Abstract
miR-155 is a regulator of immune cell development and function that is generally thought to be immunostimulatory. However, we report here that genetic ablation of miR-155 renders mice resistant to chemical carcinogenesis and the growth of several transplanted tumors, suggesting that miR-155 functions in immunosuppression and tumor promotion. Host miR-155 deficiency promoted overall antitumor immunity despite the finding of defective responses of miR-155-deficient dendritic cells and antitumor T cells. Further analysis of immune cell compartments revealed that miR-155 regulated the accumulation of functional myeloid-derived suppressive cells (MDSC) in the tumor microenvironment. Specifically, miR-155 mediated MDSC suppressor activity through at least two mechanisms, including SOCS1 repression and a reduced ability to license the generation of CD4+Foxp3+ regulatory T cells (Treg). Importantly, we demonstrated that miR-155 expression was required for MDSC to facilitate tumor growth. Thus, our results revealed a contextual function for miR-155 in antitumor immunity, with a role in MDSC support that appears to dominate in tumor-bearing hosts. Overall, the balance of these cellular effects appears to be a root determinant of whether miR-155 promotes or inhibits tumor growth.
MicroRNAs are evolutionarily conserved small non-coding RNAs that posttranscriptionally modulate the expression of multiple target genes and are hence implicated in a wide series of cellular and developmental processes (1, 2). microRNA-155 (miR-155) is processed from the B-cell integration cluster (BIC), a noncoding transcript primarily upregulated in both activated B and T cells (3) and in monocytes/macrophages upon inflammation (4, 5). Recent gene-targeting studies of miR-155 demonstrate a broad role for miR-155 in the regulation of both immune cell development and function (6, 7). Indeed, miR-155−/− mice have global immune defects due to defective B and T cell immunity and reduced dendritic cell (DC) function. Particularly, miR-155 deficient DCs fail to present antigens efficiently (6) and produce cytokines (8), whereas miR-155 in CD4+ T cells regulates differentiation into the Th1, Th2 and Th17 pathways (6, 9, 10). Furthermore, miR-155 is required for CD8+ T cell responses to acute viral and bacterial challenges (11–14). In addition to these immunostimulatory effects, miR-155 can also exert some immunosuppressive effects, such as promoting the development (15), or homeostasis and fitness (16) of Tregs, and expansion of functional MDSCs (17). Thus, miR-155 could modulate protective immune responses and inflammation through distinct mechanisms.
miR-155 dysregulation is closely related to cancer (4). miR-155 transgenic mice develop B-cell malignancy (18) and elevated miR-155 expression was reported in several types of human B-cell lymphomas (19). A correlation between increased miR-155 and development of tumors such as leukemias, glioblastoma, and breast, lung or gastric cancers has been established recently (20, 21). Therefore, targeting miR-155 has been proposed as a promising approach to treat both hematopoietic and solid cancers (22–24). However, the potent immunostimulatory effects of miR-155 have also been observed in the context of tumor. Notably, the roles of miR-155 in effector CD8+ T cells (13, 25), tumor-infiltrating DCs (26, 27) and tumor-associated macrophages (28, 29) that can be modulated to potentiate cancer immunotherapies. Thus, when cancer is treated in a immunocompetent host by inhibiting miR-155, outcomes are difficult to predict. Importantly, underlying mechanisms of host miR-155 in modulating tumor growth are still poorly understood. We show here that host miR-155 deficiency hampers the accumulaiton of functional MDSCs and inducible Treg cells in the tumor microenvironment, thereby promoting anti-tumor T cell immunity and retarding tumor growth.
Materials and Methods
Mice, cell lines and reagents
C57BL/6 miR-155−/−, CD45.1 and CD90.1 mice were purchased from the Jackson Laboratory, OT-I Rag1−/− and OT-II Rag1−/− mice from Taconic, and C57BL/6 miR-155+/+ mice from NCI-Frederick. Dr. Hans Schreiber (University of Chicago) provided the MC38, EG7, B16F10, B16-SIY cell lines, anti-Gr1 antibodies (RB6-8C5) and 2C transgenic mice. Murine Lewis Lung Carcinoma (LLC1) cells were purchased from ATCC (CRL-1642). LLC1 cells were infected with MIGR1-OVA-IRES-eGFP (30) and OVA-expressing cells (LLC1-OVA) were sorted twice based on GFP expression. OVA production was confirmed by ELISA (data not shown). All the cell lines were routinely tested for mycoplasma infections by culture and DNA stain, and maintained in complete medium composed of RPMI 1640 with 5% FBS. All animal experiments were approved by institutional animal use committees of the University of Texas Health Science Center at San Antonio and Northwestern University. The OVA-derived peptide OVA-I (SIINFEKL) was synthesized by GenScript. Dichlorofluorescin diacetate (DCFDA), azoxymethane (AOM) and 5-fluorouracil (5-FU) were purchased from Sigma-Aldrich. Dextran sulfate sodium salt (DSS) was purchased from Affymetrix, Inc. All the mAbs for flow cytometry were purchased from eBioscience and BioLegend. The Annexin V apoptosis detection kit was from BioLegend. The Kb/OVA tetramers were provided by the National Institutes of Health Tetramer Core Facility (Atlanta, GA). Depleting mAb clone GK1.5 (anti-CD4), clone 53.6.7 (anti-CD8α) and clone PK136 (anti-NK1.1) were purchased from Bio X Cell. Nω-hydroxy-nor-Arginine (Nor-NOHA) and arginase I activity kit were purchased from Cayman Chemical Company.
Analysis of cells by flow cytometry
All samples were initially incubated with 2.4G2 to block antibody binding to Fc receptors. Single cell suspensions were stained with 1 µg of relevant mAbs and then washed twice with cold PBS. ROS detection by DCFDA staining was conducted as described by Youn et al. (31). The annexin V staining, Kb/OVA tetramer staining, Foxp3 staining and intracellular IFN-γ staining were performed as previously described (32). Samples were conducted on a MACSQuant Analyzer (Miltenyi Biotec) and data were analyzed with FlowJo software.
In vivo killing assay
Analysis of tumor antigen-specific effector CTL activity in vivo was performed as previously described (32). Briefly, OVA-I (SIINFEKL) peptide-pulsed eFluor® 450high and SIY-peptide-pulsed eFluor® 450low splenocytes were mixed at a ratio of 1:1 and a total of 2×107 cells were injected i.p. into recipient animals. Draining lymph nodes (DLN) and spleen were then harvested 24 h after adoptive transfer and eFluor® 450 fluorescence intensity was analyzed by flow cytometry.
MDSC suppressive assay
Splenic MDSCs from tumor-bearing WT or miR-155−/− mice were selected using CD11b MicroBeads (Miltenyi Biotec), and tumor-infiltrating CD115+CD11b+Gr1+ or CD115−CD11b+Gr1+ MDSCs were sorted by a BD FACSAria™ cell sorter from LLC1-bearing mice. MDSCs were added at different ratios to OT-I or 2C splenocytes stimulated with 0.5 µg/ml OVA-I or SIY peptides for 3 days, and 3[H] thymidine uptake was measured. For experiments that examined the effect of arginase inhibitors, nor-NOHA (NW-hydroxyl-nor-l-arginine, 0.5 mM), were added at the beginning of the culture. To evaluate MDSC tolerogenic activity on in vivo T cell function, naive OT-1 CD90.1 cells (2×106 per mouse) were transferred to CD90.2 congeneic recipients, which were s.c. immunized, 2 days later, with 10 µg OVA-I peptides in incomplete Freund's adjuvant (IFA). MDSCs (2×106) from MC38 tumor-bearing WT or miR-155−/− mice, either pulsed or not with OVA-I peptides, were transferred on the same day of the immunization. DLNs were collected 10 days after immunization and stimulated with 0.5 µg/ml OVA-I in vitro for 3 days to measure T cell proliferation by 3[H] thymidine uptake and IFN-γ secreting CD8+ T cells by flow cytometry.
Arginase activity
Arginase activity was measured in cell lysates using the commercially available QuantiChrom Arginase Assay kit (BioAssay Systems, Hayward, USA) according to the manufacturer's instructions.
BM-derived MDSC generation
Tibias and femurs from C57BL/6 mice were removed using sterile techniques and bone marrow (BM) cells were flushed. To obtain BM-derived MDSCs, cells were cultured with GM-CSF (40 ng/ml, Biolegend) and IL-6 (40 ng/ml, Biolegend) for 4 days. BM-derived MDSCs were selected using CD11b or Gr1 MicroBeads (Miltenyi Biotec).
RNA extraction and real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer's instructions. miR-155 experssion was detected by TaqMan MicroRNA Assay kit (Applied Biosystems). The cDNA synthesis was performed using SuperScript®One-Step RT-PCR (Invitrogen). Quantitative real-time PCR was used to quantify a series of MDSC-associated genes by SYBR Green (Bio-Rad) and relative abundance of each mRNA was normalized to GAPDH mRNA.
Transfection of BM-derived MDSCs
The transfection of primary BM cells was performed according to the instructions of the manufacturer (AMAXA). BM cells were treated with GM-CSF (40 ng/ml, Biolegend) for 24 h, followed by the transfection with 1 µM pre-miR-155/BIC (P-MDSC, Ambion), 2 µM miR-155 inhibitor miRNA (I-MDSC, Dharmacon) or control oligonucleotides (C-MDSC, Dharmacon) by AMAXA. For knockdown of SOCS1, specific and respective control siRNAs used for transfection were from Santa Cruz Biotech. To recover, cells were cultured for additional 72 h in the presence of GM-CSF (40 ng/ml, Biolegend) and IL-6 (40 ng/ml, Biolegend) after transfection. After selection with CD11b or Gr1 MicroBeads (Miltenyi Biotec), these GM-CSF and IL-6-conditioned BM-derived MDSCs were tesed for suppressive assay.
