Abstract
In the biorefinery using lignocellulosic biomass as feedstock, pretreatment to breakdown or loosen lignin is important step and various approaches have been conducted. For biological pretreatment, we screened Bacillus subtilis KCTC2023 as a potential lignin-degrading bacterium based on veratryl alcohol (VA) oxidation test and the putative heme-containing dye-decolorizing peroxidase was found in the genome of B. subtilis KCTC2023. The peroxidase from B. subtilis KCTC2023 (BsDyP) was capable of oxidizing various substrates and atypically exhibits substrate-dependent optimum temperature: 30°C for dyes (Reactive Blue19 and Reactive Black5) and 50°C for high redox potential substrates (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid [ABTS], VA, and veratryl glycerol-β-guaiacyl ether [VGE]) over +1.0 V vs. normal hydrogen electrode. At 50°C, optimum temperature for high redox potential substrates, BsDyP not only showed the highest VA oxidation activity (0.13 Umg−1) among the previously reported bacterial peroxidases but also successfully achieved VGE decomposition by cleaving Cα-Cβ bond in the absence of any oxidative mediator with a specific activity of 0.086 Umg−1 and a conversion rate of 53.5%. Based on our results, BsDyP was identified as the first bacterial peroxidase capable of oxidizing high redox potential lignin-related model compounds, especially VGE, revealing a previously unknown versatility of lignin degrading biocatalyst in nature.
Owing to climate change and depletion of fossil fuels, research in the field of biotechnology has been accelerated towards a biorefinery system for producing biofuels and biochemicals from renewable resources1. In particular, lignocellulosic biomass has received great attention, because it is the most abundant bioresource in nature and does not compete with the food chain2. To develop an efficient biorefinery process, pretreatment to breakdown or loosen highly resistant lignin is one of the key steps and thus various physical, chemical, and biological approaches have been attempted3.
In nature, heme-containing peroxidases are known to be responsible for the pretreatment: lignin peroxidase (LiP, E.C. 1.11.1.14), manganese peroxidase (MnP, E.C. 1.11.1.13), versatile peroxidase (VP, E.C. 1.11.1.16), and dye-decolorizing peroxidase (DyP, E.C.1.11.1.19)4,5,6,7. They essentially require hydrogen peroxide as the electron acceptor and catalytic oxidative reactions have been usually conducted under acidic pH conditions (pH 3.0–pH 4.5) and moderate temperatures (25–37°C)6,8,9. Even if they are widely distributed in nature, most of identified peroxidases are originated from fungi such as Phanerochaete chrysosporium9,10.
Bacterial peroxidase is advantageous over fungal peroxidase because bacterial strains are more amenable to protein engineering for improving catalytic properties11. However, information about bacterial peroxidase is very limited due to a lack of identified microorganisms and enzymes exhibiting ligninolytic potential. Until now, Streptomyces viridosporus, Pseudomonas putida, and Rodococcus jostii have been researched as bacterial strains being involved in lignin degradation12,13,14. In addition, recently, Bacillus strains have been isolated as potential lignin-degrading bacteria15,16,17, even though they have not been well-known as lignin-degrading bacteria until a few years ago.
Herein, we aimed to identify a biocatalyst in Bacillus species exhibiting a potential lignin-degrading activity. Based on veratryl alcohol (VA) oxidation test9, which is usually used to assay a lignin-degrading biocatalyst, Bacillus subtilis KCTC 2023 was chosen as the potential lignin-degrading Bacillus strain and then the putative heme-containing dye-decolorizing peroxidase (BsDyP) was found in its genome. Interestingly, BsDyP exhibited substrate-dependent optimum temperature: BsDyP showed optimal oxidation activity for high redox potential substrates over +1.0 V vs. normal hydrogen electrode (NHE) such as 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid (ABTS), VA, and a β-ether dimer lignin model compound (veratryl glycerol-β-guaiacyl ether [VGE])18,19 at 50°C, but the optimum temperature for decolorizing dyes (Reactive Black5 and Reactive Blue19) requiring relatively low redox potential (< 1.0 V vs. NHE)20,21 was determined at 30°C. Furthermore, the relative activity of the high redox potential substrates at 30°C was significantly lower than that at 50°C. Thus, by discovering substrate-dependent optimum temperature profiles of BsDyP, we could identify BsDyP as the bacterial peroxidase with the highest VA oxidation activity among bacterial heme-containing peroxidases previously reported as well as the first one decomposing VGE by cleaving Cα-Cβ bond independently of oxidative mediators at 50°C. Accordingly, the unique characteristics of BsDyP presented in this study would provide an insight for elucidating the previously unknown temperature-dependent bacterial lignin-degradation process and great opportunities to realize microbial pretreatment for producing biofuels and biochemicals from lignocellulosic biomass.
