Abstract
Fanconi anemia (FA) represents a paradigm of rare genetic diseases, where the quest for cause and cure has led to seminal discoveries in cancer biology. Although a total of 16 FA genes have been identified thus far, the biochemical function of many of the FA proteins remains to be elucidated. FA is rare, yet the fact that 5 FA genes are in fact familial breast cancer genes and FA gene mutations are found frequently in sporadic cancers suggest wider applicability in hematopoiesis and oncology. Establishing the interaction network involving the FA proteins and their associated partners has revealed an intersection of FA with several DNA repair pathways, including homologous recombination, DNA mismatch repair, nucleotide excision repair, and translesion DNA synthesis. Importantly, recent studies have shown a major involvement of the FA pathway in the tolerance of reactive aldehydes. Moreover, despite improved outcomes in stem cell transplantation in the treatment of FA, many challenges remain in patient care.
Introduction
For many years, Fanconi anemia (FA) was merely acknowledged as a clinical rarity whose biological significance was not appreciated. It was understood that FA was a genetic disease of bone marrow failure, hypersensitivity to cross-linking agents, and high risk of acute myeloid leukemia. Over the last several decades, care for patients with FA has drastically improved with the advent of better blood banking, greater success in stem cell transplant, and increased recognition of the scope of care needed for FA patients. However, only with the identification of the first FA gene, FANCC, 22 years ago did the science begin to catch up with the descriptive studies of how patients and cells derived from these patients behaved.
Now, with the characterization of 16 complementation group genes, including 5 familial breast cancer genes, a better sense of the biology of FA has emerged. First, the pathway is composed of an upstream grouping of 8 proteins termed the core complex, whose primary activity appears to be an E3 ubiquitin ligase executed through one of its member subunits, FANCL. Its primary target is a heterodimer, FANCI-FANCD2 (ID2), which both become monoubiquitinated on DNA damage or during S phase of the cell cycle. Downstream members of the FA pathway are then activated in a way that is much more complex, some entailing physical interactions with FANCD2 but many resulting in coordination of direct DNA repair pathways that have a different web like character rather than the more linear pathway converging on ID2. Although classically termed a disease of interstrand cross-link hypersensitivity, FA is clearly more than that, requiring the coordination of translesion synthesis, homologous recombination, and mismatch repair in an effort to solve the Gordian knot of DNA lesion and replication fork.
In this review, we will lay out the clinical picture of FA along with the current state of care for FA patients. We will also detail the complex web of how the FA pathway orchestrates the DNA damage response to repair DNA lesions.
Clinical features of FA
Studies of FA have led to insights into bone marrow failure syndromes and common cancers. However, FA still holds many mysteries as to how mutations in the FA proteins contribute to the pathophysiology of birth defects, bone marrow failure, and cancer.1-3
Most FA patients ultimately have bone marrow failure, with ∼90% of patients exhibiting this as their first hematopoietic presentation of disease. Classically, FA patients present with congenital defects, such as malformed or absent thumbs, absent radii, short stature, and microcephaly, and subtle but abnormal facies. A much longer list of less familiar and nonspecific characteristics may be present in patients (Table 1). Strikingly, a significant percentage of all FA patients, up to a third, exhibit none of these features, which is why hematologists, especially pediatric, routinely test for FA in bone marrow failure patients despite the lack of physical findings. The literature is therefore rife with adult patients diagnosed with FA when being treated for head and neck cancer and who exhibit inordinate toxicity as a result.4,5
Table 1.
Physical abnormalities in FA patients
| Physical abnormality | Percent of FA patients |
|---|---|
| Skin discolorations (café au lait) | 55 |
| Hand, arm, and other skeletal abnormalities, including thumb (missing thumb/radius) | 51 |
| Abnormal reproductive organs (hypogenitalia, micropenis) | 35 |
| Microcephaly or microophthalmia | 26 |
| Kidney problems | 21 |
| Low birth weight | 11 |
| Heart defects | 6 |
| Gastrointestinal problems (bowel) (atresia, imperforate anus) | 5 |
Data from Alter.125
When a potential FA patient presents with evidence of a hematopoietic production defect, a bone marrow aspiration and biopsy is in order to confirm bone marrow failure. Typically, a chromosomal breakage assay is also performed using peripheral blood lymphocytes with metaphase analysis in the presence of the DNA cross-linking agent diepoxybutane or mitomycin C (MMC).4,5 FA cells would demonstrate increased breakage and radial formation in the absence and presence of drug. FA diagnosis is aided greatly by sequencing analysis using an algorithm based on the relative unequal distribution of genetic subtypes (Table 2). FA cells are prone to somatic mosaicism as a result of genetic reversion, in which formerly mutant cells acquire additional mutations, resulting in diepoxybutane resistance, most likely as a result of selective pressure in the bone marrow. Such cases along with clinical suspicion necessitate additional testing of fibroblasts obtained through skin biopsy.6,7
Table 2.
Fanconi anemia genes and proteins
| Group | Gene | Chromosome | MW (kDa) | Motifs | Percent of FA patients | Necessary for FANCD2-ub |
|---|---|---|---|---|---|---|
| A | FANCA | 16q24.3 | 163 | 2 NLSs, 5 NESs | 60-70 | Yes |
| B | FANCB | Xp22.31 | 95 | NLS | 2 | Yes |
| C | FANCC | 9q22.3 | 63 | None | 14 | Yes |
| D1 | FANCD1/BRCA2 | 13q12.13 | 380 | 8 BRC repeats, HD, 3 OBs, TD | 3 | No |
| D2 | FANCD2 | 3p25.3 | 155, 162 | None | 3 | Yes |
| E | FANCE | 6p21-22 | 60 | 2 NLSs | 3 | Yes |
| F | FANCF | 11p15 | 42 | None | 2 | Yes |
| G | FANCG/XRCC9 | 9p13 | 68 | 7 TPRs | 10 | Yes |
| I | FANCI/KIAA1794 | 15q25-26 | 146 | None | 1 | Yes |
| J | FANCJ/BRIP1/BACH1 | 17q22-24 | 130 | ATPase, 7 helicase motifs | 2 | No |
| L | FANCL/PHF9 | 2p16.1 | 43 | 3 WD40s, PHD | 0.2 | Yes |
| M | FANCM | 14q21.3 | 250 | 7 helicase motifs, degenerate endonuclease domain, ATPase | 0.2 | Yes |
| N | FANCN/PALB2 | 16p12 | 130 | 2 WD40s | 0.7 | No |
| O | FANCO/RAD51C | 17q25.1 | 42 | RAD51 paralog/recombinase | 0.2 | No |
| P | FANCP/SLX4 | 16p13.3 | 200 | Endonuclease assembly | 0.2 | No |
| Q | FANCQ/ERCC4/XPF | 16p13.12 | 101 | Endonuclease | 0.5-1.0 | No |
HD, helical domain; MW, molecular weight; NES, nuclear export signal; NLS, nuclear localization sequence; OB, oligonucleotide binding; PHD, plant homeo domain; TD, tower domain; TPR, tetratricopeptide repeat.
