Abstract
The centrosome was discovered in the late 19th century when mitosis was first described. Long recognized as a key organelle of the spindle pole, its core component, the centriole, was realized more than 50 or so years later also to comprise the basal body of the cilium. Here, we chart the more recent acquisition of a molecular understanding of centrosome structure and function. The strategies for gaining such knowledge were quickly developed in the yeasts to decipher the structure and function of their distinctive spindle pole bodies. Only within the past decade have studies with model eukaryotes and cultured cells brought a similar degree of sophistication to our understanding of the centrosome duplication cycle and the multiple roles of this organelle and its component parts in cell division and signaling. Now as we begin to understand these functions in the context of development, the way is being opened up for studies of the roles of centrosomes in human disease.
Centrosome duplication, regulated mainly by protein phosphorylation and stability, occurs in concert with cell division in metazoa. Much insight has been gained by comparing them with spindle pole bodies in yeasts.
HISTORICAL BACKGROUND
Pioneering work from Boveri, van Benenden, and others in the 1880s saw the discovery of centrosomes, descriptions of how they enlarged before mitosis, and that they were associated with multipolar mitoses in tumor cells. Only now, more than a century later, are we beginning to have an understanding of how the organelle is pieced together and how it functions as a fundamental part of the cell-division machinery.
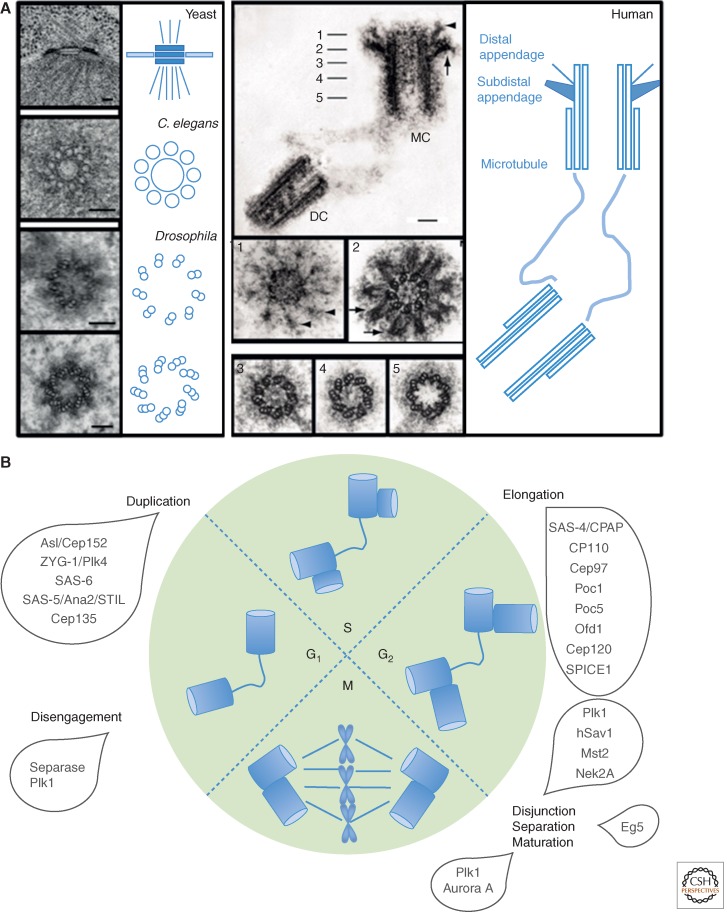
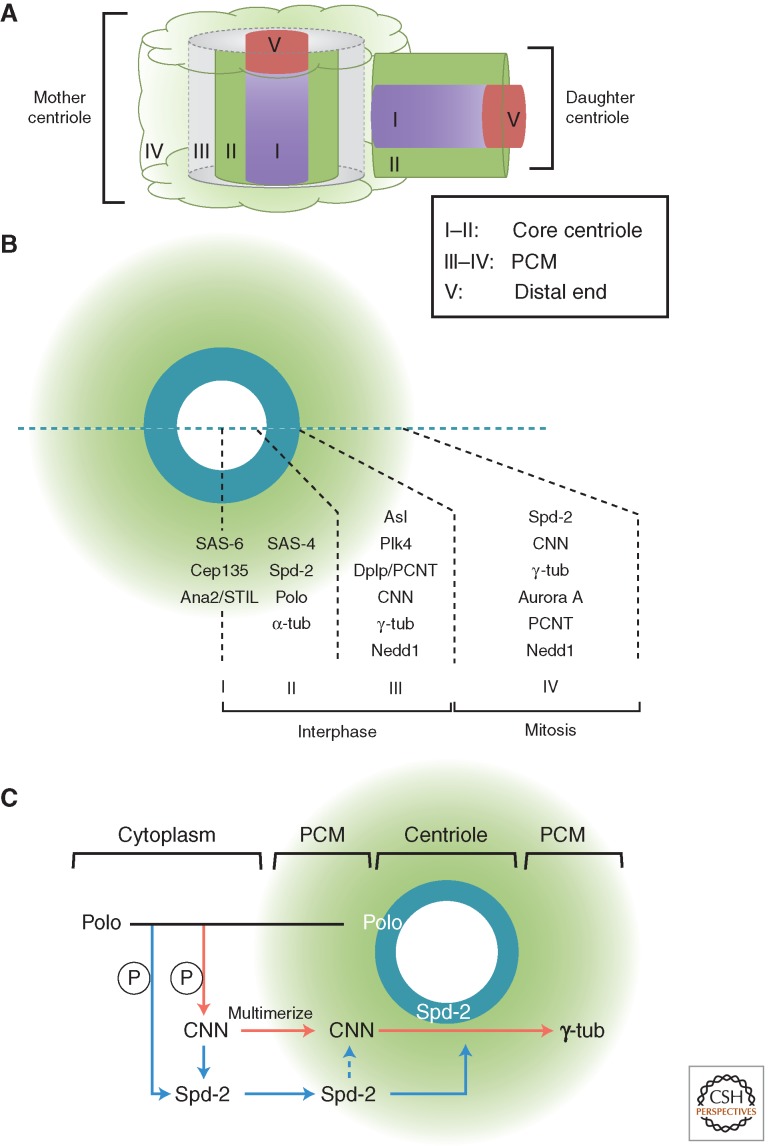
The explosion of the study of biological structures by electron microscopy (EM) in the 1950s revealed that centrosome has at its core the ninefold symmetrical centriole (Fig. 1A). A typical human centriole is a cylinder ∼200 nm in diameter and 500 nm long. At the most interior and the proximal-most part of the centriole is a cartwheel that has nine spokes, each linked to microtubule blades that form the microtubule wall (see Fig. 4B). It is surrounded by electron dense pericentriolar material (PCM) that increases in amount in mitosis providing the nucleating center for spindle and astral microtubules. In quiescent cells, a mature centriole can become associated with the plasma membrane to template cilia or flagella that function in signal transduction and cell motility. Defects in ciliogenesis lead to a group of disorders collectively known as the ciliopathies.
Figure 1.
The structure and duplication cycle of centrosomes. (A) Electron microscopy reveals the structures of the spindle pole body (SPB) centrosome with ninefold symmetrical centriole as its core. Scale bars, 100 nm. (B) C. elegans, Caenorhabditis elegans; DC, daughter centriole; MC, mother centriole. The centrosome duplication cycle occurs in concert with the cell-division cycle. Key events and players in the centrosome cycle are indicated.
Figure 4.
Centriole assembly. (A) Comparison of Caenorhabditis elegans (C. elegans) and Human/Drosophila pathways. Common elements are in the green box. (B) Structure organization of nine Sas-6 dimers (left) (Kitagawa et al. 2011c), and the relationship of cartwheel to centriole wall/microtubules (right). Molecular components are indicated.
Centrioles are present in metazoans and a variety of unicellular eukaryotes but are absent in the majority of land plants. Their ninefold symmetry is highly conserved but they do show structural differences among organisms. These differences are reflected in the molecular parts catalog for the centrioles of different organisms and give some clues to their evolution (Carvalho-Santos et al. 2010). The yeasts have evolved quite a different structure: the spindle pole body (SPB), a plate-like structure inserted into the nuclear envelope (Fig. 1A), and an ability to nucleate microtubules on its cytoplasmic and nuclear sides in the “closed” mitoses of yeast cells where the nuclear envelope does not break down. The SPBs carry much of the analogous machinery to the centriole and/or centrosome, and so it is of growing interest to compare their structure and function with centriole-containing centrosomes.
STRUCTURE AND DUPLICATION CYCLE OF YEAST SPBs
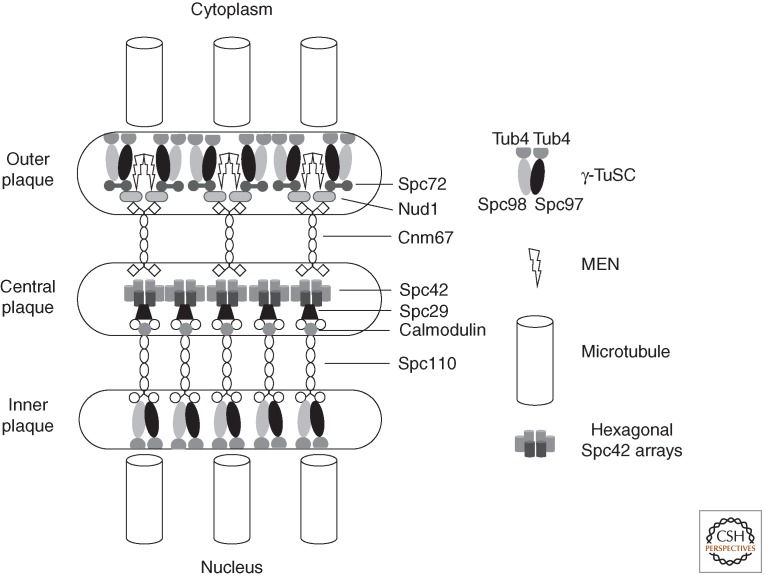
The budding yeast SPB nucleates both the nuclear spindle microtubules that segregate the genome and the cytoplasmic, astral microtubules that guide the spindle through the cytoplasm. As the nuclear envelope does not break down during mitosis, the planar trilaminar SPB is maintained within a specialized “polar fenestra” in the nuclear envelope throughout the cell-division cycle (Byers and Goetsch 1974, 1975; Heath 1980). Receptors for the γ-tubulin complexes sit on the opposing cytoplasmic and nuclear faces to nucleate the two sets of microtubules (Fig. 2). The nuclear receptor, Spc110 is a member of the pericentrin family of microtubule-nucleating proteins in which microtubule-nucleating motifs are separated from anchors by extended coiled-coil spacers (Kilmartin et al. 1993; Kilmartin and Goh 1996; Knop and Schiebel 1997; Sundberg and Davis 1997). These γ-tubulin docking motifs are highly conserved from human pericentrin and kendrin through Drosophila centrosomin (CNN) to fission yeast Mto1 and Pcp1 (Flory et al. 2002; Zhang and Megraw 2007; Fong et al. 2008; Samejima et al. 2008; Lin et al. 2014). Spc29 links Spc110 to the hexagonal crystalline lattice of Spc42 that comprises the central plaque in a coupling that relies on association of Spc110 with calmodulin (Geiser et al. 1993; Stirling et al. 1994; Donaldson and Kilmartin 1996; Spang et al. 1996; Bullit et al. 1997; Sundberg and Davis 1997; Elliott et al. 1999). On the cytoplasmic side of the central plaque, Spc42 anchors the Cnm67 linker protein that recruits Nud1 to the base of the outer plaque (Adams and Kilmartin 1999; Elliott et al. 1999; Schaerer et al. 2001). In turn, Nud1 recruits both the mitotic exit network (MEN) that regulates cell-cycle events at the end of the cycle (see the section on signaling from poles below) and the γ-tubulin complex receptor Spc72 (Knop and Schiebel 1998; Gruneberg et al. 2000).
Figure 2.
A highly schematic representation of molecular architecture of the budding yeast spindle pole body (SPB). A hexagonal crystalline array of Spc42 units associate with Spc29/Spc110 complexes on the nuclear side and cnm67 dimers on the cytoplasmic side of the SPB. These spacer proteins separate the central Spc42 plaque from the γ-TuSC microtubule-nucleating centers at the inner and outer plaques. At the inner plaque the interaction between the spacer Spc110 is direct with one Spc110 dimer associating with a single γ-TuSC (Erlemann et al. 2012). It is estimated that a functional microtubule nucleation unit comprises seven γ-TuSCs, two additional Spc98, and three extra γ-tubulins (Erlemann et al. 2012). This estimate agrees well with the reconstitution of 13-fold symmetric γ-tubulin microtubule-nucleating units in vitro (Kollman et al. 2008, 2010). At the cytoplasmic outer plaque, the association between the spacer and the γ-TuSC is mediated through the association of Nud1 with Spc72. Despite the fact that Spc72 interacts with both Spc97 and Spc98 in two hybrid assays (Knop and Schiebel 1998), in vivo measurements suggest that one Spc72 dimer interacts with a single γ-TuSC (Erlemann et al. 2012). Nud1 also acts as a scaffolding molecule for the mitotic exit network (MEN) that couples the SPB position with cell-cycle control. The stoichiometries of other associations remain to be established. The representation of Spc29 in between Spc110 and Spc42 is highly schematic, as the exact nature of its function as part of the Spc110 complex remains to be established.
γ-Tubulin recruits αβ-tubulin heterodimers to nucleate microtubules at the spindle poles of all eukaryotes (Kollman et al. 2011; Teixido-Travesa et al. 2012). Comprehensive molecular genetic analysis in budding yeast led to the characterization of the first γ-tubulin complex, the γ-tubulin small complex (γ-TuSC) (Geissler et al. 1996; Knop et al. 1997; Knop and Schiebel 1997, 1998). The γ-TuSC is conserved throughout eukaryotes and comprises two molecules of γ-tubulin and one each of the Spc97 and Spc98. Many other eukaryotes generate a larger γ-tubulin complex, the γ-tubulin ring complex (γ-TuRC) that contains Spc97/Spc98 orthologs and three further molecules that share the Grip motifs of Spc97 and Spc98 (GCP2-GCP6 [GCP2 and GCP3 being orthologous to Spc97 and Spc98, respectively]) alongside two or three additional components (Kollman et al. 2011; Teixido-Travesa et al. 2012). As its name suggests the γ-TuRC is a lock-washer-shaped ring in which the positioning of 13 γ-tubulin molecules serves as a template to recruit 13 αβ-tubulin heterodimers that seed the nucleation of 13 protofilament microtubules (Moritz et al. 1995; Kollman et al. 2011; Teixido-Travesa et al. 2012). The conserved γ-TuSC is Y shaped with Spc97/GCP2 and Spc98/GCP3 at the base of two γ-tubulin arms (Kollman et al. 2008). Because expression of the yeast γ-TuSC in baculovirus promotes the assembly of ring-like structures with 13-fold symmetry, the presence of the Grip domains in the GCP3-6 components of the γ-TuRC has been taken to infer that they act as variants of GCP2 and GCP3 to extend this core γ-TuSC complex into the larger γ-TuRC (Kollman et al. 2010). The extension of the templating function from a dimer to 13-mer that is conferred by the presence of the additional γ-TuRC components appears to be fulfilled in yeast by the γ-TuSC recruiting components of the SPB, as co-expression of the γ-TuSC with the interacting domain of the pericentrin molecule Spc110 in baculovirus generates extended γ-TuSC filaments (Kollman et al. 2010). This impact of Spc110 enhances the microtubule nucleation capacity of γ-TuSCs. Perhaps the simplicity of the budding yeast cytoskeleton with its permanent anchorage of microtubules to γ-TuSC receptors throughout the cell cycle has dispensed with the need for the complexity of the γ-TuRC that facilitates greater levels of control over microtubule nucleation.