Results
Screening of veratryl alcohol (VA) oxidation-positive Bacillus species
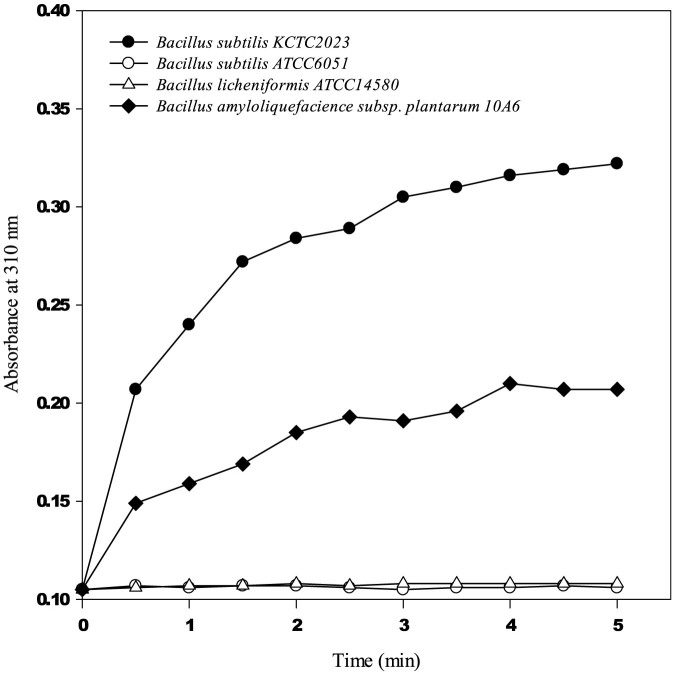
Until now, the lignin-degrading peroxidase such as LiP has been widely assayed with VA as the substrate8,22,23,24. In order to screen Bacillus species secreting the lignin-degrading biocatalyst, VA oxidation test was conducted using cell-free supernatant after two days cultivation of four Bacillus species (B. subtilis KCTC 2023, B. subtilis ATCC6051, B. licheniformis ATCC14580 and B. amyloliquefacience subsp. plantarum 10A6), because it was already reported that B. subtilis, B. licheniformis, and B. amyloliquefaciens were involved in lignin-degradation15,25. As shown in Figure 1, B. subtilis KCTC 2023 and B. amyloliquefacience subsp. plantarum 10A6 exhibited VA oxidation activity, but B. subtilis ATCC6051 and B. licheniformis ATCC14580 did not oxidize VA at all. Comparing the initial oxidation rate, B. subtilis KCTC2023 showed the highest VA oxidation activity among the tested Bacillus species, while B. amyloliquefacience subsp. plantarum 10A6 retained 52% activity of B. subtilis KCTC2023. Based on the highest ability for oxidizing VA to veratryl aldehyde, we examined the genome of B. subtilis KCTC2023 to explore the biocatalyst responsible for VA oxidation and then chose a putative heme-containing dye-decolorizing peroxidase (BsDyP, accession number:WP_003222196) as a potential VA-oxidizing biocatalyst. According to BLAST search, the putative heme-containing peroxidase is commonly discovered in the genome of Bacillus species with high identities ranging from 96% to 100% (data not shown).
Figure 1. VA oxidation activity with cell-free supernatant of four Bacillus species.
2.0 mM of VA, 0.4 mM of hydrogen peroxide, 0.5 mL of cell-free supernatant were used (total volume: 1 mL). The catalytic product veratryl aldehyde was measured at 310 nm using UV-spectrophotometer (ε310, veratryl aldehyde = 9.3 mM−1cm−1).
Expression and reconstitution of recombinant BsDyP
To obtain a sufficient amount of BsDyP for further characterization, recombinant BsDyP in Escherichia coli (E. coli) was prepared. To improve the expression level, BsDyP was induced with 1.0 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at low temperature (18°C) for 24 hours and a high expression level was achieved1. As a result, BsDyP was successfully expressed up to 2.10 mgmL−1 and purified using Ni-NTA column as a 48 kDa protein. BsDyP was expressed in two forms due to the N-terminal twin Arg sequence involving the TAT secretion machinery, commonly discovered in DyPs26 (see Supporting Information).
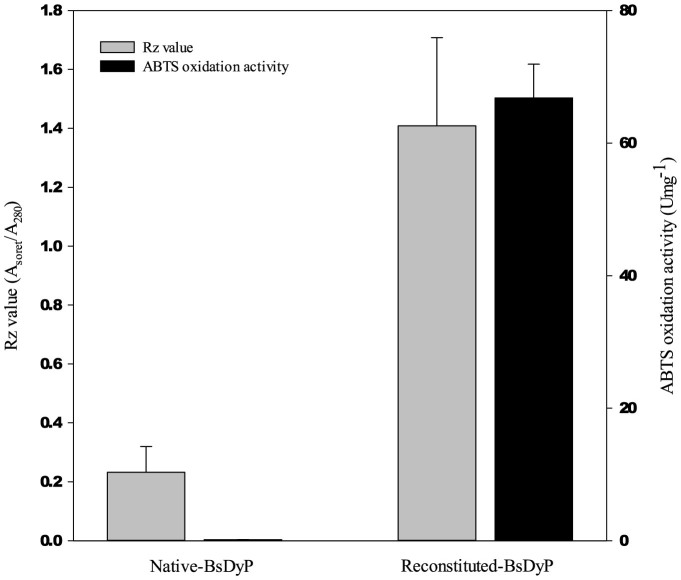
At first, BsDyP activity was assayed using ABTS as the common substrate for heme-containing peroxidase. However, the specific activity of the purified BsDyP was only 0.15 Umg−1 for ABTS, probably due to the apo-expression lacking heme in the active site. To reconstitute the apo-BsDyP to holo-BsDyP, hemin treatment was conducted and the reconstitution was evaluated by Rz value (Asoret/A280)12. After the reconstitution, a 6.1-fold higher Rz value was obtained, from 0.23 to 1.41 (Figure 2), indicating successful reconstitution of BsDyP. The specific activity of the reconstituted-BsDyP (66.8 Umg−1 with ABTS) was also 445-fold higher than the native-BsDyP (Figure 2). Accordingly, further experiments were conducted with the reconstituted-BsDyP.
Figure 2. Rz value (Asoret/A280) and ABTS oxidation activity of native- and reconstituted-BsDyP.
For ABTS oxidation, 1.0 mM of ABTS and 210 μgmL−1 of BsDyP were used (pH 3.0 and 30°C). The catalytic product was measured at 420 nm (ε420 = 36 mM−1cm−1).
Substrate spectrum of BsDyP
To investigate which substrate could be oxidized by BsDyP, various substrates including common organic dyes for a dye-decolorizing biocatalyst (Reactive Black5 and Reactive Blue19), substrates used in screening lignin-degrading biocatalyst (ABTS and VA), and β-aryl ether containing β-O-4 bond as a lignin dimer model (VGE) were tested at pH 3 and 30°C. After testing the dyes, BsDyP exhibited decolorizing activity to varying extents (data not shown). The peroxidase database (http://peroxibase.toulouse.inra.fr/index.php) classifies heme peroxidases into 6 groups: non-animal peroxidase, animal peroxidase, haloperoxidase, di-heme cytochrome c peroxidase, catalase, and DyP. It is known that the structure and reaction mechanism of DyP are definitely distinct from those of other peroxidases27,28. For example, DyP is able to oxidize anthraquinone compounds28,29, but other peroxidases do not in the least. Thus, this result demonstrates that BsDyP with the catalytic activity for Reactive Blue19, one of the anthraquinone-dyes, belongs to DyP.
