Abstract
A major challenge in the diagnosis and treatment of brain tumors is tissue heterogeneity leading to mixed treatment response. Additionally, they are often difficult or at very high risk for biopsy, further hindering the clinical management process. To overcome this, novel advanced imaging methods are increasingly being adapted clinically to identify useful noninvasive biomarkers capable of disease stage characterization and treatment response prediction. One promising technique is called functional diffusion mapping (fDM), which uses diffusion-weighted imaging (DWI) to generate parametric maps between two imaging time points in order to identify significant voxel-wise changes in water diffusion within the tumor tissue. Here we introduce serial functional diffusion mapping (sfDM), an extension of existing fDM methods, to analyze the entire tumor diffusion profile along the temporal course of the disease. sfDM provides the tools necessary to analyze a tumor data set in the context of spatiotemporal parametric mapping: the image registration pipeline, biomarker extraction, and visualization tools. We present the general workflow of the pipeline, along with a typical use case for the software. sfDM is written in Python and is freely available as an open-source package under the Berkley Software Distribution (BSD) license to promote transparency and reproducibility.
Keywords: diffusion MR, parametric mapping, brain tumors, open source
Introduction
Aggressive brain tumors are challenging to accurately diagnose and treat in both pediatric and adult populations.1 They are often characterized by a high degree of spatiotemporal tissue heterogeneity, which may not be easily interpreted on conventional imaging including magnetic resonance imaging (MRI–T1 weighting, T2 weighting or FLAIR)2 and computerized tomographic imaging. Further obstacles arise when tumors are present in regions where performing a biopsy is considered too high risk, particularly in brainstem tumors. Thus, the need for accurate quantification of noninvasive neuroimaging biomarkers is paramount. Diffusion-weighted imaging (DWI) has long been used as a qualitative supplement to tumor diagnosis.3–5 DWI is an MR technique that allows us to indirectly observe the degree of water displacement (Brownian motion) within the tissue by measuring the voxel-wise signal loss in the image following the application of an encoding gradient. This displacement can be quantitatively calculated as the apparent diffusion coefficient (ADC), measured in mm2/s.6
Quantitative DWI methods, however, have mainly been limited to region-of-interest (ROI)7–9 approaches analyzing whole tumor diffusion. While more informative than qualitative observational methods, these do not solve the problem of tissue heterogeneity. Spatial variances between aggressive tumor growth, necrosis, and inflammation may be diagnostically significant, but are averaged over the tumor volume in traditional methods.
These challenges have led to the development of many highly sophisticated advanced imaging analysis techniques. A promising approach is functional diffusion mapping (fDM).10 fDM is a method of generating parametric maps from two coregistered diffusion images by calculating the voxel-wise change in ADC. This preserves the spatial information in the diffusion changes within the tumor tissue across these two time points. Existing fDM approaches have looked only at two imaging time points, often comparing pre- and postradiation effects.10,11 While these spatial analyses seem promising in improved outcome prediction, they still provide an incomplete picture of the long-term tumor progression and treatment response, of particular importance in prolonged therapies such as tumor vaccines.12
Here, we introduce an extension of fDM called serial functional diffusion mapping (sfDM). sfDM aims to analyze the entire diffusion profile of the tumor progression through the course of the disease. This allows for (1) spatiotemporal quantitation of the diffusion properties of a tumor given a particular therapy and (2) characterization of the natural progression of specific brain tumors. sfDM is capable of quantitative and qualitative analysis of serial tumor imaging. The goal of the software is to provide the tools necessary to analyze a tumor data set in the context of spatiotemporal parametric mapping: the image registration pipeline, biomarker extraction, and visualization tools. Figure 1 outlines the general workflow when using sfDM.
Figure 1.
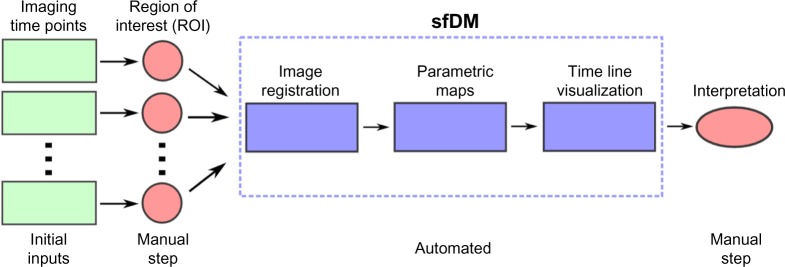
General sfDM workflow.
The typical sfDM workflow takes as inputs the patient’s diffusion and reference images for each time point, along with a user-defined tumor ROI. The sfDM pipeline automatically performs a series of image registrations and calculates parametric maps for every user-selected time point. These parametric maps are then used to generate a patient-specific fDM timeline.
