Abstract
Plasma medicine is an upcoming research area that has attracted the scientists to explore more deeply the utility of plasma. So, apart from the treating biomaterials and tissues with plasma, we have studied the effect of soft plasma with different feeding gases such as Air, N2 and Ar on modification of biomolecules. Hence, in this work we have used the soft plasma on biomolecules such as proteins ((Hemoglobin (Hb) and Myoglobin (Mb)), calf thymus DNA and amino acids. The structural changes or structural modification of proteins and DNA have been studied using circular dichroism (CD), fluorescence spectroscopy, protein oxidation test, gel electrophoresis, UV-vis spectroscopy, dynamic light scattering (DLS) and 1D NMR, while Liquid Chromatograph/Capillary Electrophoresis-Mass Spectrometer (LC/CE-MS) based on qualitative and quantitative bio-analysis have been used to study the modification of amino acids. Further, the thermal analysis of the protein has been studied with differential scanning calorimetry (DSC) and CD. Additionally, we have performed docking studies of H2O2 with Hb and Mb, which reveals that H2O2 molecules preferably attack the amino acids near heme group. We have also shown that N2 gas plasma has strong deformation action on biomolecules and compared to other gases plasma.
Supplying energy to gaseous medium leads to the dissociation of molecular bonds, resulting in the generation of fully or partially ionized gases commonly known as plasma1,2,3,4,5. Plasma is a ionized gas consisting of positive and negative ions, electrons as well as neutral atoms and molecules (e.g. radicals) and ultraviolet radiations depending on the ionized gas2. Soft plasma jet is now the center of attraction due to its significant industrial advantages over low-pressure discharge and thus offering many biomedical applications6. There are various studies on the effect of non-thermal plasmas for the inactivation of micro-organisms, wound healing, blood coagulation, skin regeneration, tooth bleaching and action against cancer cells7,8,9,10,11,12,13,14,15. These results are based on plasma created reactive agents including UV photons, reactive oxygen species (ROS), reactive nitrogen species (RNS), charged particles and electric fields1,2,3,4,5,6,7,8,9,10,11,12,13,14,15. The APPJ exposure on the DNA molecule are studied for short intervals showed that it results to the DNA damage16,17,18,19,20. Recently Han et al.19, reveals that APPJ can damage the aqueous plasmid DNA. DNA break increases as distance between the DNA and APPJ decreases and treatment time increases. Furthermore, lysozyme in the aqueous solution revealed the change in the structural information of enzyme, after plasma treatment21. Moreover, studies have been reported for the horseradish peroxidase inactivation and heme degradation induced by discharge plasma22. Hence, structural modifications in biomolecules on treatment with reactive species or free radical may vary from biomolecule to biomolecule.
In order to study the effect of soft plasma, we have studied direct effect of plasma on heme proteins such as Hemoglobin (Hb) and Myoglobin (Mb). These proteins are involved in the electron transfer and transport of oxygen in life processes. Certainly the most broadly studied allosteric protein found in nature is Hb23. Hb constitutes of seven non-helical and helical segments each in an α-chain, while each β-chain has eight helical and six non-helical segments24. While, Mb is a fairly compact globular protein whose backbone structure consists of eight α-helical segments25. Hence, it is interesting to study these protein modifications using soft plasma as model proteins.
During the study of living cells in the presence of plasma, one of the main reasons for the cell death is DNA damage26,27,28. One of the explanations of DNA damage is related to the radical attack that involves metabolic changes in the cell and results to the initiation of nucleases capable of degradation of DNA. Breaking of single and double DNA-strand26,27 a consequence of the reaction with ROS is responsible for blocking the replication process that ultimately leads to the cell death28. There might also be possibility of formation of the modified nitrogen bases on the reaction of ROS with DNA. Guanine (a component of a nucleoside, nucleotide, or polynucleotide (DNA, RNA)) in its free state, is susceptible to oxidation at C8 position, and attack of ROS generated by plasma, thus results in formation of 8-oxoguanine or 8-oxoG ultimately leading to structural changes of DNA28,29,30,31,32.
Due to oxidative stress, the modification of DNA, proteins, lipids and carbohydrates are occurred33,34. Amino acid residues in proteins are highly susceptible to oxidation by ROS which are present as by-products of normal metabolic processes, pollutants in the atmosphere and produced by exposure to different type of irradiation34,35. In order to understand the mechanisms of these ROS-mediated oxidations, many research groups explored the exposure of ionizing radiation to the solutions of amino acids, peptides, and proteins35. It was observed that OH radical dependent abstraction of a hydrogen atom from the α-carbon of amino acids, aliphatic side chains of hydrophobic amino acid residues of protein and from the protein polypeptide backbone seem to be the possible initial sites of attack31,32,36. There are some studies of the plasma action on amino acids by different research groups37,38,39,40. They observed the disulfide linkage formation and hydroxylation of amino acids after the plasma treatment. Very recently, Takai et al.40, found that APPJ can modify the 14 amino acids. They observed the hydroxylation, nitration, sulfonation, disulfide linkage formation and amidation in amino acids after the APPJ treatment using high resolution mass spectroscopy. However, there was no qualitative and quantitative analysis of the new groups originated after the plasma treatment using different type of gases.
Therefore, in this work we have tried to throw some light on the aspect that how the ROS/RNS created by the soft plasma affect the biomolecules directly. Modification of biomolecules such proteins (Hb and Mb) were studied by using circular dichroism (CD) (structural and thermal analysis), differential scanning calorimetry (DSC), fluorescence spectroscopy, protein oxidation, gel electrophoresis, UV-vis spectroscopy, dynamic light scattering (DLS) and 1D NMR. Additionally, we can locate the attacking site of H2O2 on proteins using docking studies. Modification or oxidation of calf thymus DNA was studied by CD analysis and DNA oxidation. At last, we studied the amino acid modification using the most efficient technique such as Liquid Chromatograph/Capillary Electrophoresis- Mass Spectrometer (LC/CE-MS) based qualitative and quantitative analysis.
Results
There are many studies highlighting the effect of ROS and RNS produced by plasma sources on the cells and tissues, while to gain an in-depth insight, the interaction of plasma with the involved biomolecules directly becomes necessary.
Reactive species generated using plasma in water and optical emission spectroscopy of a APPJ in the presence of different feeding gases
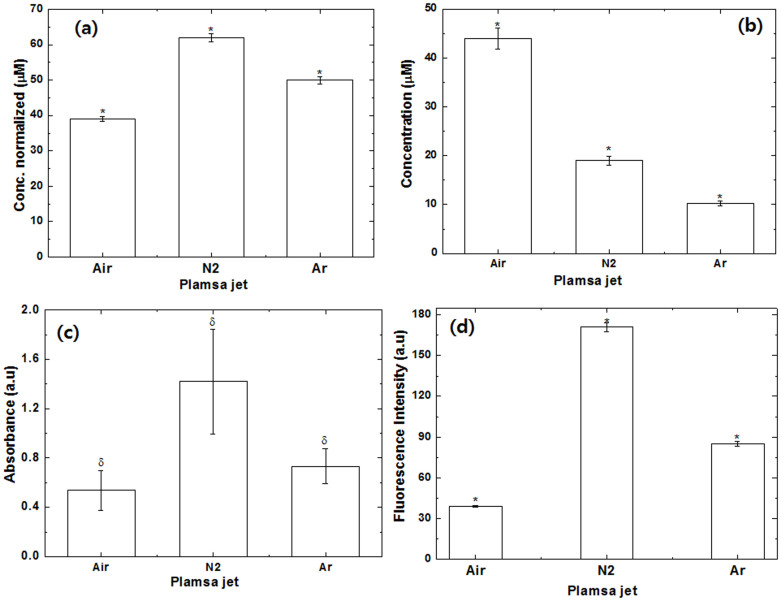
We have studied some important reactive species (RS) generated from the plasma in the presence of different feeding gases, as illustrated in Fig. 1. In the water solution, we have studied the H2O2, NO, ONOO− and OH radicals. We observed that level of H2O2 and OH radicals are highest for N2 plasma; followed by Ar plasma and least with Air plasma. From Figs. 1d and S1, we observed the generation of HTA (hydroxyterephthalic acid) from TA (terephthalic acid) in the presence of different gases, that is directly linked with the generation of OH radicals41. Fig. S1, shows that the fluorescence intensity at ≈425 nm is highest for N2 gas plasma as compared with the Ar and Air plasma, this reveals that OH radicals are generated in N2 plasma more than other gas plasma. On contrary, the concentration of NO is found out to be maximum in Air plasma, than in N2 plasma and least in the Ar plasma. Moreover, other radical also play an important role, therefore we recorded the optical emission spectra (OES) of the soft plasma.
Figure 1. H2O2, NO and OONO− concentration changes in physiological solutions exposed to Air, N2 and Ar plasma in water after 3 min treatment.
All values are expressed as ± SD in triplicates. Students't-test was performed to control (* denotes P < 0.05, ** denotes P < 0.01 and δ ± 0.4).