In addition to dye-decolorizing activity, BsDyP showed ABTS and VA oxidation activity at pH 3 and 30°C (data not shown). Considering that B. subtilis KCTC 2023 was initially screened as a potential lignin-degrading bacterium by VA oxidation assay, BsDyP might take some part in the VA oxidation reaction in the screening step. Interestingly, BsDyP also utilized VGE as a substrate, usually used as a dimeric lignin including β-O-4 linkage. This BsDyP activity on the oxidation of lignin-related substrates is very remarkable because there have been only a few reports on a VA oxidation-positive bacterial biocatalyst and even no report on a VGE-decomposing bacterial biocatalyst.
Optimum temperature of BsDyP is dependent on substrates
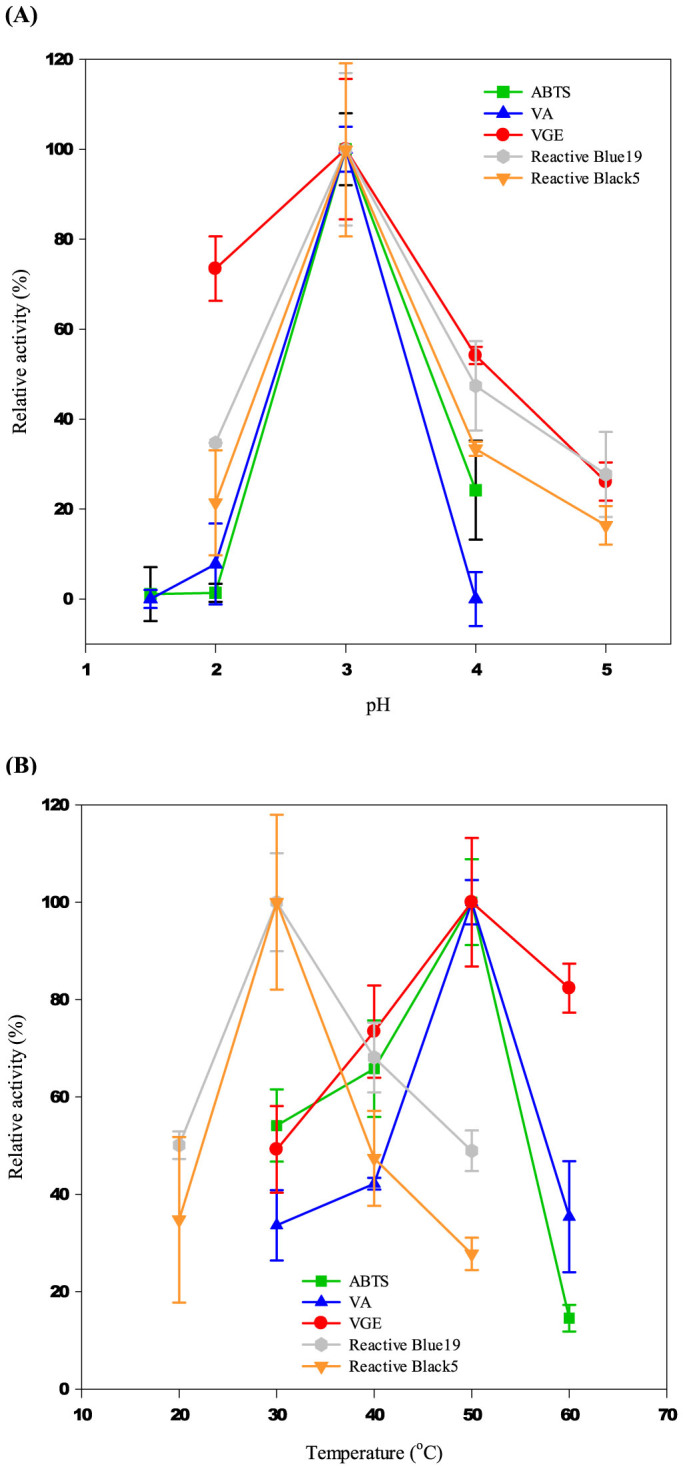
Prior to measuring the specific activity of BsDyP for each substrate, the optimum pH and temperature of BsDyP were determined. As shown in Figure 3A, the optimum pH of BsDyP was determined at pH 3.0 as usually seen in peroxidase and a narrow pH range was observed for all the substrates. The decolorization of Reactive Blue19 (anthraquinone-dye) and Reactive Black5 (azo-dye), which are organic dyes usually required relatively low redox potential (<1.0 V vs. NHS)20,21, was optimized at 30°C (Figure 3B). Notably, the optimum temperature of BsDyP for oxidizing relatively high-redox potential substrate over +1.0 V vs. NHS such as ABTS, VA, and VGE18,19 was 50°C although B. subtilis is not thermophilic bacteria. Atypical optimum temperature distribution categorized in two groups at 30°C and 50°C is clearly shown in Figure 3B. In addition, the relative activity at the other group's optimum temperature was significantly lower compared to that at its own optimum temperature. For example, in the case of ABTS, VA, and VGE, the relative activity of BsDyP at 30°C was only 30–55% of the optimum activity at 50°C.
Figure 3. Relative activity of BsDyP (157 μgmL−1) (A) at various pHs (temperature at 30°C) and (B) at various temperatures (pH at 3.0).