The vast majority of FA patients present in childhood when hematopoietic disease, be it bone marrow failure or acute myeloid leukemia, predominates. The key treatment of such patients has been stem cell transplantation (SCT).8 Because of the difficult nature of acute myelogenous leukemia therapy, especially in the wake of DNA damage hypersensitivity in FA, the best outcome results from institution of SCT prior to evolution of malignancy or neutropenia-associated infections, including bacterial and fungal. Conversely, application of SCT in the first decade is associated with significant morbidity. Nonetheless, aggressive use of SCT has resulted in an increase in survival of FA patients.9 Use of matched unrelated donors, long associated with increased risk of graft-versus-host disease, has seen marked improvement in recent years even in vulnerable populations such as FA patients. Concomitant use of milder conditioning regimens has lowered toxicity dramatically, thus ameliorating the inherent DNA damage hypersensitivity. Institution of drugs such as fludaribine has lowered the risk of graft failure, which historically has been problematic with a prevalence of up to 10%.10,11 As a result of these general improvements, outcomes for matched related donor transplants are often >80%, whereas those for matched unrelated cases are steadily improving. Late effects of SCT, namely growth delay, endocrinopathies, and second cancers, are magnified in FA patients, for whom such phenomena occur already at increased rates over the general population.
The use of modern blood banking has resulted in supportive care that enables patients to tolerate anemia and thrombocytopenia. The specter of infection remains, however, with respect to neutropenia, despite the use of granulocyte stimulating growth factor, as the use of such a growth factor promotes the evolution of clones that eventually lead to leukemia.12,13 Androgens have also been an adjunct to care and have demonstrated efficacy in FA patients, but virilization and higher risk of liver adenomas have limited their use.
There is good evidence that bone marrow failure in FA patients stems from hematopoietic stem cell (HSC) dysfunction and depletion of the HSC reservoir. The observation that CD34+ cells counts are low in FA patients supports this idea.14,15 The progressive HSC failure in FA patients is linked to the DNA damage response mediated by p53/p21.15 Reports have recently showed that knockdown of FA genes in human embryonic stem cells resulted in defective hematopoiesis, thus implicating the FA pathway in hematopoietic development.16,17 In this same study, FANCA or FANCD2 knockdown caused a significant reduction in the production of HSCs and progenitor cells on in vitro differentiation.
FA genetics
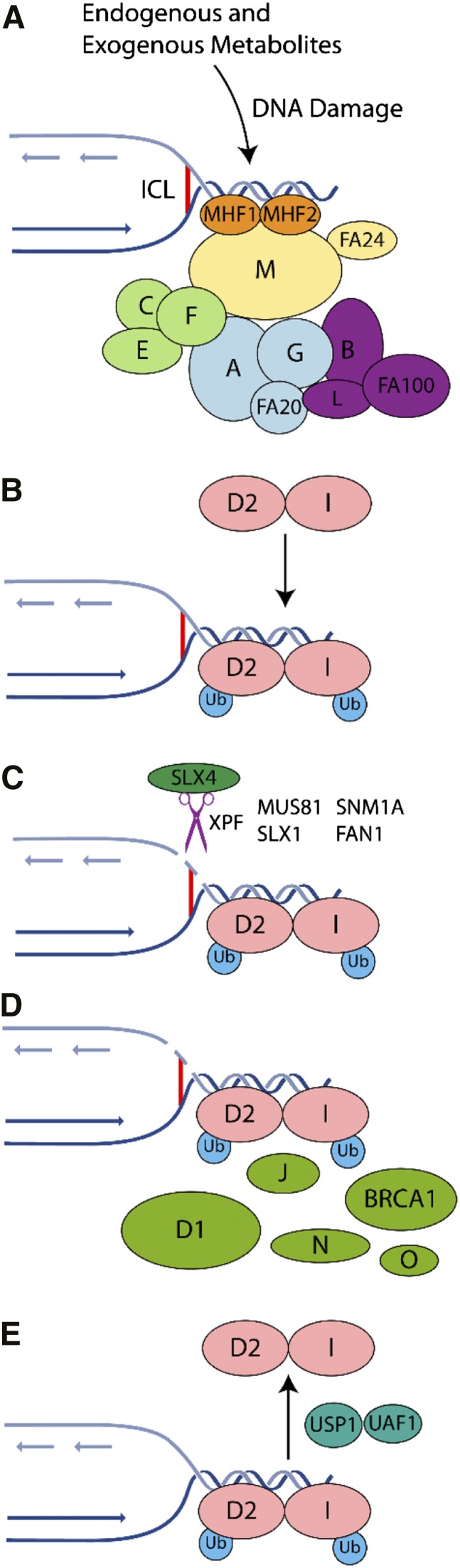
The FA pathway is genetically complex, comprised of 16 complementation groups and associated genes. The encoded proteins have been grouped into 3 categories: (1) the FA core complex, including the E3 ligase, FANCL; (2) the ID2 complex, the substrate for the E3 ubiquitin ligase activity of the core complex; and (3) downstream proteins that possess a DNA repair or damage tolerance function7,18 (Figure 1).
Figure 1.
Mechanism of ICL repair in the FA pathway on collision of a replication fork with an ICL. The FA pathway is composed of ≥16 genes (A, B, C, D1, D2, E, F, G, I, K, L, M, N, O, P, and Q). The encoded proteins can be subdivided within the FA pathway into 3 groups: (1) proteins that make up the core complex; (2) the FANCI and FANCD2 proteins, which compose the ID2 complex; and (3) downstream effector proteins. (A) The FA pathway is activated during S phase of the cell cycle or on the detection of ICLs and DNA damage caused by other agents, including endogenous acetaldehydes. The FA core complex is recruited to the damage site through its interaction with the MHF1-MHF2-FANCM complex. (B) The ID2 complex becomes monoubiquitinated and remains associated with the DNA damage. The B-L-100 complex mediates the ubiquitination reaction, with the other 2 core subcomplexes (A-G-20 and C-E-F) playing accessory roles that remain to be elucidated. (C) Specialized endonucleases, in particular XPF/FANCQ-ERCC1 in complex with SLX4/FANCP, incise the DNA. (D) Within chromatin, the monoubiquitinated ID2 complex recruits DNA repair proteins including BRCA1, BRCA2/FANCD1, FANCJ, PALB2/FANCN, and RAD51C/FANCO. (E) Following successful repair, deubiquitination of the ID2 complex by USP1-UAF1 promotes its release from chromatin.