An additional obstacle encountered in tumor research is the relative lack of transparency in the methods and algorithms employed by investigators. The use of closed-source, “in-house”– developed or expensive commercial software creates a hurdle that impedes the reproducibility and validation of promising new methods by restricting their user base. sfDM is provided as an open-source package with the intention of not only transparently broadening its user base but also to more efficiently improve and optimize its functionality through community feedback and development. This approach is critical for the use of the sfDM technique to eventually be implemented and validated for the analysis of diffusion imaging data from multi-institutional Phase II and III brain tumor therapy studies.13 In addition, this approach will also help validate the sfDM technique as a potential “point-of-care” analysis tool to be used in the clinic and help with the day-to-day management of neuro-oncology patients.
Software Details
The sfDM pipeline is described in detail below. It is designed as a series of image registrations to align every subject’s imaging time point into a common, high-resolution 3D space to allow for voxel-wise comparison across time. The multiple registrations are necessary due to the low resolution of conventionally acquired diffusion images. Beyond the created parametric maps, the pipeline outputs subject-specific diffusion timelines and calculates several fDM statistics (outline below) for each pair of time points being compared.
sfDM is written in Python and relies on a backbone built with the open-source data-processing framework Nipype.14 sfDM interfaces with several existing, commonly used neuroimage manipulation software packages and libraries including FSL,15 AFNI,16 and NIPY,17 and introduces newly developed brain tumor–specific visualization tools. sfDM is compatible with Linux and OSX operating systems and can be run on Windows using a virtual machine.
Inputs
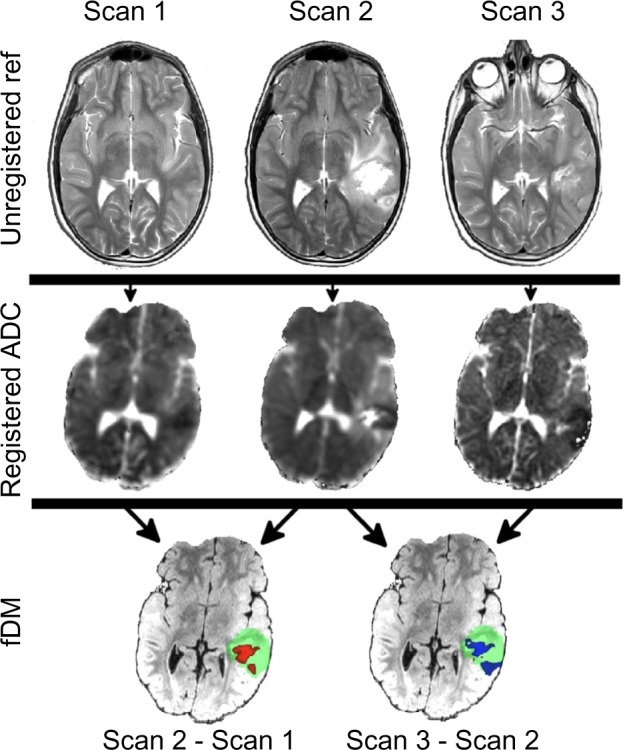
sfDM requires as input at least one high-resolution structural image (T1- or T2-weighted) to serve as the spatial reference for the registration of each imaging time point (sfDM label: Struct Brain). As its name implies, this image must be brain extracted prior to running sfDM. Brain extraction is the removal of nonbrain tissue (including skull, neck, and other image noise) from the image in order to provide a clean reference for subsequent registrations. A clean brain reference image is a pivotal point early in the processing pipeline and can be the source of misregistration if not done properly. We chose to exclude the reference brain extraction step from the pipeline in order to allow the user flexibility in choosing an adequate image without being constrained by built-in contrast or sequence-specific defaults. This also gives the opportunity for any manual corrections necessary in the generation of the Struct Brain. Conventionally, this image is from the earliest available time point, if possible, but can be arbitrarily chosen based on image quality and minimally observed mass effect. For each time point, we require as the starting set of images: (1) a reference image of medium (at most 3–4 mm slice thickness) to high resolution (sfDM label: Time Point Ref), (2) the diffusion-weighted image (sfDM label: DWI), and (3) the manually delineated tumor ROI (sfDM label: ROI) in diffusion or Time Point Ref space. An optional normal ROI can be supplied in order to calculate a patient-specific 95% confidence interval ADC change for each time point.
DWI is typically acquired at lower resolutions as a sacrifice to acquisition speed. Most modern protocols acquire a standard T2-weighted image with no diffusion gradient (the b0 image), followed by a set of images acquired using orthogonal diffusion directions. The calculated diffusion maps are created as the normalized difference in signal between the b0 image and the averaged set of diffusion images (b1), with lower signal indicating increased diffusion. Therefore, ideally the b0 image is used for registration due to its higher signal and better tissue contrast, with each resulting registration matrix applied to the ADC image. The b0 image, however, is not always consistently available in the same space as the derived ADC, particularly when analyzing retrospective or multisite data. Because of this, sfDM gives the option of directly using the available ADC image for the first registration step, in spite of its lower signal-to-noise ratio.
