Abstract
Primary brain tumors are the fourth leading cause of cancer mortality in adults under the age of 54 years and the leading cause of cancer mortality in children in the United States. Therapy for the most common type of primary brain tumors, gliomas, remains suboptimal. The development of new and more effective treatments will likely require a better understanding of the biology of these tumors. Here, we show that use of the high-density 100K single-nucleotide polymorphism arrays in a large number of primary tumor samples allows for a much higher resolution survey of the glioma genome than has been previously reported in any tumor type. We not only confirmed alterations in genomic areas previously reported to be affected in gliomas, but we also refined the location of those sites and uncovered multiple, previously unknown regions that are affected by copy number alterations (amplifications, homozygous and heterozygous deletions) as well as allelic imbalances (loss of heterozygosity/gene conversions). The wealth of genomic data produced may allow for the development of a more rational molecular classification of gliomas and serve as an important starting point in the search for new molecular therapeutic targets.
Introduction
A major problem in the quest for better therapeutics for patients with gliomas has been the relative poor understanding of glioma biology. Most of the genomic alterations described in gliomas (1, 2), with the notable exception of PTEN (3), are alterations also described in the more common epithelial tumors [epidermal growth factor receptor (EGFR), CDKN2A, TP53, and RB1]. Targeted therapies (e.g., erlotinib) directed to ubiquitously expressed targets, such as the EGFR, have met with limited success and significant toxicity (4, 5), demonstrating the need for the identification of glioma-specific genomic alterations.
Prior efforts to perform a global genomic survey of gliomas have generally used standard comparative genomic hybridization (CGH) techniques (6–9). Although useful for identifying large areas of genomic alterations, the relatively low resolution of standard CGH precludes the identification of small areas of alteration. Furthermore, it has been difficult to understand the overall significance of the findings from prior genomic surveys secondary to the relatively low number of tumor samples analyzed in these studies (8, 9). Thus, we currently have only a very general picture of the gross genomic alterations found in a small number of gliomas.
To address the problem of relatively small sample numbers in prior attempts at genetically characterizing gliomas, we have created a National Cancer Institute–funded national, multi-institutional project aimed at collecting and molecularly characterizing (at both RNA, DNA, and protein level) a large number of gliomas with corollary clinical data, which we call the Glioma Molecular Diagnostic Initiative. As part of the first phase of Glioma Molecular Diagnostic Initiative, we have analyzed 178 tumors for genomic alterations using Affymetrix 100K single-nucleotide polymorphism (SNP) arrays (10). This array relies on the detection of the alleles present in a certain sample by hybridization of enzyme-modified, PCR-amplified genomic DNA to 40 probes specific for a single SNP. By combining the results of the two chips included in the assay (one each for HindIII- and XbaI-restricted DNA), an unprecedented resolution of 25 kb can be achieved. Although these types of arrays have been primarily used in familial pedigree genotyping studies, we believe that, by using both allelic calls and signal intensity, they can be successfully used for genomic survey studies in nonfamilial diseases such as cancer.
Materials and Methods
Samples
Two hundred and forty gliomas from the Hermelin Brain Tumor Center, Departments Neurology and Neurosurgery at the Henry Ford Hospital, were analyzed in this study. The samples were provided as snap-frozen sections of areas immediately adjacent to the region used for the histopathologic diagnosis. The tumor diagnosis was kept unknown to the investigators until analysis of data was complete to avoid bias. Several tumor samples were excluded from the final analyses secondary to inconclusive pathology, reducing the total number of valid cases to 178 tumor samples (356 chips). This set included 82 glioblastomas, 33 astrocytomas, 52 oligodendrogliomas, and 11 oligoastrocytomas (mixed) tumors. Twenty-nine nontumor samples (temporal lobe resection of epileptic patients) were analyzed concurrently to provide a baseline on the accuracy of the methodology.