Treg induction
Splenic WT or miR-155−/− CD4+CD62L+ naïve T cells were selected with a CD4+CD62L+ T cell isolation kit (Miltenyi Biotec), and injected i.v. at 5 × 106 per mouse into CD45.1 mice followed by a s.c. injection of 106 LLC1-OVA cells. The conversion of transferred T cells to Foxp3+ cells (CD45.2+) in DLN and spleen from LLC1-OVA tumor-bearing mice were detected by flow cytometer 9 d after tumor cell injection. For MDSC-mediated Treg induction, splenic WT and miR-155−/− Gr1+CD11b+ MDSCs from LLC1 tumor-bearing mice were cocultured with OT-II splenocytes at a 1:4 ratio for 5 d in the absence or presence of 2 ng/ml TGF-β, and induced CD25+Foxp3+ cells among total CD4+ T cells were subsequently determined by flow cytometry.
AOM and DSS treatment
For the colitis-associated colon cancer model, mice were given 10 mg/kg AOM via i.p. injection. One week later 2.5% DSS was given in the drinking water for 7 d followed by 14 d of normal water for a total of three cycles. Colons were harvested, flushed of feces, cut longitudinally and fixed in 10% buffered formalin overnight. The colons were scored with the aid of a magnifier for the number of colonic neoplasms to determine the incidence (number of animals with at least one tumor) and multiplicity (number of tumors per animal) of neoplasms. Tumor area was also evaluated based on length and width.
Tumor challenge and treatments
B16F10, B16-SIY, LLC1, LLC1-OVA, MC38 or EG7 cells (1 × 106) in suspension were injected s.c. For MDSC depletion, 3 d after tumor cell injection mice were injected i.p. by 5-FU (50 mg/kg) or anti-Gr1 antibodies (RB6-8C5, 200 µg) once every 4 d. Depletion of CD4+ T cells, CD8+ T cells or NK cells was achieved by twice a week i.p. injection of depleting mAb clone GK1.5 (anti-CD4, 200 µg), clone 53.6.7 (anti-CD8α, 200 µg) or clone PK136 (anti-NK1.1, 200 µg) starting one day prior to tumor challenge. Flow cytometry confirmed depletion efficiency of target cells for 3 d following injections. For adoptive transfer of MDSCs, splenic Gr1+CD11b+ MDSCs from tumor-bearing WT or miR-155−/− mice were injected i.v. at 5 × 106 per mouse into LLC1-bearing mice at d7 and d14. For adoptive transfer of Tregs (33), splenic CD4+CD25+ Treg cells were selected with a CD4+CD25+ regulatory T cell isolation kit (Miltenyi Biotec) from WT or miR-155−/− mice, and i.v. injected at 2 × 106 per mouse into LLC1-bearing mice on d7, d14 and d20. The size of tumor was determined at 2–3-day intervals. Tumor volumes were measured along 3 orthogonal axes (a, b, and c) and calculated as abc/2.
Statistical analysis
Mean values were compared using an unpaired Student’s two-tailed t test. Probability values >0.05 were considered non-significant.
Results
Host miR-155 promotes tumor growth
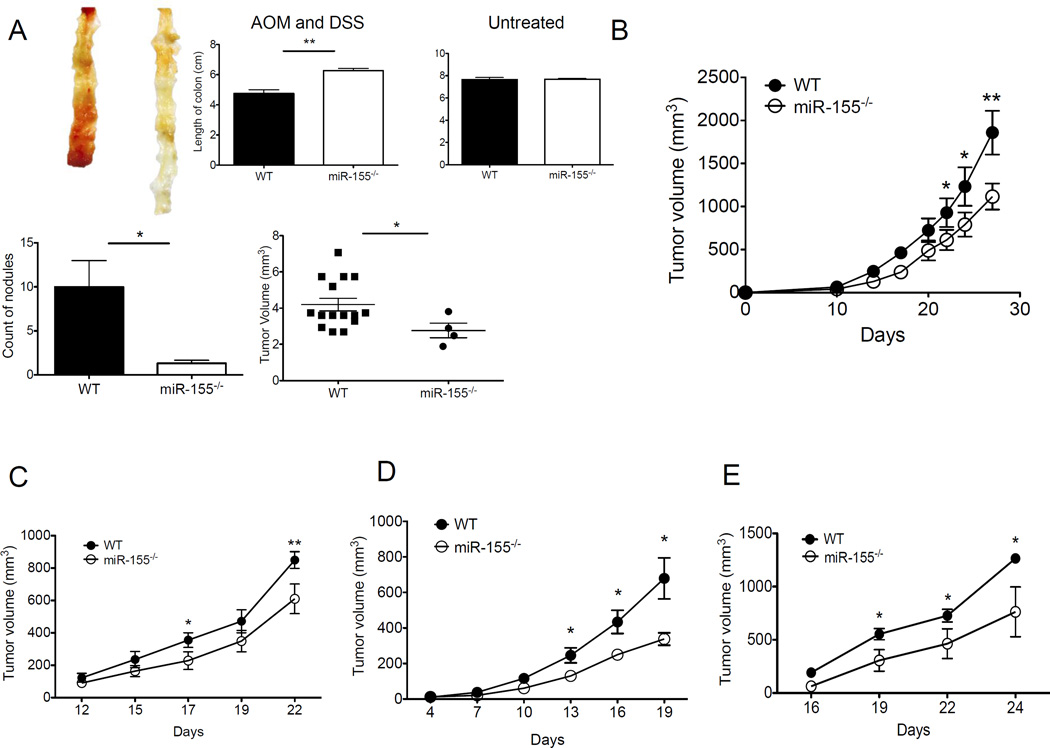
The immunoregulatory role of miR-155 has been well documented in numerous experimental settings. However, the specific contributions of endogenous miR-155 to antitumor immunity and tumorigenesis are poorly understood. We compared chemically-induced tumor and transplanted tumor growth in miR-155–deficient (miR-155−/−) versus syngeneic, wild-type (WT, miR-155+/+) mice. Mice were given AOM and DSS, as previously described to promote colorectal carcinogenesis (34). Upon AOM and DSS challenge, miR-155−/− mice exhibited less acute body weight loss comparable to WT mice (data not shown). AOM and DSS produced colonic tumors in all 8 WT mice, but in 3 of 8 miR-155−/− mice. The multiplicity of colonic neoplasms (number and size of tumors) was also significantly decreased in miR-155−/− mice. However, there was no sinificant difference in colon length between untreated WT and miR-155−/− mice (Fig. 1A). Moreover, WT and miR-155−/− mice given AOM alone or DSS alone had no macroscopic colonic tumors (data not shown). We next studied the role of host miR-155 on transplantable tumor growth in miR-155−/− mice. MC38 colon cancer cells (Fig. 1B) and LLC1 Lewis lung carcinoma cells (Fig. 1C) were s.c. inoculated into WT or miR-155−/− mice. Tumors injected into miR-155−/− mice exhibited delayed growth compared with those in control mice (Fig. 1B,C). In addition, miR-155 deficiency was also effective in inhibiting the growth of immunogenic LLC1-OVA (Fig. 1D). Similarly, the growth of lyphoma EG7 (expressing OVA antigen) tumors was inhibited in miR-155−/− mice (Fig. 1E). However, the sizes of B16-SIY melanoma (expressing SIY antigen) were comparable between the WT and miR-155−/− mice at multiple time points (Supple. Fig. 1A), suggesting that the role of host miR-155 in tumor growth is tumor-dependent.
Figure 1. Tumor growth is inhibited in miR-155−/− mice.
(A) Chemically-induced colon tumor incidence in WT and miR-155−/− mice. Mice were given no treatment, or DSS and AOM as described in Material and Methods. Length of colon, size and numbers of colon polyps were recorded (n=5). (B–E) Mice (n=3–5 per group) were inoculated s.c. with 106 MC38 (B), LLC1 (C), LLC1-OVA (D), or EG7 (E) cells. Data (mean ± SEM) are representative of at least 5 independent experiments. *, p< 0.05; **, p<0.01.