APPJ in Air, N2 and Ar gas plasma are functioning at atmospheric pressure have been shown to generate various types of radicals as shown in Figs. S2 to S5. We observed strong emission lines from a molecular NO β, γ system for N2 and Ar gas whereas weak lines between 200 and 250 nm were observed for the Air plasma. The N2 second-positive system (C3Πu–B3Πg) were observed in all gases (Air, N2 and Ar) plasma. The N2 first positive system (B3Πg–A3Πu+) observed between 400–500 nm was also observed in case of the Air plasma. However, atomic O spectra were only observed for the Air plasma at 552.91 and 776.05 nm, due to the higher gas temperature in O2 plasma. Hence, the number of electrons decreased during the formation of oxygen ions, that results in the shorter lifetime of active molecules found in Air plasma due to collisional quenching by oxygen42,43. We have also observed the Ar emission lines from 697.04 nm to 912.94 nm, as shown in Fig. S4. The OES at ≈337 nm for NO/N244 in Fig. S5, also supports the more generation of NO in Air plasma as compared to Ar and N2 plasma. Additionally, we have also studied the changes in pH and temperature of the solution after the 3 min treatment in all gases, as shown in Fig. S6.
CD spectral analysis and Melting temperature (Tm)
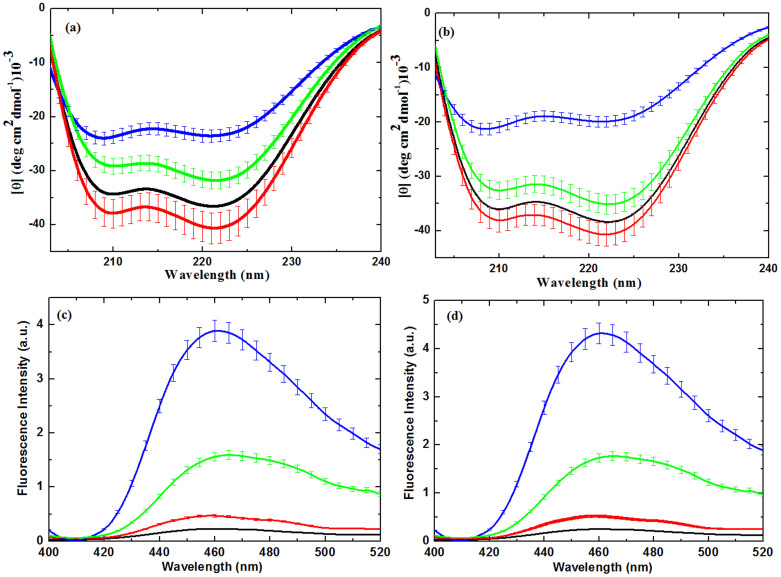
To obtain a detailed understanding of the degree of modification using soft plasma on Hb and Mb, we further performed CD experiments23. As seen from Figs. 2a, b, the far-UV CD spectra of proteins indicates the deformation of mainly secondary structure of proteins due to different feeding gases. The CD spectrum is typical for a polypeptide chain, with two well-pronounced minima at ≈210 and ≈222 nm for proteins in water suggesting that the polypeptide chain is mostly organized in α-helix conformation23. The results in this Table 1 clearly reveal that after the Air plasma treatment the α-helical structure of both Hb and Mb increases and β-sheet decreases. Whereas, after the N2 and Ar plasma treatment the decrease in α-helical structure and increase in β-sheet as compared to without treated proteins. Further, a more closer look at the observations reveals that the decrease in α-helical structure is more for Mb than Hb in the presence of N2 plasma. After the exposure of Air plasma to both proteins (Hb and Mb), their structure becomes compact and % of α-helical structure increases, whereas in case of N2 and Ar plasma modification of α-helix to β-sheet or random coil occurs. We also checked the structural changes of Hb and Mb at pH 4.8 and didn't observed any significant structural changes (data not shown), hence the observed structural changes after the plasma treatment are not due to the pH change in water. We have also measured the spectra of Hb and Mb in the presence of H2O2 at different concentration from 40 μM to 200 μM, as illustrated in Fig. S7. From the Fig. S7, we observed, slightly change in the secondary structure of both proteins at 100 and 200 μM.
Figure 2.
(a) Far-UV CD spectra analysis of Mb; (b) Far-UV CD spectra analysis of Hb; (c) Fluoresence analysis of Mb and (b) Fluoresence analysis of Hb.Where proteins without treatment (black), proteins treated with Air plasma (red), proteins treated with N2 plasma (blue) and proteins treated with Ar plasma (green).The data points are average values of at least six determinations, the error bars indicating ± mean deviation.
Table 1. Secondary structure composition of Hb and Mb, determined from Far UV CD spectra in different media at 20°C determined by K2D3.
| Sample | α-sheet (%) | β-sheet (%) |
|---|---|---|
| Hb | 67 | 8.00 |
| Hb + Air | 68 | 8.01 |
| Hb + N2 | 60 | 10.00 |
| Hb + Ar | 66 | 9.00 |
| Mb | 90 | 0.40 |
| Mb + Air | 91 | 0.30 |
| Mb + N2 | 65 | 2.00 |
| Mb + Ar | 87 | 0.40 |
Further, to investigate the thermal stability of proteins, we have studied the DSC and CD thermal analysis.The results are displayed in Figs. S8–S11 and Table S1, which reveal that the decrease in Tm values (the temperature at which 50% of the proteins are unfolded) of Hb and Mb is seen in the presence of soft plasma with different feeding gases. The observed Tm of the Hb control without treatment is found to be ≈66.37°C and 66.20°C according to DSC and CD thermal analysis, respectively, while after the Air plasma it decreases to ≈65.10°C and 65.01°C, N2 plasma has Tm ≈ 59.25°C and 59.10°C, Ar plasma has Tm ≈ 61.75°C and 61.59°C as studied by DSC and CD thermal analysis respectively. Hence, we can see clearly there is a slight difference in the Tm values from DSC and CD thermal analysis. Similarly, we observed the in Tm of Mb after the treatment with plasma, but the decrease is less for Mb as compared with Hb, as shown in Table S1.
Fluorescent analysis for Heme degradation and protein oxidation
We have studied the heme degradation using the previously reported method22,45,46, providing the excitation at 321 nm and emission at 460 nm as shown in Figs. 2c and d for Hb and Mb, respectively. The fluorescence intensity at 460 nm was used as a measurable index for heme degradation45,46. We observed that the fluorescence intensity increases in order N2 > Ar > Air plasma, which shows that N2 plasma has more influence in heme degradation. We have performed additional experiments at different concentration of H2O2 from 40 μM to 200 μM, as displayed in Fig. S12. In Fig. S12, we found an increase in fluorescence intensity as the concentration of H2O2 increases. If we compared the fluorescence intensity of N2 plasma with 200 μM of H2O2 for both proteins Hb and Mb, it is interesting that N2 plasma has high intensity than 200 μM of H2O2. Hence, the heme degradation is also affected by other radicals and parameters other than H2O2. For heme degradation the oxidation plays an important role, so we have further studied the protein oxidation using various techniques.
Protein oxidation, gel electrophoresis, UV-vis spectroscopy and DLS measurements
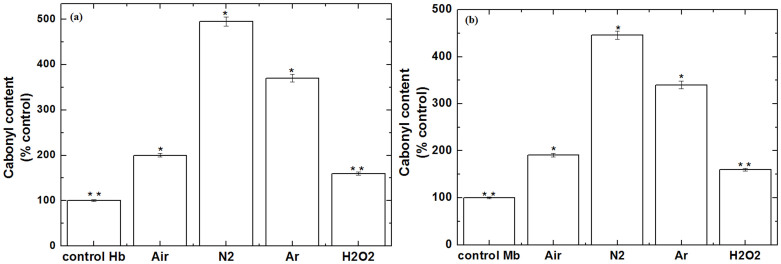
The increase in the level of ROS/RNS may consequence in oxidative stress. Protein carbonyls are generally used as an indicator of oxidative stress. Therefore, we have studied the protein oxidation in the presence of Air, N2 and Ar plasma with 2,4-dinitrophenylhydrazine (DNPH), as displayed in Fig. 3, to understand the oxidative stress due to plasma. As we have discussed earlier in the CD section that N2 and Ar plasma have a maximum decrease in α-helix structure as compared to the control. Similarly, protein oxidation levels were found to be high in the presence N2 and Ar plasma. Additionally, we have also studied the oxidation at 60 μM of H2O2 to check the influence of H2O2 on the oxidation of Hb and Mb. These results reveal that oxidation at 60 μM of H2O2 is less compared with plasma oxidation. Whereas, for both proteins Hb and Mb, the level of oxidation by plasma is approximately same, which may be possibly due to similar family proteins. Further, to see more clearly the action of soft plasma with different feeding gases on proteins, we have studied the gel electrophoresis.
Figure 3. Protein oxidation (carbonyl content) (a) Hb and (b) Mb for proteins, proteins treated with Air plasma, proteins treated with N2 plasma, proteins treated with Ar plasma for 3 min and proteins treated with 60 μM of H2O2.
All values are expressed as ± SD in triplicates. Students't-test was performed to control (*denotes P < 0.05 and ** denotes P < 0.01).