FA patient cells exhibit hypersensitivity to agents that cause interstrand DNA cross-links (ICLs), visualized as chromosomal fragility and radial formation that represent the biological hallmark of the disease. The more recent unraveling of the downstream proteins, namely, BRCA2/FANCD1, BACH1/FANCJ, PALB2/FANCN, RAD51C/FANCO, SLX4/FANCP, and XPF/FANCQ, reveals an intimate link to DNA repair and mainstream cancer biology, including breast cancer.19,20 Such links have revealed opportunities for improved cancer therapy by capitalizing on FA biology, as in the use of polyadenosine ribose polymerase inhibitors in BRCA2 mutant breast cancer.21 This is especially apropos, as FA patients who survive to adulthood display markedly higher rates of nonhematologic cancers, such as breast, head and neck, and squamous cell cancers. In addition, sporadic mutations in FA genes have been reported in many common adult cancers, including pancreatic, lung, gastrointestinal, and squamous cell cancers.22-24 Recent data indicate a direct link between FANCD2 regulation of the transcription of the Tap63 tumor suppressor and squamous cell cancer formation.25
We focus below on FA protein functions and their interacting partners, followed by a mechanistic model of DNA ICL repair. Such a model may also be more generalizable to replication fork block and collapse. We also summarize recent developments that link the FA pathway to counteracting the genotoxicity of reactive aldehydes that are byproducts of cellular metabolism.
Upstream: the FA core complex
The FA core complex is comprised of 3 subcomplexes: (1) FANCL, FANCB, and FAAP100 (FA-associated protein 100 kDa); (2) FANCA, FANCG, and FAAP20 (FA-associated protein 20 kDa); and (3) FANCC, FANCE, and FANCF26-29 (Figure 1). FANCL is an E3 ubiquitin ligase that in conjunction with the E2 conjugating enzyme UBE2T monoubiquitinates FANCD2 and FANCI at K561 and K523, respectively. Purified FANCL and UBE2T can ubiquitinate both FANCD2 and FANCI site specifically in vitro,30,31 and its activity and substrate specificity are enhanced in the context of the FANCL-FANCB-FAAP100 (the L-B-100) complex.28 In cells, all components of the core complex are required for optimal ID2 ubiquitination, although careful epistasis analysis of mutant cells has revealed that only the L-B-100 complex is absolutely required.28,29 FA proteins in general lack identifiable functional domains and are highly conserved only in vertebrates, so progress has been slow in establishing their biochemical functions. Nevertheless, multiple groups have established a preliminary network of interactions among them32 (Figure 1).
X-ray crystal structures suggest that FANCD2 binds 2 interfaces in the core complex, with contact to FANCL and FANCE.33,34 The FANCF structure suggests a flexible protein that links the FANCC-FANCE and FANCA-FANCG subcomplexes and mediates recruitment of the entire complex to sites of damage and FANCD2.29,35,36
Another core complex protein FANCA stimulates the activity of MUS81-EME1, a structure-specific nuclease involved in ICL repair37 (see below). FANCA also binds core complex members FANCG38 and FAAP20,39-41 which is necessary for normal FANCD2 ubiquitination and foci formation, mediated via its UBZ ubiquitin-binding domain. The UBZ domain also has been reported to mediate the interaction between FAAP20 and the translesion synthesis (TLS) protein REV1.42
Nexus: the FANCI-FANCD2 (ID2) complex
Monoubiquitination of FANCD2 and FANCI signals activation of the FA DNA repair network and is required for ICL resistance, which is manifest by replication fork collapse. Formation of the heterodimeric ID2 complex is necessary for FANCD2 monoubiquitination and localization of the complex to chromatin at DNA damage foci. ID2 ubiquitination occurs mainly in S-phase.43-46
The precise molecular function of FANCD2 and FANCI remains elusive, but biochemical analyses revealed that FANCD2, FANCI, and the ID2 complex possess DNA binding activity.45 Structural analysis of the ID2 complex suggests conformational changes inducible by interaction with DNA as well as core complex subcomplexes, such as L-B-100.47 An interplay of phosphorylation and presence of the entire array of core complex members is necessary for full ubiquitination of both members of the ID2 complex.30,31,48
The removal of ubiquitin from the ID2 complex on completion of replication fork repair and restart is catalyzed by the USP1-UAF1 deubiquitinase (DUB), which also targets other proteins such as ubiquitinated PCNA.49,50 USP1, the catalytic component, is stimulated by heterodimerization with the accessory protein UAF1.51,52 Negative regulation of this DUB occurs on DNA damage signaling via suppression of USP1 transcription, in addition to cleavage and proteolytic degradation of the USP1 protein.53,54 Because the timely deubiquitination of the ID2 complex is necessary for the functional integrity of the FA pathway, USP1-UAF1 has emerged as an important target for developing small molecule inhibitors that can potentiate the cytotoxic effect of ICL-inducing chemotherapeutics.55 In addition, both murine and chicken cells deficient for either UAF1 or USP1 exhibit a defect in DNA double-strand break repair by homologous recombination (HR),56-58 suggesting a HR role for USP1-UAF1 as well.
FANCM and its binding partners: regulation of replication restart
FANCM is a core complex member with an ATP-dependent DNA translocase activity. FANCM is required for resistance to ICLs via mediation of (1) recruitment of the FA core complex to chromatin59; (2) regression of stalled DNA replication forks; (3) traversal of the DNA replication machinery across an ICL60; and (4) efficient DNA damage signaling via the ATR kinase.61-63 Notably, the DNA translocase activity is not required for FANCD2 ubiquitination.64 Such activities are deemed necessary for the ability of replication to traverse an ICL using replication fork regression.65 Regression facilitates bypass of lesions in the template DNA strand,66 but how it promotes ICL removal remains unclear.