Specific activities of BsDyP for the dyes, ABTS, and VA
Specific activity of BsDyP was determined at the optimum conditions of Reactive Blue19, Reactive Black5, ABTS, and VA. BsDyP decolorized Reactive Blue19 and Reactive Black5 with a specific activity of 11.55 Umg−1, and 17.65 Umg−1, respectively (Table 1). For ABTS and VA, the specific activities of BsDyP were 66.80 Umg−1 and 0.13 Umg−1, respectively.
Table 1. Specific activities of bacterial peroxidase using various substrates.
| Origin | Biocatalyst | Substrate | Assay condition | Specific activity (U/mg protein) | Ref. | |
|---|---|---|---|---|---|---|
| pH | Temp. (°C) | |||||
| Thermobifida fusca | DyP | Reactive Blue19 | 3.5 | 25 | 4.28 | 22 |
| Reactive Black5 | 3.5 | 25 | 0.06 | |||
| Guaiacol | 3.5 | 25 | 0.03 | |||
| 2,6-dimethoxyphenol | 3.5 | 25 | 0.17 | |||
| VA | 3.5 | 25 | 0.01 | |||
| Anabaena species | DyP | Reactive Blue19 | 4.8 | 35 | 401 | 30 |
| Reactive Black5 | 4.4 | 35 | 21 | |||
| Rhodococcus jostii RHA1 | DyP | ABTS | 5.0 | 30 | 1.54a | 12 |
| ABTS | 5.0 | 30 | 13.09b | |||
| GGE | 5.5 | 30 | 0.017b | |||
| Pseudomonas putida MET94 | DyP | ABTS | 5 | 10–30 | 40 | 26 |
| VA | 5 | 10–30 | 6.2 × 10−6 | |||
| Bacillus subtilis | DyP | ABTS | 4 | 20–30 | 15 | |
| Acinetobacter calcoaceticus | LiP | VA | 1.0 | 50 | 0.043 | 23 |
| Bacillus subtilis | DyP | ABTS | 3.0 | 50 | 66.80 | This study |
| VA | 3.0 | 50 | 0.13 | |||
| VGE | 3.0 | 50 | 0.086 | |||
| Reactive Blue19 | 3.0 | 30 | 11.55 | |||
| Reactive Black5 | 3.0 | 30 | 17.65 | |||
One unit (U) corresponds to 1 μmole of product per minute.
aAssayed without MnCl2;
bAssayed with MnCl2.
The specific activity of BsDyP for the dyes, ABTS, and VA was compared with previously reported bacterial peroxidases (Table 1). Comparing specific activity of DyP from thermophilic actinomycete Thermobifida fusca (TfuDyP)22, BsDyP showed 2.7- and 294-fold higher specific activity for Reactive Blue19 and Reactive Black5, respectively. DyP from the cyanobacterium Anabaena species (AnaPX) revealed that the decolorization activity for azo-dye (Reactive Black5) was much lower than that for anthraquinone-dye (Reactive Blue19)30. Both TfuDyP and AnaPX were much more active for anthraquinone-dye (Reactive Blue19) than azo-dye (Reactive Black5), but BsDyP slightly preferred azo- than anthraquinone-dye. DyP from R. jostii showed significantly higher ABTS oxidation activity in the presence of MnCl2, implying a role of MnII as an oxidative mediator12. Compared to DyP from R. jostii, BsDyP exhibited 5.1- and 43.4-fold higher specific activity for ABTS oxidation without any oxidative mediator. Recently, Santos et al. reported two bacterial DyPs from Pseudomonas putida MET94 (PpDyP) and Bacillus subtilis (B-DyP). BsDyP possessed 1.7- and 4.5-fold higher ABTS oxidation activity than PpDyP and B-DyP. Furthermore, in case of VA oxidation activity, BsDyP retained 13.0-, 2.1 × 104- and 3.0-fold higher VA oxidation activity compared to TfuDyP, PpDyP, and LiP from Acinetobacter calcoaceticus, respectively22,23,26. Overall, as clearly shown in Table 1, BsDyP exhibited the highest ABTS and VA oxidation activity among the bacterial peroxidases previously reported.
BsDyP decomposes dimeric lignin VGE by cleaving Cα-Cβ bond
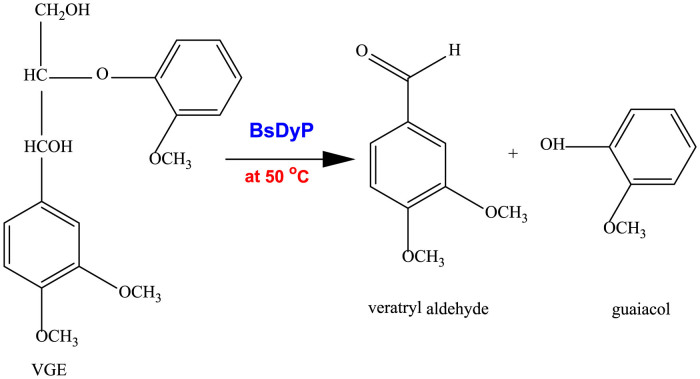
BsDyP was supposed to decompose dimeric lignin VGE by cleaving the Cα-Cβ bond and finally producing veratryl aldehyde and guaiacol as shown in Figure 412,31. By measuring veratryl aldehyde concentration, the specific activity for VGE decomposition was determined to be 0.086 Umg−1. BsDyP activity on VGE is noteworthy because there has been no report on bacterial enzymes decomposing VGE. Recently, Ahmad et al. reported that DyP from R. jostii RHA1 decomposed guaiacylglycerol-β-guaiacyl ether (GGE), a dimeric lignin model compound including the β-O-4 bond, to vanillin and guaiacol with a specific activity of 0.017 Umg−1 in the presence of MnCl2 as shown in Table 112. However, DyP from R.jostii RHA1 showed MnII –dependent activity, implying that MnII played an important role as an oxidative mediator in GGE decomposition. Compared to DyP of R.jostii RHA1, BsDyP could decompose a lignin dimer model compound without an oxidative mediator like MnII at the higher activity (0.086 Umg−1 vs. 0.017 Umg−1), implying a distinguished characteristic of BsDyP from DyP in R.jostii RHA1.