Using gel electrophoresis, we predicted the molecular weight change of the Hb and Mb proteins in the presence of soft plasma with different gases. The results are summarized in Fig. S13. Our results reveal that Hb and Mb bands are very thin in the case of N2 and Ar plasma that leads to denaturation of Hb and Mb native structure due to excess protein oxidation. These results are very well correlate with the Xiao et al.47, that excess ROS increases the BSA fragmentation that results in thinner bandwidth on SDS-PGE electrophoresis and western blotting. Further, we also detected that bandwidth is thinner for the Hb as compared to Mb. This further supports our above data that the change in the structure of the Hb is more as compared to the Mb structure after N2 and Ar plasma. Whereas, the gel bandwidth has become thicker for both Hb and Mb proteins in the presence of Air plasma. This reveals that in the presence of the Air plasma the intensity of the bandwidth increases due to the formation of higher molecular weight carbonyls, similar behaviour is observed by the Chesne et al.48, for BSA oxidation.
The heme group is submerged in a hydrophobic pocket within the protein's interior. This interaction of the heme with the protein structure results in the soret band, a strong absorbance peak in the visible spectrum at ≈409 nm for Hb and Mb, the results are shown in Figs. S14a and b. Upon denaturation of Hb or Mb, there occurs a decrease in absorbance or shift in the soret band peak owing to the exposure of heme to the polar aqueous solvent. We have observed that there is a slight red shift with quenching for the both Hb and Mb proteins in the presence of N2 plasma. Whereas, we didn't observe any change for the Air and Ar plasma, which was same as the control. In the another experiment to monitor the effect of H2O2, we have treated both proteins Hb and Mb with different concentration of H2O2, shown in Figs. S14c, d. We observed that there is no change in soret band for Hb, when the H2O2 concentration increases from 40 μM to 200 μM. While, for Mb we observed slight red shift for 100 and 200 μM of H2O2. These data reveal that soret band from Hb has less affected as compared with Mb soret band for the same concentration of H2O2.
Latter, in order to understand the change in structural size of proteins (Hb and Mb) after the plasma treatment, we have studied the DLS studies, the results are as shown in Table S2. We observed that after the treatment of N2 and Ar plasma, Hb or Mb gets denatured or oxidized, so that the particle size of Hb or Mb increased. While for the Air plasma the Hb or Mb size is approximately the same as control. The increase in size of Hb and Mb is due to protein aggregation, the other groups49,50,51 also supported this data that protein size increases due to protein aggregation that is the result of protein oxidation. Hence, this further provides an evidence that the oxidation or modification of protein are in control way, for Air plasma treatment as compared to other gases treatment.
Investigation of binding site interacting residues through molecular docking and NMR studies
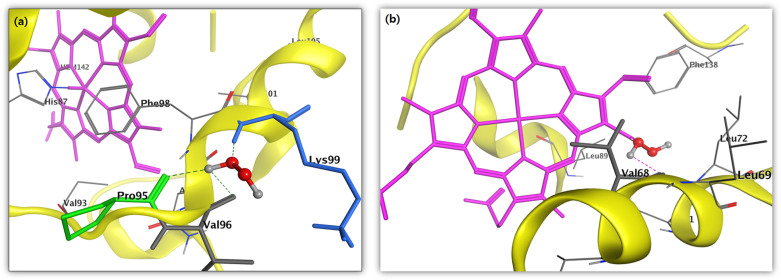
According to the previously reported work22,45,46 and our above experimental work, we can conclude that H2O2 is one of the major species for heme degradation. Therefore, in this study, we performed molecular docking study to investigate if H2O2 modulates the target, and identifies the binding site active pocket against the well-known human molecular target Hb and Mb protein receptor. This is theoretically validated by computational simulation process. The molecular docking technique is used to show the molecular insight of interacting compound with that of the target protein. This process is used to display the molecular interaction and also measured the interaction in terms of docking score or docking energy. Using molecular docking simulations, we successfully elucidate the orientations and binding affinities (in terms of docking score refer here as ‘total score') of H2O2 against molecular target Hb and Mb protein receptor.
The docking results for compound H2O2 against the target protein Hb exhibited high binding affinity docking score represented as total score of 3.1416 and also, the formation of three hydrogen bonds of length 1.9, 1.9 and 2.6 Å between backbone atoms of positively charged polar basic residue lysine (Lys)-99, backbone atom of nonpolar hydrophobic amino acid residue proline (Pro)-95 and valine (Val)-96, respectively. In this docking pose, other binding site residues within a selection radius of 4 Å from bound substrate non-polar hydrophobic residue e.g., phenylalanine (Phe)-98 and polar uncharged residue asparagine (Asn-97), as a result, the bound substrate showed a favourable interaction with Hb thus leading to more stability with H2O2, the results being displayed in Fig. 4a. Similarly, docking results for H2O2 against the target protein human Mb receptor showed a high binding affinity docking score indicated by a total score of 2.8843 and formed a hydrogen bond of length 2.0 Å to the backbone atom of non-polar hydrophobic residue e.g., valine (Val-68). In the docking pose of H2O2-Mb complex, the chemical nature of binding site residues within a radius of 4 Å was nonpolar hydrophobic residues e.g., alanine (Ala-71), leucine (Leu-69, Leu-72 and Leu-89) and isoleucine (Ile-75), as a result, the bound substrate showed a strong interaction with Mb, thus leading to more stability with H2O2 (Fig. 4b). A more deeper study of the nature of interaction of H2O2 with the binding pocket amino acid residues of the target protein, revealed that the H2O2 showed molecular interaction with conserved hydrophobic amino acid residues, in Hb and Mb active pocket (Figs. 4a, b) thus leading to more stability and potency. Hence, the presence of H2O2 due to plasma treatment might be the main source of protein modification.
Figure 4. In silico molecular docking showing the binding site residues within a selection radius of 4 Å from bound H2O2 revealing the hydrophobic pocket of active conformation.
Binding sites of human (a) Hb (PDB:1A3N) and (b) Mb (PDB:3RGK) protein receptor. The docking studies were carried out using SYBYL-X 2.0.
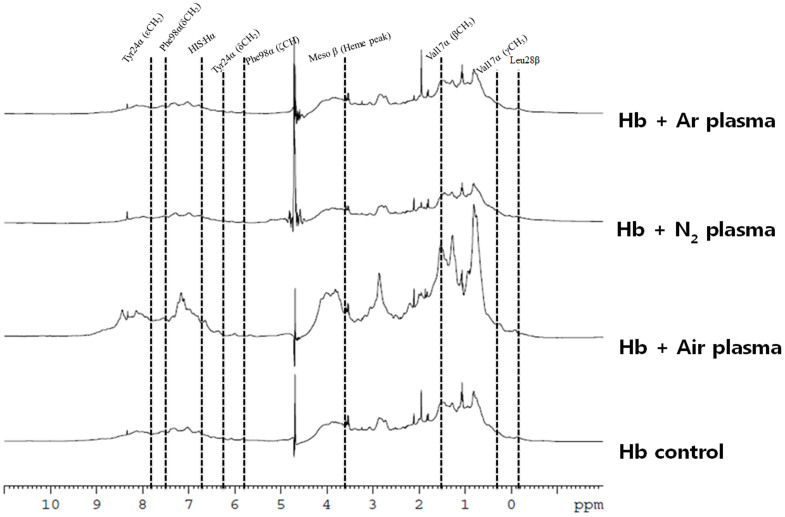
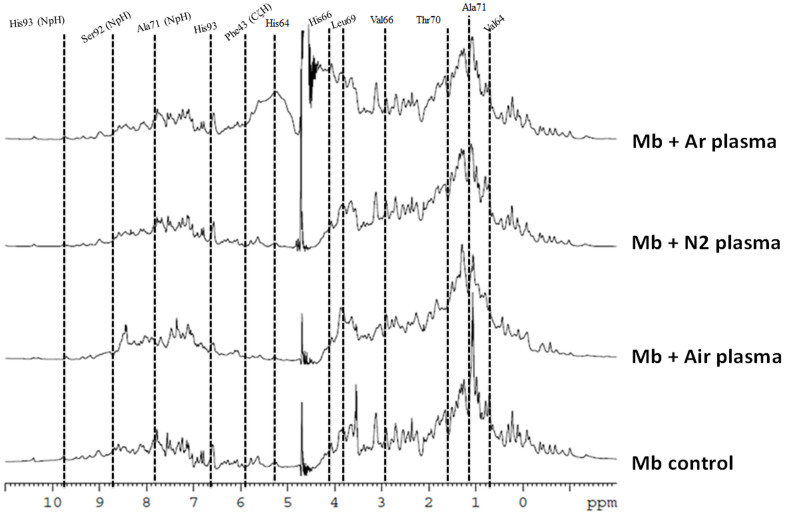
Therefore, to check the docking results at molecular level due to the H2O2, we investigated the 1D NMR for both Mb and Hb proteins, as illustrated in Figs. 5 and 6. Mb and Hb are the model system for protein folding52,53,54,55,56. For Hb NMR, we observed the peaks for Leu28β, Val17α (γCH3) Val17α (βCH3), Phe98α (ζCH), Tyr24α (δCH2), HIS:Hα, Phe98α (δCH2) and Tyr24α (εCH2) appeared at −0.81, 0.44, 1.55, 5.88, 6.19, 6.84, 7.65 and7.78 ppm in case of the Air plasma but disappeared in case of the Ar and N2 plasma. Peaks for Leu91α, Val62α, Val17α, heme group (5CH3) and Tyr42α (εCH2) were explicated at 0.94, 1.28, 1.55, 2.63 and 7.24 ppm and were found to be same in case of the N2 and Ar plasma, while the intensity was found to be enhanced in case of the Air plasma. While peaks for the Mb amino acids are following: Ala94, Val64, Ala71, Thr70, Val66, Leu69, Ser92, His66, Phe46 (CαH), His64, Phe43 (CζH), Phe33 (CεH), Phe46 (CζH), His93, Thr95 (NpH), Thr70 (NpH), Ala71 (NpH), His68 (NpH), Ser92 (NpH), Val66 (NpH)and His93 (NpH) appeared at 0.70, 0.76, 1.04, 1.59, 2.89, 3.89, 3.90, 4.05, 5.02, 5.26, 5.93,6.18, 6.27, 6.68, 7.43, 7.77, 7.96, 8.53, 8.77, 9.35 and 9.87 ppm peaks. The intensity decreases after the N2 and Ar plasma treatment, while the change is not much for Air plasma treatment. We have also studied the effect of 60 μM H2O2 on Hb and Mb, to check that the change in peaks are due to H2O2 or due to other radicals, results are displayed in Figs S15 and S16. We observed that amino acids peaks were not altered much with the presence of H2O2, hence there are contributions from the other radicals for the structural changes of Hb and Mb after the plasma treatment are very important.