Replication restart also requires the BLM helicase, which is deficient in Bloom’s syndrome, yet another way that FA interacts with a distinct repair pathway. Bloom’s syndrome is marked by cancer susceptibility and by cells that display increased sister chromatid exchange and exhibits several FA-like features. BLM regulates HR by promoting the formation of noncrossovers during homologous recombination, and it also catalyzes replication fork regression.67 Importantly, FANCM and BLM form a complex, being tethered via RMI1-RMI2, and their interaction is required for resistance to ICLs.36,68
The MHF complex (MHF1-MHF2-FAAP24), structurally similar to histones, is important for FANCM stability, recruitment of the FA core complex to damaged chromatin, FANCD2 ubiquitination, and cellular resistance to ICLs.69,70 The MHF complex also stimulates the DNA binding and translocase activities of FANCM.71,72 Based on the crystal structure of the binary complex of MHF and DNA, it has been suggested that the MHF complex serves to anchor FANCM at a DNA junction to promote replication fork regression and DNA branch migration reactions.70 FAAP24 also binds DNA and interacts with FANCM directly. The DNA binding activity of FAAP24 is required for resistance to ICLs, as well as for FANCD2 ubiquitination.73
Downstream: effectors of the FA pathway
Biallelic mutations in BRCA2/FANCD1, BACH1/FANCJ, PALB2/FANCN, RAD51C/FANCO, SLX4/FANCP, and XPF/FANCQ, which all play a role in known DNA repair reactions, can lead to FA.74 Because a deficiency in these proteins does not affect FANCD2 ubiquitination, they are characterized as downstream of FANCD2 in ICL removal.46 Conversely, FANCD2 has been found in complex and in repair foci with several of these proteins, suggesting a more direct functional link.
BRCA2, PALB2, and RAD51C (3 known familial breast cancer gene products) all have defined functions in HR.75 Specifically, BRCA2 and PALB2 associate with RAD51, the recombinase that catalyzes the HR reaction, and stimulate its activity.75,76 A paralog of RAD51, RAD51C, has been identified as a FA gene, FANCO, and is found in multiple subcomplexes, all with defined roles in HR.77-79 A subcomplex of RAD51B-RAD51C enhances RAD51-mediated DNA strand exchange in vitro.80
BACH1/FANCJ, another familial breast cancer and FA-J complementation group protein, is a DNA helicase that translocates on ssDNA with a 5′ to 3′ polarity. FANCJ also binds to the DNA mismatch repair protein MLH1,81 which interacts with FAN1 (see below) and FANCD2,82 supporting a role for mismatch repair in FA.
SLX4 acts as a nuclear scaffold to enhance the activity of 3 structure-specific nucleases previously implicated in ICL repair and other DNA repair pathways.83,84 These nucleases are (1) XPF/FANCQ-ERCC1 that functions in nucleotide excision repair85,86; (2) MUS81-EME1; and (3) SLX1. Each of these nucleases possesses a substrate specificity consistent with a role in the unhooking and removal of ICLs.87,88 Recent studies involving the use of Xenopus cell-free extracts have provided biochemical evidence for a major role of XPF-ERCC1-SLX4 in ICL removal.89
FAN1, which possesses 5′ flap endonuclease and 5′ exonuclease activities, was recently identified as a binding partner of monoubiquitinated FANCD2.90 Abrogation of FAN1 nuclease activity results in hypersensitivity to ICL agents. It has been suggested that FAN1 is recruited by ubiquitinated FANCD2 to help mediate ICL incision and removal.90 Interestingly, mutations in FAN1 do not cause FA but underlie the kidney disease karyomegalic interstitial nephritis.91
MUS81-EME1 and SLX1-SLX4 have been shown to cooperatively cleave the Holliday junction, an important DNA intermediate in HR.92 However, this function may be independent of the role of these nucleases in ICL removal.93
Translesion DNA synthesis polymerases and FA
ICL unhooking at a stalled replication fork results in a double-strand break in 1 chromatid and a single-stranded gap harboring the ICL adduct on the other (Figure 1). Repair of the strand break occurs via HR,94 whereas gap filling in the lesion-containing DNA requires TLS, being catalyzed by a specialized DNA polymerase. Several TLS polymerases have been implicated in this regard, although for replication-dependent ICL repair, the most important appears to be the REV1-Polζ complex.95 In ICL repair, a mechanism that relies on interaction of REV1 with FAAP20, an FA core complex member, is thought to mediate the recruitment of REV1-Polζ.42
Cell extracts use to examine the removal of a site-specific cross-link has led to a mechanistic model for ICL repair when 2 convergent replication forks encounter the DNA cross-link96 (Figure 1). In this system, replication fork stalling occurs momentarily ∼20 to 40 nucleotides from the lesion, followed by disassembly of the machinery, and then by polymerase stalling again at 1 nucleotide from the lesion. At this stage, unhooking incisions, requiring XPF/ERCC1 and SLX4,89 occur on the parental strand opposite to that of the approaching leading strand (Figure 1). Finally, nucleotide insertion across the adducted base by REV1-Polζ occurs. Following the unhooking and TLS events, coordinated completion of repair requires actions of HR and mismatch repair.
FA pathway and cytokinesis
Several reports have suggested that FA proteins play important roles during M phase, especially in cytokinesis.97-100 First, FANCD2 and FANCI were found to localize at ultrafine anaphase bridges between segregating sister chromatids; these bridges increase in frequency on replication stress.99,100 The number of anaphase bridges, which are normally decorated with BLM and FANCM, increases in FA-deficient cells, leading to a higher frequency of cytokinesis failure, binucleated cells, as well as supernumerary centrosomes. In addition, several FA proteins localize to centrosomes and the mitotic spindle.98,101
DNA damage response and FA: ATM, ATR, and CHK1
The FA pathway influences DNA damage signaling through the ataxia telangiectasia mutated (ATM), ataxia telangiectasia and Rad3-related (ATR),102 and Chk1 kinases. Many of the FA proteins are phosphorylated in response to DNA damage and during distinct phases of the cell cycle. Similar to ATM-mutant cells, FANCD2-deficient cells display a defect in S phase checkpoint response after ionizing radiation (IR) exposure.103 Normally, FANCD2 is phosphorylated by ATM on serine 222 on IR treatment, and this event is required for establishing the intra-S checkpoint and a proper cellular response to DSBs.103 These results thus reveal a direct interaction between the ATM and FA pathways.
The FA pathway also intersects with ATR-mediated checkpoint signaling. Following exposure to a DNA cross-linking agent, FANCD2 and ATR colocalize in nuclear foci.104 Phosphorylation of FANCD2 on threonine 691 and serine 717 by ATR is required for FANCD2 monoubiquitination and correction of MMC sensitivity in FANCD2-deficient cells.105 ATR also phosphorylates FANCG106 and FANCI.48 FANCG phosphorylation by ATR is important for the functional interaction of FANCG with BRCA2/FANCD1107. Interestingly, downregulation of FANCM or FAAP24 negatively affects ATR-mediated checkpoint signaling.62 Clinically, Seckel’s syndrome, stemming from biallelic mutations in ATR, resembles FA in the exhibition of chromosome fragility, developmental delay, growth retardation, and cancer susceptibility.