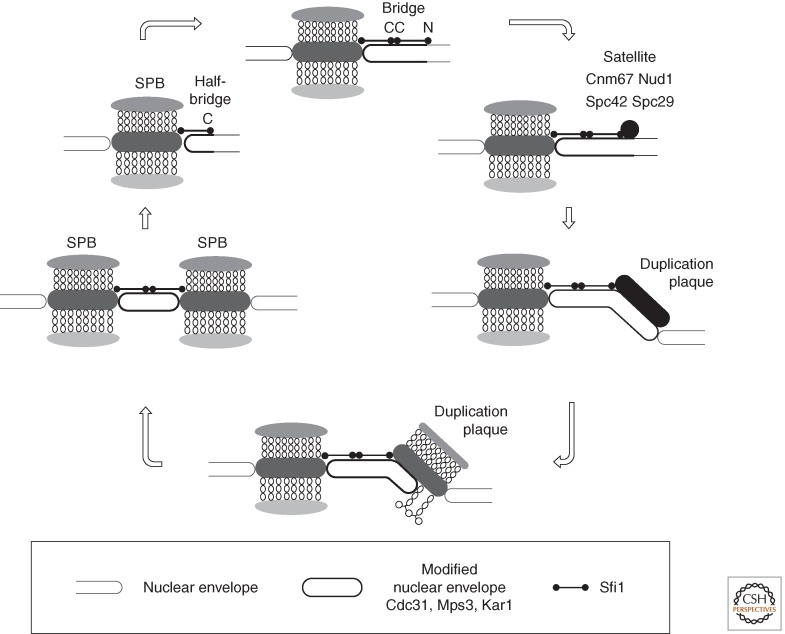
SPB duplication is conservative as a new SPB forms at the end of a “half-bridge” that extends along the inner and outer faces of the nuclear envelope from one side of the central layer of the old SPB (Fig. 3) (Byers and Goetsch 1974, 1975; Adams and Kilmartin 2000; Kilmartin 2014). The half-bridge principally comprises Cdc31, Kar1, Mps3, and Sfi1 (Baum et al. 1986; Rose and Fink 1987; Spang et al. 1993, 1995; Biggins and Rose 1994; Jaspersen et al. 2002; Kilmartin 2003; Li et al. 2006). Kar1 and the SUN domain protein Mps3 each contain a single membrane-spanning domain (Rose and Fink 1987; Jaspersen et al. 2002). Sfi1 is a long, flexible molecule principally composed of 20 Cdc31-binding repeats (Kilmartin 2003). Cdc31 is a member of the centrin family of calmodulin-related proteins that are found at spindle poles of all eukaryotes (Salisbury 2007). Sfi1-related molecules with multiple centrin-binding motifs accompany centrins at human centrosomes; however, their function remains to be determined (Kilmartin 2003; Azimzadeh et al. 2009). In budding yeast, the amino terminus of Sfi1 is anchored at the SPB, while the carboxyl terminus defines the end of the half-bridge extension. SPB duplication in the G1 phase of the cell cycle is assumed to be initiated by the end on recruitment of a second Sfi1 molecule to the free carboxyl terminus via homotypic C-C dimerization (Kilmartin 2003; Li et al. 2006). Central SPB components then bind the amino terminus of this newly docked Sfi1 to form a small satellite on the cytoplasmic face of the nuclear envelope that subsequently expands to form a complete duplication plaque on the outer face of the nuclear envelope (Adams and Kilmartin 1999). The duplication plaque is drawn into the nuclear envelope to generate side-by-side SPBs within the nuclear envelope, which are connected by an intact bridge (Byers and Goetsch 1974, 1975; Adams and Kilmartin 1999, 2000). The subsequent fission of the Sfi1–Sfi1 interface in the bridge generates two independent, half-bridge-bearing SPBs, which nucleate the microtubules of the bipolar spindle.
Figure 3.
Budding yeast spindle pole body (SPB) duplication. A highly schematic representation of SPB duplication in budding yeast. The key role played by Sfi1 C-C homotypic dimerization in establishing a point for the formation of the satellite that expands to form the duplication plaque is shown in the top panel. Immunoelectron microscopy indicates that this satellite contains at least Cnm67, Nud1, Spc42, and Spc29. (For full details, see Adams and Kilmartin 1999, 2000; Kilmartin 2014.)
STRUCTURE AND DUPLICATION CYCLES OF CENTRIOLAR CENTROSOMES
The advances in recent years of genomics and proteomics have led to identification of the multiple protein components of centrosomes and centrioles, and these, coupled with so-called superresolution light microscopy, are bringing our understanding of the functional biology of the centrosome toward the understanding that we have of yeast SPBs. Superresolution techniques overcome the limits imposed on conventional microscopy by diffraction of light and resolve what was previously seen by conventional immunostaining as an unstructured blob into a tiny cylinder (Sillibourne et al. 2011; Fu and Glover 2012; Lau et al. 2012; Lawo et al. 2012; Mennella et al. 2012; Sonnen et al. 2012). In this way, the mature Drosophila centrosome, for example, has been resolved into five major regions and three for its engaged daughter (see Fig. 6A). This places us in a position to understand precisely how the centrosome matures and how its molecular organization changes in anticipation of cell division as anticipated more than a century ago.
Figure 6.
Pericentriolar material (PCM) assembly. (A) The zones of the Drosophila centrosome (Fu and Glover 2012). (B) Expansion of the PCM in mitosis. Comparison of human and Drosophila components. (C) Pathway of PCM assembly deduced from studies in Drosophila (Fu and Glover 2012; Conduit et al. 2014a,b). tub, Tubulin.
The centrosome duplication cycle occurs in concert with the cell-division cycle. Newly born cells have a pair of centrioles, one engaged orthogonally to the other. This arrangement is lost as centrioles disengage in early G1 (Fig. 1B), and the two wander apart in G1 now linked by a loose fibrous connection. Assembly of a procentriole perpendicular to each mother begins in G1/S and the procentrioles subsequently elongate throughout G2 until a similar size to their mothers. Before mitosis, the mother centrioles begin to accumulate more PCM and are able to nucleate increased microtubules in preparation for spindle assembly. The fibrous tether among centrosomes resolves permitting centrosomes to disjoin and separate to opposite sides of the cell as the spindle poles. Preparation for centriole duplication takes place in concert with preparation for S phase.
Tying Centriole Duplication to S Phase
Centrosome duplication shares key regulators with DNA replication and yet the two processes can be uncoupled to reveal that both are dependent on Cdk2/cyclin E. Treatment of Drosophila embryos or Xenopus egg extracts with the α-DNA polymerase inhibitor, aphidicolin, for example, leads to the repeated rounds of centrosome assembly (Raff and Glover 1988; Hinchcliffe et al. 1999). If, in the latter system, Cdk2-cyclin E activity was blocked with a Cdk inhibitor derived from p57, then the multiple rounds of centrosome reproduction could be prevented and then restored by addition of purified Cdk2-cyclin E. Accordingly, injection of the Cdk inhibitor p21 or p27 into an individual blastomere of a dividing Xenopus embryo blocks centrosome duplication in that blastomere (Lacey et al. 1999). Similarly, when Chinese hamster ovary (CHO) cells are arrested in S phase by hydroxyurea (HU), then inhibition of Cdk2 activity blocks the continued centrosome duplication (Matsumoto et al. 1999; Meraldi et al. 1999). Together, these findings lead to a model in which activation of Cdk2 ensures the centrosome duplication usually in phase with DNA replication.
Although several centrosome-associated Cdk2 substrates have been identified, including NPM/B23 (nucleophosmin) (Okuda et al. 2000; Tokuyama et al. 2001), Mps1 (Fisk and Winey 2001; Fisk et al. 2003), and CP110 (Chen et al. 2002), the role of Cdk2 phosphorylation in the duplication cycle is far from clear. However, some clues are emerging from the direct involvement of proteins required to control the initiation of DNA replication in both processes. Cdk2’s partners, cyclin E and cyclin A, both interact with MCM5 and recruit it to the centrosome where they repress centrosome amplification in S phase–arrested CHO cells (Ferguson and Maller 2008; Ferguson et al. 2010). Moreover, cyclin A has been found to promote Orc1 localization to centrosomes where Orc1 prevents cyclin E–dependent reduplication of centrosomes (Hemerly et al. 2009). Finally, a direct link to the licensing of DNA replication emerges from experiments to deplete cells of Geminin, a negative regulator of the initiation of DNA replication. Geminin depletion leads to multiple DNA endoreduplication cycles at the expense of mitosis that in U2OS, HCT116 colorectal cancer cells and TIG-3 diploid fibroblasts have been shown to be accompanied with centrosome reduplication (Tachibana et al. 2005). In contrast, overexpression of Geminin inhibits centrosome reduplication in the human breast cancer cell line MDA-MB-231 (Lu et al. 2009). However, the details of the link between these licensing proteins and the centriole duplication machinery are still not clear.
Elevated levels of Cdk2/cyclin E activity have been proposed to underlie the overduplication of centrosomes seen in most p53-deficient cell lines (reviewed by Fukasawa 2008). In normal cells, this would be reflected as part of a stress response in which elevated levels of p53 would depress Cdk2 levels via the activation of p21, thus creating an environment that is not permissive for centriole duplication. However, how exactly p53 exerts its function at the centrosome is not clear. There does, however, appear to be a link between p53 and the regulation of Polo-like kinase 4 (Plk4), which, as we will see below, is a master regulator of centriole formation. The autoregulated instability of Plk4 controls its abundance, and thus preventing Plk4 autoregulation leads to centrosome amplification. This is normally associated with stabilization of p53 and loss of cell proliferation. In the absence of p53, function cells carrying amplified centrosomes are able to proliferate (Holland et al. 2012). The complexity of this regulative network is heightened by the recent report that Plk4 is directly phosphorylated and activated by stress-activated protein kinase kinase kinases (SAPKKKs) to promote centrosome duplication (Nakamura et al. 2013). However, this is balanced early in the stress response by stress-induced SAPK activation that prevents centrosome duplication. In the late stages of the stress response, however, p53 down-regulates Plk4 expression, thereby preventing sustained Plk4 activity and centrosome amplification. In cancer cells, both p53 and the SAPKK MKK4 are frequently inactivated leading to continued Plk4 activity and centrosome duplication in the absence of SAPK-mediated inhibition.
The Core Pathway of Centriole Assembly
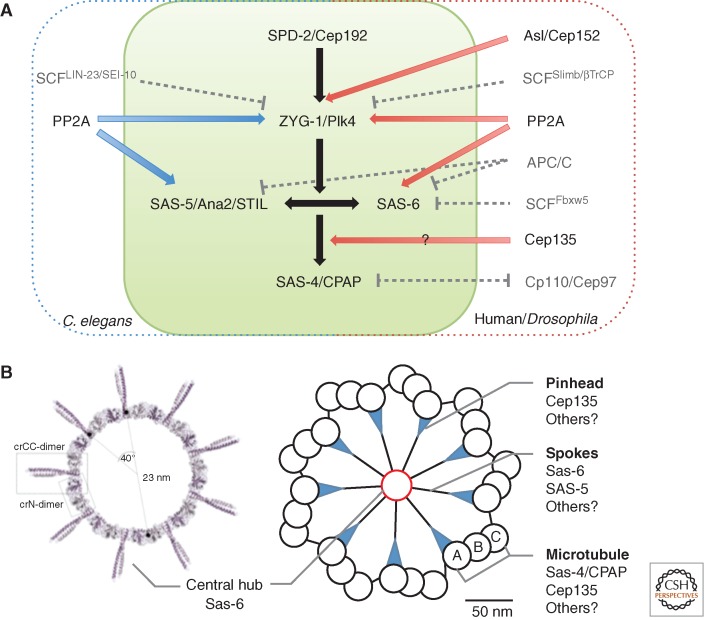
The core pathway for centriole duplication was first elucidated in Caenorhabditis elegans as a series of dependent steps. The coiled-coil protein SPD-2 (spindle defective 2) was required to recruit the ZYG-1 protein kinase that, in turn, recruits the spindle assembly abnormal proteins SAS-6 and SAS-5 as a prerequisite for procentriole assembly and, finally, SAS-4, required for the addition of centriolar microtubules (Fig. 4A) (O’Connell et al. 2001; Kirkham et al. 2003; Leidel and Gonczy 2003; Dammermann et al. 2004; Delattre et al. 2004, 2006; Kemp et al. 2004; Pelletier et al. 2004, 2006; Leidel et al. 2005). The functional homologs of these five proteins are highly conserved (Fig. 4A) (Goshima et al. 2007; Dobbelaere et al. 2008; Balestra et al. 2013).
The Cartwheel
The protein at the innermost core of the centriole is Sas-6 (zone I in Drosophila; Sonnen et al. 2012; Dzhindzhev et al. 2014). Indications for its importance in establishing ninefold symmetry first came from studies of loss of its function in Chlamydomonas, Paramecium, and from Drosophila, in which loss of, or abnormalities in, the cartwheel structure were observed (Nakazawa et al. 2007; Rodrigues-Martins et al. 2007a; Jerka-Dziadosz et al. 2010). When its structure was unveiled through crystallography of large fragments of the zebrafish and Chlamydomonas proteins, this revealed the head-to-head dimerization of its amino-terminal part and a parallel coiled-coil dimer; the ninefold symmetry could be accounted for by nine such Sas-6 homodimers interacting through adjacent amino termini to give a ring-like central hub with the carboxy-terminal coiled-coil dimers radiating outward as nine spokes (Fig. 4B) (Cottee et al. 2011; Kitagawa et al. 2011c; Schuldt 2011; van Breugel et al. 2011). Indeed, Sas-6 protein could self-assemble into ring-like structures having similar diameters to the central hub and cartwheel in solution. The cartwheel height could then be accounted for by the stacking of such structures as later revealed by electron tomography of the cartwheel in the Trichonympha basal body (Guichard et al. 2012). This may not be the only way to establish the core structure, as analogous studies revealed that the C. elegans SAS-6 also forms N-N homodimers and coiled-coil dimers but these assemble into filamentous spiral oligomers instead of rings. Such a structure could be the underlying reason for the lack of the cartwheel structure in this organism and its replacement by a central tube (Hilbert et al. 2013). However, although the detailed arrangements of Sas-6 may vary among different species, its role in dictating centriolar ninefold symmetry seems now indisputable.
The close cooperation between SAS-5 and SAS-6 in centriole duplication in C. elegans is also seen with the human and Drosophila counterparts of SAS-5, STIL, and Ana2, respectively (Stevens et al. 2010; Kitagawa et al. 2011a; Tang et al. 2011; Arquint et al. 2012; Vulprecht et al. 2012). Overexpression of SAS-6 in Drosophila syncytial embryos led to the de novo formation of tube- or vesicle-like structures that are surrounded by microtubule asters (Rodrigues-Martins et al. 2007a). However, coexpression of Sas-6 and Ana2 in Drosophila spermatocytes leads to the assembly of cartwheel-like structures (Stevens et al. 2010). Overexpression of STIL or Sas-6 in other systems leads to centriole overduplication (Kitagawa et al. 2011a; Tang et al. 2011; Arquint et al. 2012; Vulprecht et al. 2012). Both Ana2 and STIL localize to the innermost region of the centriole (Sonnen et al. 2012; Dzhindzhev et al. 2014), raising the possibility that Ana2/STIL might be part of the cartwheel structure. Indeed, the C. elegans SAS-5 physically binds a narrow central region of the SAS-6 coiled-coil domain, and is able to prevent the coiled-coil dimer from forming a tetramer in vitro (Qiao et al. 2012). Moreover, once phosphorylated by Plk4, Drosophila Ana2 becomes able to bind Sas-6 (see below) (Dzhindzhev et al. 2014).
Although SAS-6 and SAS-5/Ana2/STIL are components of the innermost part of the centriole, SAS-4 colocalizes with its microtubule wall (zone II in Drosophila cells; Fu and Glover 2012). Indeed, SAS-4 promotes polymerization of centriolar microtubules, and overexpression of its human homolog, CPAP, leads to centriole elongation (Kohlmaier et al. 2009; Schmidt et al. 2009; Tang et al. 2009). A tubulin-binding domain is critical for this function (Hsu et al. 2008; Cormier et al. 2009), and its stable incorporation into centrioles is dependent on γ-tubulin and microtubule assembly (Dammermann et al. 2008). CPAP is reported to interact with STIL and another centriole protein, Cep135 (Tang et al. 2011; Cottee et al. 2013; Hatzopoulos et al. 2013; Lin et al. 2013a), but how these molecules cooperate to regulate procentriole assembly requires further analysis. The functional importance of Cep135 was first shown by the requirement for its ortholog, Bld10p, for cartwheel formation in Chlamydomonas and Paramecium (Hiraki et al. 2007; Jerka-Dziadosz et al. 2010). Drosophila centrioles are still able to form in the absence of Cep135 but are short (Mottier-Pavie and Megraw 2009; Carvalho-Santos et al. 2010). Similarly, when Cep135 was disrupted in the chicken DT40 cell line, there was only a small decrease in the centriole number with no major defects in centrosome composition or structure (Inanc et al. 2013). However, human Cep135 is required for excessive centriole duplication following Plk4 overexpression (Kleylein-Sohn et al. 2007), and a recent report showed that depletion of Cep135 reduces the centrosome number (Lin et al. 2013a). Thus, although a common function of Cep135 seems to ensure the intact structure of centrioles, in most cases, its loss does not have a devastating effect (Roque et al. 2012; Inanc et al. 2013; Lin et al. 2013a).