Image registration module
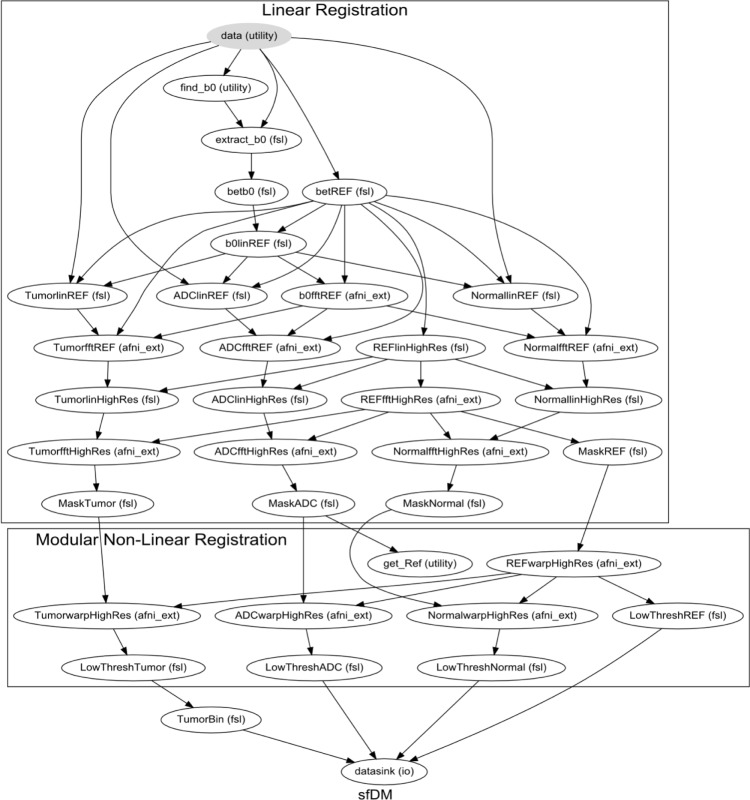
As a preprocessing step, the reference and diffusion images are brain extracted using FSL’s Brain Extraction Tool using robust brain center estimation and a low fractional intensity threshold. Figure 2 shows a typical workflow using default parameters, outlining the image registration steps performed by sfDM. This workflow is generated for each subject, outlining the specific options and parameters chosen by the user. To prevent gross misalignments, we first register the low-resolution diffusion image to the higher resolution time point reference image using a rigid-body linear registration. The time point reference image is then used as the input for registration to the high-resolution subject reference, with registration parameters applied to the diffusion and tumor ROI. The linear registration of each time point reference to subject reference space occurs in two steps: first, an affine registration using FSL’s linear registration tool, followed by a fine registration with fast Fourier transform (FFT) interpolation with AFNI’s 3dVolReg tool. An additional step following the linear registrations is to use the brain-extracted Struct Brain image as a mask to remove any nonbrain tissue or artifact from the now linearly registered ADC maps.
Figure 2.
Image registration pipeline.
The final registration node is a linear registration with a Hellinger distance metric (using AFNI) as default. sfDM also gives the option of using a nonlinear transformation using FSL’s nonlinear registration tool (FNIRT), which may be beneficial in the case of evident mass effect. As FNIRT is dependent on image intensities for registrations, only the registered ADC maps are used in this step to prevent errors due to any contrast inconsistencies in the Time Point Ref images. By default, the first time point is chosen as the reference space, but users may choose any available time point.
In a properly set up environment, sfDM can take advantage of Nipype’s built-in parallel-processing interface using Grid Engine18 to run each time point’s registration in parallel to reduce the processing time. This is of course dependent on the number of processing cores and memory available.
Image registration nodes automatically performed by the sfDM pipeline. Graph shows all the default image parameters for a typical run of sfDM. The graph is subdivided into Reg (linear registration) and Warp (modular nonlinear registration) phases to allow for modular flexibility in registration algorithms determined by the user. Each node is a processing step in the pipeline, with the used image-processing pipeline in parenthesis. A subject-specific graph outlining all user options is generated for every subject as a standard output.
sfDM generation
Functional diffusion maps are generated by calculating the voxel-wise difference between pairs of registered time points. The user must specify which time points to use to generate the fDMs for (see sfDM Usage). For ongoing studies, sfDM, the incremental addition of new time points to an existing data set, only requires the additional registration to the common space, followed by the recalculation of difference maps and voxel weighting for all time points.