Figure 4. Decomposition of veratryl glycerol-β-guaiacyl ether (VGE) to veratryl aldehyde and guaiacol.
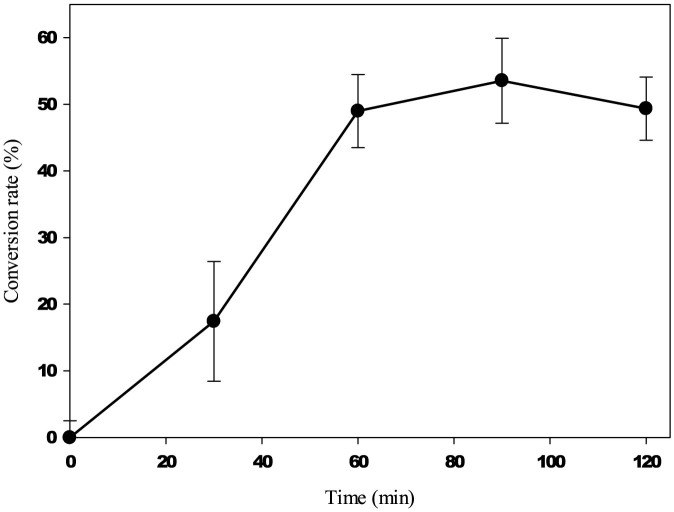
In order to further confirm BsDyP activity on VGE, we quantified VGE decomposition to veratryl aldehyde and guaiacol time-dependently. In general, heme-containing peroxidase is not stable in the presence of hydrogen peroxide because hydrogen peroxide easily attacks and destroys the heme group in the active site32. To minimize the toxic effect of hydrogen peroxide, VGE decomposition was attempted by stepwise adding hydrogen peroxide. Because guaiacol, one of the expected decomposed products, could be easily converted to guaiacol-oligomer in the presence of heme-containing peroxidase, guaiacol was not detected12,31. Thus, VGE and veratryl aldehyde were analyzed by high-performance liquid chromatography (HPLC). When using 0.2 mM of VGE and 1.9 mgmL−1 of BsDyP with stepwise feeding of 100 μM of hydrogen peroxide at every sampling point, 107 μM of VGE was consumed and 105 μM of veratryl aldehyde was formed for 2 hours, resulting in a conversion rate of up to 53.5% for 2 hours (Figure 5).
Figure 5. Decomposition of VGE (0.2 mM) to veratryl aldehyde at pH 3.0 and 50°C.
BsDyP (1.9 mgmL−1) was used with stepwise addition of hydrogen peroxide (100 μM) at every sampling points (0, 30, 60, and 90 min).
Discussion
Herein, a bacterial peroxidase from B. subtilis KCTC2023 (BsDyP) was characterized as a bacterial peroxidase capable of oxidizing various substrates including dyes (Reactive Black5 and Reactive Blue19) and relatively high redox potential substrates (ABTS, VA, and VGE) over +1.0 V vs. NHE. Atypically, BsDyP presented the substrate-dependent optimum temperature as definitely shown in Figure 3B, but optimum temperature of other bacterial peroxidase was not affected by the type of substrates. Recently, Santos et al. studied two bacterial DyPs from P. putida MET94 (PpDyP) and B. subtilis 168 (B-DyP)26. Unlike BsDyP investigated in this study, (i) B-DyP did not show VA oxidation activity; and ii) PpDyP showed flat optimum temperature profile ranging from 10 to 30°C for all substrates including VA and B-DyP presented optimal temperatures from 20 to 30°C for ABTS and dyes. Compared to PpDyP and B-DyP, the substrate-dependent optimum temperature for BsDyP is an unusual and novel feature.
Occasionally, peroxidase exhibits substrate-dependent optimum pH. For example, fungal DyP from Irpex lacteus showed substrate-dependent optimum pH: pH 2.0 for veratryl alcohol; pH 3.0 for 2,6-dimethoxyphenol and Reactive Black5; and pH 4.0 for Reactive Blue1933. LiPAc was optimized for n-propanol at pH 1.0, but the optimum pH was determined at pH 7.0 for decolorization of azo-dyes such as methyl orange and ethylene blue23. However, the substrate-dependent optimum temperature of peroxidase has not been reported so far. Exceptionally, but not in case of peroxidase, Polgár reported the substrate-dependent optimum temperature in biocatalyst for the first time; Oligopeptidase B (E.C. 3.4.21.83) from E.coli HB101, a prolyl oligopeptidase family of serine peptidase, optimally hydrolyzed benzoyl arginine p-nitroanilide and benzoyl arginine amide at 25°C, whereas the optimum temperature for benzoyl arginine ethyl ester was determined at 38°C34. Polgár aimed to thermodynamically elucidate the reason by calculating the activation entropy and enthalpy34, but the reason for the substrate-dependent temperature optimum was not completely elucidated.
It is still not clear what causes substrate-dependent temperature optimum in BsDyP. The substrates shown in Figure 3B could be categorized into two groups on the basis of the redox potential: ABTS, VA, and VGE usually required relatively high redox potential for oxidation over +1.0 V vs. NHE18,19, whereas dye-decolorization by the oxidative biocatalysis was more efficient at relatively low redox potential35: Reactive Black5 and Reactive Blue19 usually requires lower oxidation potential < +1.0 V vs. NHE in acidic condition20,21. In biocatalytic reactions, although VA, a high redox potential substrate, can be oxidized by LiP, organic dyes efficiently oxidized at low potential are not oxidized by LiP at all6,36. Distinguished from LiP, VP catalyzes both high- (e.g. VA) and low redox potential (e.g. catechol) substrates. For the biocatalytic oxidation of high redox potential substrates, it is generally known that the surface oxidation site and positively-charged environment around the surface oxidation site are necessary. In the relatively low redox potential substrates, two-independent oxidation active sites in VP are involved as shown by biphasic kinetics: the surface oxidation site as the high-specificity site and the main heme access channel as the low-specificity site5,37. As with VP, BsDyP also catalyzed both substrates requiring high- and low redox potential, implying that BsDyP might have a specific oxidation site for the low redox potential substrates different from that for the high redox potential substrates. The substrate redox potential-dependent optimum temperature of BsDyP might be resulted from a difference in the effect of temperature on each active site, but the reason could not be completely understood in this study. Further study on the structure-based catalytic mechanism will make it possible to elucidate the substrate redox potential-dependent optimum temperature in BsDyP.