Figure 5. 1H NMR of Hb before and after treatment in different feeding gases.
Figure 6. 1H NMR of Mb before and after treatment in different feeding gases.
CD analysis and DNA oxidation test for calf thymus DNA
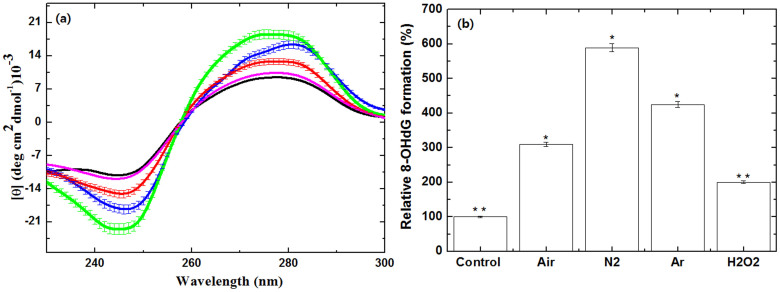
Further, to see the selective effect of the different feeding gases of soft plasma on biomolecules, we took calf thymus (ct) DNA into consideration.To check the effect of plasma on DNA, we have examined the DNA CD spectra. The measurement of ellipticity (θ) has been generally used to display the structural conformational changes of DNA28,57,58. The obtained changes in ellipticity in function of wavelength before and after plasma are shown in Fig. 7a. Before the plasma treatment, the native DNA showed positive peak at 275 nm and negative peak at 245 nm. After the Air plasma treatment, the negative peak appeared at 245 nm and positive peak at 276 nm, with enhanced negative and positive peak intensities. Whereas with the N2 and Ar plasma, the negative peaks now appeared at 247 nm and 246 nm respectively, with enhanced negative intensity. In case of the N2 and Ar plasma, the positive peaks appeared at 280 nm and 273 nm respectively, with increase in the intensity of positive peak. We have also checked the action of 60 μM H2O2 on DNA, we detected the slight change in the structure of DNA as compared to plasma treatment. That is very well correlated with the Nowicka et al.28, studied, where they observed the small change in the main positive and negative bands in presence of H2O2.
Figure 7.
(a) CD spectra of ctDNA with soft plasma treatment in the presence of different feeding gases black (control), red (Air plasma), blue (N2 plasma) and green (Ar plasma) and with 60 μM of H2O2 (magenta),The data points areaverage values of at least six determinations, the error bars indicating ± mean deviation.; (b) Relative percentage of 8-OHdG formation in the presence of different feeding gases and with 60 μM of H2O2. All values are expressed as ± SD in triplicates. Students't-test was performed to control (* denotes P < 0.05 and ** denotes P < 0.01).
This shows that the structural deformation occured in the ctDNA after the attack of plasma radicals. Further, we then studied the test for 8-oxoguanine which is a typical oxidative base lesion29,30. The presence of 8-oxoguanine in the double stranded helix leads to distortion of the double helix by disturbing the circular hydrogen bonding, when compared to the undamaged double helix (we have shown above). Also, the extent of hydration increases around the polar oxygen atom at C8. Whereas, the oxidation in the presence of 60 μM H2O2 is less as compared to plasma treated DNA. From the Fig. 7b, we observed the % of increase in the 8-OHdG formation and it was found that the maximum 8-OHdG was formed in case of the N2 plasma and the least change occurred in the presence of the Air plasma. Further, to check the effect of soft plasma in the presence of different feeding gases, we focused on amino acids.
Amino acid oxidation has studied LC/CE-MS
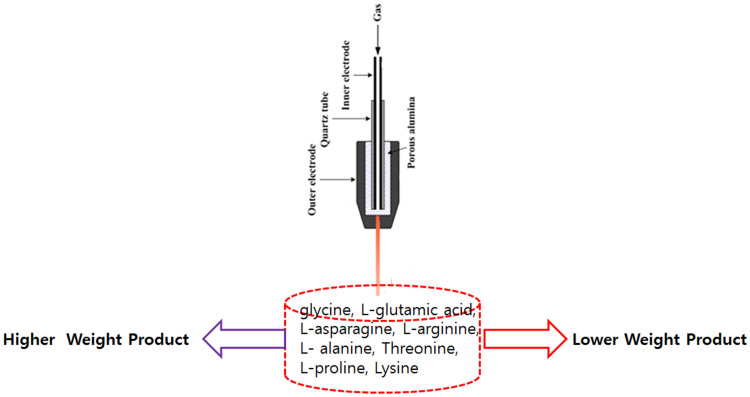
Amino acid oxidative modifications can give rise to protein carbonyls such as a direct attack by RS on certain amino acid side chains33,34,40,59,60,61. There are many pathways for the protein carbonylation, although it is very hard to visualize the exact carbonylation in proteins. Hence, we have studied the some amino acids to check the amino acid modification using LC/CE-MS. We further investigated the qualitative amino acid analysis of glycine, L-glutamic acid, L-asparagine, L-arginine, L- alanine, L-Threonine, L-proline and L-Lysine using LC/CE-MS as illustrated in Figs. S17–S24. From these Figures, we interpreted that some new compound may be generated after the plasma treatment, due to oxidation, nitration, dehydrogenation and dimerization of amino acids. While all amino acids did not respond in a similar way to the different feeding gases equally. In some cases, the oxidative products were more with some feeding gases, while in some cases, the product was not formed prominently such as L- alanine amino acid. This doesn't mean that there was no change in amino acid after the plasma treatment, while the change was not very significant to capture in spectroscopy in the given window. Therefore, to study the complete change in the amino acids after the treatment with different feeding gases, we have checked the quantitative analysis using LC/CE-MS as displayed in Table 2. We have observed that weight (μg/ml) has increased for amino acids after the treatment except the L-asparagine for N2 plasma treatment where the weight decreases. Moreover, there is no particular trend for increase in weight of amino acids for different feeding gases treatment. These studies are supported by the recently published work by Takai et al.40, that after the treatment amino acid structure modified.
Table 2. Evaluation of a LC/CE-MS method for quantitative amino acid analysis before and after the APPJ treatment in different feeding gases.
| Amino acids | Control (μg/ml) | Air plasma (μg/ml) | N2 plasma (μg/ml) | Ar plasma (μg/ml) |
|---|---|---|---|---|
| Glycine | 321.621 | 392.603 | 358.983 | 372.428 |
| L-Glutamic acid | 125.905 | 154.270 | 165.820 | 135.203 |
| L-Asparagine | 142.092 | 223.006 | 127.954 | 152.760 |
| L-Arginine | 164.200 | 216.779 | 208.316 | 180.331 |
| L-Alanine | 215.404 | 273.321 | 290.671 | 261.111 |
| L-Threonine | 235.175 | 799.575 | 785.921 | 762.577 |
| L-Proline | 81.939 | 243.887 | 249.620 | 225.251 |
| L-Lysine | 87.828 | 102.030 | 99.687 | 90.909 |
Discussion
The secondary structure of both proteins (Hb and Mb) are disturbed after the plasma treatment in all gases as compared with control. We observed that α-helix has increased for Air plasma, while decreases for N2 and Ar plasma. And among the N2 and Ar plasma, the decreases in α-helix is maximum for N2 plasma for both proteins. While in comparison of H2O2 effect with N2 plasma, we observed that N2 plasma has much more influence on the secondary structure of Hb and Mb as compared with 200 μM H2O2. From Fig. 1, we already know that the maximum concentration of H2O2 is 60 μM for N2 plasma, but modification level is much higher. This shows that other radicals (O2−, NO, OH and ONOO−) also have a strong influence on the structural modification of proteins.