Plk4
The protein kinase ZYG-1 that lies at the head of the centriole formation pathway in C. elegans is a distant member of the Plk4 family, a family of Polo-like kinases that have three Polo box domains within their carboxy-terminal domain (Slevin et al. 2012). Like ZYG-1, Plk4 lies at the head of the pathway for centriole assembly in Drosophila and human cells (Bettencourt-Dias et al. 2005; Habedanck et al. 2005; Kleylein-Sohn et al. 2007; Rodrigues-Martins et al. 2007b). When Plk4 is down-regulated, centriole duplication fails, and when it is overexpressed in Drosophila embryos, it promotes overduplication of sperm-derived centrioles (Bettencourt-Dias et al. 2005; Rodrigues-Martins et al. 2007b; Cunha-Ferreira et al. 2009). Strikingly, Plk4 was also found to drive de novo centriole formation when overexpressed in unfertilized eggs (Rodrigues-Martins et al. 2007b). Contemporaneous findings in human cells showed that Plk4 overexpression causes centrioles to overduplicate into flower-like arrays with mother centrioles in the center (Kleylein-Sohn et al. 2007). However, how Plk4 exerts this function remains largely unknown. In C. elegans, ZYG-1 binds directly to SAS-6 and recruits the SAS-6–SAS-5 complex to the centriole, independent of its kinase activity (Lettman et al. 2013). Consistently, overexpression of Plk4 can promote the recruitment of Ana2/STIL and Sas-6 to supernumerary centrioles (Kleylein-Sohn et al. 2007; Stevens et al. 2010). A number of Plk4/ZYG-1 substrates have been identified—SAS-6 (Kitagawa et al. 2009), Cep152 (Hatch et al. 2010), and a component of γ-TuRC GCP6 (Bahtz et al. 2012), but their significance is not clear. More recently, it has been shown in Drosophila that Plk4 phosphorylates Ana2 in its conserved STAN motif to enable it to bind to Sas-6. Both Ana2 and Sas-6 become recruited to the daughter centriole once it has disengaged from the mother at the end of mitosis. If the Plk4 sites in Ana2 are mutated to nonphosphorylatable residues, it can still bind to the daughter centriole but it cannot recruit Sas-6, and centriole duplication fails (Dzhindzhev et al. 2014). In contrast to Drosophila, where some Sas-6 remains at the core of the centriole once it is incorporated, in human cells, Sas-6 is destroyed during G1 (Strnad et al. 2007) and is transiently recruited to the lumen of the mother centriole in S phase (Fong et al. 2014). The human Sas-6 is then repositioned to the site of procentriole formation in a process that is dependent on STIL/Ana2) and Plk4 (Fong et al. 2014).
Since the original studies in C. elegans, it has become clear that centriole assembly requires additional factors. One of these, Asterless (Asl, Drosophila)/Cep152 (vertebrates) has particular importance in recruiting Plk4 to the centrosome. The interaction of two of the Polo boxes of Plk4 (the cryptic Polo-box domain) with Asl/Cep152 is conserved in Drosophila, human, and Xenopus cells, although their codependency for localization differs (Cizmecioglu et al. 2010; Dzhindzhev et al. 2010; Hatch et al. 2010). Asl also has additional roles in binding Sas-4 to help establish the PCM (see below). Cep192 also binds to Plk4 and so cooperates with Cep152 in recruiting Plk4 to the centriole (Sonnen et al. 2013; Firat-Karalar et al. 2014). This echoes the finding of the dependency of ZYG-1 recruitment on SPD-2 in C. elegans. However, in Drosophila, Spd-2 barely plays any role in centriole assembly but is instead required for PCM recruitment on mitotic entry (Dix and Raff 2007; Giansanti et al. 2008). In Planarians where the canonical centriole duplication pathway was abandoned during evolution and centrioles are only assembled in terminally differentiating ciliated cells through an acentriolar pathway, Spd-2/Cep192 along with CNN/CDK5RAP2 and Nek2 are all absent from the genome (Azimzadeh et al. 2012). This raises the possibility that Spd-2 is not needed for de novo centriole formation, but for canonical duplication.
To ensure that proliferating cells have only a single pair of functional centrosomes, it is crucial to tightly regulate the levels of centriolar proteins, particularly Plk4. How Plk4 becomes activated and then how its activity might be restricted to one part of the centriole at which the procentriole forms is not yet understood. In some way, licensing of duplication is linked to the disengagement of centrioles at the end of mitosis (see below) but the molecular details of this process have to be elucidated.
In contrast, we know quite a lot about the controlled proteolysis of Plk4 kinase that is achieved in both Drosophila and human cells via the SCFSlimb/βTrCP ubiquitin ligase complex (Cunha-Ferreira et al. 2009; Rogers et al. 2009; Guderian et al. 2010). If this system fails, the consequence is development of multiple centrioles both in Drosophila and human cells. Plk4 degradation first requires that Plk4 forms a homodimer through its carboxy-terminal coiled-coil region (Leung et al. 2002; Habedanck et al. 2005), where it is able to autophosphorylate a phosphodegron enabling the binding of SCFSlimb/βTrCP to promote its own destruction (Guderian et al. 2010; Holland et al. 2010, 2012; Sillibourne et al. 2010). Consistently, the ZYG-1 kinase of C. elegans is also down-regulated by the Slimb/βTrCP homolog, LIN-23, and also a second F-box protein, SEL-10 (Peel et al. 2012).
The autophosphorylation of Drosophila Plk4 is counteracted by the protein phosphatase PP2A and its B55 regulatory subunit Twins, that thereby act to stabilize Plk4 during mitosis (Brownlee et al. 2011). Plk4 is not the only centriolar protein to be targeted by PP2A. In C. elegans, PP2A’s association with the SAS-5–SAS-6 complex is instrumental in targeting SAS-5 and, hence, SAS-6 to the centriole (Kitagawa et al. 2011a), and its activity protects SAS-5 as well as ZYG-1 from degradation by the proteasome (Song et al. 2011). PP2A is similarly required for human Sas-6 to localize to centrioles (Kitagawa et al. 2011a), although it is not clear whether this is mediated through STIL.
Levels of Plk4 appear to decrease at the centrosome as cells exit mitosis, suggesting that the APC/C might also regulate Plk4 levels. Similar reductions occur for CPAP, STIL, and human Sas-6 and each of these molecules is targeted by APC/C-Cdh1 or Cdc20 (Strnad et al. 2007; Tang et al. 2009, 2011; Puklowski et al. 2011; Arquint et al. 2012; Arquint and Nigg 2014). Interplay between APC/C and SCF pathways has also been reported to regulate levels of Sas-6 (Puklowski et al. 2011). Sas-6 forms a complex with Fbxw5 to target its destruction. However, Fbxw5 is negatively regulated by Plk4 and it is also targeted for destruction by the APC/C. Thus, the Fbxw5-mediated destruction of Sas-6 would be promoted at mitotic exit and at times when Plk4 activity is minimal. However, further work is needed before we have a coordinated picture of the spatial and temporal regulation of the stability and activity of these proteins.
Centriole Elongation
Once the cartwheel of the procentriole is established, then the A, B, and C centriolar microtubules are added and begin growth (Guichard et al. 2010). Some of the components identified as essential for centriole assembly also contribute to centriolar microtubule elongation. Sas-4/CPAP, for example, promotes the polymerization of centriolar microtubules in cooperation with CEP120, which localizes preferentially to the daughter centriole (Mahjoub et al. 2010; Comartin et al. 2013; Lin et al. 2013b). Overexpression of either CPAP or CEP120 results in excessively long centrioles and their depletion abolishes this phenotype (Comartin et al. 2013; Lin et al. 2013b). CEP120 also interacts with SPICE1 that is required for centriole duplication, spindle formation, and chromosome congression (Archinti et al. 2010). Depletion of SPICE1 also results in short procentrioles, although, in contrast to its partners, overexpression of SPICE1 does not cause centriole elongation (Comartin et al. 2013). The existence of a second mechanism controlling the length of the distal part of the centriole is suggested in HeLa cells where short procentrioles or centrioles with defective distal structures accumulate in the absence of POC5 (Azimzadeh et al. 2009) and in Ofd1 mutants of mouse cells that display an abnormally elongated distal portion of centriole (Singla et al. 2010).
The elongation of centrioles brought about by overexpression of CPAP can be counteracted by another centriolar protein, CP110 (Kohlmaier et al. 2009; Schmidt et al. 2009; Tang et al. 2009). CP110 is required for the formation of supernumerary centrioles resulting from excessive Plk4 or from S-phase arrest (Chen et al. 2002; Kleylein-Sohn et al. 2007) but it also limits centriole length in HeLa and U2OS cells, where its depletion leads to abnormally long centrioles (Schmidt et al. 2009). It may act as a physical barrier for elongation as it localizes to the distal end of the centriole (zone V) as a cap (human) or plug (Drosophila) (Kleylein-Sohn et al. 2007; Fu and Glover 2012). The Cep97 protein is required to target CP110 to this distal part of the centriole and overexpression of these proteins can prevent the formation of primary cilia in RPE1 cells (Spektor et al. 2007; Tsang et al. 2008). This is suggested to occur by opposing CEP290, whose depletion prevents ciliogenesis, by interfering with Rab8a’s localization to centrosomes and cilia (Tsang et al. 2008). Depletion of a kinesin-13 subfamily member, Kif24, also induces the formation of primary cilia, but not elongated centrioles, apparently by displacing CP110 and Cep97 from the mother centriole (Kobayashi et al. 2011). Together, this indicates that the removal of CP110 and Cep97 from the centriole tip is a critical early step for ciliogenesis. A depolymerizing kinesin-like protein, Klp10A, has been found to restrict centriolar microtubule length in Drosophila cells. However, in this case, depletion of CP110 shortens centriole microtubules, apparently by exposing them to Klp10A (Delgehyr et al. 2012).
CP110 is also targeted for degradation by the SCF through the F-box protein cyclin F/Fbxo1 and if allowed to accumulate it induces centrosome overduplication in G2 (D’Angiolella et al. 2010). A deubiquitinating enzyme, USP33, antagonizes SCFcyclinF-mediated ubiquitination and stabilizes CP110 possibly during S and G2/M phases (Li et al. 2013). Consequently, depletion of USP33 inhibits centrosome amplification in S-phase-arrested U2OS cells or in cyclin-F knockdown cells.
How ubiquitin ligase and deubiquitinating enzyme counteract each other’s function as the cell cycle progresses needs further exploration. However, it seems clear that the regulated destruction of centriolar proteins plays a role not only in centriole duplication, but also in elongation. Thus, pharmacological inhibition of cellular proteolysis by Z-L3VS or MG132 not only causes assembly of multiple daughter centrioles but also their abnormal elongation. A siRNA screen for genes affecting centriole length in Z-L3VS-treated cells has revealed additional players to CPAP, CP110, and Cep97, which include the centriolar protein Sas-6, centrosomal proteins FOP and CAP350, the cohesion protein C-Nap1, as well as appendage proteins Cep170 and ninein (Korzeniewski et al. 2010). It will be fascinating to see how these fit into the regulatory network that controls centriole length.
Centrosome Disjunction at G2/M
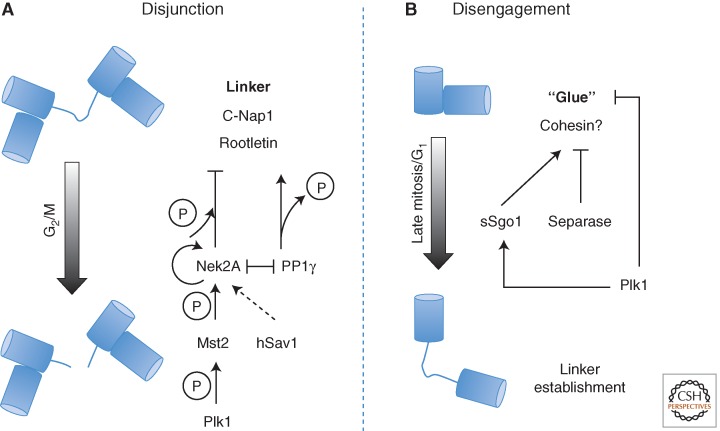
When mother and daughter centrioles lose their orthogonal arrangement during mitotic exit, a second proteinaceous linker, containing C-Nap1 and rootletin, is established that connects them at their proximal ends and persists until G2/M (Fig. 5A) (Fry et al. 1998; Mayor et al. 2000; Bahe et al. 2005; Yang et al. 2006). Depletion of C-Nap1 or rootletin causes centrosome splitting regardless of the cell-cycle stage (Mayor et al. 2000; Bahe et al. 2005). Immuno-EM revealed C-Nap1’s localization to the proximal ends of the connected centrioles but not between them, whereas rootletin is present both at ends and between the centriole pairs in nonciliated cells and, in addition, connected to basal bodies in ciliated cells (Fry et al. 1998; Mayor et al. 2000; Bahe et al. 2005; Yang et al. 2006). The overexpression of rootletin produces fibers that are able to recruit Nek2A and C-Nap1. Cep68 also decorates fibers emanating from the proximal ends of centrioles, dissociates from centrosomes during mitosis, and requires rootletin and C-Nap1 for centrosome localization. Depletion of rootletin does not affect the association of C-Nap1 with centrosomes, whereas either depletion of C-Nap1 or overexpression of its fragments affects the centrosomal localization of rootletin (Bahe et al. 2005; Yang et al. 2006). Phosphorylation of both proteins by Nek2A (Fry et al. 1998; Helps et al. 2000; Bahe et al. 2005) promotes centrosome disjunction at the G2/M transition; overexpression of wild-type Nek2A will stimulate centrosome splitting in interphase, whereas the kinase-dead mutant leads to monopolar spindles with unseparated spindle poles (Faragher and Fry 2003). Together, this has led to a model whereby C-Nap1 provides a docking site for rootletin fibers to connect the proximal ends of centrioles, Nek2A phosphorylates both C-Nap1 and rootletin, promoting their dissociation from the centrosome and leading to loss of centrosome cohesion.
Figure 5.
Centrosome disjunction and centriole disengagement. (A) Series of upstream events trigger the dissociation of C-Nap1 and rootletin from the centrosome leading to loss of centrosome cohesion. Main players are depicted. (B) Roles of Plk1 and separase in disjoining mother and daughter centrioles.
Two Hippo pathway components, Mst2 kinase (mammalian sterile 20–like kinase 2) and the scaffold protein hSav1 (scaffold protein Salvador), directly interact with Nek2A and regulate its ability to localize to centrosomes. Phosphorylation of Nek2A by Mts2 promotes its ability to induce centrosome disjunction. Depletion of Mst2, hSav1, or Nek2A results in a reduction of C-Nap1 phosphorylation and the continued association of C-Nap1 and rootletin with centrosomes that are still able to separate and form a spindle through the Eg5 pathway (Mardin et al. 2010). It seems that Plk1 functions upstream of the Mst2-Nek2A pathway. Phosphorylation of Mst2 by Plk1 blocks formation of a Nek2A–PP1γ–Mst2 complex in which Nek2 phosphorylation of C-Nap1 is counteracted by PP1 (Helps et al. 2000; Mardin et al. 2011). Nek2 also associates with two structural proteins that block its activity, namely, the focal adhesion scaffolding protein, HEF1 (Pugacheva and Golemis 2005) and pericentrin/kendrin (Matsuo et al. 2010). It has been proposed that pericentrin/kendrin serves to anchor Nek2 at the centrosome and suppress its activity.
After centrosome disjunction, the further separation of two centrosomes is mediated mainly by the kinesin-5 subfamily member, Eg5 (for review, see Ferenz et al. 2010). Eg5 homotetramers cross-link antiparallel microtubules so that when the motors walk toward microtubule plus ends, the antiparallel microtubules slide apart and centrosomes get pushed away from each other (Kashina et al. 1996). Inhibition of Eg5 by small molecule inhibitors monastrol results in prometaphase-arrested cells with monopolar spindles (Kapoor et al. 2000).
PCM Assembly
The enlargement of the centrosome that begins before mitosis results from recruitment of PCM first thought to be amorphous, but more recently revealed by 3D-structured illumination microscopy to be organized in layers that have a clear hierarchy (Fig. 6A,B) (Fu and Glover 2012; Lawo et al. 2012; Mennella et al. 2012; Sonnen et al. 2012). It is helpful to consider PCM in terms of two groups of proteins. One group of PCM proteins, including Dplp/pericentrin, Asl/Cep152, and Plk4, are associated with the mother centriole where, throughout the cell cycle, they reside in the region adjacent to the centriole microtubules. The other group, including Spd-2/Cep192, CNN/CDK5RAP2, and γ-tubulin are robustly recruited only on mitotic entry, again predominantly around the mother centriole, when the centrosome begins to nucleate increased numbers of microtubules.3
Although Asl is essential for Plk4 function (Dzhindzhev et al. 2010), pointers to another of its functions—recruiting PCM—came from early studies of asl mutants (Varmark et al. 2007). Other indications came from the discovery that CNN failed to be recruited to centrosomes following the injection of antibodies against Asl into Drosophila (Conduit et al. 2010). These findings can largely be accounted for by a direct interaction between Asl and Sas-4 and the bridging role played by Sas-4 in linking the centriole with PCM (Dzhindzhev et al. 2010). When expressed in Drosophila embryos, either Sas-4 alone or a mutant form of Asl able to bind Sas-4 but not Plk4 can promote formation of acentriolar PCM aggregates that nucleate cytoplasmic microtubules (Dzhindzhev et al. 2010). Moreover, Sas-4 null mutant flies show a reduction of PCM components in testes (Gopalakrishnan et al. 2011, 2012). This reflects the ability of Sas-4 to form complexes with CNN and Dplp; centrosomes with mutant Sas-4 unable to form such complexes have reduced PCM (Dzhindzhev et al. 2010; Gopalakrishnan et al. 2011). A double mutation in Sas-4 protein sequence that abolishes its binding to tubulin enhances centrosomal protein complex formation leading to abnormally large centrosomes and asters (Gopalakrishnan et al. 2012). Thus, tubulin binding may interfere with Sas-4-mediated PCM assembly. Sas-4 is recruited to both mother and daughter centrioles at a very early stage, and how it specifically initiates PCM assembly around mother but not daughter centrioles needs further study.
