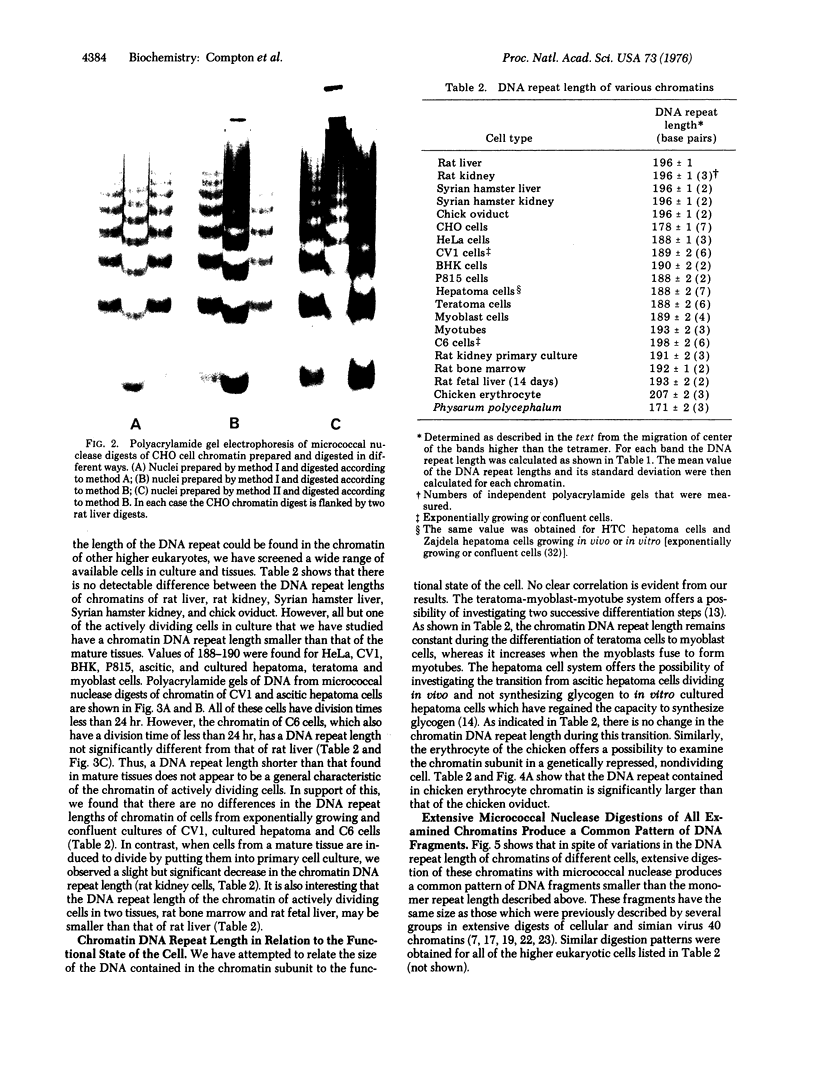
Abstract
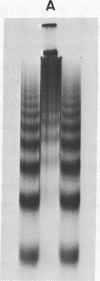
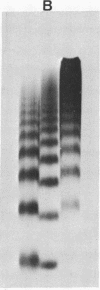
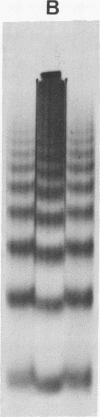
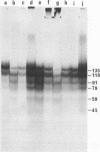
Biochemical evidence is presented which confirms that the DNA repeat length in micrococcal nuclease (spleen endonuclease, nucleate 3'-oligonucleotidohydrolase, EC 3-1-4-7) digests of Chinese hamster ovary chromatin is shorter than that of rat liver chromatin [J.L. Compton, R. Hancock, P. Oudet, and P. Chambon (1976) Eur. J. Biochem., in press]. A survey of available cells has shown that the DNA repeat length of the chromatin of higher eukaryotes varies widely. A value of 196 base pairs was found for cells of all mature tissues, regardless of the source of the tissue, whereas smaller values were found for cells of actively dividing tissues and larger values were found for a genetically inactive cell. Although the DNA repeat length of the chromatin of cells in culture was usually shorter than 196 base pairs, there was no general correlation between the size of the chromatin DNA repeat length and the rate of cell division or the functional state of the cell in culture. Examination of extensive micrococcal nuclease digests suggests that the chromatin subunits of all of the higher eukaryotic cells we have studied contain a core with approximately 140 base pairs of DNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R., Melchior W., Jr, Sollner-Webb B., Felsenfeld G. Specific sites of interaction between histones and DNA in chromatin. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4101–4105. doi: 10.1073/pnas.71.10.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R., Balhorn M., Morris H. P., Chalkley R. Comparative high-resolution electrophoresis of tumor histones: variation in phosphorylation as a function of cell replication rate. Cancer Res. 1972 Aug;32(8):1775–1784. [PubMed] [Google Scholar]
- Benda P., Lightbody J., Sato G., Levine L., Sweet W. Differentiated rat glial cell strain in tissue culture. Science. 1968 Jul 26;161(3839):370–371. doi: 10.1126/science.161.3839.370. [DOI] [PubMed] [Google Scholar]
- Blankstein L. A., Levy S. B. Changes in histone f2a2 associated with proliferation of Friend leukaemic cells. Nature. 1976 Apr 15;260(5552):638–640. doi: 10.1038/260638a0. [DOI] [PubMed] [Google Scholar]