DNA extraction and hybridization
Approximately 50 µg tissue (as recommended by the manufacturer) from each tumor were used to extract high molecular weight, genomic DNA using QIamp micro DNA extraction kit (Qiagen, Valencia, CA) following the instructions of the manufacturer. The quality of DNA was checked by electrophoresis run in a 1% agarose gel. Two hundred and fifty nanograms DNA were processed for hybridization on the Genechip Human Mapping 100K (10) arrays (Affymetrix, Inc., Santa Clara, CA), which covers 116,204 single-nucleotide polymorphism (SNP) loci in the human genome with a mean intermarker distance of 23.6 kb. The processing was done according to recommendations of the manufacturer. After hybridization, the chips were processed using fluidics station 450, high-resolution microarray scanner 3000, and GCOS workstation version 1.2. SNP calls were determined by GDAS version 3.0 (11) with 25% level of confidence. Only samples with call rates >90% were accepted for the analysis.
RNA extraction and hybridization
Approximately 50 to 80 mg of tissue from each tumor were used to total RNA using the Trizol reagent (Invitrogen, Carlsbad, CA), following the instructions of the manufacturer. The quality of RNA obtained was verified with the Bioanalyzer System (Agilent Technologies, Palo Alto, CA; ref. 12) using the RNA Pico Chips. Six micrograms of RNA were processed for hybridization on the Genechip Human Genome U133 Plus 2.0 Expression arrays (Affymetrix; ref. 13), which contains over 54,000 probe sets analyzing the expression level of over 47,000 transcripts and variants, including 38,500 well-characterized human genes. The processing was done according to recommendations of the manufacturer. After hybridization, the chips were processed using fluidics station 450, high-resolution microarray scanner 3000, and GCOS workstation version 1.3. Expression levels were determined by GCOS version 1.3, which uses the MAS5 algorithm. Expression values were scaled to a mean of 500, and only samples with a scaling factor <5, present call rates >35%, and a glyceraldehyde-3-phosphate dehydrogenase 3′/5′ ratio <3, were accepted for the analysis (14).
Loss of heterozygosity analysis
Loss of heterozygosity (LOH) scores for each SNP were calculated based on probability of LOH algorithm implemented in Affymetrix Chromosome Copy Number Tool (CCNT; ref. 15). The algorithm uses probabilities for each SNP to be homozygous derived from a reference set of 145 normal individuals. Those values are then multiplied for a stretch of homozygous SNPs giving the probability of such a stretch to happen randomly. LOH scores are −log 10 of the result. LOH scores were calculated in Matlab 7R14 and the algorithm were slightly modified to eliminate an innate error dealing incorrectly with “no calls.” For this analysis, we decreased the level of confidence for SNP calls to 0.1 to improve the scoring, by removal of less stringent heterozygous calls.4 LOH areas with a score larger or equal to 10 were considered for further analysis.
Copy number determination
To analyze the copy number changes, CCNT (Affymetrix) was used to produce a ratio between tumor samples and a normal reference set, corrected to a baseline of 2. This output was then smoothed using Hidden Markov Model algorithms in R (16, 17). This further step allowed for a clearer resolution of DNA deletions and amplifications in a high-throughput analysis, so that it could be determined which losses result from heterozygous versus homozygous deletions.
Expression/copy number correlation analysis
To correlate RNA expression with DNA copy number, each HG-U133_Plus_2 probe set was mapped against all the SNPs located in a window of 1 Mbp around the center of that probe set. The average copy number for those SNPs was calculated from the original CNAT copy numbers, and correlation coefficients and significance (P) between that average and the mRNA expression value for the associated probe set were calculated using Matlab. Positions of both expression and SNP probe sets were obtained from the NetAffx annotation website.5
EGFR immunohistochemistry
Protein expression levels for EGFR were determined by immunohistochemical staining of a tissue microarray containing all the samples used in the study. The staining was done as previously described (18). Briefly, tissue microarray sections were deparaffinized and rehydrated in PBS. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in PBS/0.05% Tween 20 for 20 minutes. Sections were then washed in PBS and blocked for 20 minutes in goat serum diluted to 10% in PBS. Pretreatment consisted of 0.025% trypsin placed on the tissue followed by a 30-minute incubation at room temperature. The EGFR mouse monoclonal (clone 528, Oncogene Science, Cambridge, MA) antibody was diluted 1:50 in PBS/10% serum, and applied to the sections in a humid chamber overnight at 4°C. After washing twice to thrice in PBS, secondary antibodies were applied using the DAKO (Carpinteria, CA) Envision kit according to the instructions from the manufacturer. Detection of bound secondary antibody was done with diaminobenzidine under microscope until optimal staining. Sections were then counterstained with light hematoxylin and mounted.