CHK1, a substrate of both ATM and ATR, phosphorylates FANCE on threonine 346 and serine 374, and these modifications are required for the cellular resistance to MMC without affecting FANCD2 monoubiquitination and focus formation.102 CHK1 is also implicated in the serine 331 phosphorylation of FANCD2, which is required for resistance to MMC and FANCD2 monoubiquitination.108 Increased CHK1 activity is associated with early marrow failure, and downregulation of such activity may promote later leukemogenesis.109
Function of the FA pathway in alleviating the genotoxicity of acetaldehyde
Recent work has implicated aldehyde metabolism as a primary cause of the FA phenotype.110,111 Chicken DT40 cells, ablated for one of several FA genes, including FANCB, FANCC, FANCL, and BRCA2/FANCD1, are hypersensitive to acetaldehyde.110 In addition, these FA gene knockouts are synthetic lethal with mutations in the formaldehyde catabolism gene ADH5, indicating that cells become sensitized to aldehyde toxicity when the FA pathway is defective.110 Notably, mice doubly deficient in Fancd2 and aldehyde dehydrogenase (Aldh2) are prone to ethanol-induced bone marrow failure compared with wild-type mice or the single mutants.110 The bone marrow failure in Fancd2−/−:Aldh2−/− mice correlates with the accumulation of damaged DNA within the hematopoietic stem and progenitor cell pool.111 A genotypic analysis of a group of Japanese FA patients revealed that ALDH2 deficiency dramatically accelerates bone marrow failure and increases the frequency of malformation in some tissues, providing additional evidence that reactive aldehydes play an important role in the pathogenesis of FA.112 However, it remains unclear if aldehydes cause FA-associated genotoxicity. Aldehydes may make FA patients clinically worse, but no evidence yet exists that antialdehyde therapy could benefit them, although the possibility is intriguing.
FA pathway, cytokine sensitivity, and oxidative stress
FA patients exhibit altered expression levels of growth factors and cytokines, including tumor necrosis factor-α (TNF-α), which is involved in the initiation of apoptosis. Studies using Fanca−/−, Fancc−/−, and Fancg−/− mouse models have demonstrated that FA cells are hypersensitive to TNF-α, and this sensitivity contributes to bone marrow failure in FA.113 Expression of Fancc cDNA in Fancc−/− stem cells prevents the formation of leukemic clonal outgrowths, implying that FANCC is crucial for proper cellular response to TNF-α. FANCD2 represses the transcription of TNF-α by binding to its promoter region,114 which may explain why FA patients have elevated TNF-α levels. Oxidative DNA damage level is persistently higher in HSC or progenitor cells from TNF-α–injected Fancc−/− mice, further supporting the notion that FA proteins protect against reactive oxygen species-induced DNA damage.115
Hematopoietic stem cells with mutant FANCC are hypersensitive to interferon-γ,116 and interferon-γ stimulates increased apoptosis in the mutant setting.117 Reconstitution of the myeloid compartment appears to depend on interferon response pathways.118 FANCC also functions as a negative regulator of cytokine-induced apoptosis by modulating the activity of PKR, a growth inhibitory kinase and key effector of apoptosis.119
Original descriptions of FA cell hypersensitivity to oxygen have led to the hypothesis that FA harbors primary defects in management of oxidative stress, and more recent evidence supports this idea. For example, FANCA and FANCG are sensitive to redox conditioning, such that hydrogen peroxide treatment triggers complex formation of these FA proteins.120 The antioxidant tempol displayed tumor onset and protective effects against oxidative damage in Fancd2 −/− mice. Low oxygen has recently been shown to stimulate ID2 ubiquitination in an ATR-dependent manner.121
The FA pathway has also been linked to mitochondrial dysfunction. A recent study has revealed excessive formation of mitochondrial reactive oxygen species in FA (FA-A, -C, and -D2) cells,122 and mice doubly deficient in Fancc and superoxide dismutase display bone marrow hypocellularity, which is not present in mice with either of the mutations.123 FA-deficient cells exhibit better growth characteristics under hypoxic conditions, and the use of low oxygen tension allows the generation of FA-deficient induced pluripotent stem cell lines.124
Summary
Although a tremendous amount of information has emerged in the 22 years since the cloning of the first FA gene, FANCC, much remains to be learned regarding how DNA repair, DNA damage checkpoints, and associated processes intersect within the FA pathway. Even though FA is rare, its general relevance to cancer biology is exemplified by ≥5 FA genes being familial breast cancer genes and the identification of somatic FA gene mutations in many common cancers.
Many challenges lie ahead in devising an effective treatment of FA. Although stem cell transplant overcomes the ravages of acute myelogenous leukemia, patients remain prone to many solid tumors, including breast, head and neck, genitourinary tumors, and second cancers after transplant. Second, stem cell transplant in FA patients is still fraught with complications, and less toxic, more efficacious modalities would be welcome. Third, as noted earlier, many common adult cancers exhibit FA gene mutations, suggesting that dysfunction in the FA pathway can contribute to oncogenic genomic instability. Such avenues of investigation promise to enhance medical care of FA and non-FA patients alike. For example, the elaboration of polyadenosine ribose polymerase inhibitors in BRCA1 and BRCA2, both central to FA biology, have led to clinical trials that help target tumors with mutations in these proteins. Understanding the biology of FA will lead to greater opportunities for therapeutic advantage.
Authorship
Contribution: All authors contributed to the writing and editing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gary M. Kupfer, LMP2073, Section of Pediatric Hematology-Oncology, Yale School of Medicine, 333 Cedar St, New Haven, CT 06510; e-mail: gary.kupfer@yale.edu.