Moreover, hemes are non-fluorescent, while the heme degradation yield fluorescent porphyrin degradation product22,45,46. We have evaluated the fluorescence intensity for heme degradation at ≈460 nm. We detected that fluorescence intensity for both proteins (Hb and Mb) in N2 plasma is higher than 200 μM H2O2 (Fig. 2 and Fig. S12). Hence, H2O2 is one of the factors for heme degradation, but for plasma, we have other factors also that increase the heme degradation. Rifkind group observed that O2−/H2O2 radicals are responsible for heme degradation45,46. In another study by Huang group22 found that H2O2, OH radicals and UV radiation generated during the plasma treatment plays role in acceleration of the heme degradation for horseradish peroxidase protein. In our present study, H2O2 and OH radicals are much higher for N2 plasma as compared with Ar and Air plasma. Hence, degradation of heme group is more for N2 plasma as compared with other gas plasma.
Further, results from protein oxidation test and gel electrophoresis experiment reveals that different feeding gases can produce different levels of efficiently oxidants that generate carbonyls into proteins. Carbonyl levels in proteins can be increased by non-oxidative and oxidative mechanism33. So, if we compare the carbonyl content in both proteins due to H2O2 in Fig. 3. We observed that carbonyl content due to 60 μM H2O2 (the highest concentration of H2O2 observed during plasma treatment in all feeding gases, Fig. 1) is much less compared to all feeding gases plasma. Among the feeding gases N2 and Ar gas plasma has a high level of carbonyl content as compared with Air plasma. On the other hand, we observed that Air plasma has thicker bandwidth for both proteins, while the bandwidth is thinner for the N2 plasma and Ar plasma. These results are explained by literature results that excess ROS increase the BSA fragmentation, that results in thin bandwidth47. While, control oxidation increase the bandwidth of BSA48. Recently published work by Ye et al.50, observed that lower level of oxidation (carbonylation) could generate soluble protein aggregates that having less particle size. Whereas, higher level of oxidation would induce insoluble aggregates of protein, that results in higher molecular size. Similar results were observed for both proteins (Hb and Mb) that Air plasma has particle size is less or approximately near the control, but for N2 and Ar plasma the particle size in more. As we have discussed above that it is due to protein carbonylation (protein oxidation).
Whereas, except in N2 plasma we have not observed the quenching and red shift in UV-vis spectroscopy for soret band. In comparison with H2O2, we observed that 60 μM H2O2 has no effect on the soret band (Figs. S14c, d). Hence, the presence of other radicals such as OH and ONOO− are the main factors responsible for the red shift in soret band for N2 plasma. Our docking results also show that H2O2 gets attached and oxidizes the amino acids near the heme group, which is one of the cause for heme degradation, as supported by other groups also45,46. However, to check the amino acid modification in proteins (Hb and Mb), we have checked the NMR. We observed that peaks of both proteins are either shifted or vanished or become broad after the plasma treatment. While, same results we are not observed for 60 μM H2O2. This further indicated that other radicals that are generated in plasma with a combination of H2O2 has influenced on protein modification.
Moreover, the main cause of cancer cell death is DNA damage62. Hence, we have also checked the structural changes in ctDNA. The CD spectra shows that plasma have much more action of structural damage of DNA as compared with 60 μM H2O2. Similar way, the 8-OHdG formation is also high for plasma treated DNA, as compared to 60 μM H2O2 treated DNA. Although, among all the treated gas plasma, N2 plasma has highest oxidation level as compared to other feeding gas plasma. Therefore, all these results of both proteins (Hb and Mb) and ctDNA show that there are some changes in the amino acid structure. Hence, we have also studied amino acids and observed that after treatment either amino acids weight increases or decreases, shown in Fig. 8. There are some cases like L-alanine has no effect of plasma, but might be in the given window, we have not observed the changes. Therefore, we have checked the quantitative analysis in Table 2, where we can observe the change in amino acids after treatment with plasma. The change in the weight of amino acids after treatment is due to oxidation, nitration, dehydrogenation and dimerization as suggested by literature37,38,39,40.
Figure 8. Change in the molecular structure of Amino acids after the treatment with soft plasma.
In order to understand the mechanism of action of the plasma in different feeding gases, we considered the reactive species (RS) generated in different feeding gases to different extent. In recent work from Keider group42, they observed that the change in the plasma parameter such as voltage, flow rate and composition of gas has various effects on cells reflected by cell viability, because these parameters changes the ROS/RNS level to medium. Similarly, in our present work we have changed the feeding gases, that results in different extent of modification for proteins, DNA and amino acids. Among all the gases N2 plasma has strong action, one of the reason is that N2 plasma are excited the N2 in metastable level of 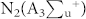 , which dissociate water molecules generating hydroxyl radicals and hydrogen atoms (detail calculation is given in supporting information) as shown in equation (1):
, which dissociate water molecules generating hydroxyl radicals and hydrogen atoms (detail calculation is given in supporting information) as shown in equation (1):
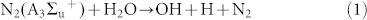 |
with dissociation coefficient63 of αHO = 5 × 10−14 cm3/s and returning back to the ground state of N2. Obviously, the water molecular dissociation due to the nitrogen molecules in the excited metastable state generates hydrogen atoms, which has some interest in the air plasma. Hydroxyl molecules meet oxygen atom by equation (2) and hydrogen atom reacts with oxygen molecules by equation (3):
 |
 |
The reaction coefficient64 of eq. 2 is αO2 = 2.4 × 10−11 exp(109/T) cm3/mole/s = 3.46 × 10−11 cm3/mole/s at T = 300 K. The main reactions of OH eliminations by equations (4) and equation (5):
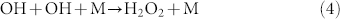 |
with its rate coefficient65 of αH2O2 = 6.83 × 10−31(Tr/T)0.8 cm6/mole2/s = 1.78 × 10−11 cm3/mole/s in air at T = 300 K and other equation (5):
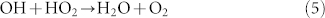 |
with its reaction coefficient64 of αOH2 = 4.8 × 10−11 exp(249/T) = 1.1 × 10−10 cm3/s. Therefore, the presence of oxygen atoms in Air plasma reduces the OH radical density, results in less concentration of H2O2 formation. Whereas, Ar plasma also reacts with water to form OH radicals and hydrogen atom, but the plasma density for the Ar is ≈1012 to 1013. While for the N2 plasma the plasma density is ≈1015, hence the generation of OH radicals in N2 plasma is much more than Ar plasma and least for the Air plasma. Due to this action, we observed from Fig. 1 that N2 plasma has highest H2O2 and OH radicals generated followed by Ar plasma and Air plasma. However, our control experiments, we observed that the structural and chemical deformation for proteins and DNA are low as compared to the concentration of H2O2 has generated during treatment. And among all the gases N2 plasma has highest modification level, this shows that OH, H2O2 and ONOO− plays an important role in the structural modification of Hb, Mb, DNA and amino acids. On the other hand, NO is maximum for Air plasma (Fig. 1 and Fig. S5), but the structural modification is very less, hence NO has no major role in structural changes. It is well that NO is removed with in second by reaction with oxyhemoglobin, during in vivo analysis66. There are many studies for binding studies of Hb and Mb with NO and their structural effects67. Whereas, the reaction between the NO and O2− produced the powerful and toxic oxidant ONOO−. Therefore, the presence of high amount of OH, H2O2 and ONOO− for N2 plasma results in high oxidation of Hb, Mb and DNA, followed by Ar plasma and least in Air plasma. As we have shown that during treatment, the pH of water becomes acidic (Fig. S6), so the peroxynitrite will be protonated to form peroxynitrous acid66. This peroxynitrous acid is very strong oxidant that can react with biomolecules and make the number of complex systems and it plays very important role in protein carbonylation (protein oxidation) and 8-OHdG product formation (DNA oxidation)66.
These results show that plasma lead oxidation can modified and oxidized the proteins, DNA and amino acids. Due to oxidation, Fe (II) oxidized to Fe (III) and also degraded the heme group. The interaction between the heme groups and peroxide cause the pseudoperoxidase activity and plays important role in pathophysiology for many diseases68. These pathologies are related to the generation of higher oxidative states of both proteins (Hb and Mb) and proteins related radical formation68,69. Moreover, oxidized forms of Hb and Mb can't be oxygenated, hence they are physiologically inactive70. The oxidative reaction of Hb protein with H2O2 plays important role in Hb mediated tissue damage71. Hence, these results are very important for the plasma medicine to understand the action of plasma on red blood cells and tissue damage studies.