Surprisingly, a whole genome siRNA screen with Drosophila cells identified only three components, Polo, Spd-2, and CNN, that are required for the second phase—the expansion of the PCM for mitosis (Fig. 6C) (Dobbelaere et al. 2008). Polo kinase or Plk1 (Polo-like kinase 1) is the major kinase required for the dramatic increase of PCM that occurs before mitotic entry (Blagden and Glover 2003; Glover 2005). Its kinase activity is needed for the normal localization of Spd-2/Cep192, CNN/CDK5RAP2, pericentrin, and Nedd1 (Haren et al. 2009; Zhang et al. 2009; Hatch et al. 2010; Lee and Rhee 2011; Fu and Glover 2012). Interestingly, Drosophila Polo is restricted to zone II (i.e., the vicinity of the microtubule wall). As Polo phosphorylates both Spd-2 and CNN, this suggests the existence of a phosphorylation gradient as PCM assembles. In accord with this notion, a FRAP experiment indicated that CNN is recruited first to the vicinity of centriole before spreading out into the PCM (Conduit et al. 2010). Continuous Plk1 activity is required during mitosis to maintain PCM structure after centrosome maturation (Mahen et al. 2011). PCM is present only around the mother centriole as cells progress through mitosis but as the mother and daughter disengage, then the daughter becomes competent to recruit PCM in the following G1 phase. This process requires Plk1, as does the accompanying process of centriole disengagement (see below), although what exactly Plk1’s substrate might be in this process remains unknown (Wang et al. 2011). In C. elegans, Plk1 is targeted to the centrosome by Spd-2, which, when overexpressed, can increase the centrosome volume (Decker et al. 2011). However, whether Plk1 is targeted by this route in other organisms is not yet clear.
There is some interdependency for recruitment of Polo’s substrates Spd-2 and CNN to the PCM; localization of Spd-2 to PCM requires CNN, whereas CNN can partially support its own recruitment in cultured Drosophila cells (Fu and Glover 2012). This may vary a little among different cell types, as in the embryo, Conduit and colleagues suggest that Asl initially helps recruit Spd-2, which, in turn, recruits CNN. CNN, on the other hand, was not required to recruit Asl or Spd-2, but was required to maintain Spd-2 in the PCM (Conduit et al. 2014a). CNN mutants show reduced γ-tubulin and Aurora A at the embryonic centrosome (Megraw et al. 1999; Zhang and Megraw 2007), and a conserved motif near the amino terminus of CNN has been identified as essential for recruitment of γ-tubulin, D-TACC (transforming acidic coiled-coil proteins), and the Ch-TOG family microtubule polymerase Msps (Minispindles) (Zhang and Megraw 2007). Spd-2 mutant flies have reduced CNN and γ-tubulin in several cell types (Dix and Raff 2007; Giansanti et al. 2008), and injection of antibodies against Spd-2 into Drosophila embryos prevents CNN recruitment in the vicinity of the centriole (Conduit et al. 2010). CDKRAP2 and Cep192, human counterparts of CNN and Spd-2, also increase on centrosome maturation as does pericentrin, and the proteins show interdependency for the recruitment of γ-tubulin (Haren et al. 2009; Lee and Rhee 2011). In part, this reflects the physical association of γ-tubulin with the amino-terminal part, and pericentrin with the carboxy-terminal part of CDKRAP2 (Fong et al. 2008; Choi et al. 2010; Wang et al. 2010).
Aurora A kinase also contributes to centrosome maturation. In part, this may be because of its role together with Bora in activating Plk1 by phosphorylating Thr210 at the G2/M transition (Macurek et al. 2008; Seki et al. 2008). Secondly, Aurora A has a role in the enrichment of multiple centrosomal factors onto the centrosome. Key to this is the phosphorylation and recruitment of the transforming acidic coiled-coil protein 3 (TACC3; D-TACC in Drosophila) and its partner proteins that influence the properties of microtubules nucleated by the PCM (Giet et al. 2002; Terada et al. 2003; Barros et al. 2005; Kinoshita et al. 2005; Mori et al. 2007; Fu et al. 2010).
Disengagement of the Mother–Daughter Linkage
At the end of S phase, the cell has two centrosomes, each comprising a mother–daughter pair of centrioles (technically, one of these mothers is a grandmother). The pairs remain tightly linked at the poles of the spindle for mitosis, after which they disengage (Fig. 5B). Centriole disengagement has been described as a licensing step that enables the duplication of centrioles purified from S-phase-arrested HeLa cells and placed in a Xenopus egg extract (Tsou and Stearns 2006). In these experiments, disengaged centrioles could duplicate during the first interphase of a cycling extract, whereas engaged centrioles required an extra cycle to become disengaged. It has been suggested that recruitment of Asl onto the daughter centriole in Drosophila embryos occurs only once mother and daughter have separated at the end of mitosis and that this provides a duplication license (Novak et al. 2014). However, it is notable that in cultured cells, the incorporation of Ana2 and Sas-6 onto the daughter’s procentriole once it has disengaged from the mother in telophase appears to mark the very first event in the duplication process (Dzhindzhev et al. 2014).
Disengagement in late mitosis requires the activity of separase, the protease that promotes sister chromatid separation, and Plk1 (Tsou et al. 2009). Although the notion that centriole disengagement and chromatid separation might share common machinery appears elegant, whether cohesin is a separase substrate in both processes remains confusing. Although one study has reported that a noncleavable cohesin subunit Scc1 would not prevent disengagement in HeLa cells (Tsou et al. 2009), another suggested otherwise in Hek293 cells (Schockel et al. 2011). If endogenous Scc1 was replaced by a variant carrying a recognition site for human rhinovirus HRV protease or if its partner protein, Smc3, carried a site for TEV protease, these molecules could be cleaved by the appropriate protease resulting in centriole disengagement. However, similar experiments in Drosophila embryos have challenged the involvement of cohesin in centriole engagement. Drosophila embryos expressing TEV-cleavable Rad21 (corresponding to human Scc1) were arrested at metaphase by the catalytically inactive E2 ubiquitin ligase, Ubch10C114S (Cabral et al. 2013; Oliveira and Nasmyth 2013). Centriole disengagement could be observed after treatment with the Cdk inhibitor, p21, to drive mitotic exit but not after by TEV protease treatment that leads to sister chromatid separation (Oliveira and Nasmyth 2013). Depletion of the C. elegans separase, sep-1, impairs the separation and consequent duplication of sperm-derived centrioles at the meiosis–mitosis transition, but subsequent cycles proceed normally (Cabral et al. 2013). To date, searches for alternative separase substrates in centriole disengagement have identified kendrin/pericentrin B (Lee and Rhee 2012; Matsuo et al. 2012). However, much remains to be done before we understand the nature of this process.
Although most engagements are eventually dissolved even without separase, the requirement for Plk1 appears absolute. Inhibition of Plk1 in a Xenopus CSF extract, for example, will block disengagement in the presence of separase (Schockel et al. 2011), and overexpression of Plk1 will promote centriole disengagement in G2 (Loncarek et al. 2010). It also seems that APC/C activity contributes to disengagement (Prosser et al. 2012), and that Plk1 and APC/C-Cdh1 activities can independently achieve this (Hatano and Sluder 2012). The duality of the regulation of sister chromatid separation and centriole disengagement is echoed in the suggestion of a role for Plk1 in localizing the small isoform of Shugoshin1 (sSgo1). Mutation of sSgo1’s putative Plk1 phosphorylation sites results in weaker localization at centrosome and promotes centriole splitting in mitosis (Wang et al. 2008). Further work is required to determine the detail and universality of this potential regulatory system and the nature of Plk1’s substrates in centriole disengagement.
Plk1 may well have multiple roles at the centrosome toward the end of mitosis that relate to the differential behavior of mother and daughter centrioles. The disengagement of the procentriole by Plk1 is hypothesized to expose a site on the mother, enabling it once again to be able to initiate formation of a new procentriole. At the same time, Plk1 is proposed to modify the daughter centriole in some way as to render it competent for the initiation for procentriole formation in the next cycle (Loncarek et al. 2010; Wang et al. 2011).
THE CILIUM CYCLE
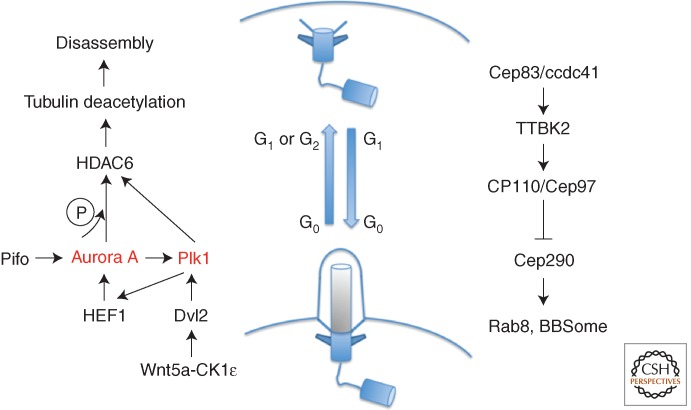
Primary cilia are present on most cell types in vertebrates where they function in signal sensing and transduction. In quiescent, G1 or G0, cells, the mother centriole associates with the ciliary vesicle at its distal end, migrates to the cell surface, and docks at the plasma membrane to become a basal body competent for cilium formation. The distal appendage proteins Cep83/ccdc41, SCLT1, FBF1 (Tanos et al. 2013), Cep89/ccdc123 (Sillibourne et al. 2011), and Cep164 (Graser et al. 2007) are all required for cilium formation (Tanos et al. 2013). CEP83 is specifically required for centriole-to-membrane docking (Joo et al. 2013; Tanos et al. 2013). The subsequent elongation of the ciliary axoneme depends on intraflagellar transport (IFT), a microtubule motor-based delivery system that transports cargos from outside the cilia to the growing tip, or retrieves them to the cell body (reviewed by Pedersen and Rosenbaum 2008; Ishikawa and Marshall 2011; Avasthi and Marshall 2012).
Ciliogenesis first requires the removal of CP110 and Cep97 from the distal end of the centriole (Fig. 7). This is promoted by Tau-tubulin kinase 2 (TTBK2); in TTBK2 null mouse embryo, E10.5 neural cilia are missing in 10.5-day-old embryos, and yet basal bodies dock to the cell cortex (Goetz et al. 2012). Cep83 appears to be upstream of this process because its depletion blocks centrosome-to-membrane association, and the undocked centrioles fail either to recruit TTBK2 or release CP110 (Riparbelli et al. 2012). It has been reported that, after cytokinesis, cilia arise considerably faster in cells inheriting the older mother centriole and that these cells are more responsive to Sonic Hedgehog signals that require receptors on the cilia (Anderson and Stearns 2009). This suggests the centriole age might transmit asymmetry to sister cells and potentially influence their ability to respond to environmental signals and so alter cell fate.
Figure 7.
Cilia assembly and disassembly cycle. Role of Plk1 and Aurora A in activating tubulin deacetylation for cilia disassembly (left). Cilia assembly pathway (right).
To regain its function as a centriole at the spindle pole, the basal body needs to dissociate from the cell cortex usually with loss of the cilium (Fig. 7). In mammalian cells, active Aurora A kinase associates with basal bodies to induce cilia disassembly by phosphorylating HDAC6 (histone deacetylase 6) to activate its tubulin deacetylase activity, necessary for primary cilia resorption (Pugacheva et al. 2007). HEF1, a transducer of integrin-initiated attachment, migration, and antiapoptotic signals at focal adhesions, appears to be required to stabilize and activate Aurora A for cilia disassembly (Pugacheva et al. 2007). In Chlamydomonas, cilia are cleaved from the basal body at the distal end of the transition zone (Parker et al. 2010) in a process requiring the scaffold protein Fa1, the NIMA family kinase Fa2 (Finst et al. 1998, 2000; Mahjoub et al. 2002), and the microtubule-severing protein katanin (Quarmby 2000). The Chlamydomonas Aurora A–like kinase, CALK, is also required for cilia excision and disassembly (Pan et al. 2004), suggesting that this is an evolutionarily conserved mechanism.
Plk1 also participates in the disassembly process by interacting with phosphorylated Dvl2 (Dishevelled 2), which is primed by both noncanonical Wnt5a signaling and CK1ε (casein kinase 1 epsilon). Depletion of Plk1, Dvl2, or disruption of their interaction leads to lower HEF1 levels and reduced Aurora A activity (Lee et al. 2012). Plk1, recruited by PCM1 to the PCM in G2, is also required for a second wave of cilia resorption (Wang et al. 2013). Plk1 appears also to interact with HDAC6 and promote tubulin deacetylation independently of Aurora A because activated Plk1 induces cilia disassembly in the presence of an Aurora A kinase inhibitor (Wang et al. 2013). This points to cooperation between Aurora A and Plk1 in centriole disassembly just as there is in centrosome maturation.
There is growing evidence that cell-cycle progression can be held up to accommodate cilia formation. When cells are depleted of Nde1 (nuclear distribution gene E homolog 1), cells develop longer cilia and cells are delayed in cell-cycle reentry (Kim et al. 2011). Cilia disassembly can also influence cycle progression; a phosphomimic mutant of the dynein-associated protein, Tctex-1, is recruited to the transition zone, where it accelerates both cilium disassembly and S-phase entry (Li et al. 2011). Suppression of Aurora A or HDAC6 activities will also inhibit both ciliary resorption and S phase (Li et al. 2011), thus pointing to an inhibitory effect of cilia on cell-cycle progression.
THE CENTROSOME AND SPB AS CONTROL CENTERS
The catalog of proteins that associate with the centrosome is very large. As the cell’s principal microtubule-organizing center (MTOC), it is a hub for microtubule-associated molecules and has great potential as a center for regulatory function.
Just as seminal advances in our understanding of cell-cycle progression came from studies with the budding yeast (Saccharomyces cerevisiae) and fission yeast (Schizosacharomyces pombe) in the 1980s, in more recent years, clues to the overriding importance of the centrosome in signaling mitotic entry have also emerged largely from studies in yeasts. To briefly recap, the universal cell-cycle controls owe much to fission yeast studies by Nurse and colleagues. Cdk1-cyclin B activity is restrained throughout interphase via phosphorylation of Cdk1 by Wee1 kinases. Removal of this phosphate from stockpiled Cdk1/cyclin B complexes by Cdc25 phosphatases then unleashes a wave of Cdk1-cyclin B activity that promotes mitotic commitment (Nurse 1990). Work in Xenopus laevis oocyte/egg extracts established that an initial impact of newly activated Cdk1-cyclin B is to use Polo kinase to enhance Cdc25 and repress Wee1 activities (Hoffmann et al. 1993; Izumi and Maller 1993, 1995; Strausfeld et al. 1994; Kovelman and Russell 1996; Kumagai and Dunphy 1996; Abrieu et al. 1998; Karaiskou et al. 1999, 2004; Lindqvist et al. 2009). These feedback loops convert mitotic commitment into a bistable, all or nothing, switch (Ferrell 2008).
It was clear from its discovery that the fission yeast Polo kinase, PoloPlo1, has roles at the SPB in both mitotic entry and exit (Ohkura et al. 1995; Tanaka et al. 2001). The first clue to PoloPlo1’s role in mitotic entry was its recruitment to the SPB with 15% of G2 phase remaining. This period of PoloPlo1 residence on the G2 spindle pole was doubled to occupy 30% of G2 phase by the cut12.s11 mutation in the SPB component Cut12 (Bridge et al. 1998; Mulvihill et al. 1999). This particular cut12 allele was originally named suppressor of cdc25, stf1.1 and, as its name suggests, is able to compensate for the loss of Cdc25 (Hudson et al. 1990). This implied that mitotic entry could be accomplished solely by the known ability of Wee1 inactivation to regulate mitotic entry in the absence of Cdc25 (Fantes 1979) when regulation of the spindle-pole-associated Cdk1/cyclin B changed (Alfa et al. 1990). The clear conservation of Cdk1/cyclin B feedback loops in fission yeast (Kovelman and Russell 1996; Tanaka et al. 2001) suggested that cut12.s11 may be exploiting these switches to inhibit Wee1 and drive mitosis.
Although the cut12.s11 mutation is an ostensibly innocuous glycine to valine change (Bridge et al. 1998), this particular glycine sits at the start of a bipartite docking site for protein phosphatase 1 (PP1) (Grallert et al. 2013a). This glycine/valine switch reduced PP1 recruitment to Cut12 leading to the revelation that a complete block to PP1 recruitment to Cut12 enabled cells to survive the otherwise lethal abolition of Cdc25 function (Grallert et al. 2013a). Thus, PP1 recruitment to Cut12 is integral to the mitotic commitment switch. The direct correlation between the degree of PP1 recruitment to Cut12 and the level of PoloPlo1 activity detected in whole cell extracts established that a primary impact of PP1 recruitment to Cut12 is to set the level of PoloPlo1 activity throughout the cell (Mulvihill et al. 1999; MacIver et al. 2003; Grallert et al. 2013a). PP1 recruitment to Cut12 also sets local PoloPlo1 activity at each individual SPB so that the temperature-sensitive loss-of-function cut12.1 mutation that enhances PP1 affinity for Cut12 blocks the conversion of the new SPB into a mitotic pole (Bridge et al. 1998; MacIver et al. 2003; Tallada et al. 2009; Grallert et al. 2013a). Consequently, cells form monopolar rather than bipolar spindles and cells die. This defect stemmed from deficient PoloPlo1-based feedback controls because a simple elevation of Cdc25 levels activated the new SPB to drive cut12.1 cells through a functional mitosis (Tallada et al. 2007, 2009). This evidence for spindle pole–driven mitotic commitment was consolidated by the ability of ectopic activation of either PoloPlo1 or Cdk1 at interphase SPBs to inappropriately promote mitotic commitment (Grallert et al. 2013b). These data categorically establish that events on the spindle pole form an integral part of the switch that regulates mitotic commitment.