Results
Validation of the technology
We first set out to validate our methodology given that the Affymetrix 100K SNP chip is a new and unproven technology for genomic surveying. As a first step in this validation process, we tried to determine if the pool of normal samples provided with Affymetrix CCNT (15) was providing a stable baseline for our calculations. To this end, we hybridized a set of 29 nontumor brain tissue samples to the arrays and analyzed their genomic integrity by means of the aforementioned algorithm. With some recently reported exceptions (19), it is widely accepted that most of the genome of a “normal” sample should be diploid. Our analysis of the copy numbers for every SNP in the nontumor set showed that this assumption was correct in most cases; however, some discreet SNPs (~ 30 single and grouped) were consistently showing alterations in our nontumor controls at the same high levels as the tumor samples, suggesting either a hybridization specificity problem or a tissue-specific amplification event. In any case, we decided to exclude these aberrant SNPs from our analysis of the tumor samples because the copy number alterations (CNA) detected were also represented in the nontumor controls, suggesting that they were not relevant to the tumor biology, thus avoiding the possibility of reporting false positives.
Although the CNA we found were specific for the tumor samples and were not seen in the nontumor tissues, we decided to perform a second level of validation of the results by determining whether the SNP-derived alterations could be detected by established and accepted methodologies. To this end, real-time PCR copy number analysis (20) was done using the genomic DNA from a number of samples in three separate regions (EGFR, CDKN2A, and KIF21B). The results obtained (Supplementary Table S1) show consistency between the two techniques, with the SNP analysis and the real-time PCR techniques revealing nearly identical copy number levels.
Although the results from the real-time PCR and the SNP arrays we examined were consistent, it was not possible to perform this type of analysis for every area where alterations were detected. Thus, to further validate the results obtained by our SNP analyses, we analyzed the correlation found between the SNP-derived CNAs and mRNA expression levels for probe sets located in the vicinity of the altered SNPs. Overall, we found a significantly high degree of correlation between genomic alterations and mRNA expression of the neighboring genes (Fig. 1). In addition to validating the findings from the SNP analysis, the correlation between CNA and mRNA expression of neighboring genes suggests the possibility of genomic changes with biological significance.
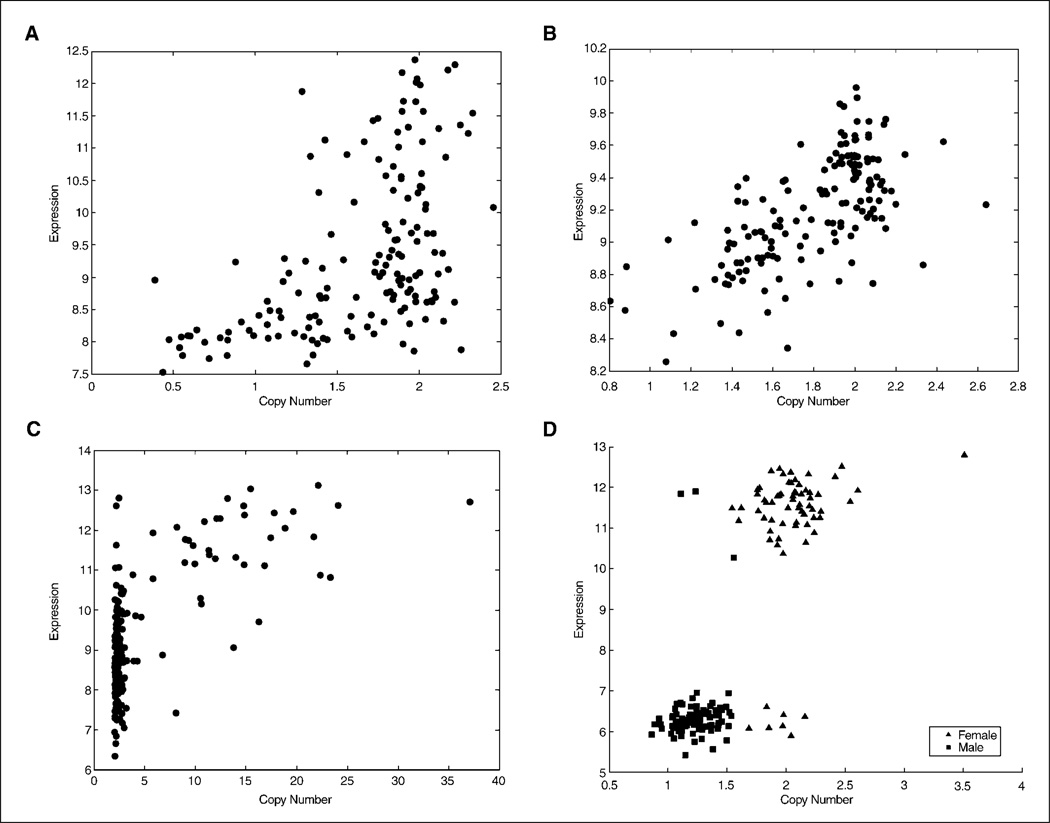
Figure 1.