Experiment section
Materials
The Myoglobin, Hemoglobin proteins, calf thymus DNA, amino acids (glycine, L-glutamic acid (Glu), L-asparagine (Asn), L-arginine (Arg), L- alanine (Ala), Threonine (Thr), L-proline andLysine (Lys)) were supplied by Aldrich Chemical Co. (USA). All chemicals andreagents were used without any further purification. The 8-OHdG quantitation was analyzed using Cell biolabs, inc, OxiSelect™ oxidative DNA damage ELISA kit. The concentrations of H2O2 were measured using an Amplex H2O2 assay kit (Invitrogen, GrandIsland, NY, USA) and NO detection assay kit (Biovision, Milpitas,CA, USA), respectively, following the manufacturer's protocols. For calculation of peroxynitrite we prepared a solution by treating acidified H2O2 with a solution of sodium nitrite followed by addition of NaOH. The concentration is indicated by the absorbance at 302 nm at pH 12, having ε = 1670 M−1 cm−1, through this we can check the concentration of the standard66. Then, pH of solution was made 12 with the addition of NaOH and treated with plasma and monitored at 302 nm (δ denotes ± 0.4).
Measurements
Quantitative and qualitative analysis of amino acids was studied using Liquid Chromatograph/Capillary Electrophoresis- Mass Spectrometer (LC/CE-MS) of Model Quattro LC Triple quadrupole mass spectrometer (Micromass & Waters), HP-1100 High Performance Liquid Chromatography (Hewlett Packard), HP-3D Capillary Electrophoresis (Hewlett Packard). Dynamic light scattering (DLS) is of Photalostuka of Zeta-potential & Particle size Analyzer ELSZ-2. They have wide size range (0.6 nm ~ 7 μm), wide concentration range (0.001% ~ 40%), for DLS we have used 1 mg/ml concentration of proteins (Hb and Mb). UV-Vis S-3100 Spectrophotometer having wavelength resolution of 0.95 nm, wavelength accuracy ±0.5 nm and wavelength reproducibility of ±0.02 nm was utilized for the analysis. The 0.5 mg/ml concentration of sample, we have used for UV-vis spectroscopy.
Fluorescence spectroscopy
The fluorescence spectroscopy instrument used for measuring fluorescence intensity in the present investigation is similar to that depicted in our earlier articles23,72,73,74. Steady-state fluorescence measurements were carried out in a Perkin Elmer LS 55 fluorescence spectrometer. The excitation wavelength was fixed at 321 nm to obtain the contribution of the degradation of heme group from the overall fluorescence emission. Slit width of excitation and emissions were set at 10 and 10 nm, respectively. The concentration for this experiment is 1 mg/ml for both proteins (Hb and Mb).
Circular dichroism spectroscopy
CD spectroscopic studies23,72,73,74 were performed using a J-815 spectrophotometer (Jasco, Japan) equipped with a Peltier system for controlling the temperature. (1S)-(+)-10-camphorsulfonic acid (Aldrich, Milwaukee, WI) was utilised for CD calibrations, exhibiting molar extinction coefficient 34.5 M/cm at 285 nm, and 2.36 M/cm molar ellipticity (θ) at 295 nm. Samples were pre-equilibrated at the desired temperature for 15 min and the scan speed was fixed for adaptative sampling (error F 0.01) with a response time of 1 s and 1 nm bandwidth. The secondary structures of Hb and Mb were monitored by using 1.0 cm path length cuvette. The concentrations for secondary structures of Hb and Mb were 0.1 mg/ml, each spectrum being an average of six spectra. Each sample spectrum was obtained by subtracting appropriate blank media without Hb and Mb from the experimental proteins spectrum. The percentages of secondary structures were then calculated by using K2D3 online software. To measure the Tm value (The higher the transition midpoint (Tm) when 50% of the biomolecules are unfolded, the more stable the molecule) from CD, we have used 1 mg/ml concentration of the Hb and Mb.
Differential scanning calorimeter (DSC)
A differential scanning calorimeter VP-DSC was used to measure the melting point (Tm) for Hb and Mb in the water. A biomolecules in solution is in equilibrium between the native (folded) and its denatured (unfolded) state. The bubble-free solution was placed into a reference and sample cells are fixed in place lollipop shaped vessels with effective volumes of approximately 0.5 ml and sealed using a press. The approximate volume of each cell stem was 0.5 ml. The concentration of the proteins were 2 mg/ml of Hb and Mb. The DSC operates in the temperature range of 1°C to 110°C. The cells were allowed to stabilize at 20°C inside the calorimeter before heating up to 100°C with a scanning rate of 1.0°C/min. All DSC measurements were carried out in triplicate. The uncertainties of temperature and heat flow readings are ±0.02°C and ±0.1%, respectively. To obtain accurate results, the instrument was calibrated with pure water and sapphire. Each value is the average over three measurements. The error in Tm does not exceed ±0.1°C.
Determination of Hb and Mb protein oxidation
Oxidation of proteins (Hb and Mb) were determined using methods reported earlier75. Cold trichloroacetic acid was used to precipitate Hb and Mb and the final concentration being 10% (v/v). Samples were centrifuged at 11,000 g for 3 min after a 10 min incubation period at 4°C, followed by resuspension of the Hb and Mb pellet in 0.5 ml of 10 mM 2,4-dinitrophenylhydrazine (DNPH)/2 M HCl. After that, samples were placed in a sample holder, vortexed continuously at room temperature for 1 h and then precipitated with 0.5 ml of 20% TCA followed by centrifugation at11,000 g for 3 min. To remove free DNPH reagent, the pellet was washed with 1 ml of ethanol-ethyl acetate (1:1(v/v)) and then allowed to stand for 10 min. Further, the sample was centrifuged for 5 min at 11,000 g and the supernatant was then discarded. The washing procedure was repeated twice. After completing the above steps, the resulting Hb and Mb pellets were resuspended in 1.0 ml of 6 M guanidine with 2 mM potassium phosphate buffer (pH 2.3, adjusted with trifluoroacetic acid) and now samples were incubated at 37°C for 15–30 min. Samples having difficulties going into solutions were briefly sonicated and incubated at an increased temperature of upto 70°C. In order to remove any insoluble material remaining in the suspension, all samples were then centrifuged. The concentration of DNPH was determined at its maximum wavelength of 360 nm and the levels of proteins oxidation were quantified by using the molar absorption coefficient of 22, 000 M−1 cm−1. Protein oxidation content was expressed as % in comparison to control.
In silico molecular modeling and docking parameters
To explore the bioactive site of H2O2 with amino acids of Hb and Mb, the Sybyl × 2.0 interfaced with Surflex-Dock module was exploited for molecular docking. The ligand is automatically docked into binding pocket of a target protein by using protomol-based algorithm and empirically produced scoring function. The X-ray crystallographic structures of Hb (PDB:1A3N)76 and Mb (PDB:3RGK)77 protein receptor were taken from the protein data bank (PDB) and modified further for docking calculations. During protein preparation, co-crystallized ligand, water molecules were removed from the structure, and H atoms were added without disturbing the side chains. Protein structure minimization was performed by applying Tripos force field and Gasteiger-Huckel method was used to calculate the partial atomic charges. In the reasonable binding pocket, all the compounds were docked into the binding pocket and 20 possible active docking conformations with different scores were obtained for each compound. During the docking process, default values were taken for all of the other parameters78,79.
Atmospheric Pressure Plasma Device (APPJ)
The soft plasma comprised of components mainly, electrodes, dielectrics, and a high-voltage power supply72,73,74. Voltage of 60 Hz at 60 V was input into a commercial transformer, resulting in an output frequency of approximately 8 kHz. The Air, N2 and Ar gas (99.99% pure) flow rate was 1.0 lpm, and a plasma jet plume was ejected into the open air through a 1 mm hole. The OES spectra of the APPJ emission were recorded by the use of HR4000CG-UV-NIR (Ocean Optics, FL, USA) and optical fiber (QP400-2-SR) with a diameter of 400 mm, in the humid free atmosphere (The vacuum chamber is fabricated to study the activity in a closed environment). UV over a wide wavelength range 200–1100 nm without solution. The signal was accumulated for 3 min, and the data was analyzed using the Origin 8.0 software package. The emission spectra was recorded as illustrated in Figs. S2–S5.
pH and temperature measurement
After exposure of plasma for 3 min in water, the pH and temperature of the water was measured using a pH meter (Eutech Instruments, Singapore) and Infrared (IR) camera (Fluke Ti100 Series Thermal Imaging Cameras, UK). All measurements were carried out in triplicate.
NMR studies
Spectra were recorded on a Bruker DRX 500 MHz NMR spectrometer equipped with a cryoprobe at 25°C. The spectra are the result of 1000 scans, 1 s (delay time) at 25°C, and were processed with Bruker Topspin version 2.1. The concentration of Hb and Mb in all solutions used for NMR spectroscopy was 4 mg/ml. About 10% of 2H2O was added to provide an internal-field-frequency lock signal80,81.
Sample Preparation
Protein stability was analysed by incubating 2 ml screw-capped vials in deionized water, at 25°C for 4 h to attain complete equilibrium. The samples were treated at 6 mm distances from the soft plasma tip for 3 min, then incubated for 4 h at room temperature. The concentration of proteins after the plasma treatment is determined by the Bradford method82. Three samples were treated for each condition to minimize the error.