The Cut12/PP1/PoloPlo1 switch can be altered either at the level of Cut12 or PoloPlo1. Phosphorylation within Cut12’s bipartite PP1 docking motif by Cdk1-cyclin B or the NIMA kinase Fin1 reduces PP1 affinity for Cut12, whereas simultaneous phosphorylation by both kinases blocks it completely (Grallert et al. 2013a). Cdk1/cyclin B phosphorylation of Cut12 clearly constitutes another feedback control as the enhancement of Plo1 activity it will generate will, in turn, accelerate activation of further Cdk1-cyclin B reserves. It is unclear whether Fin1 is also another mode of feedback control, or whether it coordinates division with specific external cues. In contrast, direct control of PoloPlo1’s SPB affinity clearly does couple division timing to environmental cues (Petersen and Hagan 2005; Petersen and Nurse 2007; Hartmuth and Petersen 2009; Halova and Petersen 2011). Phosphorylation of the conserved serine 402 between the kinase and Polo-box domains enhances SPB recruitment of PoloPlo1 in response to signaling flux through the Sty1 MAP kinase stress-response pathway (equivalent to p38 of higher eukaryotes) (Petersen and Hagan 2005; Halova and Petersen 2011). Heat, pressure, and nutritional signaling from the TOR network compromise the function of the protein phosphatase Pyp2 (equivalent to human DUSP65) toward the MAP kinase Sty1 (George et al. 2007; Petersen and Nurse 2007; Hartmuth and Petersen 2009; Halova and Petersen 2011). This reduction in Pyp2 function enhances MAP kinase signaling to boost serine 402 phosphorylation, thereby driving PoloPlo1 onto the SPB (Petersen and Nurse 2007) to couple division to at least three external cues: nutrition, heat, and pressure stresses.
Similar pathways may operate in animal cells but are less well characterized. It is long known that Cdk1/cyclin B associates with the centrosome (Bailly et al. 1989) and that its initial activation occurs here (Jackman et al. 2003). This appears to require the activity of the Cdc25B isoform (Gabrielli et al. 1996; De Souza et al. 2000; Lindqvist et al. 2005) whose association with centrosomes is promoted by Aurora A (Dutertre et al. 2004). Specific phosphorylation events on centrosomal Cdc25C in late G2 phase could also play some role (Franckhauser et al. 2010). Although the relationship to the spindle pole is less well developed than in fission yeast, Plk1 activity levels do determine the timing of mitotic commitment in human cells (Gavet and Pines 2010). In a further echo, the role played by PoloPlo1. S402 phosphorylation following heat stress in fission yeast; human Plk1 drives the first division after recovery from DNA damage (Macurek et al. 2008; Seki et al. 2008). A firmer link to centrosomal activities sits at the heart of the promotion of mitotic commitment by Aurora A in C. elegans (Hachet et al. 2007), the acceleration of mitotic commitment in Xenopus egg extracts following the addition of centrosomes (Perez-Mongiovi et al. 2000), and in humans on removal of the centrosomal component MCPH1 (Gruber et al. 2011). Modeling experiments in Xenopus egg extracts now bring these diverse threads together into a coherent reiteration of the fission yeast data by supporting the concept of waves of Cdk1/cyclin B feedback loop activities emanating from the centrosome (Chang and Ferrell 2013).
A final twist to centrosomal control has been provided by the demonstration that pericentrin mutations alter the DNA damage checkpoint response (Griffith et al. 2008). This link had been anticipated by reports that centrosomal Chk1 played a critical role in determining the timing of mitotic commitment. However, concerns about the specificity of the antibody used for immunolocalization and the expression levels of centrosomally targeted Chk1 fusion proteins in the early studies has questioned whether Chk1 does indeed act at centrosomes (Matsuyama et al. 2011). Thus, the link between pericentrin and cell-cycle control remains enigmatic, although it is tempting to speculate that the potential coupling between fission yeast pericentrin and Cut12/PoloPlo1 controls could hold the key (Fong et al. 2010).
In summary, although less developed than the fission yeast data, the indications are that centrosomal triggering of mitotic commitment will be a universal phenomenon that may well involve the same players that operate in fission yeast.
CENTROSOME ASYMMETRIES
Cellular Asymmetry and Spindle Alignment in Budding Yeast
The two mitotic SPBs can have strikingly different impacts on yeast cell fate, even though they both reside within the same cytoplasm at the same point in the cell-division cycle. This distinctive behavior can arise from intrinsic differences in SPB maturation or extrinsic cues that influence the recruitment of SPB components or the modification of resident molecules. In the budding yeast S. cerevisiae, an initial intrinsic distinction between the old and new SPB is overwhelmed by extrinsic factors that arise from inherent cellular asymmetry to impose asymmetric behavior that is entirely dependent on SPB position rather than age. In contrast, intrinsic control predominates in the fission yeast S. pombe.
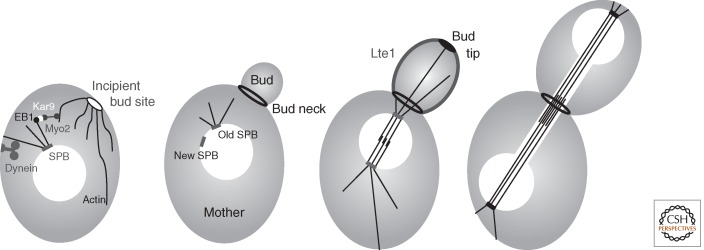
The intrinsic asymmetry of budding yeast cells is generated through expansive growth at the tip of a bud that emerges from a defined zone of the mother cell cortex. The narrow channel connecting the mother and bud (the bud neck) presents a unique challenge for genome segregation as the mitotic spindle must be aligned to promote passage of one set of the duplicated chromosomes through the neck (Fig. 8). Two redundant pathways guide this alignment by directing the astral microtubules that emerge from the cytoplasmic face of one of the two SPBs through the narrow aperture. In one, Kar9 associates with both the microtubule plus end-binding protein EB1 and the myosin motor protein Myo2 to guide astral microtubules along actin filaments that emanate from the incipient bud site and growing bud tips (Beach et al. 2000; Lee et al. 2000; Yin et al. 2000). In the second pathway, dynein both guides the astral microtubules along the cell cortex and promotes instability of the plus ends to ensure end-on association of microtubules with the cortex (McMillan and Tatchell 1994; Carminati and Stearns 1997; Adames and Cooper 2000; Heil-Chapdelaine et al. 2000; Yeh et al. 2000). When astral guidance fails, the ensuing nuclear division within the mother cell blocks cytokinesis and mitotic exit through activation of the spindle orientation checkpoint (SPOC) (Yeh et al. 1995; Miller and Rose 1998; Miller et al. 1998; Bardin et al. 2000; Pereira et al. 2000; Caydasi and Pereira 2012).
Figure 8.
Key landmarks in the control of asymmetric spindle pole body (SPB) function in budding yeast. The figure shows the key landmarks that are referred to in the text imposed on depictions of bud growth from a mother yeast cell. See text for full details on how each of these molecules/features contributes to asymmetries of SPB function that ensure each daughter cell will inherit one genome.
SPB Asymmetry and Spindle Alignment in Budding Yeast
The strong tendency for the old SPB to lead the spindle into the bud (Pereira et al. 2001) stems from a delay in the recruitment of the Spc72 γ-tubulin docking component of the cytoplasmic outer plaque of the new SPB (Juanes et al. 2013). Because microtubules cannot be nucleated until γ-tubulin is recruited to Spc72 (Knop and Schiebel 1998), the inherent pause in Spc72 docking delays microtubule nucleation from the new SPB to allow the microtubules extending from the old SPB to be guided into the bud (Shaw et al. 1997; Juanes et al. 2013). However, if the connection between astral microtubules and the cortex is broken before the old SPB enters the bud (via transient dissolution of the microtubule cytoskeleton), then microtubules extending from either the old or the new SPB can lead spindle orientation into the bud (Pereira et al. 2001). Thus, although intrinsic asymmetry in SPB maturation drives spindle orientation in unperturbed divisions, extrinsic controls are equally competent to drive asymmetry of SPB function.
Extrinsic Control of Asymmetric SPB Behavior Drives the End of the Budding Yeast Cell Cycle
Exit from budding yeast mitosis is driven by the Hippo-signaling-related MEN that releases the protein phosphatase Cdc14 from sequestration in the nucleolus (Visintin et al. 1998; Pereira and Schiebel 2001; Stegmeier and Amon 2004; Hergovich et al. 2006). Newly released Cdc14 can then dephosphorylate sites that were initially phosphorylated by Cdk1/cyclins to promote the mitotic state. This impact of the MEN on Cdc14 release, and its coincident control of the cytokinetic machinery (Meitinger et al. 2012), acts at sites that are remote from the heart of MEN control on the SPBs. Whether or not an SPB resides within the mother or the bud determines its competence to promote MEN signaling.
MEN activity is controlled by a GTPase of the Ras superfamily called Tem1. Tem1 activation recruits the Pak kinase Cdc15 that then recruits the NDR kinase Dbf2/Mob1 kinase to drive Cdc14 from the nucleolus. Tem1 activation is promoted by PoloCdc5 phosphorylation of its Bfa1/Bub2 GAP complex (Hu et al. 2001; Pereira and Schiebel 2001; Geymonat et al. 2003; Simanis 2003; Stegmeier and Amon 2004; Maekawa et al. 2007). Once the GAP is inhibited, the intrinsic GTP exchange activity of Tem1 family GTPases (Furge et al. 1998; Geymonat et al. 2003) then flips Tem1 into an active GTP-bound form that drives MEN activation.
The primary mode of spatial control of MEN activity is through modulation of Kin4 kinase activity (D’Aquino et al. 2005; Pereira and Schiebel 2005). Kin4 phosphorylation of Bfa1 both blocks its ability to be inhibited through PoloCdc5 phosphorylation and promotes docking of 14-3-3 proteins to drive Bfa1 turnover at SPBs (Caydasi and Pereira 2009; Caydasi et al. 2014). Bfa1 phosphorylation by Kin4, therefore, locks MEN signaling in the off state. Kin4 is inhibited by Lte1 (Bertazzi et al. 2011). Lte1 accumulates on the bud cortex until one SPB enters the bud where upon it is released into the cytoplasm (Bardin et al. 2000; Pereira et al. 2000). Kin4, on the other hand, associates with both the cortex and SPB of the mother cell (D’Aquino et al. 2005; Pereira and Schiebel 2005). This partitioning of the kinase and its inhibitor ensures high levels Kin4 activity in the mother and very low levels in the daughter (Bertazzi et al. 2011). Spindle misalignment errors that result in the retention of both SPBs in the mother cell place these two SPBs in the zone of high Kin4 activity that drives Kin4 onto both SPBs to block mitotic exit. If one SPB eventually enters the bud, Kin4 activity will be inhibited on that SPBbud by Lte1 to promote Kin4 departure and so drive MEN activation from this SPBbud (Bertazzi et al. 2011; Falk et al. 2011; Caydasi et al. 2014).
Intrinsic Controls Set by SPB Maturation Establishes One SPB as the Signaling Center for Mitotic Exit in Fission Yeast
Although the asymmetric competence for signaling of two otherwise indistinguishable SPBs arises primarily from extrinsic, spatial context cues to drive mitotic exit of budding yeast, it is primarily the age of the SPB and not external cues that determines which of the two anaphase SPBs will host the active version of the fission yeast equivalent of the MEN (Simanis 2003; Grallert et al. 2004). Thus, even in the simple single-celled yeasts, different strategies can turn one of the two spindle poles into a unique signaling platform from which to determine cell fate.
Asymmetry of Centrioles, Centrosomes, and Fate Decisions
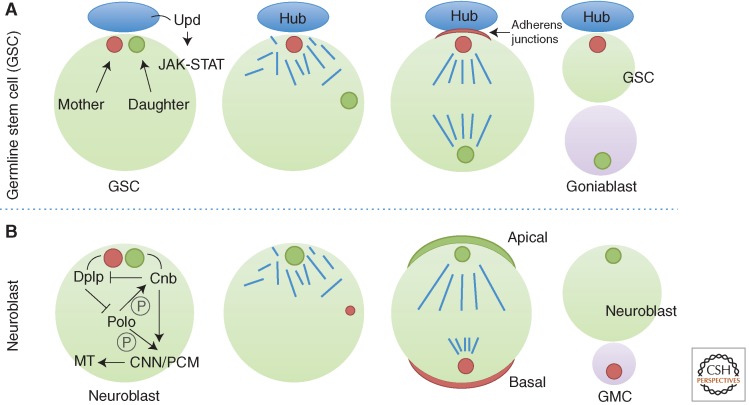
In animal cells, there is increasing evidence that stereotypical centrosome inheritance correlates with cell fate decisions. The Drosophila male germline stem cell (GSC) divides asymmetrically to produce one stem cell that is able to self-renew and one gonioblast that subsequently undergoes differentiation (Fig. 9A). GSCs attach to a cluster of support cells called the hub that secrete the signaling ligand Upd (Unpaired), which activates the JAK-STAT (Janus kinase-signal transducer and activator of transcription) pathway in adjacent germ cells to specify stem cell identity (Kiger et al. 2001; Tulina and Matunis 2001). During cell division, the spindle is orientated perpendicular to the hub so that one daughter cell remains attached to the hub and the other is placed outside the niche. Using a heat shock–Gal4 control promoter, only the newly assembled centriole is labeled on pulse-chase heat shock by transient expression of a centrosomal marker, GFP-PACT (Yamashita et al. 2007). The mother centrosome was observed to be normally inherited by the GSC, whereas the daughter centrosome moves away from the hub and is inherited by the cell that commits to differentiation (Yamashita et al. 2007).
Figure 9.
Inheritance of centrosomes in asymmetric cell division in Drosophila. The stem cells of the male germline inherit the mother centrosome (A) in contrast to the stem cell of the neuroblasts that inherit the daughter (B). GMC, ganglion mother cell; MT, microtubule; PCM, pericentriolar material.
A similar finding was made in mouse radial glia progenitors that produce self-renewing radial glia in the ventricular zone and differentiating cells for the future neocortex (Fig. 9B) (Wang et al. 2009). Centrosomes were marked with photoconvertible Centrin1 that would change color from green to red when pulsed with violet light. Original centrioles (red) could then be distinguished from newly formed centrioles (green). During the peak phases of neurogenesis, mother centrosomes are preferentially inherited by radial glia progenitors that stay in the ventricular zone, whereas the daughter centrosomes mostly leave the ventricular zone and are associated with differentiating cells. Removal of the subdistal appendage protein, ninein, disrupts the asymmetric segregation and inheritance of the centrosome and causes premature depletion of progenitors from the ventricular zone.
The reverse pattern of inheritance is seen in Drosophila neuroblasts that function as stem cells for brain development, where the mother centrosome is inherited by the differentiating daughter cell. Mother and daughter centrosomes could be traced either by photoconvertible PACT and the daughter centriole marker Cnb (centrobin) (Januschke et al. 2011) or distinguished by the different fluorescence intensity of PACT (Conduit and Raff 2010). Early in the cell cycle, the daughter centrosome organizes an aster that remains by the apical cortex, whereas the mother loses PCM and microtubule-organizing activity and moves extensively throughout the cell (Rebollo et al. 2007). Before mitosis, the mother centrosome begins to recruit PCM and organizes a mitotic aster as it settles near the basal cortex. When the daughter centrosome-specific protein, Centrobin (Cnb), is depleted, both centrosomes are free to wander in the cells, and they lose their interphase association with centrosomal proteins Asl and CNN. Strikingly, when Cnb is ectopically targeted to both centrosomes by fusing to PACT, the mother centrosome is able to retain PCM and microtubule organizing activity. Cnb coprecipitates with a set of centrosomal proteins, including γ-tubulin, Ana2, CNN, Sas-4, Sas-6, Asl, DGrip71, and Polo. These data suggest that Cnb is both necessary and sufficient for centrosomes to retain microtubule organizing activity probably through interaction with PCM components (Januschke et al. 2013). Cnb is regulated by Polo in this process; following chemical inhibition of Polo or mutation of three Polo phosphorylation sites, the interphase microtubule aster is lost (Januschke et al. 2013). The centriolar protein, Cep135, is required to establish centrosome asymmetry in Drosophila neuroblasts through shedding of Polo from the mother centrosome (Singh et al. 2014). Dplp (pericentrin-like protein), on the other hand, is more enriched on the mother centrosome (Lerit and Rusan 2013). In Dplp mutant neuroblasts, the mother centrosome retains γ-tubulin and Polo levels comparable to the daughter. Targeting Cnb to both centrosomes abolishes the asymmetric distribution of Dplp, suggesting that Dplp is one of Cnb’s targets to establish centrosome asymmetry.