Representative diagrams of correlation between SNP copy numbers and mRNA expression for neighboring gene probe sets. A, CDKN2A (homozygous and heterozygous deletion). B, PTEN (heterozygous deletion). C, EGFR (amplified). D, XIST (gender specific). mRNA expression units are arbitrary.
In addition to validating our SNP analysis–derived CNA through comparisons with findings published in the literature, real-time PCR data, and microarray gene expression data, we also evaluated the correlation between the level of tumor tissue protein expression of EGFR and the corresponding CNA of the EGFR gene. For this analysis, we created a tissue microarray consisting of all of the gliomas used in this study and stained for the EGFR receptor as previously described (18). All 38 tumors showing genomic amplification in the EGFR region in our SNP analysis also showed strong immunostaining of the EGFR protein in the tissue array, further substantiating the validity of the CNA assessment. A number of samples also showed EGFR staining without genomic alterations, consistent with previous reports indicating that EGFR overexpression can be produced by transcriptional control. Representative images of the immunohistochemical staining are shown in Supplementary Fig. S2.
Data smoothing and preparation
Despite removal of the aforementioned aberrant SNPs, we found the output from CCNT is fraught with high background noise. To identify the regions of CNAs more clearly, we decided to smooth the data using a common tool in CGH analysis, namely the Hidden Markov Model algorithms (16, 21). As shown in Supplementary Fig. S1, this process is necessary for optimal data interpretation. Without the smoothing process, small deletions like the ones shown in Supplementary Fig. S1B would likely be dismissed in view of the high level of background noise. Furthermore, although larger deletions or amplifications (Supplementary Fig. S1A and C) can be detected with the original CCNT output, our ability to determine the region boundaries was greatly improved after Hidden Markov Model processing.
Although the smoothing process greatly reduces the innate noise level of the technology, the high resolution of the arrays still produces such a large number of CNA/LOH hits that minor changes may obscure more relevant findings (Figs. 2–4). In fact, in the case of heterozygous deletions and LOH analysis, this phenomenon reaches a level such that the alterations are seen as almost a continuum across the genome. Thus, we used thresholding as a way to remove this background layer of noise to reveal the relevant genomic alterations. We chose to use a 10% threshold (a common cutoff used for LOH analysis) of the samples (both as a whole and segregated by tumor histologic subtype) as our minimal threshold for identification of an alteration.
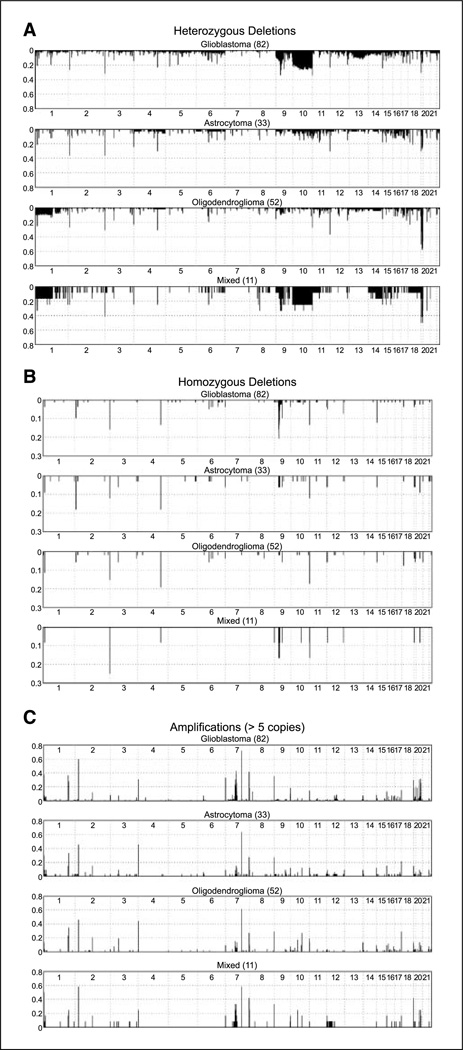
Figure 2.