Outputs
In addition to the calculated fDMs, the pipeline outputs three-plane slice images at a user-defined co ordinate, such as the center of the tumor or any other ROI, as well as scatter plot of voxel intensity changes (Fig. 3) for every generated map. Currently, the default threshold for a significant change in ADC is 0.4 mm2/s, as empirically derived by Ellingson et al.19 However, future development will include the option of calculating patient-specific values by using an ROI of normal tissue at each time point.
Figure 3.
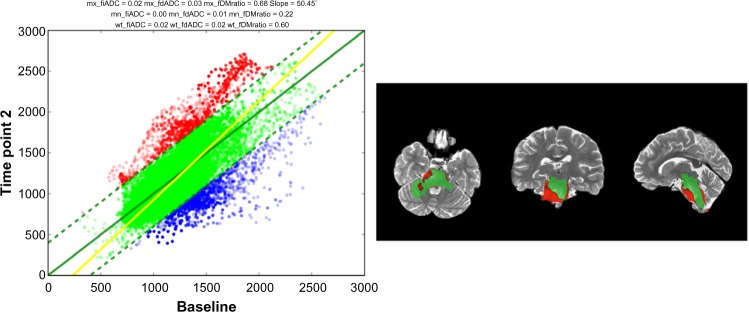
Sample time point output images.
Notes: Left: Scatter plot of ADC changes derived from fDM. Right: Three-plane view of fDM at a user-defined coordinate.
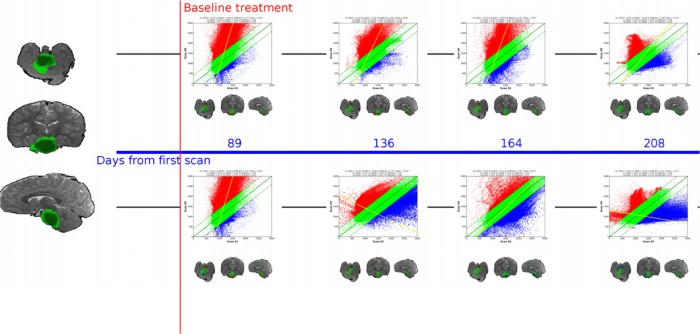
Optionally, sfDM will generate a subject-specific fDM timeline (Fig. 4). The fDM timeline provides a comprehensive view of the tumor progression from baseline through the subsequent time points.
Figure 4.
Sample patient timeline.
Notes: sfDM generates an fDM timeline allowing comprehensive visualization of the entire tumor progression through the course of treatment. Left image shows weighted tumor volume. Spacing between fDM’s is proportional to elapsed time.
Table 1 lists the metrics that are calculated for each map generated. Previous fDM studies have proposed multiple approaches for the calculation of fDM metrics. When comparing pre- and postradiation images, for example, including only voxels classified as tumor tissue at both time points (AND operation) was shown to have superior predictive power.11 Other studies suggest including any tissue classified as tumor in either time points (OR operation).20 sfDM generates fractional increase in ADC (fiADC), fractional decrease in ADC (fdADC), and the ratio of fiADC/(fdADC + 0.01) (fDM Ratio – the addition of 0.01 to the denominator avoids a potential division by 0) using both approaches. Additionally, sfDM generates a weighted metric, where each voxel’s contribution to the extracted metric is weighted by the proportion of time points in which the voxel is classified as tumor (ie, a voxel present in all time points is given a weight of 1, whereas a voxel only present in 10% of time points is given a weight of 0.1). This novel approach presents a compromise between excluding potentially explanatory voxels from the metric extraction (AND function) and inevitably including healthy tissue in the image subtraction, which may obfuscate desired within-tumor ADC changes (OR function).
Table 1.
sfDM output metrics.
| METRIC | DESCRIPTION | APPROACH | |
|---|---|---|---|
| Time point specific | Mean ADC | Mean ADC at specific time point | Time Point ROI |
| Tumor Volume | Volume in Reference space (mm3 and voxels) | ||
| fDM specific | mx_fiADC | fractional increase in ADC | Maximum ROI (OR) |
| mx_fdADC | fractional decrease in ADC | ||
| mx_fdm Ratio | mx_fiADC/(mx_fdADC + 0.01) | ||
| mn_fiADC | fractional increase in ADC | Minimum ROI (AND) | |
| mn_fdADC | fractional decrease in ADC | ||
| mn_fdm Ratio | mn_fiADC/(mn_fdADC + 0.01) | ||
| wt_fiADC | fractional increase in ADC | Weighted ROI (OR) | |
| wt_fdADC | fractional decrease in ADC | ||
| wt_fdm Ratio | wt_fiADC/(wt_fdADC + 0.01) |
Notes: Maximum ROI approach uses all voxels classified as tumor over any time point used in the analysis (OR operation). Minimum ROI approach uses only tissue classified as tumor in all time points (AND operation). Weighted ROI weighs each voxel’s contribution to the metric by the proportion of time points in which it is classified as tumor.