Defects in Division Symmetry in Microcephaly
In contrast to the above examples of asymmetric centriolar behavior, the reasons why aberrant centrosome behavior might lead to the various forms of inherited microcephaly are less clear. Microcephaly develops as the result of a failure of neuroprogenitor cells to undertake sufficient symmetric divisions to generate sufficient numbers of cells before asymmetric, differentiating divisions to generate the cerebral cortex (Jeffers et al. 2008; Rai et al. 2008). It is generally thought that proper centrosomal function is required to ensure correct spindle orientation to prevent asymmetric divisions from occurring before the progenitor cell population has been sufficiently expanded (reviewed in Morrison and Kimble 2006). The most commonly affected gene in microcephaly is aspm (abnormal spindle-like microcephaly associated, MCPH5) (Thornton and Woods 2009). In cultured human cells, ASPM localizes to centrosomes and spindle poles, similar to its fly and worm ortholog (Saunders et al. 1997; Kouprina et al. 2005; van der Voet et al. 2009; Noatynska et al. 2012). Neuroprogenitors depleted of ASPM fail to orient the mitotic spindle perpendicular to the ventricular surface of the neuroepithelium giving asymmetric, rather than symmetric, divisions (Fish et al. 2006). Mutations in CPAP (MCPH6) and STIL (MCPH7) also affect spindle orientation (Kitagawa et al. 2011b; Brito et al. 2012). Other genes for centrosomal proteins that are mutated in microcephaly include the CNN homolog, CDK5RAP2 (MCPH3), required for centrosome maturation and DNA damage-induced G2 arrest (Barr et al. 2010; Lizarraga et al. 2010), and CEP63 with its partner Cep152, which participate in centriole assembly (Sir et al. 2011). Centriole amplification has also been shown to be one cause of microcephaly in human patients that is triggered by mutants that stabilize STIL by removing a KEN destruction box (Arquint and Nigg 2014).
The link tying microcephaly mutations to spindle orientation in brain development may not be quite as straightforward as it seems. Not all forms of microcephaly are associated with defects in spindle orientation; a mutant encoding a truncated version of ASPM results in microcephaly without interfering with spindle orientation (Pulvers et al. 2010). Conversely, division orientation can be completely randomized in neuroprogenitor cells by mutations in LGN, a protein that helps orient the spindle with respect to the axis of cell polarity, and yet this does not lead to smaller brains. One of the genes mutated in microcephaly patients encodes MCPH1, which regulates several DNA damage response proteins. MCPH1 appears in radiation-induced nuclear foci (Rai et al. 2006) and specifically regulates levels of both BRCA1 and Chk1 (Xu et al. 2004; Lin et al. 2005; Tibelius et al. 2009). MCPH1 knockdown cells show increased radiosensitivity (Peng et al. 2009; Liang et al. 2010) as well as recombination and chromosome condensation defects (Wood et al. 2008, Liang et al. 2010; Trimborn et al. 2010). MCPH1 also participates in altering the chromatin structure in response to DNA damage to allow DNA repair (Peng et al. 2009).
We, therefore, seem still not to fully understand the underlying causes of these defects in brain development and all of the roles of the centrosome in this process.
CONCLUDING REMARKS
We have come a long way in understanding centrosome structure and function, particularly with the explosion of molecular detail that has been added to the picture over the past decade. We are now beginning to piece together the molecular components of the organelle and their dynamic interactions during cell-cycle progression. As with many cell-cycle processes, the principal events of the centrosome cycle are regulated by a combination of protein phosphorylation/dephosphorylation and stability. We now have to find out exactly how and when centrosomal proteins become modified and how this affects their function. Only then will we be able to bring our level of understanding of the metazoan centrosome up to what we already have for yeast SPBs. We also have to fully appreciate the diversity of centrosomal function in different cell types of metazoans and their roles in development. Perhaps only when we have a greater understanding of these functions in normal cells will we be able to properly address how these processes go awry in the multiple types of tumor cells that show centrosomal abnormalities. Many challenges still lie ahead before we can come to grips with the many roles of the centrosome uncovered at the end of the 19th century.
This is in addition to Spd-2 present within zone II throughout the cell cycle.
Editors: Mitsuhiro Yanagida, Anthony A. Hyman, and Jonathon Pines
Additional Perspectives on Mitosis available at www.cshperspectives.org
REFERENCES
- Abrieu A, Brassac T, Galas S, Fisher D, Labbe JC, Doree M 1998. The Polo-like kinase Plx1 is a component of the MPF amplification loop at the G2/M-phase transition of the cell cycle in Xenopus eggs. J Cell Sci 111: 1751–1757. [DOI] [PubMed] [Google Scholar]
- Adames NR, Cooper JA 2000. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol 149: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams IR, Kilmartin JV 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol 145: 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams IR, Kilmartin JV 2000. Spindle pole body duplication: A model for centrosome duplication? Trend Cell Biol 10: 329–335. [DOI] [PubMed] [Google Scholar]
- Alfa CE, Ducommun B, Beach D, Hyams JS 1990. Distinct nuclear and spindle pole body populations of cyclin-cdc2 in fission yeast. Nature 347: 680–682. [DOI] [PubMed] [Google Scholar]
- Anderson CT, Stearns T 2009. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr Biol 19: 1498–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archinti M, Lacasa C, Teixido-Travesa N, Luders J 2010. SPICE—A previously uncharacterized protein required for centriole duplication and mitotic chromosome congression. J Cell Sci 123: 3039–3046. [DOI] [PubMed] [Google Scholar]
- Arquint C, Nigg EA 2014. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr Biol 24: 351–360. [DOI] [PubMed] [Google Scholar]
- Arquint C, Sonnen KF, Stierhof YD, Nigg EA 2012. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J Cell Sci 125: 1342–1352. [DOI] [PubMed] [Google Scholar]
- Avasthi P, Marshall WF 2012. Stages of ciliogenesis and regulation of ciliary length. Differentiation 83: S30–S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Hergert P, Delouvee A, Euteneuer U, Formstecher E, Khodjakov A, Bornens M 2009. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol 185: 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Wong ML, Downhour DM, Sanchez Alvarado A, Marshall WF 2012. Centrosome loss in the evolution of planarians. Science 335: 461–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA 2005. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol 171: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahtz R, Seidler J, Arnold M, Haselmann-Weiss U, Antony C, Lehmann WD, Hoffmann I 2012. GCP6 is a substrate of Plk4 and required for centriole duplication. J Cell Sci 125: 486–496. [DOI] [PubMed] [Google Scholar]
- Bailly E, Doree M, Nurse P, Bornens M 1989. p34cdc2 is located in both nucleus and cytoplasm; part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J 8: 3985–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra FR, Strnad P, Fluckiger I, Gonczy P 2013. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev Cell 25: 555–571. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102: 21–31. [DOI] [PubMed] [Google Scholar]
- Barr AR, Kilmartin JV, Gergely F 2010. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J Cell Biol 189: 23–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros TP, Kinoshita K, Hyman AA, Raff JW 2005. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J Cell Biol 170: 1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P, Furlong C, Byers B 1986. Yeast gene required for spindle pole body duplication—Homology of its product with Ca2+-binding proteins. Proc Natl Acad Sci 83: 5512–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr Biol 10: 1497–1506. [DOI] [PubMed] [Google Scholar]
- Bertazzi DT, Kurtulmus B, Pereira G 2011. The cortical protein Lte1 promotes mitotic exit by inhibiting the spindle position checkpoint kinase Kin4. J Cell Biol 193: 1033–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol 15: 2199–2207. [DOI] [PubMed] [Google Scholar]
- Biggins S, Rose MD 1994. Direct interaction between yeast spindle pole body components—Kar1p is required for Cdc31p localization to the spindle pole body. J Cell Biol 125: 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden SP, Glover DM 2003. Polar expeditions—Provisioning the centrosome for mitosis. Nat Cell Biol 5: 505–511. [DOI] [PubMed] [Google Scholar]
- Bridge AJ, Morphew M, Bartlett R, Hagan IM 1998. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev 12: 927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Gouveia SM, Bettencourt-Dias M 2012. Deconstructing the centriole: Structure and number control. Curr Opin Cell Biol 24: 4–13. [DOI] [PubMed] [Google Scholar]
- Brownlee CW, Klebba JE, Buster DW, Rogers GC 2011. The Protein Phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J Cell Biol 195: 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullit E, Rout MP, Kilmartin JV, Akey CW 1997. The yeast spindle pole body is assembled around a central crystal of Spc42. Cell 89: 1077–1087. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L 1974. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol 38: 123–131. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol 124: 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral G, Sans SS, Cowan CR, Dammermann A 2013. Multiple mechanisms contribute to centriole separation in C. elegans. Curr Biol 23: 1380–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol 138: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M 2010. Stepwise evolution of the centriole-assembly pathway. J Cell Sci 123: 1414–1426. [DOI] [PubMed] [Google Scholar]
- Caydasi AK, Pereira G 2009. Spindle alignment regulates the dynamic association of checkpoint proteins with yeast spindle pole bodies. Dev Cell 16: 146–156. [DOI] [PubMed] [Google Scholar]
- Caydasi AK, Pereira G 2012. SPOC alert—When chromosomes get the wrong direction. Exp Cell Res 318: 1421–1427. [DOI] [PubMed] [Google Scholar]
- Caydasi AK, Micoogullari Y, Kurtulmus B, Palani S, Pereira G 2014. The 14-3-3 protein Bmh1 functions in the spindle position checkpoint by breaking Bfa1 asymmetry at yeast centrosomes. Mol Biol Cell 25: 2143–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JB, Ferrell JE Jr. 2013. Mitotic trigger waves and the spatial coordination of the Xenopus cell cycle. Nature 500: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD 2002. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell 3: 339–350. [DOI] [PubMed] [Google Scholar]
- Choi YK, Liu P, Sze SK, Dai C, Qi RZ 2010. CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex. J Cell Biol 191: 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol 191: 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comartin D, Gupta GD, Fussner E, Coyaud E, Hasegan M, Archinti M, Cheung SW, Pinchev D, Lawo S, Raught B, et al. 2013. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr Biol 23: 1360–1366. [DOI] [PubMed] [Google Scholar]
- Conduit PT, Raff JW 2010. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr Biol 20: 2187–2192. [DOI] [PubMed] [Google Scholar]
- Conduit PT, Brunk K, Dobbelaere J, Dix CI, Lucas EP, Raff JW 2010. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr Biol 20: 2178–2186. [DOI] [PubMed] [Google Scholar]
- Conduit PT, Feng Z, Richens JH, Baumbach J, Wainman A, Bakshi SD, Dobbelaere J, Johnson S, Lea SM, Raff JW 2014a. The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev Cell 28: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Richens JH, Wainman A, Holder J, Vicente CC, Pratt MB, Dix CI, Novak ZA, Dobbie IM, Schermelleh L, et al. 2014b. A molecular mechanism of mitotic centrosome assembly in Drosophila. eLife 3: e03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier A, Clement MJ, Knossow M, Lachkar S, Savarin P, Toma F, Sobel A, Gigant B, Curmi PA 2009. The PN2-3 domain of centrosomal P4.1-associated protein implements a novel mechanism for tubulin sequestration. J Biol Chem 284: 6909–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee MA, Raff JW, Lea SM, Roque H 2011. SAS-6 oligomerization: The key to the centriole? Nat Chem Biol 7: 650–653. [DOI] [PubMed] [Google Scholar]
- Cottee MA, Muschalik N, Wong YL, Johnson CM, Johnson S, Andreeva A, Oegema K, Lea SM, Raff JW, van Breugel M 2013. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. Elife 2: e01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbelli M, Zhang W, Laue E, Callaini G, Glover DM, Bettencourt-Dias M 2009. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol 19: 43–49. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell 7: 815–829. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Maddox PS, Desai A, Oegema K 2008. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the γ-tubulin-mediated addition of centriolar microtubules. J Cell Biol 180: 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, Dynlacht B, Pagano M 2010. SCF (cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature 466: 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aquino KE, Monje-Casas F, Paulson J, Reiser V, Charles GM, Lai L, Shokat KM, Amon A 2005. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol Cell 19: 223–234. [DOI] [PubMed] [Google Scholar]
- Decker M, Jaensch S, Pozniakovsky A, Zinke A, O’Connell KF, Zachariae W, Myers E, Hyman AA 2011. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr Biol 21: 1259–1267. [DOI] [PubMed] [Google Scholar]
- Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gonczy P 2004. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol 6: 656–664. [DOI] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gonczy P 2006. Sequential protein recruitment in C. elegans centriole formation. Curr Biol 16: 1844–1849. [DOI] [PubMed] [Google Scholar]
- Delgehyr N, Rangone H, Fu J, Mao G, Tom B, Riparbelli MG, Callaini G, Glover DM 2012. Klp10A, a microtubule-depolymerizing kinesin-13, cooperates with CP110 to control Drosophila centriole length. Curr Biol 22: 502–509. [DOI] [PubMed] [Google Scholar]
- De Souza CP, Ellem KA, Gabrielli BG 2000. Centrosomal and cytoplasmic Cdc2/cyclin B1 activation precedes nuclear mitotic events. Exp Cell Res 257: 11–21. [DOI] [PubMed] [Google Scholar]
- Dix CI, Raff JW 2007. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr Biol 17: 1759–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol 6: e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Kilmartin JV 1996. Spc42p—A phosphorylated component of the Saccharomyces cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J Cell Biol 132: 887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, Cazales M, Quaranta M, Froment C, Trabut V, Dozier C, Mirey G, Bouche JP, Theis-Febvre N, Schmitt E, et al. 2004. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J Cell Sci 117: 2523–2531. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, Glover DM 2010. Asterless is a scaffold for the onset of centriole assembly. Nature 467: 714–718. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Tzolovsky G, Lipinszki Z, Schneider S, Lattao R, Fu J, Debski J, Dadlez M, Glover DM 2014. Plk4 phosphorylates Ana2 to Trigger Sas6 recruitment and procentriole formation. Curr Biol 24: 2526–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S, Knop M, Schlenstedt G, Schiebel E 1999. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc Natl Acad Sci 96: 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlemann S, Neuner A, Gombos L, Gibeaux R, Antony C, Schiebel E 2012. An extended γ-tubulin ring functions as a stable platform in microtubule nucleation. J Cell Biol 197: 59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk JE, Chan LY, Amon A 2011. Lte1 promotes mitotic exit by controlling the localization of the spindle position checkpoint kinase Kin4. Proc Natl Acad Sci 108: 12584–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes P 1979. Epistatic gene interactions in the control of division in fission yeast. Nature 279: 428–430. [DOI] [PubMed] [Google Scholar]
- Faragher AJ, Fry AM 2003. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell 14: 2876–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Gable A, Wadsworth P 2010. Mitotic functions of kinesin-5. Semin Cell Dev Biol 21: 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson RL, Maller JL 2008. Cyclin E–dependent localization of MCM5 regulates centrosome duplication. J Cell Sci 121: 3224–3232. [DOI] [PubMed] [Google Scholar]
- Ferguson RL, Pascreau G, Maller JL 2010. The cyclin A centrosomal localization sequence recruits MCM5 and Orc1 to regulate centrosome reduplication. J Cell Sci 123: 2743–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE Jr. 2008. Feedback regulation of opposing enzymes generates robust, all-or-none bistable responses. Curr Biol 18: R244–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finst RJ, Kim PJ, Quarmby LM 1998. Genetics of the deflagellation pathway in Chlamydomonas. Genetics 149: 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finst RJ, Kim PJ, Griffis ER, Quarmby LM 2000. Fa1p is a 171 kDa protein essential for axonemal microtubule severing in Chlamydomonas. J Cell Sci 113: 1963–1971. [DOI] [PubMed] [Google Scholar]
- Firat-Karalar EN, Rauniyar N, Yates JR III, Stearns T 2014. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol 24: 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB 2006. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci 103: 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk HA, Winey M 2001. The mouse Mps1p-like kinase regulates centrosome duplication. Cell 106: 95–104. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Mattison CP, Winey M 2003. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc Natl Acad Sci 100: 14875–14880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory MR, Morphew MM, Joseph JD, Means AR, Davis TR 2002. Pcp1, an Spc110p related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ 13: 47–58. [PubMed] [Google Scholar]
- Fong KW, Choi YK, Rattner JB, Qi RZ 2008. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the γ-tubulin ring complex. Mol Biol Cell 19: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong CS, Sato M, Toda T 2010. Fission yeast Pcp1 links Polo kinase-mediated mitotic entry to γ-tubulin-dependent spindle formation. EMBO J 29: 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong CS, Kim M, Yang TT, Liao JC, Tsou MF 2014. SAS-6 assembly templated by the lumen of cartwheel-less centrioles precedes centriole duplication. Dev Cell 30: 238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franckhauser C, Mamaeva D, Heron-Milhavet L, Fernandez A, Lamb NJ 2010. Distinct pools of cdc25C are phosphorylated on specific TP sites and differentially localized in human mitotic cells. PloS ONE 5: e11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA 1998. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol 141: 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Glover DM 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol 2: 120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Tao W, Zheng P, Fu J, Bian M, Jiang Q, Clarke PR, Zhang C 2010. Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J Cell Sci 123: 3645–3651. [DOI] [PubMed] [Google Scholar]
- Fukasawa K 2008. P53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim Biophys Acta 1786: 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furge KA, Wong K, Armstrong J, Balasubramanian M, Albright CF 1998. Byr4 and Cdc16 form a two component GTPase activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr Biol 8: 947–954. [DOI] [PubMed] [Google Scholar]
- Gabrielli BG, De Souza CP, Tonks ID, Clark JM, Hayward NK, Ellem KA 1996. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J Cell Sci 109: 1081–1093. [DOI] [PubMed] [Google Scholar]