Compared to bacterial peroxidases previously reported, BsDyP discussed in this study exhibited the highest ABTS and VA oxidation activity. Notably, BsDyP cleaved Cα-Cβ bond and then decomposed a dimeric lignin VGE including β-O-4 linkage dominant in lignin (> 50%) without any oxidative mediator. Furthermore, this is the first report to present BsDyP showing a significant shift of optimum temperature in catalyzing high redox potential substrates, revealing a previously unknown versatility of bacterial lignin-degrading biocatalyst in nature and opening up the possibility for efficient biocatalytic pretreatment of lignocellulosic biomass to produce biofuels and biochemical from lignocellulosic biomass.
Methods
Chemicals and bacterial strain
Four Bacillus species, Bacillus subtilis KCTC 2023, B. subtilis ATCC6051, B. licheniformis ATCC14580 and B. amyloliquefacience subsp. plantarum 10A6 were obtained from Korea Collection for Type Culture (KCTC), American Type Culture Collection (ATCC) and Bacillus Genetic Stock Center (BGSC). Phusion High-Fidelity DNA polymerase, pET21a(+) vector, hydrogen peroxide, E.coli BL21 (DE3) competent cell, and VGE were purchased from New England Biolabs (Ipswich, MA, USA), Novagen (Bilerica, MA, USA), Junsei (Tokyo, Japan), RBC Bioscience (Taipei, Taiwan), and AstaTech (Bristol, PA, USA), respectively. BugBuster® reagent for cell lysis and fast column kit for Ni-NTA purification were bought from Quiagen (Valencia, CA, USA). Hemin, ABTS, VA, Reactive Black5, Reactive Blue19, and veratryl aldehyde were purchased from Sigma-Aldrich (St. Louis, MO, USA).
VA oxidation test using Bacillus species
Lignin-degrading activity was screened by oxidation of VA to veratryl aldehyde. Four Bacillus species, B. subtilis KCTC 2023, B. subtilis ATCC6051, B. licheniformis ATCC14580 and B. amyloliquefacience subsp. plantarum 10A6 were grown in Luria-Bertani (LB) media for two days (optical density at 600 nm = 1.5–1.9) and centrifuged (13,000 rpm, 10 min, and 4°C) for obtaining cell-free supernatant including secreted proteins. For VA oxidation test, 2.0 mM of VA and 0.4 mM of H2O2 were added into 0.5 ml of cell-free supernatant in total volume of 1 ml (pH 3.0 and 30°C). The catalytic product veratryl aldehyde (ε310, veratryl aldehyde = 9.3 mM−1cm−1) was detected at 310 nm using UV-spectrophotometer (Cary60, Agilent, CO, USA).
Cloning and construction of expression vector
After growing B.subtilis KCTC2023 in LB media, cells were harvested by centrifugation (13,000 rpm, 10 min, and 4°C) and genomic DNA was extracted using Genomic DNA purification kit (iNtRON Biotechnology, Korea). The PCR primers were designed based on the genome sequence of B. subtilis KCTC2023 (NZ_ADGS01000000). Gene encoding BsDyP (accession number:WP_003222196) was amplified with primers: 5′-AAAACATATGAGCGATGAACAGAAAAAGC-3′ (forward, NdeI site underlined) and 5′-AAAAGAATTCCTAGTGATGATGATGATGATGTGATTCCAGCAAACGCTG-3′ (reverse, EcoRI site underlined, His tag coding sequence in italic). The PCR condition included an initial denaturation step at 98°C for 5 min, followed by 25 cycles of 98°C for 30 second, 54°C for 30 second, and 72°C for 1 min, with a final step of 72°C for 5 min using Phusion High-Fidelity DNA Polymerase. The PCR product was cloned into the pET-21a(+) vector.
Expression, purification, and reconstitution of BsDyP
E.coli BL21 (DE3) with pET-21a(+) containing the gene for BsDyP was grown in 5 mL of LB medium containing 50 μgmL−1 ampicillin and incubated at 37°C shaking 180 rpm. After overnight incubation, the culture was inoculated into 100 mL of fresh LB medium including 50 μgmL−1 of ampicillin and incubated at 37°C shaking at 200 rpm until the optical density at 600 nm was reached at 0.8. For induction, IPTG was added to a final concentration of 1.0 mM and further cultivation was conducted at 18°C for 24 hours, shaking 150 rpm to improve the expression level1. Then, the cell was harvested by centrifugation (8000 rpm, 20 min, and 4°C) and the pellet was stored at −20°C until purification.
For cell lysis, the harvested cell pellet was re-suspended in 4 mL BugBuster® reagent and incubated on ice for 1 hour, shaking 100 rpm. The lysate was purified using Ni-NTA fast column kit and finally 1 mL of purified BsDyP was obtained.
In order to reconstitute the native-BsDyP, 10 μL of hemin solution (10 mgmL−1) in dimethylsulfoxide was added to the purified BsDyP and then incubated on ice for 1 hour, shaking 100 rpm12. For calculating the Rz value (Asoret/A280)12, native- and reconstituted-BsDyP were scanned using a UV-spectrophotometer (Cary60, Agilent, CO, USA) from 500 nm to 250 nm. The concentration of the native- and the reconstituted-BsDyP was quantified by Bradford reagent using bovine serum albumin as a standard protein.