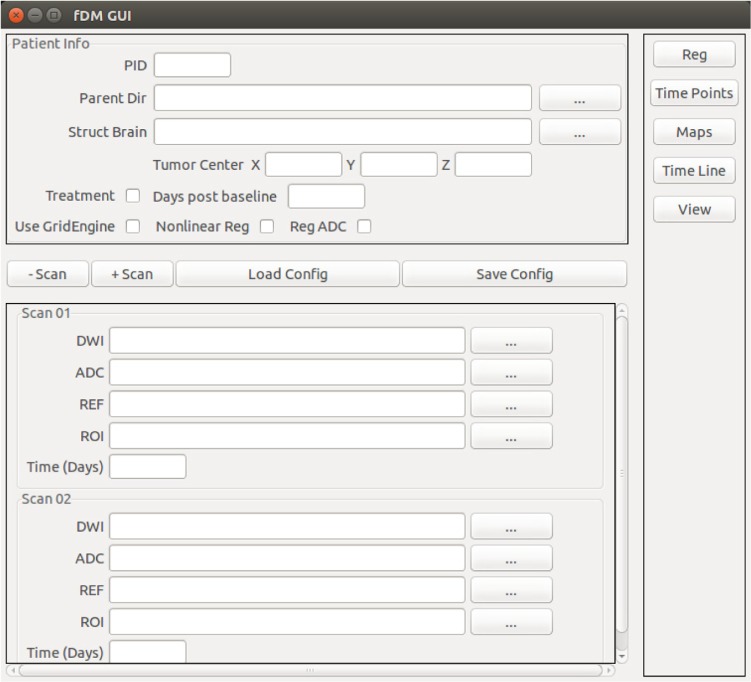
sfDM usage
sfDM is run using a graphical user interface (Fig. 5). The user must specify the path of each input, along with the days after each scan from the first available imaging time point (referenced as baseline). Finally, the user supplies the coordinates of the ROI to be used for image generation (in structural reference space), along with the option to include a treatment marker to signal the start of a treatment. This is only used in the fDM timeline image generation. The selected image location and options are conveniently saved in JavaScript Object Notation format to allow easy recall and record keeping. The processing modules are then run sequentially by using the buttons on the right-hand side, allowing the user to perform a data check upon each module completion.
Figure 5.
Graphical user interface for sfDM.
Figure 6 shows the graphical user interface for generating fDM timelines. This step is performed following registration and allows the user to select the time point pairs to be used in fDM generation. The timeline generation takes as input a “Serial” list, which consists of sequential parametric maps, and a “Baseline” list, which consists of subsequent time points compared to a specific time point of interest. These, however, are only by convention and may be freely modified by the user.
Figure 6.
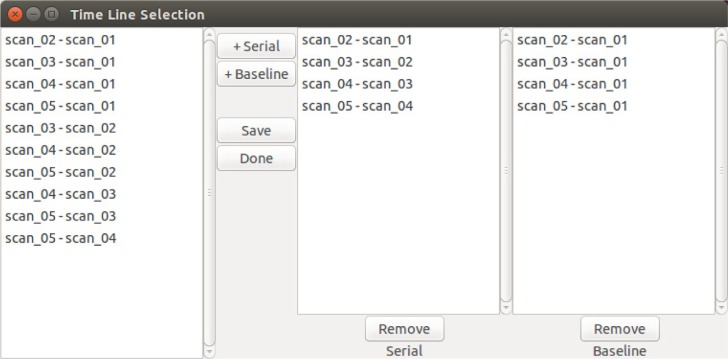
Timeline generation Graphical User Interface (GUI).
Notes: Example of user-defined time points that are used to generate subsequent fDMs and patient timeline.
Testing
To test for potential variation in the output as a result of different registration methods, we ran diffusion-weighted images from a newly diagnosed pediatric brainstem glioma patient enrolled in a Phase I tumor vaccine trial at our institution21 through our software using various settings. We ran five time points starting with the postradiation scan as baseline, using three registration methods: linear registration using b0 as initial source, linear registration using ADC as initial source, and nonlinear registration using b0 as initial source. All tumor ROIs were manually drawn using FSLView, and all additional parameters were set as default (see software documentation for full list of parameters at https://pypi.python.org/pypi/sfDM/). The patient was treated with concurrent radiation and chemotherapy prior to the start of vaccine, and the baseline scan was chosen as the time point at initial vaccine dose.
Additionally, we tested the nonlinear registration on a pediatric subject with a supratentorial mass that demonstrated a large calcification with mass effect distorting the adja left lateral ventricle, which later subsided. The large tissue distortion and proximity to the ventricle can be a challenge to image registration. We chose the scan prior to the observed calcification as the baseline and ran the pipeline with the default parameters using the nonlinear registration option.