- Gavet O, Pines J 2010. Progressive activation of cyclin B1-Cdk1 coordinates entry to mitosis. Dev Cell 18: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Sundberg HA, Chang BH, Muller EGD, Davis TN 1993. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. Mol Cell Biol 13: 7913–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E 1996. The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J 15: 5124–5124. [PMC free article] [PubMed] [Google Scholar]
- George VT, Brooks G, Humphrey TC 2007. Regulation of cell cycle and stress responses to hydrostatic pressure in fission yeast. Mol Biol Cell 18: 4168–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Walker PA, Johnston LH, Sedgwick SG 2003. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J Biol Chem 278: 14591–14594. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M 2008. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr Biol 18: 303–309. [DOI] [PubMed] [Google Scholar]
- Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, Glover DM 2002. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol 156: 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover DM 2005. Polo kinase and progression through M phase in Drosophila: A perspective from the spindle poles. Oncogene 24: 230–237. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Liem KF Jr, Anderson KV 2012. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell 151: 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan J, Mennella V, Blachon S, Zhai B, Smith AH, Megraw TL, Nicastro D, Gygi SP, Agard DA, Avidor-Reiss T 2011. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat Commun 2: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan J, Chim YC, Ha A, Basiri ML, Lerit DA, Rusan NM, Avidor-Reiss T 2012. Tubulin nucleotide status controls Sas-4-dependent pericentriolar material recruitment. Nat Cell Biol 14: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Krapp A, Bagley S, Simanis V, Hagan IM 2004. Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev 18: 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Chan KY, Alonso-Nunez ML, Madrid M, Biswas A, Alvarez-Tabares I, Connolly Y, Tanaka K, Robertson A, Ortiz JM, et al. 2013a. Removal of centrosomal PP1 by NIMA kinase unlocks the MPF feedback loop to promote mitotic commitment in S. pombe. Curr Biol 23: 213–222. [DOI] [PubMed] [Google Scholar]
- Grallert A, Patel A, Tallada VA, Chan KY, Bagley S, Krapp A, Simanis V, Hagan IM 2013b. Centrosomal MPF triggers the mitotic and morphogenetic switches of fission yeast. Nat Cell Biol 15: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith E, Walker S, Martin CA, Vagnarelli P, Stiff T, Vernay B, Al Sanna N, Saggar A, Hamel B, Earnshaw WC, et al. 2008. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet 40: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ 2011. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol 13: 1325–1334. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E 2000. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J 19: 6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian G, Westendorf J, Uldschmid A, Nigg EA 2010. Plk4 trans-autophosphorylation regulates centriole number by controlling βTrCP-mediated degradation. J Cell Sci 123: 2163–2169. [DOI] [PubMed] [Google Scholar]
- Guichard P, Chretien D, Marco S, Tassin AM 2010. Procentriole assembly revealed by cryo-electron tomography. EMBO J 29: 1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gonczy P 2012. Cartwheel architecture of Trichonympha basal body. Science 337: 553. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA 2005. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7: 1140–1146. [DOI] [PubMed] [Google Scholar]
- Hachet V, Canard C, Gonczy P 2007. Centrosomes promote timely mitotic entry in C. elegans embryos. Dev Cell 12: 531–541. [DOI] [PubMed] [Google Scholar]
- Halova L, Petersen J 2011. Aurora promotes cell division during recovery from TOR-mediated cell cycle arrest by driving spindle pole body recruitment of Polo. J Cell Sci 124: 3441–3449. [DOI] [PubMed] [Google Scholar]
- Haren L, Stearns T, Luders J 2009. Plk1-dependent recruitment of γ-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS ONE 4: e5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth S, Petersen J 2009. Fission yeast Tor1 functions as part of TORC1 to control mitotic entry through the stress MAPK pathway following nutrient stress. J Cell Sci 122: 1737–1746. [DOI] [PubMed] [Google Scholar]
- Hatano T, Sluder G 2012. The interrelationship between APC/C and Plk1 activities in centriole disengagement. Biol Open 1: 1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol 191: 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzopoulos GN, Erat MC, Cutts E, Rogala KB, Slater LM, Stansfeld PJ, Vakonakis I 2013. Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure 21: 2069–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath I 1980. Variant mitosis in lower eukaryotes: Indicators of the evolution of mitosis? Int Rev Cytol 64: 1–40. [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Oberle JR, Cooper JA 2000. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol 151: 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helps NR, Luo X, Barker HM, Cohen PT 2000. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem J 349: 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Prasanth SG, Siddiqui K, Stillman B 2009. Orc1 controls centriole and centrosome copy number in human cells. Science 323: 789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Stegert MR, Schmitz D, Hemmings BA 2006. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol 7: 253–264. [DOI] [PubMed] [Google Scholar]
- Hilbert M, Erat MC, Hachet V, Guichard P, Blank ID, Fluckiger I, Slater L, Lowe ED, Hatzopoulos GN, Steinmetz MO, et al. 2013. Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry. Proc Natl Acad Sci 110: 11373–11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283: 851–854. [DOI] [PubMed] [Google Scholar]
- Hiraki M, Nakazawa Y, Kamiya R, Hirono M 2007. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol 17: 1778–1783. [DOI] [PubMed] [Google Scholar]
- Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G 1993. Phosphorylation and activation of human cdc25-C by cdc2—Cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J 12: 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol 188: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Zhu Q, Bauer M, Verma IM, Nigg EA, Cleveland DW 2012. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev 26: 2684–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu WB, Hung LY, Tang CJ, Su CL, Chang Y, Tang TK 2008. Functional characterization of the microtubule-binding and -destabilizing domains of CPAP and d-SAS-4. Exp Cell Res 314: 2591–2602. [DOI] [PubMed] [Google Scholar]
- Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ 2001. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107: 655–665. [DOI] [PubMed] [Google Scholar]
- Hudson JD, Feilotter H, Young PG 1990. stf1: Non-wee mutations epistatic to cdc25 in the fission yeast Schizosaccharomyces pombe. Genetics 126: 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanc B, Putz M, Lalor P, Dockery P, Kuriyama R, Gergely F, Morrison CG 2013. Abnormal centrosomal structure and duplication in Cep135-deficient vertebrate cells. Mol Biol Cell 24: 2645–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF 2011. Ciliogenesis: Building the cell’s antenna. Nat Rev Mol Cell Biol 12: 222–234. [DOI] [PubMed] [Google Scholar]
- Izumi T, Maller JL 1993. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol Biol Cell 4: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Maller J 1995. Phosphorylation and activation of Xenopus Cdc25 phosphatase in the absence of Cdc2 and Cdk2 kinase activity. Mol Biol Cell 6: 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Lindon C, Nigg EA, Pines J 2003. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat Cell Biol 5: 143–148. [DOI] [PubMed] [Google Scholar]
- Januschke J, Llamazares S, Reina J, Gonzalez C 2011. Drosophila neuroblasts retain the daughter centrosome. Nat Commun 2: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Reina J, Llamazares S, Bertran T, Rossi F, Roig J, Gonzalez C 2013. Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat Cell Biol 15: 241–248. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Giddings TH Jr, Winey M 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol 159: 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers LJ, Coull BJ, Stack SJ, Morrison CG 2008. Distinct BRCT domains in Mcph1/Brit1 mediate ionizing radiation-induced focus formation and centrosomal localization. Oncogene 27: 139–144. [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz M, Gogendeau D, Klotz C, Cohen J, Beisson J, Koll F 2010. Basal body duplication in Paramecium: The key role of Bld10 in assembly and stability of the cartwheel. Cytoskeleton (Hoboken) 67: 161–171. [DOI] [PubMed] [Google Scholar]
- Joo K, Kim CG, Lee MS, Moon HY, Lee SH, Kim MJ, Kweon HS, Park WY, Kim CH, Gleeson JG, et al. 2013. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc Natl Acad Sci 110: 5987–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juanes MA, Twyman H, Tunnacliffe E, Guo Z, ten Hoopen R, Segal M 2013. Spindle pole body history intrinsically links pole identity with asymmetric fate in budding yeast. Curr Biol 23: 1310–1319. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol 150: 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaiskou A, Jessus C, Brassac T, Ozon R 1999. Phosphatase 2A and Polo kinase, two antagonistic regulators of Cdc25 activation and MPF auto-amplification. J Cell Sci 112: 3747–3756. [DOI] [PubMed] [Google Scholar]
- Karaiskou A, Lepretre AC, Pahlavan G, Du Pasquier D, Ozon R, Jessus C 2004. Polo-like kinase confers MPF autoamplification competence to growing Xenopus oocytes. Development 131: 1543–1552. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM 1996. A bipolar kinesin. Nature 379: 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF 2004. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell 6: 511–523. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT 2001. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science 294: 2542–2545. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol 162: 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV 2014. Lessons from yeast: The spindle pole body and the centrosome. Phil Trans R Soc Lond B Biol Sci 369: pii: 20130456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Goh PY 1996. Spc110p—Assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J 15: 4592–4602. [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT 1993. A spacer protein in the Saccharomyces cerevisiae spindle pole body whose transcript is cell-cycle regulated. J Cell Biol 123: 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Zaghloul NA, Bubenshchikova E, Oh EC, Rankin S, Katsanis N, Obara T, Tsiokas L 2011. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat Cell Biol 13: 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Noetzel TL, Pelletier L, Mechtler K, Drechsel DN, Schwager A, Lee M, Raff JW, Hyman AA 2005. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J Cell Biol 170: 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112: 575–587. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Busso C, Fluckiger I, Gonczy P 2009. Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev Cell 17: 900–907. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Fluckiger I, Polanowska J, Keller D, Reboul J, Gonczy P 2011a. PP2A phosphatase acts upon SAS-5 to ensure centriole formation in C. elegans embryos. Dev Cell 20: 550–562. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Kohlmaier G, Keller D, Strnad P, Balestra FR, Fluckiger I, Gonczy P 2011b. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J Cell Sci 124: 3884–3893. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, et al. 2011c. Structural basis of the 9-fold symmetry of centrioles. Cell 144: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA 2007. Plk4-induced centriole biogenesis in human cells. Dev Cell 13: 190–202. [DOI] [PubMed] [Google Scholar]
- Knop M, Schiebel E 1997. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J 16: 6985–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E 1998. Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J 17: 3952–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E 1997. The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J 16: 1550–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD 2011. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell 145: 914–925. [DOI] [PubMed] [Google Scholar]
- Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gonczy P 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol 19: 1012–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Zelter A, Muller EG, Fox B, Rice LM, Davis TN, Agard DA 2008. The structure of the γ-tubulin small complex: Implications of its architecture and flexibility for microtubule nucleation. Mol Biol Cell 19: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA 2010. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466: 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA 2011. Microtubule nucleation by γ-tubulin complexes. Nat Rev Mol Cell Biol 12: 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniewski N, Cuevas R, Duensing A, Duensing S 2010. Daughter centriole elongation is controlled by proteolysis. Mol Biol Cell 21: 3942–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Collins NK, Nakano M, Noskov VN, Ohzeki J, Mochida GH, Risinger JI, Goldsmith P, Gunsior M, et al. 2005. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum Mol Genet 14: 2155–2165. [DOI] [PubMed] [Google Scholar]
- Kovelman R, Russell P 1996. Stockpiling of Cdc25 during a DNA-replication checkpoint arrest in Schizosaccharomyces pombe. Mol Cell Biol 16: 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG 1996. Purification and molecular-cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science 273: 1377–1380. [DOI] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T 1999. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci 96: 2817–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L, Lee YL, Sahl SJ, Stearns T, Moerner WE 2012. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys J 102: 2926–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol 14: 1148–1158. [DOI] [PubMed] [Google Scholar]
- Lee K, Rhee K 2011. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J Cell Biol 195: 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Rhee K 2012. Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle 11: 2476–2485. [DOI] [PubMed] [Google Scholar]
- Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science 287: 2260–2262. [DOI] [PubMed] [Google Scholar]
- Lee KH, Johmura Y, Yu LR, Park JE, Gao Y, Bang JK, Zhou M, Veenstra TD, Yeon Kim B, Lee KS 2012. Identification of a novel Wnt5a-CK1varepsilon-Dvl2-Plk1-mediated primary cilia disassembly pathway. EMBO J 31: 3104–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Gonczy P 2003. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell 4: 431–439. [DOI] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol 7: 115–125. [DOI] [PubMed] [Google Scholar]
- Lerit DA, Rusan NM 2013. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J Cell Biol 202: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettman MM, Wong YL, Viscardi V, Niessen S, Chen SH, Shiau AK, Zhou H, Desai A, Oegema K 2013. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev Cell 25: 284–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung GC, Hudson JW, Kozarova A, Davidson A, Dennis JW, Sicheri F 2002. The Sak Polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat Struct Biol 9: 719–724. [DOI] [PubMed] [Google Scholar]
- Li S, Sandercock AM, Conduit P, Robinson CV, Williams RL, Kilmartin JV 2006. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J Cell Biol 173: 867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Saito M, Chuang JZ, Tseng YY, Dedesma C, Tomizawa K, Kaitsuka T, Sung CH 2011. Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat Cell Biol 13: 402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, D’Angiolella V, Seeley ES, Kim S, Kobayashi T, Fu W, Campos EI, Pagano M, Dynlacht BD 2013. USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature 495: 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Gao H, Lin SY, Peng G, Huang X, Zhang P, Goss JA, Brunicardi FC, Multani AS, Chang S, et al. 2010. BRIT1/MCPH1 is essential for mitotic and meiotic recombination DNA repair and maintaining genomic stability in mice. PLoS Genet 6: e1000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Rai R, Li K, Xu ZX, Elledge SJ 2005. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc Natl Acad Sci 102: 15105–15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chang CW, Hsu WB, Tang CJ, Lin YN, Chou EJ, Wu CT, Tang TK 2013a. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J 32: 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YN, Wu CT, Lin YC, Hsu WB, Tang CJ, Chang CW, Tang TK 2013b. CEP120 interacts with CPAP and positively regulates centriole elongation. J Cell Biol 202: 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TC, Neuner A, Schlosser YT, Scharf AN, Weber L, Schiebel E 2014. Cell-cycle dependent phosphorylation of yeast pericentrin regulates γ-TuSC-mediated microtubule nucleation. eLife 3: e02208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, Kallstrom H, Lundgren A, Barsoum E, Rosenthal CK 2005. Cdc25B cooperates with Cdc25A to induce mitosis but has a unique role in activating cyclin B1-Cdk1 at the centrosome. J Cell Biol 171: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, Rodriguez-Bravo V, Medema RH 2009. The decision to enter mitosis: Feedback and redundancy in the mitotic entry network. J Cell Biol 185: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga SB, Margossian SP, Harris MH, Campagna DR, Han AP, Blevins S, Mudbhary R, Barker JE, Walsh CA, Fleming MD 2010. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development 137: 1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Khodjakov A 2010. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr Biol 20: 1277–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Lan R, Zhang H, Jiang Q, Zhang C 2009. Geminin is partially localized to the centrosome and plays a role in proper centrosome duplication. Biol Cell 101: 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver FH, Tanaka K, Robertson AM, Hagan IM 2003. Physical and functional interactions between Polo kinase and the spindle pole component Cut12 regulate mitotic commitment in S. pombe. Genes Dev 17: 1507–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH 2008. Polo-like kinase-1 is activated by Aurora A to promote checkpoint recovery. Nature 455: 119–123. [DOI] [PubMed] [Google Scholar]
- Maekawa H, Priest C, Lechner J, Pereira G, Schiebel E 2007. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J Cell Biol 179: 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahen R, Jeyasekharan AD, Barry NP, Venkitaraman AR 2011. Continuous Polo-like kinase 1 activity regulates diffusion to maintain centrosome self-organization during mitosis. Proc Natl Acad Sci 108: 9310–9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Montpetit B, Zhao L, Finst RJ, Goh B, Kim AC, Quarmby LM 2002. The FA2 gene of Chlamydomonas encodes a NIMA family kinase with roles in cell cycle progression and microtubule severing during deflagellation. J Cell Sci 115: 1759–1768. [DOI] [PubMed] [Google Scholar]
- Mahjoub MR, Xie Z, Stearns T 2010. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J Cell Biol 191: 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardin BR, Lange C, Baxter JE, Hardy T, Scholz SR, Fry AM, Schiebel E 2010. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat Cell Biol 12: 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardin BR, Agircan FG, Lange C, Schiebel E 2011. Plk1 controls the Nek2A-PP1γ antagonism in centrosome disjunction. Curr Biol 21: 1145–1151. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Hayashi K, Nishida E 1999. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr Biol 9: 429–432. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Nishimura T, Hayakawa A, Ono Y, Takahashi M 2010. Involvement of a centrosomal protein kendrin in the maintenance of centrosome cohesion by modulating Nek2A kinase activity. Biochem Biophys Res Commun 398: 217–223. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Ohsumi K, Iwabuchi M, Kawamata T, Ono Y, Takahashi M 2012. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr Biol 22: 915–921. [DOI] [PubMed] [Google Scholar]