Detailed histogram representation of heterozygous DNA (A) and homozygous (B) deletions as well as amplifications at the level of five copies or more (C) found segregated by histopathologic diagnosis. Y axis, ratio of samples showing the alteration at a given SNP. Numbers below each subpanel, chromosome number.
Figure 4.
Detailed histogram representing LOH with a pLOH > 10 segregated by histopathologic diagnosis. Y axis, ratio of samples showing the alteration at a given SNP. Numbers below, chromosome number.
Detection of copy number alterations
As shown in Fig. 2, the 100K SNP arrays proved useful in detecting both homozygous (B, threshold at copy number < − 1.3) and heterozygous (A, copy number < −0.7) deletions in the 178 gliomas assayed. Homozygous and heterozygous deletions of CDKN2A were found in 21% and 34% of glioblastoma samples, respectively. These rates are consistent with those reported in the literature using standard genetic approaches (i.e., PCR, Southern analysis; refs. 22, 23), thereby adding validity to both our technique and the data we derived from our sample set. Other regions of deletion commonly described in the literature using standard genomic methodology, including loss of 10q23.2 in glioblastoma (24–26) and loss of 1p and/or 19q (9, 20, 27) in oligodendrogliomas were also detected in our analyses at rates consistent with those previously reported. A sample of the genes located in the affected areas can be found in Table 1; a complete list of other regions we discovered to be deleted by CNA analysis can be found in Supplementary Tables S2 and S3.
Table 1.
Sample of regions found altered by copy number analysis
| Chromosome no. | SNP, n | Staring band |
Ending band |
Gene(s) | All max (%) |
GBM max (%) |
Astrocytoma max (%) |
Oligodendroglioma max (%) |
Mixed max (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | p35.2 | p35.2 | PTP4A2 | 7 | 4 | 9 | 12 | 8 |
| 2 | 1 | p25.1 | p25.1 | E2F6/GREB1 | 10 | 10 | 18 | 6 | 0 |
| 9 | 72 | p21.3 | p21.3 | CDKN2B | 12 | 21 | 6 | 2 | 17 |
| 10 | 1 | q26.3 | q26.3 | CYP2E1 | 14 | 13 | 12 | 17 | 8 |
| 11 | 1 | p15.4 | p15.4 | ZNF195 | 2 | 1 | 0 | 0 | 17 |
| 19 | 1 | q13.12 | q13.12 | ZNF566/ZNF545 | 11 | 9 | 12 | 12 | 17 |
| 1 | 1 | p13.2 | p13.2 | CAPZA1 | 16 | 20 | 12 | 13 | 17 |
| 6 | 1 | q13 | q13 | COL12A1 | 11 | 13 | 6 | 12 | 8 |
| 9 | 93 | p23 | p23 | TYRP1/PTPRD | 7 | 12 | 3 | 2 | 8 |
| 9 | 4 | q22.2 | q22.2 | GADD45 | 8 | 12 | 0 | 4 | 17 |
| 10 | 7 | q23.2 | q23.2 | GRID1/PTEN | 12 | 22 | 3 | 0 | 25 |
| 12 | 1 | q24.11 | q24.11 | PPP1CC | 18 | 20 | 15 | 17 | 17 |
| 18 | 1 | p11.21 | p11.21 | PTPN2 | 19 | 18 | 18 | 23 | 8 |
| 4 | 1 | p16.1 | p16.1 | PPP2R2C | 37 | 30 | 45 | 44 | 25 |
| 7 | 32 | p11.2 | p11.2 | EGFR | 26 | 43 | 15 | 6 | 33 |
| 8 | 1 | q24.3 | q24.3 | PTP4A3 | 32 | 35 | 27 | 29 | 33 |
| 13 | 1 | q34 | q34 | ING1 | 9 | 7 | 9 | 13 | 0 |
| 20 | 1 | q13.13 | q13.13 | PTPN1 | 10 | 18 | 6 | 0 | 8 |
NOTE: Type of alteration found is as follows: Top six rows, homozygous deletions; mid seven rows, heterozygous deletions; bottom five rows, amplifications.