Activity assay
BsDyP activity was assayed using a UV-spectrophotometer (Cary60, Agilient, USA) at the maximum absorption wavelength of each substrate or product. The assay was initiated by adding 100 μM of hydrogen peroxide and 157 μgmL−1 of BsDyP into the substrate solution and conducted at the optimum condition; ABTS, VA, and VGE at pH 3.0 and 50°C; Reactive Black5 and Reactive Blue19 at pH 3.0 and 30°C. The wavelength and molar extinction coefficient for each substrate or product were as following: ABTS, ε420 = 36 mM−1cm−1; veratryl aldehyde, ε310 = 9.3 mM−1cm−1; Reactive Black5, ε597 = 37 mM−1cm−1; Reactive Blue19, ε595 = 10 mM−1cm−1 12,22.
For determining the optimum pH, 50 mM of KCl-HCl buffer for pH 1.5-pH 2.0 and 50 mM of acetate buffer for pH 3.0-pH 5.0 were used at 30°C. For the temperature optimum, 50 mM of acetate buffer was used for all substrate. The specific activities shown in Table 1 were assayed at the optimum pH and temperature for each substrate.
VGE decomposition with stepwise addition of hydrogen peroxide
For VGE decomposition, 0.2 mM of VGE in 1.0% ethanol and 1.9 mgmL−1 of reconstituted-BsDyP were used. The reaction initiated by adding 100 μM of hydrogen peroxide. The total reaction volume was of 2.0 mL. During the reaction, the pH and temperature were maintained at pH 3.0 (50 mM acetate buffer) and 50°C. At every sampling point, 100 μM of hydrogen peroxide was added after sampling. VGE and veratryl aldehyde were analyzed by HPLC (Agilent, CO, USA). The HPLC procedure was performed by injecting fractions using an Agilent 1200 HPLC system onto a reverse-phase Eclipse XDB-C18 column (4.6 × 150 mm, 5 μm, Agilent). Gradient separation was performed from 0.1% aqueous trifluoroacetic acid (solvent A) to methanol-acetonitrile (25:75; v/v; solvent B) using the following conditions: flow 1.5 mLmin−1; column temperature 30°C; time 0 min–15% B, time 2 min–30% B, time 11 min–60% B, time 11.5 min–100% B, time 13 min–0% B. Using the diode array detector, VGE and veratryl aldehyde were detected at 280 nm and 310 nm, respectively. Before injection, the sample was filtered through a PTFE hydrophilic 0.2 μm membrane filter. The conversion rate was calculated from the consumption of VGE31.
Author Contributions
K.M., H.M.W., Y. K. and Y.U. conceived and coordinated the experiments. K.M. performed the experiments. K.M. and Y.U. wrote the manuscript. G.G. conducted VA oxidation test using Bacillus species and analyzed the genome of B. subtilis KCTC2023.
Supplementary Material
Figure S1.
Acknowledgments
The authors thank to Dr. Seil Kim at the Korea Research Institute of Standards and Science for helping cloning. This work was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2009-C1AAA001-0093286) and the KIST Institutional Program (Project No. 2E24500).
References
- Min K. et al. Conversion of levulinic acid to 2-butanone by acetoacetate decarboxylase from Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 97, 5627–5634(2013). [DOI] [PubMed] [Google Scholar]
- FitzPatrick M., Champagne P., Cunningham M. F. & Whitney R. A. A biorefinery processing perspective: Treatment of lignocellulosic materials for the production of value-added products. Bioresour. Technol. 101, 8915–8922 (2010). [DOI] [PubMed] [Google Scholar]
- Agbor V. B., Cicek N., Sparling R., Berlin A. & Levin D. B. Biomass pretreatment: Fundamentals toward application. Biotechnol. Adv. 29, 675–685 (2011). [DOI] [PubMed] [Google Scholar]
- Rothschild N., Novotný Č., Šašek V. & Dosoretz C. G. Ligninolytic enzymes of the fungus Irpex lacteus (Polyporus tulipiferae): isolation and characterization of lignin peroxidase. Enzyme Microb. Technol. 31, 627–633 (2002). [Google Scholar]
- Ruiz-Dueñas F. J. et al. Substrate oxidation sites in versatile peroxidase and other basidiomycete peroxidases. J. Exp. Bot. 60, 441–452 (2009). [DOI] [PubMed] [Google Scholar]
- Brown M. E., Barros T. & Chang M. C. Y. Identification and Characterization of a Multifunctional Dye Peroxidase from a Lignin-Reactive Bacterium. ACS Chem. Biol. 7, 2074–2081 (2012). [DOI] [PubMed] [Google Scholar]
- Hofrichter M. Review: lignin conversion by manganese peroxidase (MnP). Enzyme Microb. Technol. 30, 454–466 (2002). [Google Scholar]
- Jing D. Improving the simultaneous production of laccase and lignin peroxidase from Streptomyces lavendulae by medium optimization. Bioresour. Technol. 101, 7592–7597 (2010). [DOI] [PubMed] [Google Scholar]
- Arora D. S. & Gill P. K. Comparison of two assay procedures for lignin peroxidase. Enzyme Microb.Technol. 28, 602–605 (2001). [DOI] [PubMed] [Google Scholar]
- Ürek R. Ö. & Pazarlioğlu N. K. Purification and partial characterization of manganese peroxidase from immobilized Phanerochaete chrysosporium. Process Biochem. 39, 2061–2068 (2004). [Google Scholar]
- Bugg T. D. H., Ahmad M., Hardiman E. M. & Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22, 394–400 (2011). [DOI] [PubMed] [Google Scholar]
- Ahmad M. et al. Identification of DypB from Rhodococcus jostii RHA1 as a Lignin Peroxidase. Biochemistry 50, 5096–5107 (2011). [DOI] [PubMed] [Google Scholar]
- Ahmad M. et al. Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders. Mol. BioSyst. 6, 815–821 (2010). [DOI] [PubMed] [Google Scholar]