Host miR-155 deficiency enhances antigen-specific antitumor T cell immunity
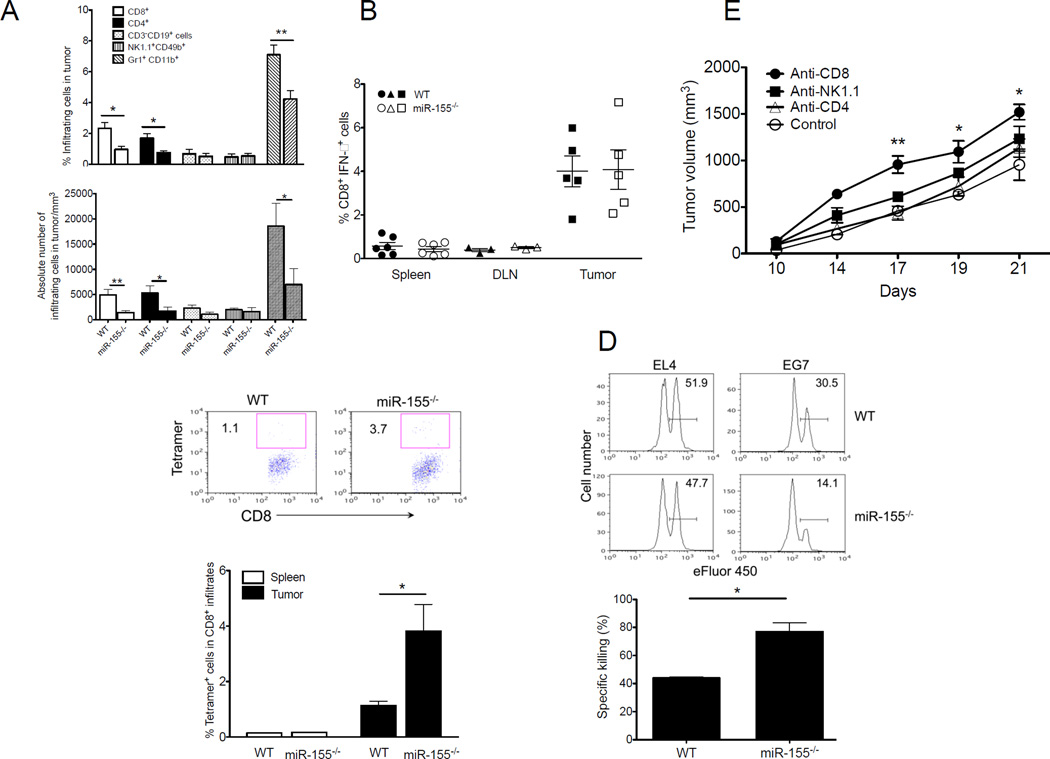
Given the importance of miR-155 in immune regulation, we next examined the phenotype and cytokine profile of tumor-infiltrating immune cells in tumor-bearing mice. At 19 days after tumor inoculation, we found no significant alterations in the percentages of B cells (CD19+), NK cells (NK1.1+), or myeloid DCs (CD11b+CD11c+) in local infiltrates of EG7 (Fig. 2A) or LLC1-OVA (Supple. Fig. 2A) tumors in miR-155−/− versus WT mice. Interestingly, remarkably fewer tumor-infiltrating CD8+, CD4+ lymphocytes were found in miR-155−/− versus WT mice (Fig. 2A and Supple. Fig. 2A). Although percentages of tumor-infiltrating IFN-γ+CD8+ T cells were comparable between groups, tetramer staining showed a greater number of OVA-reactive (tumor-specific) CD8+ T cells in EG7-bearing (Fig. 2B, C) or LLC1-OVA-bearing (Supple. Fig. 2B,C) miR-155−/− mice than WT mice. We next examined the cytolytic function of tumor antigen-specific T cells. Target cell lysis in vivo was remarkably improved in DLN of EG7 tumor-bearing miR-155−/− mice compared to tumor-bearing WT mice (Fig. 2D). To assess the roles of CD4+, CD8+ and NK cells in the tumor-inhibiting effects observed in miR-155−/− mice, mice were inoculated with EG7 cells, and subsequently received depleting anti-CD4, anti-CD8α or anti-NK1.1 antibodies against CD4+ or CD8+ T cells, or NK cells, respectively. Notably, the tumor-inhibiting advantage of host miR-155 deficiency was primarily dependent on CD8+ cells, but independent of CD4+ cells or NK cells (Fig. 2E). These data suggest that loss of miR-155 expression in mice results in the enhanced antitumor T cell immunity that contributes to the inhibition of immunogenic tumor growth.
Figure 2. Host miR-155 deficiency enhances antigen-specific antitumor T cell immunity.
(A) Percentage and absolute number of CD4+CD3+, CD8+CD3+, Gr1+CD11b+, CD3−CD19+ and CD49b+NK1.1+ cells in tumor infiltrates of WT or miR-155−/− mice collected 21 days after inoculation with EG7 tumor cells (n=5). (B) CD8+IFN-γ+ T cell frequency in spleen, DLN and tumor from EG7-bearing WT or miR-155−/− mice 21 days after tumor inoculation (n=3–6). (C) Representative flow cytometric analysis of tumor antigen-specific CD8+ T cells from EG7-bearing WT or miR-155−/− mice. Frequency of tetramer+ cells specific for the OVA epitope SIINFEKL in CD8+ infiltrates from mice in B, was summarized. (D) Representative flow cytometric analyses of in vivo antigen-specific killing capacity of antitumor T cells from EL4- or EG7-bearing WT and miR-155−/− mice. Equal numbers of eFluor® 450high SIINFEKL peptide-pulsed and eFluor® 450low SIY–peptide-pulsed WT splenocytes were adoptively transferred into tumor-bearing mice. Numbers denote percentage of SIINFEKL peptide-pulsed target cell killing in DLN. Percent killing for EG7-bearing mice in DLN was calculated as described in Methods (n=3). (E) In miR-155−/− mice (n= 5), depletion of CD4+ T cells, CD8+ T cells, or NK cells was achieved by twice weekly i.p. injection of control Ig, anti-CD4, anti-CD8, or anti–NK1.1 depleting Abs, respectively, beginning 1 day prior to tumor challenge. Data are representative of 2 independent experiments. *, p< 0.05; **, p<0.01.
Previous studies have demonstrated the involvement of miR-155 in the DCs (26, 27) and T cells (13, 25) in controlling tumor growth. As expected, we found that tumor-associated miR-155−/− DCs expressed less MHC-II (Supple. Fig. 3A), and induced less antigen-specific CD8+ T cell proliferation compared to WT DCs (Supple. Fig. 3B). Similarly, tumor-infiltrated miR-155−/− CD8+ T cells sorted from LLC1-OVA-tumors displayed reduced response to DCs pulsed with OVA-I peptides in vitro (Supple. Fig. 3C). In this immune cell-specific context, it is of interest that we observed intrinsic defects in miR-155−/− tumor-associated DCs and antitumor T cells.
miR-155 is required for MDSC accumulation in tumor-bearing mice
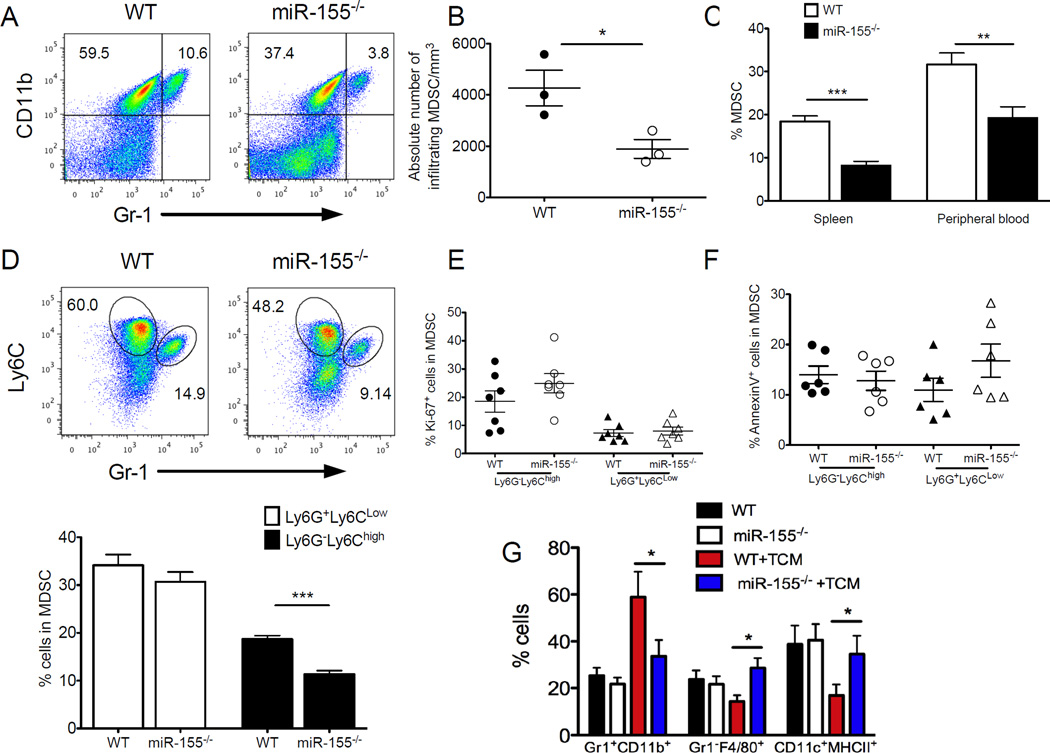
Although above data suggest a cell-intrinsic role of miR-155 in tumor-associated DCs and antitumor T cells, host miR-155 deficiency promoted overall antitumor T cell immunity and inhibited tumor growth. In search of a cellular mechanism for the miR-155-mediated tumor-promoting effect we investigated the well-defined immunosuppressive immune cell subsets in tumor, including MDSCs and Tregs. We observed that intratumoral Gr1+CD11b+ MDSCs were consistently decreased in LLC1-OVA-bearing miR-155−/− mice in comparison to WT mice (Fig. 3A,B). Further analysis revealed significant reductions in the percentage of CD11b+Gr1+ cells from miR-155−/− mice compared with WT controls in spleen and peripheral blood (Fig. 3C). We also tested other tumor models including EL4, B16F10 and LLC1, and found that miR-155−/− mice have much fewer splenic MDSCs than WT mice (Supple. Fig. 4A). These results confirmed that miR-155 is required for MDSC accumulation under tumor-bearing conditions because no significant differences were noted in the percentages of splenic CD11b+Gr1+ cells between tumor-free miR-155−/− and WT mice (data not shown). MDSC consists of ly6G−ly6Chigh (monocytic) and ly6G+ly6Clow (granulocytic) subpopulations (31, 35). Of note, the preferential reduction of the splenic (Supple. Fig. 4B,C) and tumor-infiltrating (Fig. 3D) monocytic ly6G−ly6Chigh subset was observed in LLC1-OVA-bearing miR-155−/− mice compared with WT mice. These results suggest miR-155 is required for tumor-associated MDSC accumulation particularly with a monocytic phenotype. To dissect the role of miR-155 further in regulating MDSC accumulation, we stained with Ki67 (Fig. 3E) and annexin V (Fig. 3F) to test the proliferative ability and apoptotic status of MDSCs within the tumor microenvironment, respectively. No significant differences in both granulocytic and monocytic MDSC subsets were found between WT mice and miR-155−/− mice.