- Boon T., Buckingham M. E., Dexter D. L., Jakob H., Jacob F. Tératocarcinome de la souris: isolement et propriétés de deux lignées de myoblastes. Ann Microbiol (Paris) 1974 Jul-Aug;125B(1):13–28. [PubMed] [Google Scholar]
- Burgoyne L. A., Hewish D. R., Mobbs J. Mammalian chromatin substructure studies with the calcium-magnesium endonuclease and two-dimensional polyacrylamide-gel electrophoresis. Biochem J. 1974 Oct;143(1):67–72. doi: 10.1042/bj1430067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L. H., Newrock K. M., Zweidler A. Stage-specific switches in histone synthesis during embryogenesis of the sea urchin. Science. 1975 Dec 5;190(4218):994–997. doi: 10.1126/science.1237932. [DOI] [PubMed] [Google Scholar]
- Elgin S. C., Weintraub H. Chromosomal proteins and chromatin structure. Annu Rev Biochem. 1975;44:725–774. doi: 10.1146/annurev.bi.44.070175.003453. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Howze G. B., Hsie A. W., Olins A. L. nu-Bodies in mitotic chromatin. Exp Cell Res. 1976 Jul;100(2):424–428. doi: 10.1016/0014-4827(76)90173-7. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Yeast chromatin subunit structure. Science. 1975 Apr 11;188(4184):165–166. doi: 10.1126/science.1090006. [DOI] [PubMed] [Google Scholar]
- Morris N. R. Nucleosome structure in Aspergillus nidulans. Cell. 1976 Jul;8(3):357–363. doi: 10.1016/0092-8674(76)90147-1. [DOI] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Branch A., Hamkalo B. A. Electron microscopy of whole mount metaphase chromosomes. Chromosoma. 1975 Nov 11;52(4):329–338. doi: 10.1007/BF00364017. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P., Jr Chemical evidence that chromatin DNA exists as 160 base pair beads interspersed with 40 base pair bridges. Nucleic Acids Res. 1976 Jan;3(1):117–127. doi: 10.1093/nar/3.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Spadafora C., Bellard M., Compton J. L., Chambon P. The DNA repeat lengths in chromatins from sea urchin sperm and gastrule cells are markedly different. FEBS Lett. 1976 Oct 15;69(1):281–285. doi: 10.1016/0014-5793(76)80704-1. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINOCOUR E. Purification of polyoma virus. Virology. 1963 Feb;19:158–168. doi: 10.1016/0042-6822(63)90005-9. [DOI] [PubMed] [Google Scholar]
- Woodcock C. L., Safer J. P., Stanchfield J. E. Structural repeating units in chromatin. I. Evidence for their general occurrence. Exp Cell Res. 1976 Jan;97:101–110. doi: 10.1016/0014-4827(76)90659-5. [DOI] [PubMed] [Google Scholar]