References
- 1.Bagby GC, Alter BP. Fanconi anemia. Semin Hematol. 2006;43(3):147–156. doi: 10.1053/j.seminhematol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 2.D’Andrea AD, Dahl N, Guinan EC, Shimamura A. Marrow failure. Hematology (Am Soc Hematol Educ Program) 2002 doi: 10.1182/asheducation-2002.1.58. 58-72. [DOI] [PubMed] [Google Scholar]
- 3.Chirnomas SD, Kupfer GM. The inherited bone marrow failure syndromes. Pediatr Clin North Am. 2013;60(6):1291–1310. doi: 10.1016/j.pcl.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green AM, Kupfer GM. Fanconi anemia. Hematol Oncol Clin North Am. 2009;23(2):193–214. doi: 10.1016/j.hoc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alter BP. Diagnosis, genetics, and management of inherited bone marrow failure syndromes. Hematology (Am Soc Hematol Educ Program) 2007 doi: 10.1182/asheducation-2007.1.29. 29-39. [DOI] [PubMed] [Google Scholar]
- 6.Khincha PP, Savage SA. Genomic characterization of the inherited bone marrow failure syndromes. Semin Hematol. 2013;50(4):333–347. doi: 10.1053/j.seminhematol.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kee Y, D’Andrea AD. Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Invest. 2012;122(11):3799–3806. doi: 10.1172/JCI58321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gluckman E, Wagner JE. Hematopoietic stem cell transplantation in childhood inherited bone marrow failure syndrome. Bone Marrow Transplant. 2008;41(2):127–132. doi: 10.1038/sj.bmt.1705960. [DOI] [PubMed] [Google Scholar]
- 9.Farzin A, Davies SM, Smith FO, et al. Matched sibling donor haematopoietic stem cell transplantation in Fanconi anaemia: an update of the Cincinnati Children’s experience. Br J Haematol. 2007;136(4):633–640. doi: 10.1111/j.1365-2141.2006.06460.x. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhury S, Auerbach AD, Kernan NA, et al. Fludarabine-based cytoreductive regimen and T-cell-depleted grafts from alternative donors for the treatment of high-risk patients with Fanconi anaemia. Br J Haematol. 2008;140(6):644–655. doi: 10.1111/j.1365-2141.2007.06975.x. [DOI] [PubMed] [Google Scholar]
- 11.Bitan M, Or R, Shapira MY, et al. Fludarabine-based reduced intensity conditioning for stem cell transplantation of Fanconi anemia patients from fully matched related and unrelated donors. Biol Blood Marrow Transplant. 2006;12(7):712–718. doi: 10.1016/j.bbmt.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Myers KC, Davies SM. Hematopoietic stem cell transplantation for bone marrow failure syndromes in children. Biol Blood Marrow Transplant. 2009;15(3):279–292. doi: 10.1016/j.bbmt.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 13.MacMillan ML, Wagner JE. Haematopoeitic cell transplantation for Fanconi anaemia - when and how? Br J Haematol. 2010;149(1):14–21. doi: 10.1111/j.1365-2141.2010.08078.x. [DOI] [PubMed] [Google Scholar]
- 14.Kelly PF, Radtke S, von Kalle C, et al. Stem cell collection and gene transfer in Fanconi anemia. Mol Ther. 2007;15(1):211–219. doi: 10.1038/sj.mt.6300033. [DOI] [PubMed] [Google Scholar]
- 15.Ceccaldi R, Parmar K, Mouly E, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11(1):36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tulpule A, Lensch MW, Miller JD, et al. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood. 2010;115(17):3453–3462. doi: 10.1182/blood-2009-10-246694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamimae-Lanning AN, Goloviznina NA, Kurre P. Fetal origins of hematopoietic failure in a murine model of Fanconi anemia. Blood. 2013;121(11):2008–2012. doi: 10.1182/blood-2012-06-439679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garaycoechea JI, Patel KJ. Why does the bone marrow fail in Fanconi anemia? Blood. 2014;123(1):26–34. doi: 10.1182/blood-2013-09-427740. [DOI] [PubMed] [Google Scholar]
- 19.Masserot C, Peffault de Latour R, Rocha V, et al. Head and neck squamous cell carcinoma in 13 patients with Fanconi anemia after hematopoietic stem cell transplantation. Cancer. 2008;113(12):3315–3322. doi: 10.1002/cncr.23954. [DOI] [PubMed] [Google Scholar]
- 20.Lowy DR, Gillison ML. A new link between Fanconi anemia and human papillomavirus-associated malignancies. J Natl Cancer Inst. 2003;95(22):1648–1650. doi: 10.1093/jnci/djg125. [DOI] [PubMed] [Google Scholar]
- 21.Lee JM, Ledermann JA, Kohn EC. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann Oncol. 2014;25(1):32-40. [DOI] [PMC free article] [PubMed]
- 22.Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19(24):2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 23.Levitus M, Joenje H, de Winter JP. The Fanconi anemia pathway of genomic maintenance. Cell Oncol. 2006;28(1-2):3–29. doi: 10.1155/2006/974975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valeri A, Martínez S, Casado JA, Bueren JA. Fanconi anaemia: from a monogenic disease to sporadic cancer. Clin Transl Oncol. 2011;13(4):215–221. doi: 10.1007/s12094-011-0645-6. [DOI] [PubMed] [Google Scholar]
- 25.Park E, Kim H, Kim JM, et al. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Mol Cell. 2013;50(6):908–918. doi: 10.1016/j.molcel.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling C, Ishiai M, Ali AM, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26(8):2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medhurst AL, Laghmani H, Steltenpool J, et al. Evidence for subcomplexes in the Fanconi anemia pathway. Blood. 2006;108(6):2072–2080. doi: 10.1182/blood-2005-11-008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajendra E, Oestergaard VH, Langevin F, et al. The genetic and biochemical basis of FANCD2 monoubiquitination. Mol Cell. 2014;54(5):858–869. doi: 10.1016/j.molcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang YL, Justin WC, Lowery M, et al. Modularized functions of the Fanconi anemia core complex. Cell Reports. 2014;7(6):1849–1857. doi: 10.1016/j.celrep.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato K, Toda K, Ishiai M, Takata M, Kurumizaka H. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res. 2012;40(10):4553–4561. doi: 10.1093/nar/gks053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longerich SKY, Tsai M-S, Hlaing A, Kupfer GM, Sung P. Regulation of FANCD2 and FANCI ubiquitination by their interaction and by DNA. Nucleic Acids Res. 2014;42(9):5657–5670. doi: 10.1093/nar/gku198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodson C, Walden H. Towards a molecular understanding of the fanconi anemia core complex. Anemia. doi: 10.1155/2012/926787. [DOI] [PMC free article] [PubMed]
- 33.Hodson C, Cole AR, Lewis LP, Miles JA, Purkiss A, Walden H. Structural analysis of human FANCL, the E3 ligase in the Fanconi anemia pathway. J Biol Chem. 2011;286(37):32628–32637. doi: 10.1074/jbc.M111.244632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nookala RK, Hussain S, Pellegrini L. Insights into Fanconi Anaemia from the structure of human FANCE. Nucleic Acids Res. 2007;35(5):1638–1648. doi: 10.1093/nar/gkm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kowal P, Gurtan AM, Stuckert P, D’Andrea AD, Ellenberger T. Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex. J Biol Chem. 2007;282(3):2047–2055. doi: 10.1074/jbc.M608356200. [DOI] [PubMed] [Google Scholar]
- 36.Deans AJ, West SC. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol Cell. 2009;36(6):943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 37.Benitez A, Yuan F, Nakajima S, et al. Damage-dependent regulation of MUS81-EME1 by Fanconi anemia complementation group A protein. Nucleic Acids Res. 2014;42(3):1671–1683. doi: 10.1093/nar/gkt975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Higuera I, Kuang Y, Denham J, D’Andrea AD. The fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood. 2000;96(9):3224–3230. [PubMed] [Google Scholar]