Figure 3. miR-155 is required for MDSC accumulation in the tumor microenvironment.
(A) Percent tumor-infiltrating Gr1+CD11b+ MDSCs were determined by flow cytometry from LLC1-OVA tumor-bearing mice. (B) The absolute number of tumor-infiltrating MDSCs (n=3). (C) Percent MDSC in spleen (n=9) and peripheral blood (n=5) from LLC1-OVA tumor-bearing mice were summarized. (D) Percent CD11b+Ly6G+Ly6Clow (granulocytic) and CD11b+Ly6G−Ly6Chigh (monocytic) MDSCs were indicated within plots and summarized (n=9). Flow cytometry anlaysis of expression of ki-67 (E) and annexin-V (F) on both granulocytic and monocytic tumor-infiltrating MDSCs (n=6). (G) Bone marrow cells were cultured with GM-CSF and IL-4 for 5 d in complete culture medium or in the tumor cell conditioned medium (TCM). The cell phenotypes were examined by flow cytometry. Data are given as means ± SEM. *, p< 0.05; **, p<0.01. Data are representative of 2 independent experiments.
Given the critical function of miR-155 in promoting myeloid lineage commitment in hematopoietic stem cells and myeloid progenitors (36), we asked whether MDSC differentiation requires miR-155. To evaluate differentiation of myeloid cells in the presence of tumor-derived factors, BM cells from miR-155−/− mice and their WT littermates were cultured with GM-CSF for 5 d in tumor cell conditioned medium (TCM). As expected, tumor-derived factors significantly reduced the differentiation of DCs and macrophages and increased the generation of Gr1+CD11b+ MDSCs in WT populations (Fig. 3G), consistent with previous observation (37). In contrast, TCM failed to inhibit the differentiation of myeloid progenitor cells appreciably from miR-155−/− mice (Fig. 3G), suggesting an important role of miR-155 in MDSC differentiation in the tumor microenvironment.
miR-155 is required for MDSC suppressive function during tumor growth
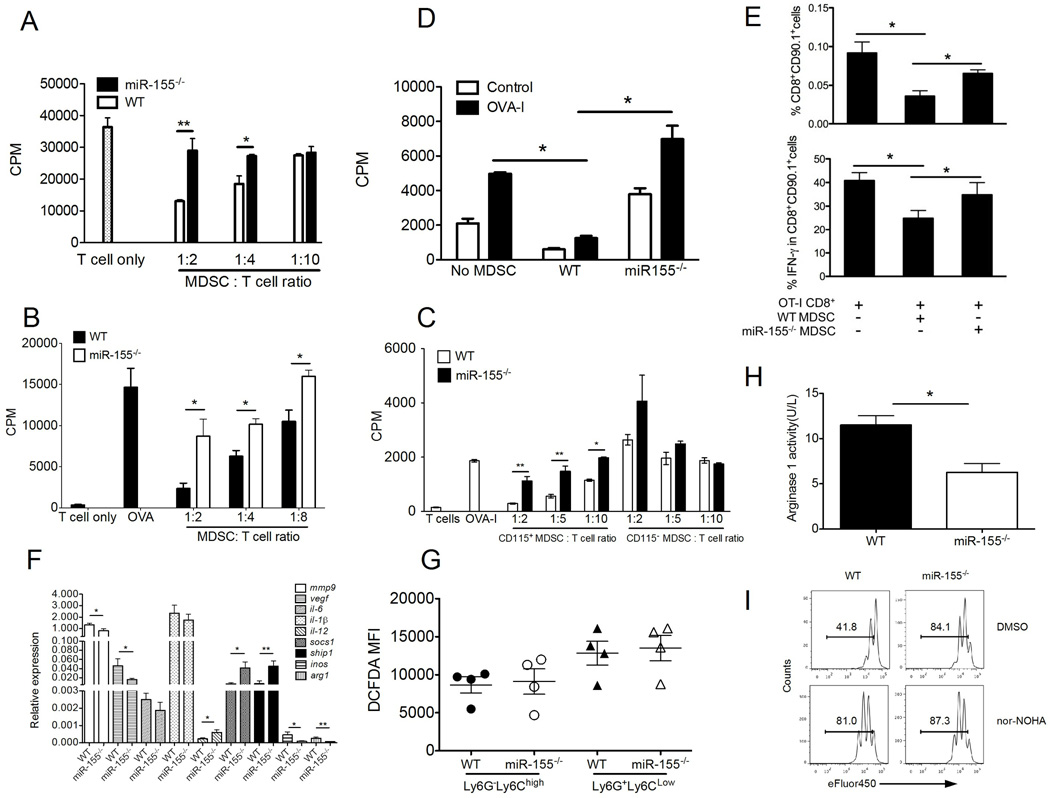
To determine whether miR-155 is required for MDSC suppressive function, we purified CD11b+ cells from MC38-bearing mice and co-cultured with OT-I splenocytes. Notably, the miR-155−/− MDSC appreciably lost their capacity to suppress proliferation of antigen-specific T cell in vitro, whereas WT MDSC remained strongly suppressive (Fig. 4A). To corroborate these findings further, splenic MDSCs were isolated from EG7 (Fig. 4B) or B16-SIY (Supple. Fig. 1B) tumor-bearing WT or miR-155−/− mice. As expected, miR-155−/− MDSCs were unable to inhibit antigen-specific T cell proliferation in vitro compared with WT MDSCs. Emerging data show that the degree of immunosuppression varies among populations of MDSCs isolated from different organs, and intratumoral MDSCs are the most immunosuppressive (38). In the LLC1 tumor model, CD115 acts as a function marker for MDSCs (39). Thus, we sorted CD115+CD11b+ cells and CD115−CD11b+ cells from LLC1 tumor tissues, and compared their suppressive activity between WT and miR-155−/− mice (Fig. 4C). Consistent with previous results, intratumoral WT CD115+CD11b+ cells but not CD115−CD11b+ cells were inhibitory. In contrast, miR-155−/− CD115+CD11b+ cells were unable to suppress T cell proliferation (Fig. 4B). To evaluate MDSC tolerogenic activity on antigen-specific CD8+ T cells in vivo, MDSCs from tumor-bearing WT or miR-155−/− mice, either pulsed or not with OVA-I peptides, were transferred on the same day of the immunization. DLNs were collected 10 days after immunization and stimulated in vitro to measure T cell proliferation (Fig. 4D) and enumerate CD8+ T cells producing IFN-γ (Fig. 4E). Both the number of transferred CD90.1+ cells and number of IFN-γ secreting CD8+ T cells in DLNs were significantly reduced in mice that received MDSCs derived from WT tumor-bearing mice, but not miR-155−/− tumor-bearing mice.
Figure 4. miR-155 is required for MDSC suppressive function during tumor growth.
(A) Suppressive activity of MDSCs from MC38-bearing WT versus miR-155−/− mice. Splenic Gr1+CD11b+ MDSCs from either MC38-bearing WT or miR-155−/− mice were added at different ratios to OT-I splenocytes stimulated with OVA-I peptides for 3d, and 3[H] thymidine uptake was measured. The suppressive activities of tumor-infiltrating MDSCs from EG7-bearing (B) or LLC1-bearing (C) WT and miR-155−/− mice were assessed in a similar manner as described in A. (D) To evaluate MDSC tolerogenic activity on in vivo T cell function, naive OT-1 CD90.1 cells were transferred to CD90.2 congeneic recipients, which were s.c. immunized, 2 days later, with OVA-I peptides in IFA. MDSCs from MC38 tumor- bearing WT or miR-155−/− mice, either pulsed or not with OVA-I peptides, were transferred on the same day of the immunization. DLNs were collected 10 days after immunization and stimulated with OVA-I in vitro to measure T cell proliferation by 3[H] thymidine uptake. (E) Frequencies of CD8+CD90.1+ cells and IFN-γ secreting CD8+ T cells as determined by flow cytometry were summarized. (F) Real-time quantitative RT-PCR analysis of different gene expression in WT and miR-155−/− MDSCs from LLC1-OVA-bearing mice (n=5–14). (G) ROS production was measured with DCFDA staining by flow cytometry and summarized within the granulocytic and monocytic tumor-infiltrating MDSCs. (H) Arginase I activity of WT versus miR-155−/− MDSCs. (I) Arginase I inhibitor nor-NOHA was able to blunt the suppressive activity of WT MDSC but not miR-155−/− MDSCs. All samples had MDSC, and the ratio of T cell/MDSC was 2:1. *, p<0.05; **, p<0.01. Data are representative of 2 independent experiments.
miR-155 is upregulated and functions in cytokine-induced MDSCs
It is generally accepted that MDSCs are elicited by tumor-derived factors (e.g. GM-CSF, IL-6) from precursors present in hematopoietic organs such as the BM and possibly spleen (at least in mice) (40–42). GM-CSF alone (43) or the combination of GM-CSF plus IL-6 (44) has been used successfully to generate MDSCs in short term culture in vitro from BM precursor cells. Interestingly, GM-CSF alone upregulated miR-155 expression during the induction of BM-derived MDSCs. Moreover, a combination of GM-CSF and IL-6 induced higher levels of miR-155 expression (Supple. Fig. 5A). We next analyzed whether miR-155 affected cytokine-induced MDSC function as observed in tumor-bearing mice. As shown in Supple. Fig. 5B, miR-155−/− MDSCs failed to suppress antigen-specific T cell proliferation in vitro compared with WT MDSCs. To examine further the functional contribution of miR-155 expression to the immunoregulatory activity of MDSCs, we transfected BM cells with a miR-155-specific inhibitor or a respective pre-miR-155 (precursor) and analyzed the proliferative capacity of antigen-specific T cells in the presence of cytokine-induced MDSCs (Supple. Fig. 5C). To this end, pre-miR-155, miR-155 inhibitor or control-transfected MDSCs were co-cultured with responder T cells at different ratios. As expected, control-transfection in MDSCs did not alter their suppressive capacity. In sharp contrast, treatment with miR-155 inhibitors abrogated MDSC suppressive activity. Consistent with theses, overexpression of miR-155 resulted in stronger suppression of T cell proliferation versus control-transfected MDSCs.