Additionally, we tested the nonlinear registration on a pediatric subject with a supratentorial mass that demonstrated a large calcification with mass effect distorting the adjacent left lateral ventricle, which later subsided. The large tissue distortion and proximity to the ventricle can be a challenge to image registration. We chose the scan prior to the observed calcification as the baseline and ran the pipeline with the default parameters using the nonlinear registration option.
Results
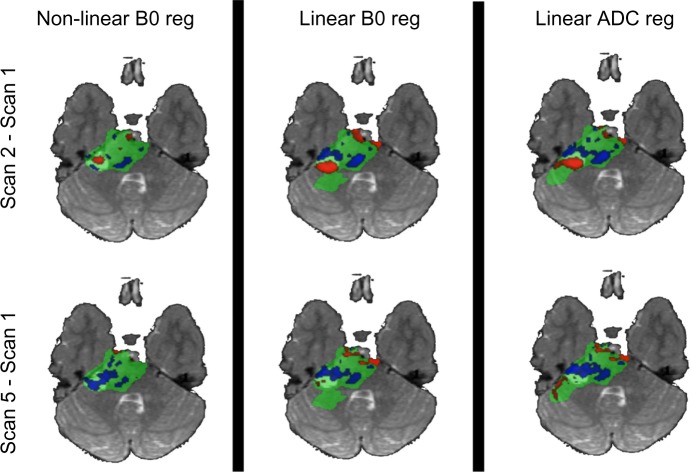
The provided defaults performed generally well with the data set used. However, using only ADC maps for initial registration required some manual removal of noise from the initial image on two of the five time points. Figure 7 shows the comparison between fDMs using three different registration methods at two different time points compared to baseline. As previously observed by Ellingson et al.22, nonlinear registration shows a decrease in the volume of tissue classified as significantly different (Fig. 8). While a small difference is observed between the linear registration methods, particularly in the fdADC, the overall trend is consistent. Finally, the nonlinear registration on the subject presenting with a calcification and mass effect was successful in spite of the large tissue distortion and intensity variation (Fig. 9). It should be noted that the diagnostic utility of a voxel-wise comparison between such large tissue distortion needs to be validated.
Figure 7.
fDM comparison between registration methods.
Notes: Nonlinear registration using b0 image as initial source shows the lowest number of significantly different voxels, as previously observed in the literature. Linear registrations using b0 and ADC maps as initial target show comparable parametric maps, with subtle differences around EPI distortion-susceptible regions.
Figure 8.
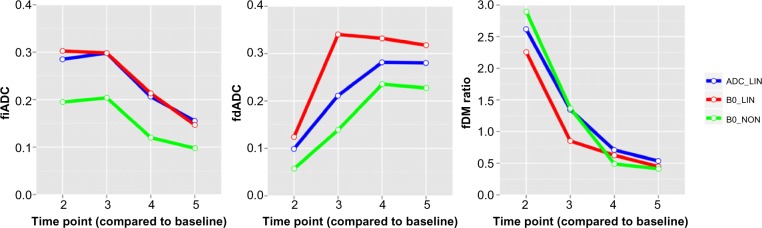
Comparison of extracted weighted fDM metrics using different image registration methods for five time points using the same patient. While some differences are observed, the overall trend is consistent.
Notes: ADC_LIN: Linear registrations only using ADC map as initial source image. B0_LIN: Linear registrations only using b0 image as initial source. B0_NON: final nonlinear registration, with b0 image as initial source.
Figure 9.
Nonlinear registration on a supratentorial mass that presented with calcification with mass effect on the adjacent left lateral ventricle. Despite large tissue distortion and intensity variation, the image registration was successful as evident by the consistent spatial location of the lateral ventricles.
Image registration required the longest amount of processing time, generally estimated at 20–30 minutes per image time point when using a linear pipeline approach (ie, no parallel processing). The final nonlinear registration option requires an additional 20 minutes per time point. Parallelizing the registrations is able to significantly reduce processing time. Care must be taken with the number of cores required, however, as this creates a bottleneck in memory with each parallel registration utilizing roughly 300 MB of RAM. All subsequent steps required less than 10 minutes per patient.
Discussion
When applied to fDMs, nonlinear registration algorithms have been previously shown to improve image registration and outcome prediction. However, care must be taken with assumptions made about corresponding tissue mapping across time points when doing so.22 When analyzing patient cohorts, a consistent registration method should be enforced whenever possible. Even though the observed registration methods showed similar trends, the subtle difference in extracted metrics could become a confounding factor.
sfDM is a promising tool for the generation of serial parametric maps. While the test cases we presented were pediatric brain tumors, sfDM can easily be applied to adult populations. Additionally, this approach can be extended to include multimodal quantitative imaging data beyond DWI including diffusion tensor MRI and also perfusion MR (which included permeability MR imaging and arterial spin labeling).23 Conveniently, sfDM provides the direct ADC registration method, which eliminates the need for a b0 image as the initial source of registration, which can be generalized to these additional imaging modalities.