- 39.Ali AM, Pradhan A, Singh TR, et al. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood. 2012;119(14):3285–3294. doi: 10.1182/blood-2011-10-385963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung JW, Wang Y, Fong KW, Huen MS, Li L, Chen J. Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc Natl Acad Sci USA. 2012;109(12):4491–4496. doi: 10.1073/pnas.1118720109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan Z, Guo R, Paramasivam M, et al. A ubiquitin-binding protein, FAAP20, links RNF8-mediated ubiquitination to the Fanconi anemia DNA repair network. Mol Cell. 2012;47(1):61–75. doi: 10.1016/j.molcel.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H, Yang K, Dejsuphong D, D’Andrea AD. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol. 2012;19(2):164–170. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22(1):106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang LC, Stone S, Hoatlin ME, Gautier J. Fanconi anemia proteins stabilize replication forks. DNA Repair (Amst) 2008;7(12):1973–1981. doi: 10.1016/j.dnarep.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garner E, Smogorzewska A. Ubiquitylation and the Fanconi anemia pathway. FEBS Lett. 2011;585(18):2853–2860. doi: 10.1016/j.febslet.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joo W, Xu G, Persky NS, et al. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science. 2011;333(6040):312–316. doi: 10.1126/science.1205805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishiai M, Kitao H, Smogorzewska A, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15(11):1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nijman SM, Huang TT, Dirac AM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17(3):331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 50.Smogorzewska A, Matsuoka S, Vinciguerra P, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129(2):289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohn MA, Kowal P, Yang K, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28(5):786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 52.Villamil MA, Liang Q, Chen J, et al. Serine phosphorylation is critical for the activation of ubiquitin-specific protease 1 and its interaction with WD40-repeat protein UAF1. Biochemistry. 2012;51(45):9112–9123. doi: 10.1021/bi300845s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang TT, Nijman SM, Mirchandani KD, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8(4):339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 54.Piatkov KI, Colnaghi L, Békés M, Varshavsky A, Huang TT. The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol Cell. 2012;48(6):926–933. doi: 10.1016/j.molcel.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang Q, Dexheimer TS, Zhang P, et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10(4):298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murai J, Yang K, Dejsuphong D, Hirota K, Takeda S, D’Andrea AD. The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol Cell Biol. 2011;31(12):2462–2469. doi: 10.1128/MCB.05058-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park E, Kim JM, Primack B, et al. Inactivation of Uaf1 causes defective homologous recombination and early embryonic lethality in mice. Mol Cell Biol. 2013;33(22):4360–4370. doi: 10.1128/MCB.00870-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim JM, Parmar K, Huang M, et al. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell. 2009;16(2):314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JM, Kee Y, Gurtan A, D'Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111(10):5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang J, Liu S, Bellani MA, et al. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell. 2013;52(3):434–446. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blackford AN, Schwab RA, Nieminuszczy J, Deans AJ, West SC, Niedzwiedz W. The DNA translocase activity of FANCM protects stalled replication forks. Hum Mol Genet. 2012;21(9):2005–2016. doi: 10.1093/hmg/dds013. [DOI] [PubMed] [Google Scholar]
- 62.Collis SJ, Ciccia A, Deans AJ, et al. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell. 2008;32(3):313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 63.Huang M, Kim JM, Shiotani B, Yang K, Zou L, D’Andrea AD. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell. 2010;39(2):259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ. The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res. 2009;37(13):4360–4370. doi: 10.1093/nar/gkp365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ray Chaudhuri A, Hashimoto Y, Herrador R, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19(4):417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 66.Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol. 2013;5(5):a012815. doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manthei KA, Keck JL. The BLM dissolvasome in DNA replication and repair. Cell Mol Life Sci. 2013;70(21):4067–4084. doi: 10.1007/s00018-013-1325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoadley KA, Xue Y, Ling C, Takata M, Wang W, Keck JL. Defining the molecular interface that connects the Fanconi anemia protein FANCM to the Bloom syndrome dissolvasome. Proc Natl Acad Sci USA. 2012;109(12):4437–4442. doi: 10.1073/pnas.1117279109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tao Y, Jin C, Li X, et al. The structure of the FANCM-MHF complex reveals physical features for functional assembly. Nat Commun. 2012;3:782. doi: 10.1038/ncomms1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Q, Saro D, Sachpatzidis A, et al. The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes. Nat Commun. 2014;5:2987. doi: 10.1038/ncomms3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh TR, Saro D, Ali AM, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37(6):879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan Z, Delannoy M, Ling C, et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell. 2010;37(6):865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 74.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11(7):467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 76.Dray E, Etchin J, Wiese C, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol. 2010;17(10):1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Somyajit K, Subramanya S, Nagaraju G. RAD51C: a novel cancer susceptibility gene is linked to Fanconi anemia and breast cancer. Carcinogenesis. 2010;31(12):2031–2038. doi: 10.1093/carcin/bgq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42(5):406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 79.Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin Cell Dev Biol. 2011;22(8):898–905. doi: 10.1016/j.semcdb.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 80.Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15(24):3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peng M, Litman R, Xie J, Sharma S, Brosh RM, Jr, Cantor SB. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007;26(13):3238–3249. doi: 10.1038/sj.emboj.7601754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Williams SA, Wilson JB, Clark AP, et al. Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Hum Mol Genet. 2011;20(22):4395–4410. doi: 10.1093/hmg/ddr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43(2):142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stoepker C, Hain K, Schuster B, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43(2):138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 85.Bogliolo M, Schuster B, Stoepker C, et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92(5):800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kashiyama K, Nakazawa Y, Pilz DT, et al. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet. 2013;92(5):807–819. doi: 10.1016/j.ajhg.