miR-155 deficiency down-regulates tumor-associated MDSC suppressive pathways
To identify the factors by which miR-155 regulated MDSCs, we analyzed gene expression profiles in WT versus miR-155−/− MDSCs from LLC1-OVA-bearing mice. We used real-time PCR to evaluate mRNA levels of genes related to tumor angiogenesis, immune responses and immune suppression (Fig. 4F). We found that mmp9, vegf, inos and arg1 were down regulated, whereas socs1 and ship1 were upregulated in miR-155−/− MDSCs. Based on previous observations (45–47), VEGF and MMP-9 expressed by MDSCs contributes to the proangiogenic tumor microenvironment. Thus, our results raised the possibility that miR-155 expressed by MDSCs could promote tumor growth by stimulating tumor angiogenesis. Because iNOS and arginase-I in MDSCs are essential for their immunosuppressive function, we asked whether down-regulation of inos or arg1 was implicated in the link between MDSC suppressive activity and miR-155. We evaluated reactive oxygen species (ROS) levels within the population of tumor-infiltrating MDSCs. No significant differences in ROS production from MDSCs were found between miR-155−/− and WT mice (Fig. 4G). However, miR-155−/− MDSCs has a lower level of arginase activity than WT counterparts (Fig. 4H). Moreover, inhibition of arginase-I with specific inhibitor nor-NOHA completely abrogated suppressive activity of WT MDSCs, whereas the nor-NOHA treatment did not affect the miR-155−/− MDSCs (Fig. 4I), suggesting that miR-155 modulates arginase-dependent suppressive activity of MDSCs. Given the importance of CD115 and CD124 (IL-4Rα) (48) in MDSCs, we also compared the expression of CD115 and CD124 in both MDSC subsets from tumor-bearing mice. There were no significant difference between miR-155−/− and WT mice (Supple. Fig. 4D). Taken together, our data indicate that miR-155 is likely required for MDSC-mediated tumor angiogenesis and immunosuppression.
miR-155 targets SOCS1 in MDSCs
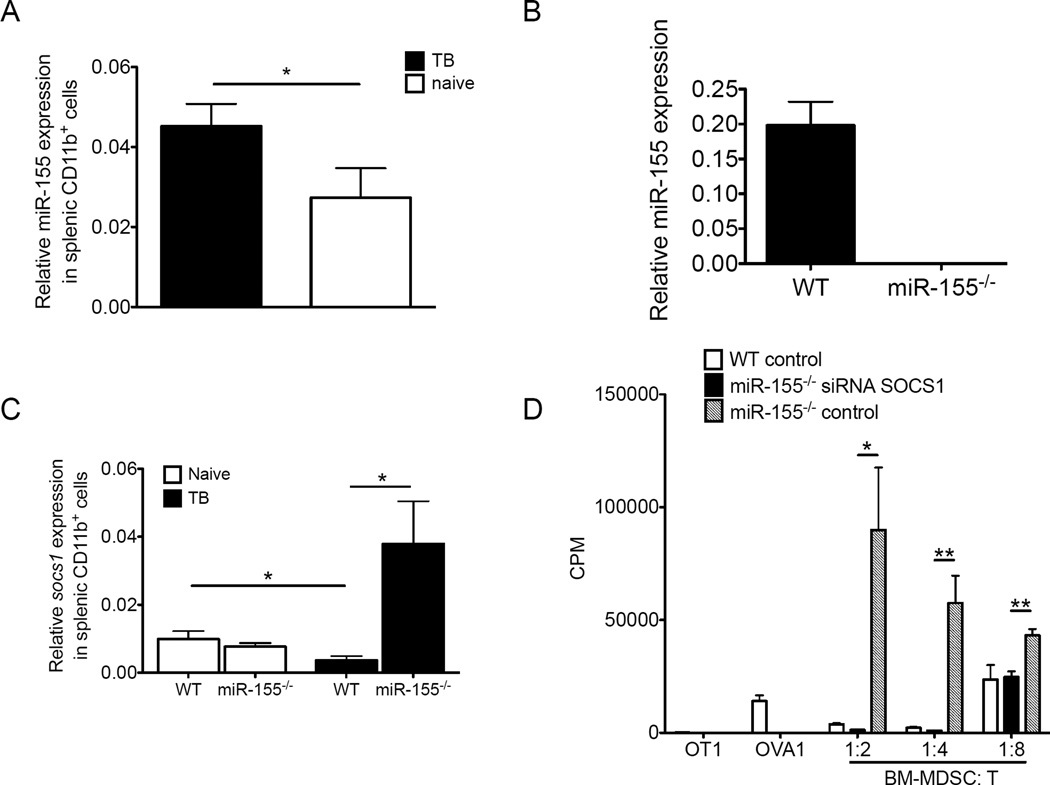
We initially confirmed that splenic CD11b+ cells from tumor-bearing mice had higher levels of miR-155 expression than those counterparts from tumor-free mice (Fig. 5A), whereas no detectable miR-155 expression was found in tumor-bearing miR-155−/− mice (Fig. 5B), suggesting a link between miR-155 up-regulation and MDSC induction upon tumor-bearing conditions. Interestingly, we detected enhanced levels of socs1 in miR-155−/− MDSCs compared to WT MDSCs in the tumor-bearing mice but not tumor-free mice (Fig. 5C). To test the functional consequence of elevated socs1 expression (as observed in the absence of miR-155) on MDSC suppressive activity, we utilized siRNAs to knock down socs1 expression in activated miR-155−/− MDSCs. We found that socs1 knockdown by specific siRNAs completely restored suppressive activity of miR-155−/− MDSCs compared to cells given a scrambled control (Fig. 5D). Thus, miR-155 targets SOCS1 to regulate the suppressive function of MDSCs.
Figure 5. miR-155 targets socs1 within MDSCs.
(A) miR-155 expression was measured by the real-time quantitative RT-PCR in splenic Gr1+CD11b+ cells from WT naïve mice, LLC1 tumor-bearing WT mice, and (B) miR-155−/− tumor bearing mice (n=3). (C) Socs1 gene expression was measured by the real-time quantitative RT-PCR in splenic WT or miR-155−/− Gr1+CD11b+ cells from naïve mice and LLC1 tumor-bearing mice (n=5). (D) To identify the function of SOCS1 within Gr1+CD11b+ MDSC, miR-155−/− MDSCs were transfected with siRNAs against SOCS1 or control oligos, and WT MDSCs were also transfected with control oligos by AMAXA. MDSCs 48 h after transfection were added at different ratios to OT-I splenocytes stimulated with OVA-I peptides for 3d, and 3[H] thymidine uptake was measured. Data are given as means ± SEM. Data are representative of 2 independent experiments. *, p<0.05; **, p<0.01.
miR-155 is required for MDSC-mediated Treg induction
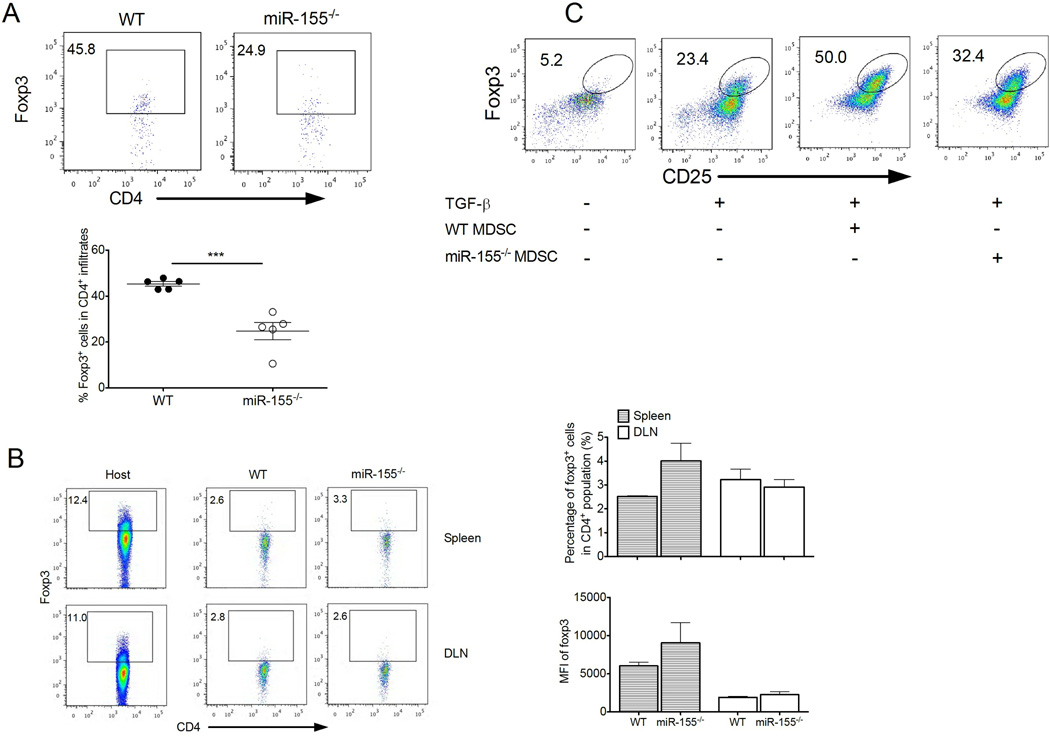
Because fewer tumor-infiltrating CD4+Foxp3+ Tregs were found in tumor-bearing miR-155−/− mice than in WT mice (Fig. 6A and Supple. Fig. 6), we tested a role for miR-155 in Treg in immune suppression. As shown in Supple. Fig. 7A, Tregs from either WT or miR-155−/− mice potently suppressed proliferation of CD4+ T cells in vitro, consistent with previous results (15, 16). Moreover, miR-155−/− Tregs from tumor-bearing mice had similar levels of CD39, CD73, CTLA-4, GITR, CD44 and CD62L expression compared with WT Treg (Supple. Fig. 7B). To test further the tumor-promoting role of Tregs, we performed adoptive transfer of WT or miR-155−/− Tregs into LLC1-bearing mice. There was a subtle difference in tumor growth between mice receiving WT and miR-155−/− Tregs (Supple. Fig. 7C). In addition, the role of miR-155 in tumor-mediated conversion of Tregs was also evaluated. We found similar conversion of CD4+ Foxp3+ cells in spleen and DLNs after tranfer of WT or miR-155−/− CD62L+CD4+ naive T cells into the tumor-bearing mice (Fig. 6B). Thus, these data exclude a direct contribution of miR-155 to Treg-mediated suppressive function and tumor promotion, and tumor-mediated conversion of Tregs.
