Abstract
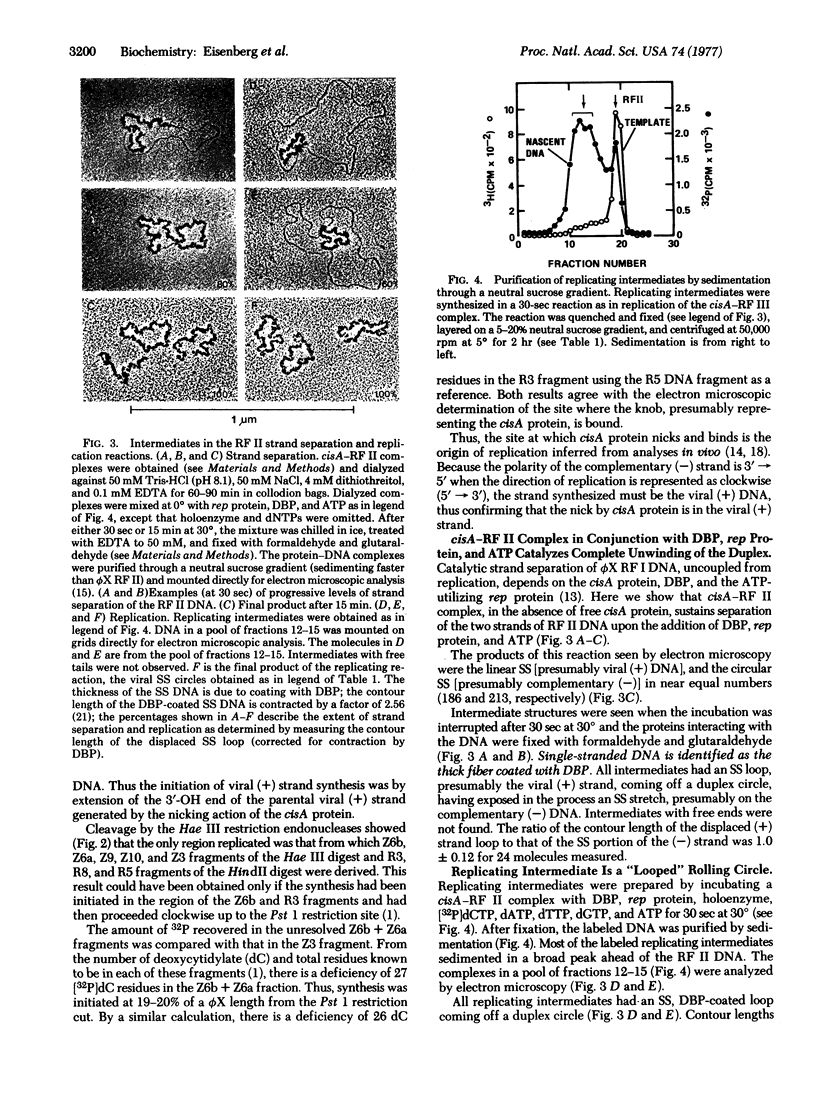
The cistron A protein induced by phage ϕX174 nicks (produces a single-strand break in) the viral strand of the superhelical ϕX duplex DNA, thereby forming a complex with the DNA. The protein, seen bound to the DNA in the electron microscope, was located in the restriction endonuclease fragment between nucleotides 4290 and 4330 on the ϕX map [Sanger, F., Air, G. M., Barrel, B. G., Brown, N. L., Coulson, A. R., Fiddes, J. C., Hutchison, C. A., III, Slocomb, P. M. Y. & Smith, M. (1977) Nature 265, 687-695]. Replication also was initiated at this point, thus identifying the site of cistron A protein nicking and binding as the origin of replication.
The cisA-DNA complex (separated from free cistron A protein), upon the addition of Escherichia coli rep protein, ATP, and DNA binding protein, is unwound to generate a single-stranded linear [presumably the nicked (+) strand] and a circular [presumably the (-) strand] molecule. The cisA-DNA complex, upon the further addition of DNA polymerase III holoenzyme and deoxynucleoside triphosphates, supports replication to generate viral, single-stranded circles, as many as 15 circles per cisA-DNA complex.