- Yang Y. S., Zhou J. T., Lu H., Yuan Y. L. & Zhao L. H. Isolation and characterization of Streptomyces spp. strains F-6 and F-7 capable of decomposing alkali lignin. Environ. Technol. 33, 2603–2609 (2012). [DOI] [PubMed] [Google Scholar]
- Chang Y.-C., Choi D., Takamizawa K. & Kikuchi S. Isolation of Bacillus sp. strains capable of decomposing alkali lignin and their application in combination with lactic acid bacteria for enhancing cellulase performance. Bioresour. Technol. 152, 429–436 (2014). [DOI] [PubMed] [Google Scholar]
- Bandounas L., Wierckx N., de Winde J. & Ruijssenaars H. Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotech. 11, 94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Krishna Reddy M. M. & Chandra R. Identification of low molecular weight aromatic compounds by gas chromatography–mass spectrometry (GC–MS) from kraft lignin degradation by three Bacillus sp. Int. Biodeterior. Biodegrad. 59, 292–296 (2007). [Google Scholar]
- Smith A. T., Doyle W. A., Dorlet P. & Ivancich A. Spectroscopic evidence for an engineered, catalytically active Trp radical that creates the unique reactivity of lignin peroxidase. Proc. Natl. Acad. Sci. 106, 16084–16089 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbonnais R., Leech D. & Paice M. G. Electrochemical analysis of the interactions of laccase mediators with lignin model compounds. Biochim. Biophys. Acta, Gen. Subj. 1379, 381–390 (1998). [DOI] [PubMed] [Google Scholar]
- Esteves F. & Cunha E. P. Voltametric study and electrochemical degradation of reactive dyes. 5th World Textile Conference AUTEX 2005 (2005). [Google Scholar]
- Radi A., Mostafa M. R., Hegazy T. A. & Elshafey R. M. Electrochemical study of vinylsulphone azo dye Reactive Black 5 and its determination at a glassy carbon electrode. J. Anal. Chem. 67, 890–894 (2012). [Google Scholar]
- van Bloois E., Torres Pazmiño D., Winter R. & Fraaije M. A robust and extracellular heme-containing peroxidase from Thermobifida fusca as prototype of a bacterial peroxidase superfamily. Appl. Microbiol. Biotechnol. 86, 1419–1430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodake G., Kalme S., Jadhav J. & Govindwar S. Purification and Partial Characterization of Lignin Peroxidase from Acinetobacter calcoaceticus NCIM 2890 and Its Application in Decolorization of Textile Dyes. Appl. Biochem. Biotechnol. 152, 6–14 (2009). [DOI] [PubMed] [Google Scholar]
- Yang Y. S., Zhou J. T., Lu H., Yuan Y. L. & Zhao L. H. Isolation and characterization of a fungus Aspergillus sp. strain F-3 capable of degrading alkali lignin. Biodegradation 22, 1017–1027 (2011). [DOI] [PubMed] [Google Scholar]
- Li H. Y., Li S. N., Wang S. X., Wang Q. & Zhu B. C. Screening, Identification of Lignin-Degradating Bacillus MN-8 and Its Characteristics in Degradation of Maize Straw Lignin. China Agric.Sci. 47, 324–333 (2014). [Google Scholar]
- Santos A., Mendes S., Brissos V. & Martins L. New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: towards biotechnological applications. Appl. Microbiol. Biotechnol. 98, 2053–2065 (2014). [DOI] [PubMed] [Google Scholar]
- Yoshida T., Tsuge H., Hisabori T. & Sugano Y. Crystal structures of dye-decolorizing peroxidase with ascorbic acid and 2,6-dimethoxyphenol. FEBS Lett. 586, 4351–4356 (2012). [DOI] [PubMed] [Google Scholar]
- Yoshida T., Tsuge H., Konno H., Hisabori T. & Sugano Y. The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue. FEBS J. 278, 2387–2394 (2011). [DOI] [PubMed] [Google Scholar]
- Kim S. J. & Shoda M. Purification and Characterization of a Novel Peroxidase from Geotrichum candidum Dec 1 Involved in Decolorization of Dyes. Appl.Environ.Microbiol. 65, 1029–1035 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogola H. J. O. et al. Molecular Characterization of a Novel Peroxidase from the Cyanobacterium Anabaena sp. Strain PCC 7120. Appl. Environ. Microbiol. 75, 7509–7518 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh Mai Pham L., Eom M.-H. & Kim Y. H. Inactivating effect of phenolic unit structures on the biodegradation of lignin by lignin peroxidase from Phanerochaete chrysosporium. Enzyme Microb. Technol. 61–62, 48–54 (2014). [DOI] [PubMed] [Google Scholar]
- Valderrama B., Ayala M. & Vazquez-Duhalt R. Suicide Inactivation of Peroxidases and the Challenge of Engineering More Robust Enzymes. Chem. Biol. 9, 555–565 (2002). [DOI] [PubMed] [Google Scholar]
- Salvachúa D., Prieto A., Martínez Á. T. & Martínez M. J. Characterization of a Novel Dye-Decolorizing Peroxidase (DyP)-Type Enzyme from Irpex lacteus and Its Application in Enzymatic Hydrolysis of Wheat Straw. Appl. Environ. Microbiol. 79, 4316–4324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgár L. Oligopeptidase B: A New Type of Serine Peptidase with a Unique Substrate-Dependent Temperature Sensitivity. Biochemistry 38, 15548–15555 (1999). [DOI] [PubMed] [Google Scholar]
- Zille A. et al. Predicting Dye Biodegradation from Redox Potentials. Biotechnol.Progr. 20, 1588–1592 (2004). [DOI] [PubMed] [Google Scholar]
- Heinfling A. et al. A study on reducing substrates of manganese-oxidizing peroxidases from Pleurotus eryngii and Bjerkandera adusta. FEBS Lett. 428, 141–146 (1998). [DOI] [PubMed] [Google Scholar]
- Morales M. et al. Two Oxidation Sites for Low Redox Potential Substrates: A directed mutagenesis, kinetic, and crystallographic study on Pleurotus eryngii versatile peroxidase. J. Biol. Chem. 287, 41053–41067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.