Figure 6. miR-155 is required for MDSC-mediated Treg induction.
(A) Representative dot plots of Foxp3 expression in LLC1-OVA tumor-infiltrating CD4+ cells. Percent Foxp3+ cells is indicated within plots and summarized. (n= 5). (B) WT or miR-155−/− CD4+CD62L+ naïve T cells were transferred into CD45.1 mice followed by a s.c. injection of LLC1-OVA cells. The conversion of transferred T cells to Foxp3+ cells (CD45.2+) in DLN and spleen from LLC1-OVA tumor-bearing mice were detected by flow cytometer 9 d after tumor cell injection. The levels of converted Foxp3 expression are determined by mean fluorescent intensity (MFI). Endogenous Foxp3+ cells (CD45.1+) from host mice were shown as controls. (C) WT and miR-155−/− MDSCs from LLC1 tumor-bearing mice were cocultured with OT-II splenocytes at a 1:4 ratio for 5 days in the absence or presence of TGF-β, and induced CD25+Foxp3+ cells among total CD4+ T cells were subsequently determined by flow cytometry. ***, P<0.001. Data are representative of 2 independent experiments.
MDSCs induce Treg expansion in tumor-bearing mice (39, 49). To determine whether miR-155 mediates MDSC-mediated Treg induction, miR-155−/− or WT MDSCs were cultured with OT-II T cells plus OVA-II peptides. As expected, MDSCs were ineffective to induce antigen-specific Treg in the absence of TGF-β, but decreased Treg cell induction was observed when comparing miR-155−/− to WT MDSCs in the presence of TGF-β (Fig. 6C), indicating a role for miR-155 in MDSC-mediated Treg induction.
miR-155 expression by MDSC facilitates tumor growth
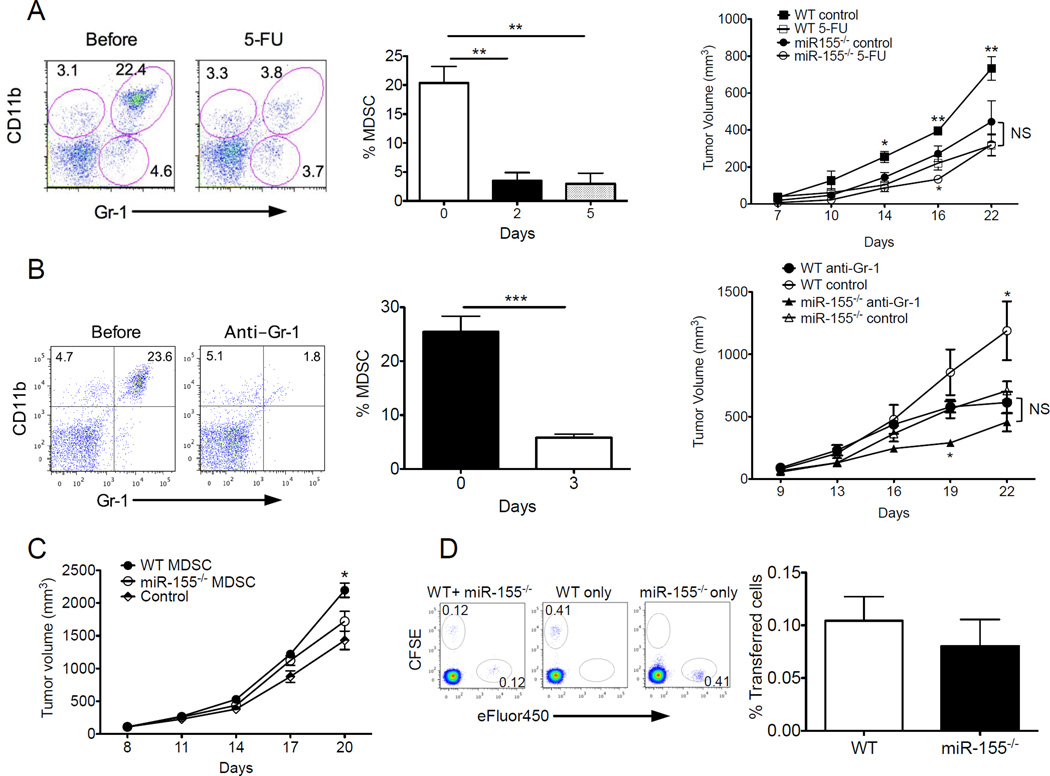
As miR-155 was required for MDSC accumulation and function, we tested whether miR-155 promoted tumor growth in an MDSC-dependent manner. We performed MDSC depletion in WT and miR-155−/− mice using either 5-FU (Fig. 7A) or depleting anti-Gr1 antibodies (Fig. 7B) after LLC1 tumor challenge. Consistent with prior published data (50, 51), both 5-FU and anti-Gr1 efficiently depleted CD11b+Gr1+ populations, especially the Gr1hi population within tumor-bearing mice (Fig. 7A,B). Importantly, MDSC depletion greatly inhibited tumor growth in WT mice, indicating a tumor-promoting role for MDSCs. By contrast, MDSC depletion minimally affected tumor growth in miR-155−/− mice compared to WT mice at later time points (starting from day 22) (Fig. 7A,B). Moreover, adoptive transfer of WT MDSCs into miR-155−/− mice resulted in faster tumor growth than transfer of miR-155−/− MDSCs (Fig. 7C), further consistent with the direct role of miR-155 on MDSCs in tumor growth. We did not expect miR-155−/− MDSCs to have migration defects. This notion is supported by the fact that miR-155−/− MDSCs displayed equal ability to traffic to the tumor site as WT MDSC (Fig. 7D), excluding the possibility miR-155−/− MDSC may not reach the tumor to exert their effect. These results indicate that miR-155 expression is required for MDSCs to facilitate tumor growth.
Figure 7. miR-155 expression by MDSCs facilitates tumor growth.
(A) WT or miR-155−/− mice were injected s.c. with 106 LLC1 tumor cells. Three days later, mice were injected i.p. by (A) 5-FU or PBS (control) or (B) anti-Gr1 antibodies once every 4 days. MDSC depletion by 5-FU or anti-Gr1 depleting antibodies in vivo were determined by flow cytometry and summarized (n=5). Tumor volume was measured and plotted at indicated times. NS: No Significant. (C) Splenic Gr1+CD11b+ MDSCs from tumor-bearing WT or miR-155−/− mice were injected i.v. into LLC1-bearing mice at d7 and d14. Mice receiving PBS without MDSCs were controls. (D) MDSC homing to tumors. Equal numbers of splenic WT Gr1+CD11b+ MDSCs labeled with CFSE and miR-155−/− MDSCs labeled with eFluro450 were mixed and transferred i.v. into LLC1-tumor bearing mice. Either WT MDSCs labeled with CFSE alone or miR-155−/− MDSCs labeled with eFluro450 alone were used as controls. Representative flow cytometric analysis of CFSE+ cells versus eFluro+ cells in the tumor 24 hours after transfer. Frequencies of CFSE+ MDSCs (WT) and eFluro+ MDSCs (miR-155−/−) among tumor tissues were summarized (n=5). *, p<0.05; **, p<0.01; ***, p<0.001. Data (mean ± SEM) are representative of 2 independent experiments.
Discussion
miR-155 is required for development and function of both innate and adaptive immune cells, and is thought to be largely immune stimulatory (52–54). However, to our surprise, chemically-induced tumor incidence and transplanted tumor growth were decreased in miR-155−/− mice. This was associated with a number of immune phenotypic and functional alterations.
MDSCs and Treg cells are important immunosuppressive cells in the tumor microenvironment. Notably, there were significantly more MDSCs and Treg cells in tumor-bearing WT mice than in tumor-bearing miR-155−/− mice. However, the prevalence of MDSCs and Tregs was similar in tumor-free WT versus miR-155−/− mice. Our results indicate that miR-155 could regulate the development of MDSCs and Treg cells in the context of tumor, and in turn affect anti-tumor immune responses. In line with our observations, a recent study showed that miR-155 was upregulated in cytokine-induced MDSCs from BM cultures and spleen MDSCs isolated from tumor-bearing mice, and promoted expansion of functional MDSCs (17). However, it remained unclear whether miR-155 mediates inhibtion of tumor growth in a MDSC-dependent manner despite its defined immune-stimulatory functions. In addition to regulating immunosuppressive factors, we do not rule out the contribution of miR-155 to tumor immunity through other immune components. Indeed, we observed the intrinsic defects in miR-155−/− tumor-associated DCs and antitumor T cells. Consistent with this concept, miR-155 is required for activation of tumor-associated DCs (26, 27) and effector CD8+ T cell (13, 25) responsed to cancer. Moreover, ectopic miR-155 expression repolarized pro-tumoral M2 macrophages towards an antitumor M1 phenotype (27), and increasing miR-155 levels in tumor-associated DCs by microRNA mimetics increased antitumor responses (26). We do not exclude, however, that miR-155 insufficiency in other immune compartments may have similar pro-tumoral effects, as recently proposed for NK cells (55).