Statistical analysis
All values are represented by the mean ± S.D of the indicated number of replicates. Statistical analyses of the data were performed using student's t-test to establish significance between data points, and significant differences have been based on the P < 0.05 or P < 0.01.
Author Contributions
P.A. conceived and designed the experiments, analyzed the data,wrote the manuscript. P.A., N.K. and J.H.P. performed research. D.K.Y. attempted insilico drug design approaches. W.T. and S.C. attempted and analyzed the NMR data. W.T., H.S.U., I.T.K. and E.H.C. supervised the study/project, provided assistance in design the plasma setup and provides the OES spectra measurement characterization.
Supplementary Material
Supporting Information
Acknowledgments
We gratefully acknowledge SRC program of National Research Foundation of Korea (NRF) Grant funded by the Korean Government (MEST) (No. 2010-0027963) and in part by Kwangwoon University 2014. This work was also supported by Mid-career Researcher Program (NRF-2013R1A2A2A01068963) through NRF grant funded by the MEST and in part by the Brain Korea 21(BK21) PLUS program (Sooho Choi, Yonsei U.).
References
- Volotskova O. et al. Targeting the cancer cell cycle by cold atmospheric plasma. Sci Rep 2, 636 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratovitski E. A. et al. Anti-Cancer Therapies of 21st Century: Novel Approach to Treat Human Cancers Using Cold Atmospheric Plasma. Plasma Process Polym 10.1002/ppap.201400071 (2014). [DOI] [Google Scholar]
- Kirson E. D. et al. Alternating electric fields arrest cell proliferation in animal tumor models and human brain tumors. Proc Natl Acad Sci USA 104, 10152–10157 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staack D. et al. Nanoscale Corona Discharge in Liquids, Enabling Nanosecond Optical Emission Spectroscopy. Angew Chem Int Ed 47, 8020–8024 (2008). [DOI] [PubMed] [Google Scholar]
- Ling L. et al. Effects of cold plasma treatment on seed germination and seedling growth of soybean. Sci Rep 4, 5859 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmann K. D. & von Woedtke T. Campus PlasmaMed-From Basic Research to Clinical Proof. IEEE Trans Plasma Sci 39, 1015–1025 (2011). [Google Scholar]
- Yan X. et al. Plasma-Induced Death of HepG2 Cancer Cells: Intracellular Effects of Reactive Species. Plasma Process Polym 9, 59–66 (2012). [Google Scholar]
- Ehlbeck J. et al. Low temperature atmospheric pressure plasma sources for microbial decontamination. J Phys D: Appl Phys 44, 013002–013020 (2011). [Google Scholar]
- Kang S. K. et al. Reactive hydroxyl radical-driven oral bacterial inactivation by radio frequency atmospheric plasma. Appl Phys Lett 98, 143702 (2011). [Google Scholar]
- Bai N. et al. Inactivation of Staphylococcus aureus in Water by a Cold, He/O2 Atmospheric Pressure Plasma Micro jet. Plasma Process Polym 8, 424–431 (2011). [Google Scholar]
- Stoffels E. et al. Delayed effects of cold atmospheric plasma on vascular cells. Plasma Process Polym 5, 599–605 (2008). [Google Scholar]
- Bogle M. A. et al. Evaluation of Plasma Skin Regeneration Technology in Low-Energy Full-Facial Rejuvenation. Arch Dermatol 143, 168–174 (2007). [DOI] [PubMed] [Google Scholar]
- Attri P. et al. Influence of nanosecond pulsed plasma on the non-enzymatic pathway for the generation of nitric oxide from L-arginine and the modification of graphite oxide to increase the solar cell efficiency. Phys Chem Chem Phys 16, 18375–18382 (2014). [DOI] [PubMed] [Google Scholar]
- Brulle L. et al. Effects of a Non Thermal Plasma Treatment Alone or in Combination with Gemcitabine in a MIA PaCa2-luc Orthotopic Pancreatic Carcinoma Model. PLoS ONE 7, e52653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme M. et al. ROS implication in a new antitumor strategy based on non-thermal plasma. Int J Cancer 130, 2185–2194 (2012). [DOI] [PubMed] [Google Scholar]
- O'Connel D. et al. Cold Atmospheric Pressure Plasma Jet Interactions with Plasmid DNA. Appl Phys Lett 98, 043701–043703 (2011). [Google Scholar]
- Ptasinska S. et al. DNA strand scission induced by a non-thermal atmospheric pressure plasma jet. Phys Chem Chem Phys 28, 7779–7781 (2010). [DOI] [PubMed] [Google Scholar]
- Li G. et al. Genetic effects of radio-frequency, atmospheric-pressure glow discharges with helium. Appl Phys Lett 92, 221504–221507 (2008). [Google Scholar]
- Han X. et al. Plasmid DNA damage induced by helium atmospheric pressure plasma jet. Eur Phys J D 10.1140/epjd/e2014-40753-y (2014). [DOI] [Google Scholar]
- Yan X. et al. Effect of the atmospheric pressure nonequilibrium plasmas on the conformational changes of plasmid DNA Appl. Phys. Lett. 95, 083702 (2009). [Google Scholar]
- Takai E. et al. Protein Inactivation by Low-temperature Atmospheric Pressure Plasma in Aqueous Solution. Plasma Processes Polym 9, 77–82 (2012). [Google Scholar]
- Ke Z. & Huang Q. Inactivation and Heme Degradation of Horseradish Peroxidase Induced by Discharge. Plasma Plasma Processes Polym 10, 731–739 (2013). [Google Scholar]
- Jha I. et al. Unexpected effects of the alteration of structure and stability of myoglobin and hemoglobin in ammonium-based ionic liquids. Phys Chem Chem Phys 16, 5514–5526 (2014). [DOI] [PubMed] [Google Scholar]
- Katz D. S. et al. Structure determination of aquomet porcine hemoglobin at 2.8 Å resolution. J Mol Biol 244, 541–553 (1994). [DOI] [PubMed] [Google Scholar]
- Kendrew J. C. et al. Structure of Myoglobin: A Three-Dimensional Fourier Synthesis at 2 Å. Resolution. Nature 185, 422–427 (1960). [DOI] [PubMed] [Google Scholar]
- Olive P. L. The role of DNA single- and double-strand breaks in cell killing by ionizing radiation. Radiat Res 150, S42–S51 (1998). [PubMed] [Google Scholar]
- McKinnon P. J. & Caldecott K. W. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet 8, 37–55 (2007). [DOI] [PubMed] [Google Scholar]
- Nowicka A. M. et al. Oxidation of DNA Followed by Conformational Change after OH Radical Attack. Anal Chem 85, 355–361 (2013). [DOI] [PubMed] [Google Scholar]
- Wei H. et al. Tamoxifen reduces endogenous and UV light-induced oxidative damage to DNA, lipid and protein in vitro and in vivo. Carcinogenesis 19, 1013–1018 (1998). [DOI] [PubMed] [Google Scholar]
- Gao D. et al. Benzo[a]pyrene and its metabolites combined with ultraviolet A synergistically induce 8-hydroxy-2V-deoxyguanosine via reactive oxygen species. Free Radical Biol Med 39, 1177–1183 (2005). [DOI] [PubMed] [Google Scholar]
- Abolfath R. M. et al. Multiscale QM/MM Molecular Dynamics Study on the First Steps of Guanine Damage by Free Hydroxyl Radicals in Solution. J Phys Chem A 116, 3940–3945 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abolfath R. M. et al. A molecular dynamics simulation of DNA damage induction by ionizing radiation. Phys Med Biol 58, 7143–7157 (2013). [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I. et al. Protein carbonylation, cellular dysfunction, and disease progression. J Cell Mol Med 10, 389–406 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsrud P. A. et al. Oxidative Stress and Covalent Modification of Protein with Bioactive Aldehydes. J Biol Chem 283, 21837–21841 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman E. R. Role of Oxidant Species in Aging. Curr Med Chem 11, 1105–1112 (2004). [DOI] [PubMed] [Google Scholar]
- Stadtman E. R. & Levine R. L. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 25, 207–218 (2003). [DOI] [PubMed] [Google Scholar]
- Ke Z. G. et al. A paradigm study for assessment of phenylalanine's damage under arc-discharge irradiation. Nucl InstrumMethods Phys Res B 268, 1618–1625 (2010). [Google Scholar]
- Ke Z. G. et al. A study of low-energy ion induced radiolysis of thiol-containing amino acid cysteine in the solid and aqueous solution states. Nucl Instrum Methods Phys Res B 268, 2729–2734 (2010). [Google Scholar]
- Ke Z. G. et al. Assessment of damage of glutathione by glow discharge plasma at the gas solution interface through Raman spectroscopy. Plasma Process Polym 10, 181–188 (2013). [Google Scholar]
- Takai E. et al. Chemical modification of amino acids by atmospheric-pressure cold plasma in aqueous solution. J Phys D: Appl Phys 47, 285403 (2014). [Google Scholar]