Figure 2C shows the distribution of amplification events for all the samples assayed segregated by histopathology. For this analysis, we required that the SNP be amplified by five or more copies for the amplification to be considered significantly above background noise. The transition from a continuous spectrum of CNA to one of discrete areas of amplification is shown in Fig. 3. This adjustment was necessary to avoid the formation of a quasi-continuum of alterations that would have obscured the regions of clear relevance.
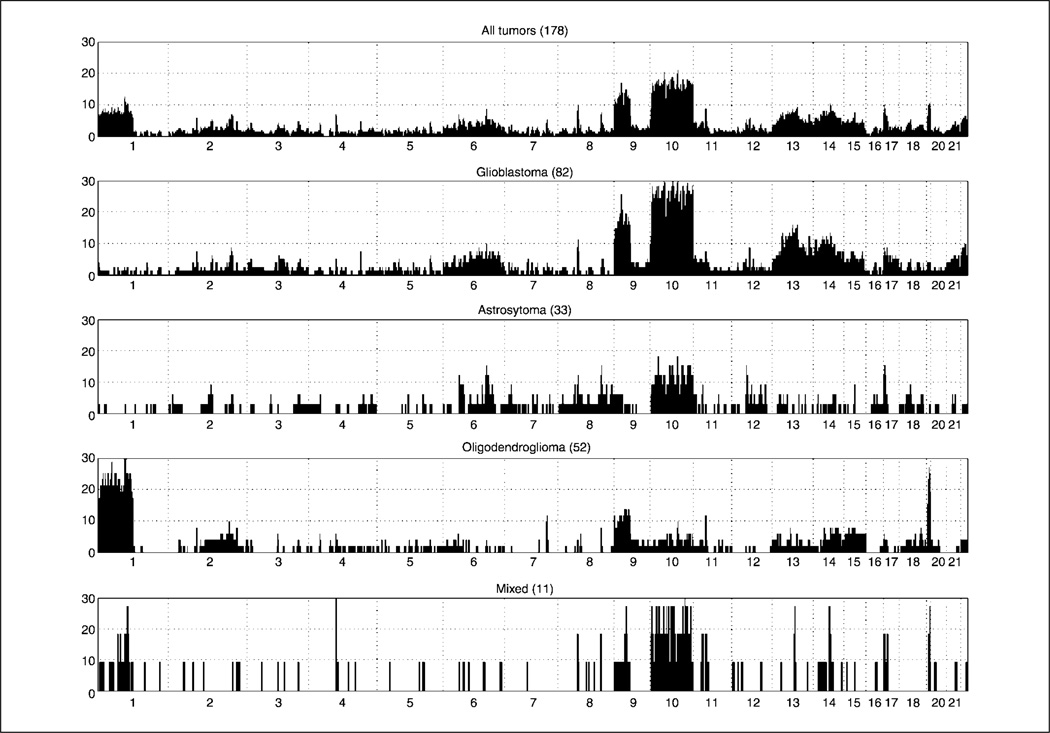
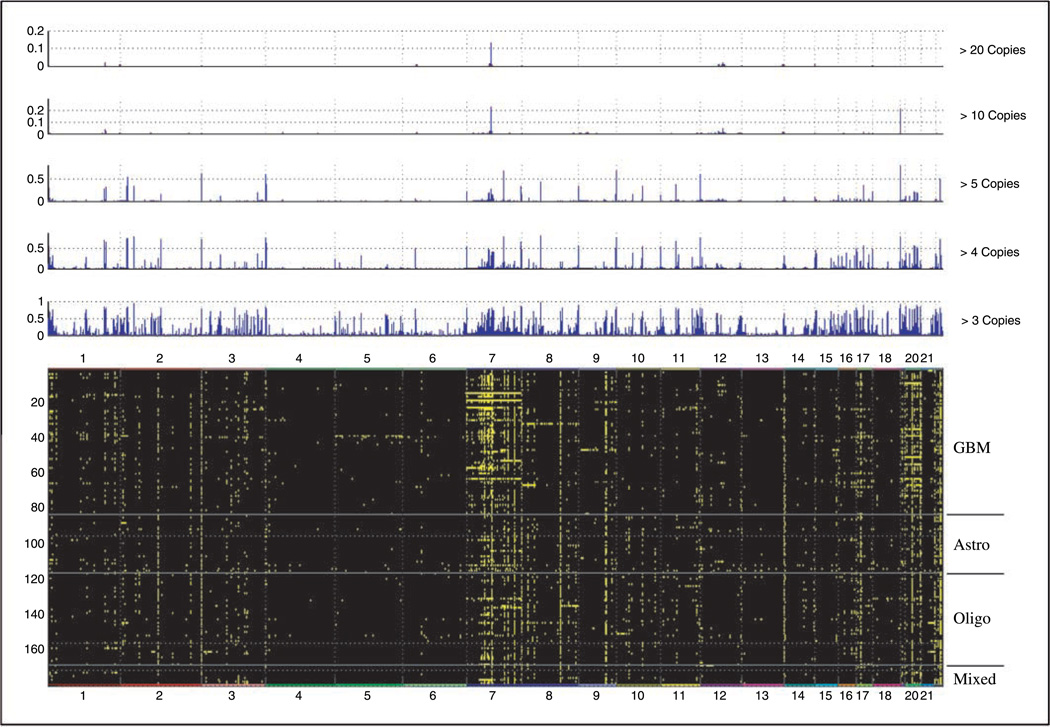
Figure 3.
Top, histogram representing DNA amplifications found in all samples by analyzing copy number alterations. Different thresholding conditions are shown. X axis, all somatic chromosomes; the length of the segment is proportional to the size of chromosome. Y axis, ratio of samples showing a particular amplification. Bottom, heat map showing the altered SNP (amplified at three copies or more) as a green dot against the background. Numbers on top, chromosome numbers. Different types of tumors are noted to the right (Astro, astrocytoma; Oligo, oligodendroglioma) and separated by straight line.
As in the case of deletions, we found areas of amplification in our data sets that have been previously reported, further supporting the validity of our methodology. Most notable among these is the 7p11.2 amplicon (EGFR) that we found altered in 43% glioblastoma multiforme (GBM) and 15% astrocytomas, rates consistent with those in the literature (26, 28, 29). The reminder of the regions found amplified (Supplementary Table S4) are novel, and, therefore, cannot