Notably, our results on host miR-155 deficiency and tumor growth differ from a recent study (25) using the EL4 tumor model. Reasons for this discrepancy could include differences in the tumor cell lines that could alter the accumulation of distinct immune cell subsets in the tumor microenvironment. We also used EG7 cells expressing the surrogate antigen OVA, rather than parental (OVA-negative) EL4 cells in the prior report (25). These immune differences could alter in vivo outcomes. Finally, given that miR-155 regulation of one immune cell type can antagonize the function of other cells, the balance of these effects may determine whether miR-155 is beneficial or detrimental to tumor growth. In this regard, the prior study (25) focused on the intrinsic role of miR-155 in effector T cells, but did not analyze other distinct cellular subsets within the tumor such as MDSCs and Treg cells that promote tumor growth, as we showed in our study. Despite defects in immunostimulatory activities observed in miR-155−/− effector T cells and DCs, they are still able to mount antitumor responses. Increased miR-155 could play a critical role in balancing anti- and pro-tumor immune components within the tumor. In a given tumor model system, miR-155 could preferentially promote MDSCs and Treg cells before potent antitumor T cell immunity is established. Furthermore, the extent and regulation of tumor-induced immunosuppression including MDSC and Tregs could vary in different tumor types and/or tumor stages. In support, we showed that host miR-155 dificiency inhibited the growth of MC38 and LLC1 tumors rather than B16 tumors. Thus, it is likely that miR-155 plays dominant, MDSC-intrinsic roles in impairing antitumor T cell immunity in these tumor models. Our data suggest that the immune regulation of miR-155 is highly context-dependent, and varies in the presence of different cells, phases of immune responses and tumor model systems. Our studies highlight the importance of evaluating the intrinsic contribution of miR-155 carefully in major immune cell subsets, where miR-155 could be either protective or deleterious to antitumor immunity.
Although miR-155 is required for Treg cell homeostasis in the presence of limiting amounts of IL-2, it is dispensable in noncompetitive lymphopenic settings (16). Indeed, we showed no impaired ability of miR-155-deficient T cells to induce Foxp3 in tumor-bearing hosts. Moreover, intact suppressive activity of miR-155-deficient Tregs was observed, consistent with previous results (15, 16). In addition to the direct inhibiton of T cell proliferation, MDSCs can induce Treg expasion in tumor-bearing mice. Considering the importance of miR-155 for functional MDSC development, we tested whether miR-155 is required for MDSC-mediated Treg induction. It appears that loss of miR-155 results in the reduced accumulation of MDSCs that not only can inhibit clonal expansion of activated effector T cells but also induce tumor-specific Tregs to establish and maintain T cell suppression in tumor-bearing mice. Therefore, our results indicate that miR-155 is likely involved in a close interaction of MDSCs and Treg development during tumor progression.
In an effort to unravel the molecular basis for miR-155’s function in the MDSCs, we found the miR-155 targeted SOCS1 to retain the suppressive activity of MDSCs. SOCS1 is defined as an important mechanism for the negative regulation of the cytokine–JAK–STAT pathway (56). Several studies have demonstrated that the expansion and suppressive function of MDSC is mediated via the STATs (40–42). A recent study reported that miR-155 deficiency in Treg cells resulted in increased SOCS1 expression accompanied by impaired activation of STAT5 transcription factor in response to limiting amounts of IL-2, and suggested that Foxp3-dependent regulation of miR-155 maintains competitive fitness of Treg cells by targeting SOCS1 (16). In line with these findings, our SOCS1 shRNA experiments showed that defective suppressive activity by miR-155−/− MDSCs could be complemented by knockdown of SOCS1 expression, which was elevated in these MDSCs. Our data indicate that SOCS1 could impair the suppressive function of MDSCs when miR-155 is absent, and at least partially explain why miR-155 helps maintain MDSC activity. We also noted increased SHIP-1 expression in miR-155−/− MDSCs. Interestingly, SHIP-1 was recently reported as a target of miR-155 specifically in MDSC expansion (17), consistent with the previous observation that myeloid-specific ablation of SHIP resulted in an increase in MDSC numbers (57). However, the prior study did not examine the importance of MDSC miR-155 status in tumor growth (17). To our knowledge, our data clearly provide the first evidence that cell-intrinsic MDSC miR-155 is required for MDSCs to facilitate tumor growth, using both adoptive transfer and MDSC depletion analyses. We showed inverse correlations between MDSC SHIP-1/SOCS1 and miR-155, suggesting both SHIP-1 and SOCS1 as target genes of miR-155 during functional MDSC generation. As down-regulation of either SHIP-1 or SOCS1 could increase STAT3 activation (17, 58), which promotes functional MDSC expansion (37, 45), targeting both SHIP-1 and SOCS1 by miR-155 would enhance STAT3 activity and MDSC accumulation. However, the biology of miRNA signaling in MDSC development is likely to be more complex. At this stage, we cannot exclude the involvement of additional targets other than SHIP-1 and SOCS1 or even miRNAs other than miR-155 in regulation of functional MDSC induction.
miR-155 expression is controlled by a wide range of inflammatory factors, and transgenic over-expression of miR-155 results in cancer (18). Being oncogenic and pertinent to inflammation, miR-155 is considered as prototypical microRNA bridging inflammation and cancer development (4, 59). In support, we found that miR-155 deficiency inhibited carcinogenesis in the AOM and DSS-induced colorectal cancer model. MiR-155 deficiency could reduce colon inflammation that is known to drive carcinogenesis in this model (60). Moreover, miR-155 might promote tumor growth in an intrinsic manner as this is an induced and not transplanted model. Nevertheless, miR-155 positively regulates myeloid cell development by acting on BM progenitors during inflammatory stress. Particularly, our and other data (17) show that miR-155 is upregulated in MDSC either from tumor-bearing hosts or generated from BM progenitors by GM-CSF and IL-6. Overexpression of miR-155 enhanced, whereas depletion of miR-155 reduced the suppressive function of cytokine-induced MDSCs. Moreover, MDSC accumulation was impaired in tumor-bearing mice lacking miR-155 and miR155-deficient MDSCs failed to inhibit T cell functions. Thus, the induction of MDSC by proinflammatory mediators led to the novel hypothesis that inflammation promotes the accumulation of functional MDSC by increased miR-155 that down-regulates immune surveillance and antitumor immunity, thereby facilitating tumor growth. MDSCs also promote tumor progression through non-immune mechanisms. Their release of MMP-9 and VEGF contributes to tumor angiogenesis. Given the decreased production of MMP-9 and VEGF from miR-155−/− MDSCs, further studies will determine whether miR-155 mediates MDSC-dependent tumor angiogenesis.
Extensive evidence indicates that miR-155 functions as an oncomiR in many solid as well as hematologic tumors, and it is often associated with poor prognosis (61, 62). Thus, it has been suggested that therapeutic inhibition of miR-155 could be an effective strategy to treat cancer (22–24). However, in these studies, the contributions of immune regulation by miR-155 to tumor progression were unappreciable (23, 63, 64). Although our data expand the role of miR-155 to MDSC-mediated tumor protection, the cancer cell-intrinsic roles of miR-155 in both immune and non-immune conditions need further investigation. On the other hand, miR-155 activation in effector T cells and DCs boosts antitumor immunity, demonstrating a potential beneficial role for this miRNA during tumor progression. In this regard, besides its oncogenic activity, miR-155 functions as a cell context dependent "immunomiR" in orchestrating pro-tumor or anti-tumor immune responses. Thus, our results suggest additional investigations before considering miR-155 manipulation for cancer therapy. For example, cell-specific targeting of miR-155, and consideration of tumor effects on miR-155-mediated outcomes merit additional attention.
In summary, we investigated the role of host miR-155 in AOM and DSS-induced colon carcinogenesis and multiple transplantable tumor models. Our study identified a crucial cell-intrinsic role of miR-155 and its target SOCS-1 in MDSCs and demonstrated that this miRNA is required by MDSCs to limit antitumor T cell immunity. Despite the evidence for an established role of miR-155 in effector T cells and DCs, this miRNA is closely linked to the development of MDSCs and Treg cells, triggers tumor immune suppression, and thereby facilitates tumor growth. Our data indicate that the biological activities of miR-155 are highly cell context-dependent, including tumor dependent. Further studies will also be necessary to determine if host miR-155 affects tumor angiogenesis and metastasis.
Supplementary Material
Acknowledgments
This research was in part supported by National Institutes of Health grant CA149669, Ovarian Cancer Research Foundation Funds (LT/UTHSC/01.2011), the Northwestern University RHLCCC Flow Cytometry Facility, Cancer Center Support Grants (NCI CA060553 and CA054174), the Owens Foundation, the Skinner Endowment the Holly Beach Public Library Association. We thank the National Institutes of Health Tetramer Facility for providing the Kb/OVA tetramers.