This software does have limitations. Image registration is highly dependent on the quality of image acquisition, and careful quality control must be established in order to obtain meaningful results. Diffusion imaging can also suffer from susceptibility, gradient, and physiological artifacts, and ultimately, the results will only be as good as the images used to derive them. These artifacts can be particularly apparent in sensitive areas such as the brainstem, and we recommend a careful image acquisition protocol design in order to attenuate these problems in prospective studies (such as cardiac gating and proper shimming).
Furthermore, the presence of resection cavities, excessive mass effect, or tumor recurrence can cause the registrations to fail. To attenuate potential interpretation errors due to misregistration, we have incorporated break points at the end of each module that require visual inspection of the pipeline outputs. This does require the user to have previous experience with image registration methods in order to accurately identify any existing problems. Future iterations of the software will aim to incorporate more robust and automated methods for detecting and alerting users of any errors. This will have the added benefit of expanding the potential user base to include researchers and clinicians who may not be as familiar with the underlying methodology, with the goal of developing it into a true “point-of-care” tool. Finally, fDM is a useful and promising technique, but it is also very context-specific and requires clinical validation across several populations. Interpretation is highly dependent on tumor type, treatment, and patient population.24
This software is still young, and future work will be aimed at improving usability from user feedback. Additional features will include the ability to differentially weigh tissue types such as necrosis or contrast enhancement in order to increase fDM sensitivity, as well as allowing users to more easily alter patient-specific parameters directly from the graphical user interface.
Availability
sfDM is available for download from https://pypi.python.org/pypi/sfDM/with documentation and installation instructions. The latest source code can be found at http://github.com/PIRCImagingTools/sfDM. sfDM is distributed under the BSD license, which allows anyone to freely use, distribute, and modify the source code.
Conclusion
We have introduced sfDM: a new open-source package capable of generating serial functional diffusion maps and extracting meaningful tumor diffusion-weighted MR biomarkers. sfDM is an efficient way to standardize and process serial functional diffusion maps in large cohorts, emphasizing transparency and reproducibility.
Glossary
Abbreviations
- DWI
Diffusion Weighted Imaging
- ROI
Region of Interest
- fDM
functional Diffusion Mapping
- sfDM
serial functional Diffusion Mapping
- ADC
Apparent Diffusion Coefficient
- fiADC
fractional increase in ADC
- fdADC
fractional decrease in ADC
- fdm Ratio
ratio of fiADC/fdADC
Footnotes
Author Contributions
Conceived and designed the experiments: RC, AP, VG. Analyzed the data: RC. Wrote the first draft of the manuscript: RC. Contributed to the writing of the manuscript: RC, AP, VG. Agree with manuscript results and conclusions: RC, AP, VG. Jointly developed the structure and arguments for the paper: RC, AP, VG. Made critical revisions and approved final version: RC, AP, VG. All authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: J.T. Efird, Editor in Chief
FUNDING: This work is supported by National Library of Medicine (NLM) Training grant 5T15LM007059-27 and Ian’s Friend Foundation Grant. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETIN G INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
REFERENCES
- 1.Pollack IF, Jakacki RI. Childhood brain tumors: epidemiology, current management and future directions. Nat Rev Neurol. 2011;7(9):495–506. doi: 10.1038/nrneurol.2011.110. [DOI] [PubMed] [Google Scholar]
- 2.Panigrahy A, Blüml S. Neuroimaging of pediatric brain tumors: from basic to advanced magnetic resonance imaging (MRI) J Child Neurol. 2009;24(11):1343–65. doi: 10.1177/0883073809342129. [DOI] [PubMed] [Google Scholar]
- 3.Chenevert TL, Stegman LD, Taylor JM, et al. Diffusion magnetic resonance imaging: an early surrogate marker of therapeutic efficacy in brain tumors. J Natl Cancer Inst. 2000;92(24):2029–36. doi: 10.1093/jnci/92.24.2029. [DOI] [PubMed] [Google Scholar]
- 4.Chenevert TL, McKeever PE, Ross BD. Monitoring early response of experimental brain tumors to therapy using diffusion magnetic resonance imaging. Clin Cancer Res. 1997;3(9):1457–66. [PubMed] [Google Scholar]
