and snap-frozen with liquid nitrogen. Total RNA were first isolated from these tissues with Trizol (Invitrogen), and subsequently used for the synthesis of cDNA according to manufacturer's protocol (MMLV reverse-transcriptase kit, Invitrogen). Quantitative real-time PCR was performed using iQ SYBR green supermix with the iCycler iQTM multicolor real-time detection system (Bio-Rad). All quantitative RT-PCRs (qRT-PCRs) were performed at 60°C annealing temperature with standard conditions for 40 cycles. Relative expression values of each gene were normalized by the Ct (threshold cycle) values of an internal control, β-actin. All expression values from qRTPCR were analyzed according to their ΔΔCt values [23].
ChIP (Chromatin Immuno-Precipitation) experiments
Thymus tissues of one-month-old C57BL/6N mice (Taconic) were used for ChIP experiments as previously described [8]. This particular protocol has one additional step compared to other existing ChIP protocols. The two four base pair (bp) cutter enzymes, AluI and Tsp509, were used to shorten the average length of ChIP DNA right before the elution step from the AEBP2 antibody. The eluted DNA was further processed for the construction of a library according to the manufacturer's protocol (NEBNext Fast DNA Library Prep Set for Ion Torrent, NEB cat. No. E6270S). The constructed library was sequenced using a NGS sequencer (Ion Torrent PGM, Life Technologies). This series of ChIP-Seq experiments were also repeated using an input DNA as a negative control. The sequence reads from both libraries were mapped with Bowtie2 [24], and the mapped reads were subsequently used for generating peaks with MACS2 [25]. The Aebp2 ChIP sequenced data is available in GEO under accession number GSE62680. Several candidates loci predicted through peak calling were further confirmed through three independent ChIP experiments. This series of qPCR-based ChIP assays were also performed using iQ SYBR green supermix with the iCycler iQTM multicolor real-time detection system (Bio-Rad). All qPCRs conditions were same as described for cDNA qRT-PCR except that each ChIP DNA was normalized by the Ct value of input DNA.
Construction of expression and promoter assay vectors
The promoter regions of Jarid2 (mm9, chr13:44826128-44827446), Snai2 (mm9, chr16: 14705572-14706495) and Aebp2 (mm9, chr6:140570728-140573129) were amplified with PCR (iStar Master mix, Intron). Each promoter region was individually cloned into the NotI site of the promoterless β-Geo expression vector [26]. The two ORFs (Open Reading Frames) corresponding to the isoforms of AEBP2 were also individually cloned into the NotI site of pcDNA (-) 3.1 hygro (Invitrogen): the somatic form (GenBank accession no. NM_001005605.1, NP_001005605) and the embryonic form (GenBank accession no. NM_178803, NP_848918.1). After cloning, all constructs were sequenced to verify their orientation and integrity. The following primers used for cloning: Jarid2-promoter-F/-R, 5’-AGCCATTTTGTAGTCAAGGGAC-3’ and 5’-ACTAGGCAGACACGACTTTGC-3’; Snai2-promoter-F/-R, 5’-CCAAATATAGACTCTCTGGCCAC-3’ and 5-TTCTAGCTGTACCGTGCCTGT-3’; Aebp2-promoter-E-F/-S-R, 5’-TCCCTTCTAGCCTCATACTACAT-3’ and 5’-GGAATCTACAGAGCAAGGGATC-3’; Aebp2 somatic form-F 5’-ATGGCCGCCGCGCTCGCCGACATG-3’ Aebp2 somatic form-R 5’-ATTGCAAATGTCGTTCACTGTTTGCT-3’ and Aebp2 embryonic form-F 5’-ATGGACATAGACAGCACAATTTCCAG-3’; Aebp2 embryonic form-R 5’- ATTGCAAATGTCGTTCACTGTTTGCT-3’.
Promoter assay
HEK 293T cells (1×106) were transfected with a series of the promoterless β-Geo vectors containing individual promoters (2 μg) along with the pcDNA vectors expressing the embryonic and somatic forms of AEBP2 (500 ng) using Lipofectamine 2000 (Invitrogen). All cells were grown in 10% Fetal Bovine Serum in glutamax DMEM (Gibco BRL) with 1% antibiotic-antimycotic (Gibco BRL) in 5% CO2 humidified incubator at 37°C. The transfected cells were harvested 48 hours post-transfection, and used for measuring the β-Galactosidase activity at 405 nm with Wallac 1420 multilable counter VICTOR (PerkinElmer). Transfection efficiency of each well was also monitored by measuring the luciferase activity that had been derived from the co-transfected pGL3-Luc (Promega). Luminescence was measured with Wallac 1420 multilabel counter VICTOR (Perkin Elmer).