Footnotes
Authors' Contributions
Conception and design: S. Chen, Y. Zhang, B. Zhang
Development of methodology: S. Chen, L. Wang, J. Fan, T.J. Curiel, B. Zhang
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S. Chen, L. Wang, J. Fan, D. Dominguez, C. Ye, B. Zhang
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S. Chen, Y. Zhang, B. Zhang
Writing, review, and/or revision of the manuscript: S. Chen, D. Fang, T.J. Curiel, T. Kuzel, B. Zhang
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): L. Wang, J. Fan, D. Dominguez, C. Ye, T. Kuzel, B. Zhang
Study supervision: B. Zhang
The authors have no conflicting financial interests.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 4.Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 5.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 8.Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI, et al. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117:4293–4303. doi: 10.1182/blood-2010-12-322503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai CY, Allie SR, Zhang W, Usherwood EJ. MicroRNA miR-155 affects antiviral effector and effector Memory CD8 T cell differentiation. J Virol. 2013;87:2348–2351. doi: 10.1128/JVI.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lind EF, Elford AR, Ohashi PS. Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J Immunol. 2013;190:1210–1216. doi: 10.4049/jimmunol.1202700. [DOI] [PubMed] [Google Scholar]
- 13.Dudda JC, Salaun B, Ji Y, Palmer DC, Monnot GC, Merck E, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38:742–753. doi: 10.1016/j.immuni.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J, et al. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 16.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Zhang J, Diao W, Wang D, Wei Y, Zhang CY, et al. MicroRNA-155 and MicroRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J Immunol. 2014;192:1034–1043. doi: 10.4049/jimmunol.1301309. [DOI] [PubMed] [Google Scholar]
- 18.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2011;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 21.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 23.Poltronieri P, D'Urso PI, Mezzolla V, D'Urso OF. Potential of anti-cancer therapy based on anti-miR-155 oligonucleotides in glioma and brain tumours. Chem Biol Drug Des. 2013;81:79–84. doi: 10.1111/cbdd.12002. [DOI] [PubMed] [Google Scholar]
- 24.Chen Z, Ma T, Huang C, Hu T, Li J. The pivotal role of microRNA-155 in the control of cancer. J Cell Physiol. 2014;229:545–550. doi: 10.1002/jcp.24492. [DOI] [PubMed] [Google Scholar]
- 25.Huffaker TB, Hu R, Runtsch MC, Bake E, Chen X, Zhao J, et al. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep. 2012;2:1697–1709. doi: 10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cubillos-Ruiz JR, Baird JR, Tesone AJ, Rutkowski MR, Scarlett UK, Camposeco-Jacobs AL, et al. Reprogramming Tumor-Associated Dendritic Cells In Vivo Using miRNA Mimetics Triggers Protective Immunity against Ovarian Cancer. Cancer Res. 2012;72:1683–1693. doi: 10.1158/0008-5472.CAN-11-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zonari E, Pucci F, Saini M, Mazzieri R, Politi LS, Gentner B, et al. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood. 2013;122:243–252. doi: 10.1182/blood-2012-08-449306. [DOI] [PubMed] [Google Scholar]
- 28.He M, Xu Z, Ding T, Kuang DM, Zheng L. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPbeta. Cell Mol Immunol. 2009;6:343–352. doi: 10.1038/cmi.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu F, Jia X, Du F, Wang J, Wang Y, Ai W, et al. miR-155-Deficient Bone Marrow Promotes Tumor Metastasis. Mol Cancer Res. 2013;11:923–936. doi: 10.1158/1541-7786.MCR-12-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin D, Fan J, Wang L, Thompson LF, Liu A, Daniel BJ, et al. CD73 on tumor cells impairs antitumor T-cell responses: a novel mechanism of tumor-induced immune suppression. Cancer Res. 2010;70:2245–2255. doi: 10.1158/0008-5472.CAN-09-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang B, Karrison T, Rowley DA, Schreiber H. IFN-gamma- and TNF-dependent bystander eradication of antigen-loss variants in established mouse cancers. J Clin Invest. 2008;118:1398–1404. doi: 10.1172/JCI33522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin PY, Sun L, Thibodeaux SR, Ludwig SM, Vadlamudi RK, Hurez VJ, et al. B7-H1-dependent sex-related differences in tumor immunity and immunotherapy responses. J Immunol. 2010;185:2747–2753. doi: 10.4049/jimmunol.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka T, Kohno H, Suzuki R, Yamada Y, Sugie S, Mori H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 2003;94:965–973. doi: 10.1111/j.1349-7006.2003.tb01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Movahedi K, Guilliams M, Van den Bossche J, Van den Bergh R, Gysemans C, Beschin A, et al. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 36.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng P, Corzo CA, Luetteke N, Yu B, Nagaraj S, Bui MM, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205:2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serafini P, Meckel K, Kelso M, Noonan K, Califano J, Koch W, et al. Phosphodiesterase-5 inhibition augments endogenous antitumor immunity by reducing myeloid-derived suppressor cell function. J Exp Med. 2006;203:2691–2702. doi: 10.1084/jem.20061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang B, Pan PY, Li Q, Sato AI, Levy DE, Bromberg J, et al. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 2006;66:1123–1131. doi: 10.1158/0008-5472.CAN-05-1299. [DOI] [PubMed] [Google Scholar]
- 40.Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32:19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonda N, Chioda M, Zilio S, Simonato F, Bronte V. Transcription factors in myeloid-derived suppressor cell recruitment and function. Curr Opin Immunol. 2011;23:279–285. doi: 10.1016/j.coi.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. 2009;182:4499–4506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossner S, Voigtlander C, Wiethe C, Hanig J, Seifarth C, Lutz MB. Myeloid dendritic cell precursors generated from bone marrow suppress T cell responses via cell contact and nitric oxide production in vitro. Eur J Immunol. 2005;35:3533–3544. doi: 10.1002/eji.200526172. [DOI] [PubMed] [Google Scholar]
- 44.Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 2011;32:790–802. doi: 10.1016/j.immuni.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Kujawski M, Kortylewski M, Lee H, Herrmann A, Kay H, Yu H. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J Clin Invest. 2008;118:3367–3377. doi: 10.1172/JCI35213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, et al. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell. 2004;6:409–421. doi: 10.1016/j.ccr.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 47.Jodele S, Chantrain CF, Blavier L, Lutzko C, Crooks GM, Shimada H, et al. The contribution of bone marrow-derived cells to the tumor vasculature in neuroblastoma is matrix metalloproteinase-9 dependent. Cancer Res. 2005;65:3200–3208. doi: 10.1158/0008-5472.CAN-04-3770. [DOI] [PubMed] [Google Scholar]
- 48.Gallina G, Dolcetti L, Serafini P, De Santo C, Marigo I, Colombo MP, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J Clin Invest. 2006;116:2777–2790. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan PY, Ma G, Weber KJ, Ozao-Choy J, Wang G, Yin B, et al. Immune stimulatory receptor CD40 is required for T-cell suppression and T regulatory cell activation mediated by myeloid-derived suppressor cells in cancer. Cancer Res. 2010;70:99–108. doi: 10.1158/0008-5472.CAN-09-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomihara K, Guo M, Shin T, Sun X, Ludwig SM, Brumlik MJ, et al. Antigen-specific immunity and cross-priming by epithelial ovarian carcinoma-induced CD11b(+)Gr-1(+) cells. J Immunol. 2010;184:6151–6160. doi: 10.4049/jimmunol.0903519. [DOI] [PubMed] [Google Scholar]
- 51.Vincent J, Mignot G, Chalmin F, Ladoire S, Bruchard M, Chevriaux A, et al. 5-Fluorouracil selectively kills tumor-associated myeloid-derived suppressor cells resulting in enhanced T cell-dependent antitumor immunity. Cancer Res. 2010;70:3052–3061. doi: 10.1158/0008-5472.CAN-09-3690. [DOI] [PubMed] [Google Scholar]
- 52.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253:146–157. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 53.Lind EF, Ohashi PS. Mir-155, a central modulator of T-cell responses. Eur J Immunol. 2014;44:11–15. doi: 10.1002/eji.201343962. [DOI] [PubMed] [Google Scholar]
- 54.Seddiki N, Brezar V, Ruffin N, Levy Y, Swaminathan S. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology. 2014;142:32–38. doi: 10.1111/imm.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trotta R, Chen L, Costinean S, Josyula S, Mundy-Bosse BL, Ciarlariello D, et al. Overexpression of miR-155 causes expansion, arrest in terminal differentiation and functional activation of mouse natural killer cells. Blood. 2013;121:3126–3134. doi: 10.1182/blood-2012-12-467597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 57.Collazo MM, Paraiso KH, Park MY, Hazen AL, Kerr WG. Lineage extrinsic and intrinsic control of immunoregulatory cell numbers by SHIP. Eur J Immunol. 2012;42:1785–1795. doi: 10.1002/eji.201142092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao XD, Zhang W, Liang HJ, Ji WY. Overexpression of miR-155 promotes proliferation and invasion of human laryngeal squamous cell carcinoma via targeting SOCS1 and STAT3. PLoS One. 2013;8:e56395. doi: 10.1371/journal.pone.0056395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tili E, Michaille JJ, Wernicke D, Alder H, Costinean S, Volinia S, et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A. 2011;108:4908–4913. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Robertis M, Massi E, Poeta ML, Carotti S, Morini S, Cecchetelli L, et al. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J Carcinog. 2011;10:9. doi: 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han ZB, Chen HY, Fan JW, Wu JY, Tang HM, Peng ZH. Up-regulation of microRNA-155 promotes cancer cell invasion and predicts poor survival of hepatocellular carcinoma following liver transplantation. J Cancer Res Clin Oncol. 2012;138:153–161. doi: 10.1007/s00432-011-1076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang S, Wang RH, Akagi K, Kim KA, Martin BK, Cavallone L, et al. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat Med. 2011;17:1275–1282. doi: 10.1038/nm.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109:E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Roccaro AM, Rombaoa C, Flores L, Obad S, Fernandes SM, et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–1686. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.