- Matsuyama M, Goto H, Kasahara K, Kawakami Y, Nakanishi M, Kiyono T, Goshima N, Inagaki M 2011. Nuclear Chk1 prevents premature mitotic entry. J Cell Sci 124: 2113–2119. [DOI] [PubMed] [Google Scholar]
- Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA 2000. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol 151: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JN, Tatchell K 1994. The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J Cell Biol 125: 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Li K, Kao LR, Kaufman TC 1999. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126: 2829–2839. [DOI] [PubMed] [Google Scholar]
- Meitinger F, Palani S, Pereira G 2012. The power of MEN in cytokinesis. Cell Cycle 11: 219–228. [DOI] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol 14: 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol 1: 88–93. [DOI] [PubMed] [Google Scholar]
- Miller RK, Rose MD 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol 140: 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Heller KK, Frisen L, Wallack DL, Loayza D, Gammie AE, Rose MD 1998. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol Biol Cell 9: 2051–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori D, Yano Y, Toyo-oka K, Yoshida N, Yamada M, Muramatsu M, Zhang D, Saya H, Toyoshima YY, Kinoshita K, et al. 2007. NDEL1 phosphorylation by Aurora-A kinase is essential for centrosomal maturation, separation, and TACC3 recruitment. Mol Cell Biol 27: 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA 1995. Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature 378: 638–640. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J 2006. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441: 1068–1074. [DOI] [PubMed] [Google Scholar]
- Mottier-Pavie V, Megraw TL 2009. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell 20: 2605–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill DP, Petersen J, Ohkura H, Glover DM, Hagan IM 1999. Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol Biol Cell 10: 2771–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Saito H, Takekawa M 2013. SAPK pathways and p53 cooperatively regulate PLK4 activity and centrosome integrity under stress. Nat Commun 4: 1775. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M 2007. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol 17: 2169–2174. [DOI] [PubMed] [Google Scholar]
- Noatynska A, Gotta M, Meraldi P 2012. Mitotic spindle (DIS)orientation and DISease: Cause or consequence? J Cell Biol 199: 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak ZA, Conduit PT, Wainman A, Raff JW 2014. Asterless licenses daughter centrioles to duplicate for the first time in Drosophila embryos. Curr Biol 24: 1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503–508. [DOI] [PubMed] [Google Scholar]
- O’Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105: 547–558. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Hagan IM, Glover DM 1995. The conserved Schizosaccharomyces pombe kinase Plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev 9: 1059–1073. [DOI] [PubMed] [Google Scholar]
- Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, et al. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103: 127–140. [DOI] [PubMed] [Google Scholar]
- Oliveira RA, Nasmyth K 2013. Cohesin cleavage is insufficient for centriole disengagement in Drosophila. Curr Biol 23: R601–R603. [DOI] [PubMed] [Google Scholar]
- Pan J, Wang Q, Snell WJ 2004. An Aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev Cell 6: 445–451. [DOI] [PubMed] [Google Scholar]
- Parker JD, Hilton LK, Diener DR, Rasi MQ, Mahjoub MR, Rosenbaum JL, Quarmby LM 2010. Centrioles are freed from cilia by severing prior to mitosis. Cytoskeleton (Hoboken) 67: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL 2008. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol 85: 23–61. [DOI] [PubMed] [Google Scholar]
- Peel N, Dougherty M, Goeres J, Liu Y, O’Connell KF 2012. The C. elegans F-box proteins LIN-23 and SEL-10 antagonize centrosome duplication by regulating ZYG-1 levels. J Cell Sci 125: 3535–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA 2004. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol 14: 863–873. [DOI] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T 2006. Centriole assembly in Caenorhabditis elegans. Nature 444: 619–623. [DOI] [PubMed] [Google Scholar]
- Peng G, Yim EK, Dai H, Jackson AP, Burgt I, Pan MR, Hu R, Li K, Lin SY 2009. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol 11: 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Schiebel E 2001. The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr Opin Cell Biol 13: 762–769. [DOI] [PubMed] [Google Scholar]
- Pereira G, Schiebel E 2005. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol Cell 19: 209–221. [DOI] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell 6: 1–10. [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth K, Schiebel E 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J 20: 6359–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mongiovi D, Beckhelling C, Chang P, Ford CC, Houliston E 2000. Nuclei and microtubule asters stimulate maturation/M phase promoting factor (MPF) activation in Xenopus eggs and egg cytoplasmic extracts. J Cell Biol 150: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Hagan IM 2005. Polo kinase links the stress pathway to cell cycle control and tip growth in fission yeast. Nature 435: 507–512. [DOI] [PubMed] [Google Scholar]
- Petersen J, Nurse P 2007. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat Cell Biol 9: 1263–1272. [DOI] [PubMed] [Google Scholar]
- Prosser SL, Samant MD, Baxter JE, Morrison CG, Fry AM 2012. Oscillation of APC/C activity during cell cycle arrest promotes centrosome amplification. J Cell Sci 125: 5353–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EN, Golemis EA 2005. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol 7: 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129: 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklowski A, Homsi Y, Keller D, May M, Chauhan S, Kossatz U, Grunwald V, Kubicka S, Pich A, Manns MP, et al. 2011. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat Cell Biol 13: 1004–1009. [DOI] [PubMed] [Google Scholar]
- Pulvers JN, Bryk J, Fish JL, Wilsch-Brauninger M, Arai Y, Schreier D, Naumann R, Helppi J, Habermann B, Vogt J, et al. 2010. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci 107: 16595–16600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao R, Cabral G, Lettman MM, Dammermann A, Dong G 2012. SAS-6 coiled-coil structure and interaction with SAS-5 suggest a regulatory mechanism in C. elegans centriole assembly. EMBO J 31: 4334–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby L 2000. Cellular samurai: Katanin and the severing of microtubules. J Cell Sci 113: 2821–2827. [DOI] [PubMed] [Google Scholar]
- Raff JW, Glover DM 1988. Nuclear and cytoplasmic mitotic-cycles continue in Drosophila embryos in which DNA-synthesis is inhibited with aphidicolin. J Cell Biol 107: 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Dai H, Multani AS, Li K, Chin K, Gray J, Lahad JP, Liang J, Mills GB, Meric-Bernstam F, et al. 2006. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell 10: 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Phadnis A, Haralkar S, Badwe RA, Dai H, Li K, Lin SY 2008. Differential regulation of centrosome integrity by DNA damage response proteins. Cell Cycle 7: 2225–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C 2007. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell 12: 467–474. [DOI] [PubMed] [Google Scholar]
- Riparbelli MG, Callaini G, Megraw TL 2012. Assembly and persistence of primary cilia in dividing Drosophila spermatocytes. Dev Cell 23: 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM 2007a. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr Biol 17: 1465–1472. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M 2007b. Revisiting the role of the mother centriole in centriole biogenesis. Science 316: 1046–1050. [DOI] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL 2009. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol 184: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque H, Wainman A, Richens J, Kozyrska K, Franz A, Raff JW 2012. Drosophila Cep135/Bld10 maintains proper centriole structure but is dispensable for cartwheel formation. J Cell Sci 125: 5881–5886. [DOI] [PubMed] [Google Scholar]
- Rose MD, Fink GR 1987. Kar1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell 48: 1047–1060. [DOI] [PubMed] [Google Scholar]
- Salisbury JL 2007. A mechanistic view on the evolutionary origin for centrin-based control of centriole duplication. J Cell Physiol 213: 420–428. [DOI] [PubMed] [Google Scholar]
- Samejima I, Miller VJ, Groocock LM, Sawin KE 2008. Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the γ-tubulin complex in vivo. J Cell Sci 121: 3971–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RD, Avides MC, Howard T, Gonzalez C, Glover DM 1997. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J Cell Biol 137: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerer F, Morgan G, Winey M, Philippsen P 2001. Cnm67p is a spacer protein of the Saccharomyces cerevisiae spindle pole body outer plaque. Mol Biol Cell 12: 2519–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA 2009. Control of centriole length by CPAP and CP110. Curr Biol 19: 1005–1011. [DOI] [PubMed] [Google Scholar]
- Schockel L, Mockel M, Mayer B, Boos D, Stemmann O 2011. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol 13: 966–972. [DOI] [PubMed] [Google Scholar]
- Schuldt A 2011. Cytoskeleton: SAS-6 turns a cartwheel trick. Nat Rev Mol Cell Biol 12: 137. [DOI] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Jang CY, Yates JR, Fang G 2008. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 320: 1655–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K 1997. Astral microtubule dynamics in yeast: A microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol 139: 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne JE, Tack F, Vloemans N, Boeckx A, Thambirajah S, Bonnet P, Ramaekers FC, Bornens M, Grand-Perret T 2010. Autophosphorylation of Polo-like kinase 4 and its role in centriole duplication. Mol Biol Cell 21: 547–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne JE, Specht CG, Izeddin I, Hurbain I, Tran P, Triller A, Darzacq X, Dahan M, Bornens M 2011. Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton (Hoboken) 68: 619–627. [DOI] [PubMed] [Google Scholar]
- Simanis V 2003. Events at the end of mitosis in the budding and fission yeasts. J Cell Sci 116: 4263–4275. [DOI] [PubMed] [Google Scholar]
- Singh P, Ramdas Nair A, Cabernard C 2014. The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr Biol 24: 1548–1555. [DOI] [PubMed] [Google Scholar]
- Singla V, Romaguera-Ros M, Garcia-Verdugo JM, Reiter JF 2010. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev Cell 18: 410–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir JH, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D’Santos C, Woods CG, Gergely F 2011. A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet 43: 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin LK, Nye J, Pinkerton DC, Buster DW, Rogers GC, Slep KC 2012. The structure of the Plk4 cryptic Polo box reveals two tandem Polo boxes required for centriole duplication. Structure 20: 1905–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MH, Liu Y, Anderson DE, Jahng WJ, O’Connell KF 2011. Protein phosphatase 2A-SUR-6/B55 regulates centriole duplication in C. elegans by controlling the levels of centriole assembly factors. Dev Cell 20: 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open 1: 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Gabryjonczyk AM, Anselm E, Stierhof YD, Nigg EA 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J Cell Sci 126: 3223–3233. [DOI] [PubMed] [Google Scholar]
- Spang A, Courtney I, Fackler U, Matzner M, Schiebel E 1993. The calcium-binding protein cell-division cycle 31 of Saccharomyces cerevisiae is a component of the half-bridge of the spindle pole body. J Cell Biol 123: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Courtney I, Grein K, Matzner M, Schiebel E 1995. The Cdc31p-binding protein Kar1p is a component of the half-bridge of the yeast spindle pole body. J Cell Biol 128: 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Grein K, Schiebel E 1996. The spacer protein Spc110p targets calmodulin to the central plaque of the yeast spindle pole body. J Cell Sci 109: 2229–2237. [DOI] [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD 2007. Cep97 and CP110 suppress a cilia assembly program. Cell 130: 678–690. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Amon A 2004. Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet 38: 203–232. [DOI] [PubMed] [Google Scholar]
- Stevens NR, Roque H, Raff JW 2010. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev Cell 19: 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling DA, Welch KA, Stark MJR 1994. Interaction with calmodulin is required for the function of Spc110p, an essential component of the yeast spindle pole body. EMBO J 13: 6180–6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld U, Fernandez A, Capony JP, Girard F, Lautredou N, Derancourt J, Labbe JC, Lamb NJ 1994. Activation of p34cdc2 protein kinase by microinjection of human cdc25C into mammalian cells. Requirement for prior phosphorylation of cdc25C by p34cdc2 on sites phosphorylated at mitosis. J Biol Chem 269: 5989–6000. [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell 13: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg HA, Davis TN 1997. A mutational analysis identifies three functional regions of the spindle pole component Spc110p in Saccharomyces cerevisiae. Mol Biol Cell 8: 2575–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana KE, Gonzalez MA, Guarguaglini G, Nigg EA, Laskey RA 2005. Depletion of licensing inhibitor geminin causes centrosome overduplication and mitotic defects. EMBO Rep 6: 1052–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallada VA, Bridge AJ, Emery PE, Hagan IM 2007. Suppression of the S. pombe cut12.1 cell cycle defect by mutations in cdc25 and genes involved in transcriptional and translational control. Genetics 176: 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallada VA, Tanaka K, Yanagida M, Hagan IM 2009. The S. pombe mitotic regulator Cut12 promotes spindle pole body activation and integration into the nuclear envelope. J Cell Biol 185: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Petersen J, MacIver F, Mulvihill DP, Glover DM, Hagan IM 2001. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J 20: 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK 2009. CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol 11: 825–831. [DOI] [PubMed] [Google Scholar]
- Tang CJ, Lin SY, Hsu WB, Lin YN, Wu CT, Lin YC, Chang CW, Wu KS, Tang TK 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J 30: 4790–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MF 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev 27: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixido-Travesa N, Roig J, Luders J 2012. The where, when and how of microtubule nucleation—One ring to rule them all. J Cell Sci 125: 4445–4456. [DOI] [PubMed] [Google Scholar]
- Terada Y, Uetake Y, Kuriyama R 2003. Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J Cell Biol 162: 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton GK, Woods CG 2009. Primary microcephaly: Do all roads lead to Rome? Trends Genet 25: 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibelius A, Marhold J, Zentgraf H, Heilig CE, Neitzel H, Ducommun B, Rauch A, Ho AD, Bartek J, Kramer A 2009. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J Cell Biol 185: 1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K 2001. Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J Biol Chem 276: 21529–21537. [DOI] [PubMed] [Google Scholar]
- Trimborn M, Ghani M, Walther DJ, Dopatka M, Dutrannoy V, Busche A, Meyer F, Nowak S, Nowak J, Zabel C, et al. 2010. Establishment of a mouse model with misregulated chromosome condensation due to defective Mcph1 function. PLoS ONE 5: e9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD 2008. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell 15: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T 2006. Mechanism limiting centrosome duplication to once per cell cycle. Nature 442: 947–951. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell 17: 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulina N, Matunis E 2001. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science 294: 2546–2549. [DOI] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science 331: 1196–1199. [DOI] [PubMed] [Google Scholar]
- van der Voet M, Berends CW, Perreault A, Nguyen-Ngoc T, Gonczy P, Vidal M, Boxem M, van den Heuvel S 2009. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Gα. Nat Cell Biol 11: 269–277. [DOI] [PubMed] [Google Scholar]
- Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C 2007. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr Biol 17: 1735–1745. [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of CDK-dependent phosphorylation. Mol Cell 2: 709–718. [DOI] [PubMed] [Google Scholar]
- Vulprecht J, David A, Tibelius A, Castiel A, Konotop G, Liu F, Bestvater F, Raab MS, Zentgraf H, Izraeli S, et al. 2012. STIL is required for centriole duplication in human cells. J Cell Sci 125: 1353–1362. [DOI] [PubMed] [Google Scholar]
- Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, Dai W 2008. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell 14: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH 2009. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461: 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wu T, Shi L, Zhang L, Zheng W, Qu JY, Niu R, Qi RZ 2010. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J Biol Chem 285: 22658–22665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Soni RK, Uryu K, Tsou MF 2011. The conversion of centrioles to centrosomes: Essential coupling of duplication with segregation. J Cell Biol 193: 727–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Chen Q, Zhang X, Zhang B, Zhuo X, Liu J, Jiang Q, Zhang C 2013. PCM1 recruits Plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J Cell Sci 126: 1355–1365. [DOI] [PubMed] [Google Scholar]
- Wood JL, Liang Y, Li K, Chen J 2008. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J Biol Chem 283: 29586–29592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Lee J, Stern DF 2004. Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J Biol Chem 279: 34091–34094. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315: 518–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Adamian M, Li T 2006. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol Biol Cell 17: 1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K 1995. Spindle dynamics and cell-cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol 130: 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K 2000. Dynamic positioning of mitotic spindles in yeast: Role of microtubule motors and cortical determinants. Mol Biol Cell 11: 3949–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pruyne D, Huffaker TC, Bretscher A 2000. Myosin V orientates the mitotic spindle in yeast. Nature 406: 1013–1015. [DOI] [PubMed] [Google Scholar]
- Zhang J, Megraw TL 2007. Proper recruitment of γ-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires centrosomin motif 1. Mol Biol Cell 18: 4037–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen Q, Feng J, Hou J, Yang F, Liu J, Jiang Q, Zhang C 2009. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the γTuRC to the centrosome. J Cell Sci 122: 2240–2251. [DOI] [PubMed] [Google Scholar]