The replicating intermediates seen in the electron microscope are a novel form of “rolling circle” [Gilbert, W. & Dressler, D. H. (1969) Cold Spring Harbor Symp. Quant. Biol. 33, 473-485]. The 5′ end (presumably with the cistron A protein bound to it) is locked in the replication fork and loops back to accompany the strand-separation and replication fork around the template [(-) strand] circle. Thus, the multiple functions of cistron A protein include: (i) nicking the viral strand at the origin of replication to initiate a round of replication, (ii) participating in a complex which supports fork movement in strand separation and replication, (iii) nicking again at the regenerated origin to produce a unit-length DNA, and (iv) ligating the newly generated 3′-OH end to the 5′-phosphate-complexed end to form a circular viral molecule.
Keywords: cisA-RF II complex, origin, strand separation, rep protein
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. D., Jansz H. S. Bacteriophage phiX174 DNA synthesis in a replication-deficient host: determination of the origin of phiX DNA replication. J Mol Biol. 1976 Apr 15;102(3):633–656. doi: 10.1016/0022-2836(76)90339-9. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. Evidence for an intermediate with a single-strand break in the reaction catalyzed by the DNA untwisting enzyme. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3488–3491. doi: 10.1073/pnas.73.10.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T., Iwaya M., Larison L. L. The rep mutation. II. Its effect on Escherichia coli and on the replication of bacteriophage phi X174. Virology. 1972 Aug;49(2):486–496. doi: 10.1016/0042-6822(72)90500-4. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Harbers B., Hours C., Denhardt D. T. The mechanism of replication of phiX174 DNA. XII. Non-random location of gaps in nascent phiX174 RF II DNA. J Mol Biol. 1975 Nov 25;99(1):107–123. doi: 10.1016/s0022-2836(75)80162-8. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Scott J. F., Kornberg A. An enzyme system for replication of duplex circular DNA: the replicative form of phage phi X174. Proc Natl Acad Sci U S A. 1976 May;73(5):1594–1597. doi: 10.1073/pnas.73.5.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Scott J. F., Kornberg A. Enzymatic replication of viral and complementary strands of duplex DNA of phage phiX174 proceeds by seprate mechanisms. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3151–3155. doi: 10.1073/pnas.73.9.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke B., Ray D. S. Formation of the parental replicative form DNA of bacteriophage phi-X174 and initial events in its replication. J Mol Biol. 1971 Nov 14;61(3):565–586. doi: 10.1016/0022-2836(71)90065-9. [DOI] [PubMed] [Google Scholar]
- Fujisawa H., Hayashi M. Gene A product of phi X174 is required for site-specific endonucleolytic cleavage during single-stranded DNA synthesis in vivo. J Virol. 1976 Aug;19(2):416–424. doi: 10.1128/jvi.19.2.416-424.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Henry T. J., Knippers R. Isolation and function of the gene A initiator of bacteriophage phi-chi 174, a highly specific DNA endonuclease. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1549–1553. doi: 10.1073/pnas.71.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda J. E., Yudelevich A., Hurwitz J. Isolation and characterization of the protein coded by gene A of bacteriophage phiX174 DNA. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2669–2673. doi: 10.1073/pnas.73.8.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Scott J. F., Eisenberg S., Bertsch L. L., Kornberg A. A mechanism of duplex DNA replication revealed by enzymatic studies of phage phi X174: catalytic strand separation in advance of replication. Proc Natl Acad Sci U S A. 1977 Jan;74(1):193–197. doi: 10.1073/pnas.74.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal N., Delius H., Kornberg T., Gefter M. L., Alberts B. A DNA-unwinding protein isolated from Escherichia coli: its interaction with DNA and with DNA polymerases. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3537–3541. doi: 10.1073/pnas.69.12.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsheimer R. L. Bacteriophage phi-X174 and related viruses. Prog Nucleic Acid Res Mol Biol. 1968;8:115–169. [PubMed] [Google Scholar]
- Sumida-Yasumoto C., Yudelevich A., Hurwitz J. DNA synthesis in vitro dependent upon phiX174 replicative form I DNA. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1887–1891. doi: 10.1073/pnas.73.6.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessman E. S. Mutants of bacteriophage S13 blocked in infectious DNA synthesis. J Mol Biol. 1966 May;17(1):218–236. doi: 10.1016/s0022-2836(66)80104-3. [DOI] [PubMed] [Google Scholar]
- Tessman E. S., Peterson P. K. Bacterial rep- mutations that block development of small DNA bacteriophages late in infection. J Virol. 1976 Nov;20(2):400–412. doi: 10.1128/jvi.20.2.400-412.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]