- Kanazawa S. et al. Observation of OH radicals produced by pulsed discharges on the surface of a liquid. Plasma Sources Sci. Technol. 20, 034010 (2011). [Google Scholar]
- Baik K. Y. et al. Feeding-gas effects of plasma jets on escherichia coli in physiological solutions. Plasma Process Polym 10, 235–242 (2013). [Google Scholar]
- Machala Z. et al. Emission spectroscopy of atmospheric pressure plasmas for bio-medical and environmental application. J Mol Spectrosc 243, 194–201 (2007). [Google Scholar]
- Cheng X. et al. The Effect of Tuning Cold Plasma Composition on Glioblastoma Cell Viability. PLoS ONE 9, e98652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagababu E. & Rifkind J. M. Formation of Fluorescent Heme Degradation Products during the Oxidation of Hemoglobin by Hydrogen Peroxide. Biochem Biophys Res Commun 247, 592–596 (1998). [DOI] [PubMed] [Google Scholar]
- Nagababu E. & Rifkind J. M. Reaction of Hydrogen Peroxide with Ferrylhemoglobin: Superoxide Production and Heme Degradation. Biochemistry 39, 12503–12511 (2000). [DOI] [PubMed] [Google Scholar]
- Xiao H. et al. Novel physiological properties of ethanol extracts from Eremurus chinensis Fedtsch. roots: in vitro antioxidant and anticancer activities. Food Funct 3, 1310–1318 (2012). [DOI] [PubMed] [Google Scholar]
- Chesne S. et al. Effects of oxidative modifications induced by the glycation of bovine serum albumin on its structure and on cultured adipose cells. Biochimie 88, 1467–1477 (2006). [DOI] [PubMed] [Google Scholar]
- Bee J. S. et al. Aggregation of a monoclonal antibody induced by adsorption to stainless steel. Biotechnol Bioeng 105, 121–129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L. et al. Effect of Protein Oxidation on the Conformational Properties of Peanut Protein Isolate. Journal of Chemistry 2013, Article ID 423254 (2013). [Google Scholar]
- Coelho F. R. et al. Oxidation of the Tryptophan 32 Residue of Human Superoxide Dismutase 1 Caused by Its Bicarbonate dependent Peroxidase Activity Triggers the Non-amyloid Aggregation of the Enzyme. J Biol Chem 289, 30690–30701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. E. et al. Monitoring fast reactions by spatially-selective and frequency-shifted continuous NMR spectroscopy: application to rapid-injection protein unfolding. Chem Commun 49, 3155–3157 (2013). [DOI] [PubMed] [Google Scholar]
- Sogbein O. O. et al. Effects of pH on the kinetic reaction mechanism of myoglobin unfolding studies by time-resolved electrospray ionization mass spectrometry. J Am Soc Mass Spectrom 11, 312–319 (2000). [DOI] [PubMed] [Google Scholar]
- Uzawa T. et al. Hierarchical folding mechanism of apomyoglobin revealed by ultra-fast H/D exchange coupled with 2D NMR. Proc Natl Acad Sci USA 105, 13859–13864 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juillard S. et al. Dynamics of heme in hemoproteins: proton NMR study of myoglobin reconstituted with iron 3-ethyl-2-methylporphyrin. Biochimica et Biophysica Acta 1814, 1188–1194 (2011). [DOI] [PubMed] [Google Scholar]
- Juillard S. et al. Proton NMR study of myoglobin reconstituted with 3,7-diethyl-2,8-dimethyl iron porphyrin: Remarkable influence of peripheral substitution on heme rotation. J Inorg Biochem 100, 1441–1448 (2006). [DOI] [PubMed] [Google Scholar]
- De Bont R. & Van Larebeke N. Endogenous DNA damage in humans: A review of quantitative data. Mutagenesis 19, 169–185 (2004). [DOI] [PubMed] [Google Scholar]
- Maheswari P. U. & Palaniandavar M. DNA binding and cleavage properties of certain tetrammineruthenium(II) complexes of modified 1,10 phenenthrolines-effect of hydrogen-bonding on DNA-binding affinity. J Inorg Biochem 98, 219–230 (2004). [DOI] [PubMed] [Google Scholar]
- Amici A. et al. Conversion of amino acid residues in proteins and amino acid homopolymers to carbonyl derivatives by metal-catalyzed oxidation reactions. J Biol Chem 264, 3341–3346 (1989). [PubMed] [Google Scholar]
- Dalle-Donne I. et al. Protein carbonylation in human diseases. Trends Mol Med 9, 169–176 (2003). [DOI] [PubMed] [Google Scholar]
- Berger P. et al. Ozone and Hydroxyl Radicals Induced Oxidation of Glycine. Wat Res 33, 433–441 (1999). [Google Scholar]
- Kumar N. et al. Induced apoptosis in melanocytes cancer cell and oxidation in biomolecules through deuterium oxide generated from atmospheric pressure non-thermal plasma jet. Sci Rep (2014) In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Herron J. T.
Evaluated chemical kinetics data for reactions of N(2D), N(2P), and
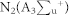 in the gas phase. J Phys Chem Ref Data
28, 1453–1483 (1999). [Google Scholar]
in the gas phase. J Phys Chem Ref Data
28, 1453–1483 (1999). [Google Scholar] - Atkinson R. et al. Evaluated kinetic and photochemical data for atmospheric chemistry: Volume I - gas phase reactions of Ox, HOx, NOx and SOx species. Atmos Chem Phys 4, 1461–1738 (2004). [Google Scholar]
- Forster R. et al. High pressure range of the addition of HO to HO, NO, NO2, and CO. I. Saturated laser induced fluorescence measurements at 298 K. J Chem Phys 103, 2949 (1995). [Google Scholar]
- Beckman J. S. & Koppenol W. H. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J Physiol 271, C1424–C1437 (1996). [DOI] [PubMed] [Google Scholar]
- Kim S. & Lim M. Protein conformation-controlled rebinding barrier of NO and its binding trajectories in myoglobin and hemoglobin at room temperature. J Phys Chem B 24, 5819–5830 (2012). [DOI] [PubMed] [Google Scholar]
- Lu N. et al. Anti- and pro-oxidant effects of (+)-catechin on hemoglobin-induced protein oxidative damage. Toxicology in Vitro 25, 833–838 (2011). [DOI] [PubMed] [Google Scholar]
- Alayash A. I. et al. Redox reactions of hemoglobin and myoglobin: biological and toxicological implications. Antioxid Redox Signaling 3, 313–327 (2001). [DOI] [PubMed] [Google Scholar]
- Shikama K. et al. The molecular mechanism of autoxidation for myoglobin and hemoglobin: A venerable puzzle. Chem Rev 98, 1357–1374 (1998). [DOI] [PubMed] [Google Scholar]
- Alayash A. I. et al. Reactions of Sperm Whale Myoglobin with Hydrogen Peroxide: Effects of distal pocket mutations on the formation and stability of the ferryl intermediate. J Biol Chem 274, 2029–2037 (1999). [DOI] [PubMed] [Google Scholar]
- Attri P. et al. Activity and Stability of α-Chymotrypsin in Biocompatible Ionic Liquids: Enzyme Refolding by Triethyl Ammonium Acetate. Phys Chem Chem Phys 13, 2788–2796 (2011). [DOI] [PubMed] [Google Scholar]
- Attri P. & Choi E. H. Influence of Reactive Oxygen Species on the Enzyme Stability and Activity in the Presence of Ionic Liquids. PloS One 8, e75096 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attri P. et al. TMAO and Sorbitol Attenuate the Deleterious Action of Atmospheric Pressure Non-Thermal Jet Plasma on α-Chymotrypsin. RSC Adv 2, 7146–7155 (2012). [Google Scholar]
- Yan L. J. & Forster M. J. Chemical probes for analysis of carbonylated proteins: a review. J Chromatogr B Analyt Technol Biomed Life Sci 879, 1308–1315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tame J. R. & Vallone B. The structures of deoxy human haemoglobin and the mutant Hb Tyralpha42His at 120 K. ActaCrystallogr D BiolCrystallogr 56, 805–811 (2000). [DOI] [PubMed] [Google Scholar]
- Hubbard S. R. et al. X-ray crystal structure of a recombinant human myoglobin mutant at 2.8 Å resolution. J Mol Biol 213, 215–218 (1990). [DOI] [PubMed] [Google Scholar]
- Yadav D. K. et al. QSAR and docking based semi-synthesis and in vivo evaluation of artemisinin derivatives for antimalarial activity. Current Drug Target 15, 753–761 (2014). [DOI] [PubMed] [Google Scholar]
- Yadav D. K. et al. QSAR and docking studies on Chalcone derivatives for anti-tubercular activity against M. tuberculosis H37Rv. Journal of Chemometrics 28, 499–507 (2014). [Google Scholar]
- Sung H. J. et al. Inhibition of human neutrophil activity by an RNA aptamer bound to interleukin-8. Biomaterials 35, 578–589 (2014). [DOI] [PubMed] [Google Scholar]
- Yun J. H. et al. Solution Structure and Rpn1 Interaction of the UBL Domain of Human RNA Polymerase II CTerminalDomain Phosphatase. PLoS ONE 8, e62981 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information