Scratch assay
HeLa cells were transfected with the linearized pcDNA constructs expressing Aebp2. The transfected cells were selected under hygromycin (300 μg per 12 well) for two weeks. These cells were re-plated to 100% confluence in 6 well plates. Scratch assay was performed as described by Valster et al [27] with some modifications. In brief, after serum starvation treatment (0.5% FBS) for 18 hours, a scratch was made on the 100% confluent cells with a p10 pipet tip. The migrating cells were observed every two hours, and picture images were captured using the Lycia DM2500 microscope. All scratch assays were repeated at least three independent times.
Agarose droplet assay
The overall procedure was performed as described by Varani et al with some modifications [25]. In brief, HEK293T cells were transfected with the pcDNA constructs expressing both AEBP2 and eGFP simultaneously as a bicistronic transcript through an IRES (internal ribosomal entry site). The cells were first trypsinized 24 hours post-transfection, and mixed with agarose to a final concentration of 0.25% agarose in 0.5% FBS. The melted agarose droplets (50 μl) containing the transfected cells were placed onto a petri dish and left at room temperature for 10 minutes to be solidified. Later, 5 ml of DMEM containing 10% FBS was added to the petri dish to submerge the solidified droplets. The cells were examined for one week to check for their migration out of the agarose droplets.
Supplementary Material
Highlights.
Aebp2 has three alternative promoters responsible for two protein isoforms.
AEBP2 functions as a transcriptional activator or repressor.
The somatic form of AEBP2 is involved in cell migration.
Acknowledgements
We like to thank the members of JooKim Lab, Wesley Frey and Suman Lee for discussion and reading the manuscript. This work was supported by the National Institute of Health (R01GM066225 and R01GM097074).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatic structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 3.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 4.Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by Polycomb and Trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz YB, Pirrotta V. A new world of Polycomb: unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14:853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 8.Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009;37:2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji Coordinates Control of PRC2 Enzymatic Activity and Target Gene Occupancy in Pluripotent Cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Kim H. Recruitment and biological consequences of histone modification of H3K27me3 and H3K9me3. ILAR J. 2012;53:232–239. doi: 10.1093/ilar.53.3-4.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E. Molecular architecture of human polycomb repressive complex 2. eLife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He GP, Kim S, Ro HS. Cloning and characterizing of a novel zinc finger transcriptional repressor. J Biol Chem. 1999;274:14678–14684. doi: 10.1074/jbc.274.21.14678. [DOI] [PubMed] [Google Scholar]
- 13.Cao R, Zhang Y. Suz12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Montell DJ. Jing: a downstream target of slbo required for developmental control of border cell migration. Development. 2001;128:321–330. doi: 10.1242/dev.128.3.321. [DOI] [PubMed] [Google Scholar]
- 15.Herz HM, Mohan M, Garrett AS, Miller C, Casto D, Zhang Y, Seidel C, Haug JS, Florens L, Washburn MP, Yamaguchi M, Shiekhattar R, Shilatifard A. Polycomb Repressive Complex 2-Dependent and –Independent Functions of Jarid2 in Transcriptional Regulation in Drosophila. Mol Cell Biol. 2012;32:1683–1693. doi: 10.1128/MCB.06503-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H, Kang K, Ekram MB, Roh TY, Kim J. Aebp2 as an epigenetic regulator for neural crest cells. PLoS One. 2011;6:e25174. doi: 10.1371/journal.pone.0025174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 18.Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 19.Sigler PB. Acid blobs and negative noodles. Nature. 1988;333:210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- 20.Hope IA, Struhl K. Functional dissection of a eukaryotic transcriptional activator, GCN4 of yeast. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 21.Hahn Structure(?) and Function of Acidic Transcription Activators. Cell. 1993;72:481–483. doi: 10.1016/0092-8674(93)90064-w. [DOI] [PubMed] [Google Scholar]
- 22.Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 23.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myosytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 24.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JD, Hinz AK, Bergmann A, Huang JM, Ovcharenko I, Stubbs L, Kim J. Identification of clustered YY1 binding sites in imprinting control region. Genome Research. 2006;16:901–911. doi: 10.1101/gr.5091406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valster A, Tran NL, Nakada M, Berens ME, Chan AY, Symons M. Cell migration and invasion assays. Methods. 2005;37:208–215. doi: 10.1016/j.ymeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Varani J, Orr W, Ward PA. A Comparison of the Migration Patterns of Normal and Malignant Cells in Two Assay Systems. American Journal of Pathology. 1977;90:159–172. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.