- 5.Ross BD, Moffat BA, Lawrence TS, et al. Evaluation of cancer therapy using diffusion magnetic resonance imaging. Mol Cancer Ther. 2003;2(6):581–7. [PubMed] [Google Scholar]
- 6.Le Bihan D. The “wet mind”: water and functional neuroimaging. Phys Med Biol. 2007;52(7):R57–90. doi: 10.1088/0031-9155/52/7/R02. [DOI] [PubMed] [Google Scholar]
- 7.Tsougos I, Svolos P, Kousi E, et al. Differentiation of glioblastoma multiforme from metastatic brain tumor using proton magnetic resonance spectroscopy, diffusion and perfusion metrics at 3T. Cancer Imaging. 2012;12:423–36. doi: 10.1102/1470-7330.2012.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvar JA, Meli FJ, Romero C, et al. Characterization of brain tumors by MRS, DWI and Ki-67 labeling index. J Neurooncol. 2005;72(3):273–80. doi: 10.1007/s11060-004-3342-2. [DOI] [PubMed] [Google Scholar]
- 9.Holodny AI, Makeyev S, Beattie BJ, Riad S, Blasberg RG. Apparent diffusion coefficient of glial neoplasms: correlation with fluorodeoxyglucose-positron-emission tomography and gadolinium-enhanced MR imaging. Am J Neuroradiol. 2010;31(6):1042–8. doi: 10.3174/ajnr.A1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moffat BA, Chenevert TL, Lawrence TS, et al. Functional diffusion map: a noninvasive MRI biomarker for early stratification of clinical brain tumor response. Proc Natl Acad Sci USA. 2005;102(15):5524–9. doi: 10.1073/pnas.0501532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamstra DA, Galbán CJ, Meyer CR, et al. Functional diffusion map as an early imaging biomarker for high-grade glioma: correlation with conventional radiologic response and overall survival. J Clin Oncol. 2008;26(20):3387–94. doi: 10.1200/JCO.2007.15.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada H, Pollack IF, Lieberman F, et al. Gene therapy of malignant gliomas: a pilot study of vaccination with irradiated autologous glioma and dendritic cells admixed with IL-4 transduced fibroblasts to elicit an immune response. Hum Gene Ther. 2001;12(5):575–95. doi: 10.1089/104303401300042528. [DOI] [PubMed] [Google Scholar]
- 13.Galbán S, Brisset J-C, Rehemtulla A, Chenevert TL, Ross BD, Galbán CJ. Diffusion-weighted MRI for assessment of early cancer treatment response. Curr Pharm Biotechnol. 2010;11(6):701–8. doi: 10.2174/138920110792246627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorgolewski K, Burns CD, Madison C, et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in Python. Front Neuroinform. 2011;5:13. doi: 10.3389/fninf.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM, FSL Neuroimage. 2012;62(2):782–90. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162–73. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 17.Millman KJ, Brett M. Analysis of functional magnetic resonance imaging in Python. Comput Sci Eng. 2007;9(3):52–5. [Google Scholar]
- 18.Open Grid Scheduler: The official Open Source Grid Engine. 2014. [Accessed July 24, 2014]. Available from: http://gridscheduler.sourceforge.net/
- 19.Ellingson BM, Malkin MG, Rand SD, et al. Validation of functional diffusion maps (fDMs) as a biomarker for human glioma cellularity. J Magn Reson Imaging. 2010;31(3):538–48. doi: 10.1002/jmri.22068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellingson BM, Cloughesy TF, Zaw T, et al. Functional diffusion maps (fDMs) evaluated before and after radiochemotherapy predict progression-free and overall survival in newly diagnosed glioblastoma. Neuro Oncol. 2012;14(3):333–43. doi: 10.1093/neuonc/nor220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollack IF, Jakacki RI, Butterfield LH, et al. Antigen-specific immune responses and clinical outcome after vaccination with glioma-associated antigen peptides and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethyl-cellulose in children with newly diagnosed malignant brainstem and n. J Clin Oncol. 2014;32(19):2050–8. doi: 10.1200/JCO.2013.54.0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellingson BM, Cloughesy TF, Lai A, Nghiemphu PL, Pope WB. Nonlinear registration of diffusion-weighted images improves clinical sensitivity of functional diffusion maps in recurrent glioblastoma treated with bevacizumab. Magn Reson Med. 2011;67(1):237–45. doi: 10.1002/mrm.23003. [DOI] [PubMed] [Google Scholar]
- 23.Tsien C, Galbán CJ, Chenevert TL, et al. Parametric response map as an imaging biomarker to distinguish progression from pseudoprogression in high-grade glioma. J Clin Oncol. 2010;28(13):2293–9. doi: 10.1200/JCO.2009.25.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grech-Sollars M, Saunders DE, Phipps KP, et al. Challenges for the functional diffusion map in pediatric brain tumors. Neuro Oncol. 2013;16(3):449–56. doi: 10.1093/neuonc/not197. [DOI] [PMC free article] [PubMed] [Google Scholar]