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cybulski KE, Howlett NG. FANCP/SLX4: a Swiss army knife of DNA interstrand crosslink repair. Cell Cycle. 2011;10(11):1757–1763. doi: 10.4161/cc.10.11.15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sengerová B, Wang AT, McHugh PJ. Orchestrating the nucleases involved in DNA interstrand cross-link (ICL) repair. Cell Cycle. 2011;10(23):3999–4008. doi: 10.4161/cc.10.23.18385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klein Douwel D, Boonen RA, Long DT, et al. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54(3):460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang M, D’Andrea AD. A new nuclease member of the FAN club. Nat Struct Mol Biol. 2010;17(8):926–928. doi: 10.1038/nsmb0810-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou W, Otto EA, Cluckey A, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44(8):910–915. doi: 10.1038/ng.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brill SJ. Linking the Enzymes that Unlink DNA. Mol Cell. 2013;52(2):159–160. doi: 10.1016/j.molcel.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Castor D, Nair N, Déclais AC, et al. Cooperative control of holliday junction resolution and DNA repair by the SLX1 and MUS81-EME1 nucleases. Mol Cell. 2013;52(2):221–233. doi: 10.1016/j.molcel.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Long DT, Räschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333(6038):84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sharma S, Helchowski CM, Canman CE. The roles of DNA polymerase ζ and the Y family DNA polymerases in promoting or preventing genome instability. Mutat Res. 2013;743-744:97–110. doi: 10.1016/j.mrfmmm.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nalepa G, Clapp DW. Fanconi anemia and the cell cycle: new perspectives on aneuploidy. F1000Prime Rep. 2014;6:23. doi: 10.12703/P6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vinciguerra P, Godinho SA, Parmar K, Pellman D, D'Andrea AD. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. J Clin Invest. 2010;120(11):3834–3842. doi: 10.1172/JCI43391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11(6):761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 100.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11(6):753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 101.Nalepa G, Enzor R, Sun Z, et al. Fanconi anemia signaling network regulates the spindle assembly checkpoint. J Clin Invest. 2013;123(9):3839–3847. doi: 10.1172/JCI67364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang X, Kennedy RD, Ray K, Stuckert P, Ellenberger T, D'Andrea AD. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2007;27(8):3098–3108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Taniguchi T, Garcia-Higuera I, Xu B, et al. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109(4):459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 104.Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18(16):1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ho GPH, Margossian S, Taniguchi T, D'Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol. 2006;26(18):7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wilson JB, Yamamoto K, Marriott AS, et al. FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. Oncogene. 2008;27(26):3641–3652. doi: 10.1038/sj.onc.1211034. [DOI] [PubMed] [Google Scholar]
- 107.Wilson JB, Blom E, Cunningham R, Xiao Y, Kupfer GM, Jones NJ. Several tetratricopeptide repeat (TPR) motifs of FANCG are required for assembly of the BRCA2/D1-D2-G-X3 complex, FANCD2 monoubiquitylation and phleomycin resistance. Mutat Res. 2010;689(1-2):12–20. doi: 10.1016/j.mrfmmm.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhi G, Wilson JB, Chen X, et al. Fanconi anemia complementation group FANCD2 protein serine 331 phosphorylation is important for fanconi anemia pathway function and BRCA2 interaction. Cancer Res. 2009;69(22):8775–8783. doi: 10.1158/0008-5472.CAN-09-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ceccaldi R, Briot D, Larghero J, et al. Spontaneous abrogation of the G2DNA damage checkpoint has clinical benefits but promotes leukemogenesis in Fanconi anemia patients. J Clin Invest. 2011;121(1):184–194. doi: 10.1172/JCI43836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol. 2011;18(12):1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- 111.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489(7417):571. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 112.Hira A, Yabe H, Yoshida K, et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122(18):3206–3209. doi: 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li XX, Le Beau MM, Ciccone S, et al. Ex vivo culture of Fancc(−/−) stem/progenitor cells predisposes cells to undergo apoptosis, and surviving stem/progenitor cells display cytogenetic abnormalities and an increased risk of malignancy. Blood. 2005;105(9):3465–3471. doi: 10.1182/blood-2004-06-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Matsushita N, Endo Y, Sato K, et al. Direct inhibition of TNF-alpha promoter activity by Fanconi anemia protein FANCD2. PLoS ONE. doi: 10.1371/journal.pone.0023324. doi: 10.1371/journal.pone.0023324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang X, Sejas DP, Qiu Y, Williams DA, Pang Q. Inflammatory ROS promote and cooperate with the Fanconi anemia mutation for hematopoietic senescence. J Cell Sci. 2007;120(9):1572–1583. doi: 10.1242/jcs.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fagerlie S, Lensch MW, Pang QS, Bagby GC. The Fanconi anemia group C gene product: Signaling functions in hematopoietic cells. Exp Hematol. 2001;29(12):1371–1381. doi: 10.1016/s0301-472x(01)00755-x. [DOI] [PubMed] [Google Scholar]
- 117.Rathbun RK, Christianson TA, Faulkner GR, et al. Interferon-gamma-induced apoptotic responses of Fanconi anemia group C hematopoietic progenitor cells involve caspase 8-dependent activation of caspase 3 family members. Blood. 2000;96(13):4204–4211. [PubMed] [Google Scholar]
- 118.Hu L, Huang W, Hjort E, Eklund EA. Increased Fanconi C expression contributes to the emergency granulopoiesis response. J Clin Invest. 2013;123(9):3952–3966. doi: 10.1172/JCI69032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pang QS, Christianson TA, Keeble W, Koretsky T, Bagby GC. The anti-apoptotic function of Hsp70 in the interferon-inducible double-stranded RNA-dependent protein kinase-mediated death signaling pathway requires the Fanconi anemia protein, FANCC. J Biol Chem. 2002;277(51):49638–49643. doi: 10.1074/jbc.M209386200. [DOI] [PubMed] [Google Scholar]
- 120.Park SJ, Ciccone SLM, Beck BD, et al. Oxidative stress/damage induces multimerization and interaction of Fanconi anemia proteins. J Biol Chem. 2004;279(29):30053–30059. doi: 10.1074/jbc.M403527200. [DOI] [PubMed] [Google Scholar]
- 121.Scanlon SE, Glazer PM. Hypoxic stress facilitates acute activation and chronic downregulation of Fanconi anemia proteins. Mol Cancer Res. 2014 doi: 10.1158/1541-7786.MCR-13-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kumari U, Ya Jun W, Huat Bay B, Lyakhovich A. Evidence of mitochondrial dysfunction and impaired ROS detoxifying machinery in Fanconi anemia cells. Oncogene. 2014;33(2):165–172. doi: 10.1038/onc.2012.583. [DOI] [PubMed] [Google Scholar]
- 123.Hadjur S, Ung K, Wadsworth L, et al. Defective hematopoiesis and hepatic steatosis in mice with combined deficiencies of the genes encoding Fancc and Cu/Zn superoxide dismutase. Blood. 2001;98(4):1003–1011. doi: 10.1182/blood.v98.4.1003. [DOI] [PubMed] [Google Scholar]
- 124.Muller LU, Milsom MD, Harris CE, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119(23):5449–5457. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Alter B. Diagnostic evaluation of FA. In: Owen J, Frohnmayer L, Eiler ME, eds. Fanconi Anemia Standards for Clinical Care. 2nd Ed. London: The Fanconi Anemia Research Fund; 2003:3-17.