be compared with results in the literature. Of note is the observation that a number of genes found within the deleted/amplified regions seem to have the potential for being candidate oncogenes and/or have been implicated in the biology of other cancer types (e.g., protein phosphatase and protein tyrosine phosphatase superfamilies; Table 1; Supplementary Tables S2–S4). A sampling of the regions found amplified can be seen in Table 1, with a complete listing in Supplementary Table S4.
Loss of heterozygosity analysis
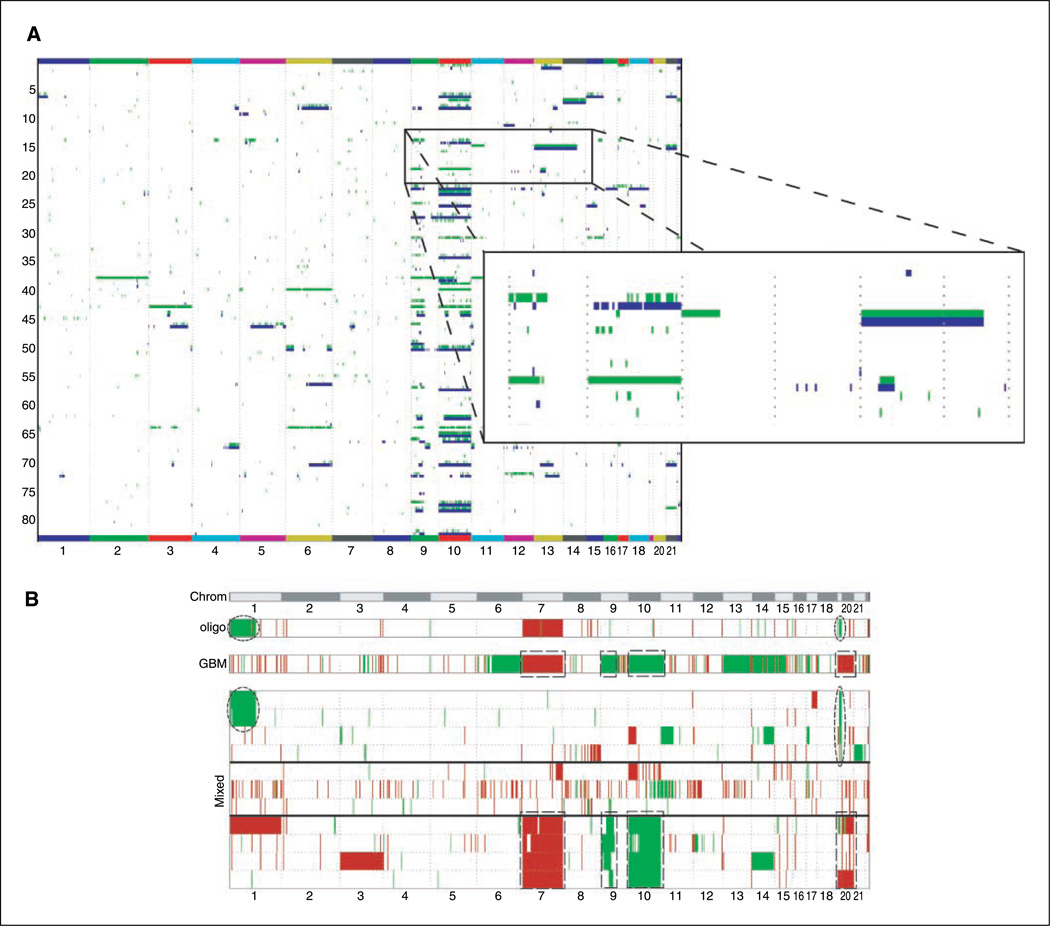
LOH analysis of the sample gliomas (Fig. 4; Supplementary Table S5) yielded a pattern similar to the one obtained by CNA although each analysis uses independent data types gathered from the arrays (signal intensity in the case of CNA, and allelic calls in the case of LOH). The similar findings using two different data types further show the robustness of the methodology and our data analysis. Figure 5 shows an example of several discrete areas where LOH is found without a corresponding deletion, pointing to a recombination event leading to allelic imbalance (gene conversion events).
Figure 5.
A, correlation heat map showing the altered SNP as calculated by LOH analysis (green) or deletions by copy number alteration (blue). Numbers below, chromosome numbers. Shown here are only the glioblastoma samples. B, heat map showing the alterations found in mixed tumors: amplified SNPs (red) and deletions/LOH (green). Numbers on top, chromosome numbers. For comparison purposes, two panels showing the consensus alterations (defined as changes seen in more than five samples) in all glioblastomas and oligodendrogliomas are shown on top of the main panel. The alterations found on oligodendrogliomas and GBMs that are mirrored in mixed tumors are highlighted by an ellipsoidal or rectangular marquee, respectively.
Discussion
In the present study, we showed that the use of Affymetrix 100K SNP chip allows one to scan the genome of a large number of samples at an unprecedented 25 kb average resolution. Several novel regions of copy number alterations and allelic imbalance were detected by the use of this methodology in 179 glioma samples.
There are, however, some limitations that need to be taken into account when using these arrays and data derived from the analysis. First, a series of tissue-matched “normal” samples need to be run in parallel with the disease subjects to make sure that the copy numbers obtained by the CCNT tool are correct. This is necessary because, as our analysis of the copy numbers for every SNP in the normal set shows, some discreet SNPs (~ 30 single and grouped) were consistently showing alterations at high levels in our nontumor controls. Although it has been recently recognized that a “normal” genotype can contain local regions of amplification/ deletions (19), it is accepted that most of the genome of a normal sample should be diploid. Because the normal baseline for copy number determination by CCNT is a set of DNA from the Coriell panels (30), derived from peripheral blood lymphocytes, it is possible that these aberrant SNPs represent either a brain-specific alteration or, more likely, a SNP with unspecific hybridization characteristics. We are at present conducting additional experiments to elucidate which of these two explanations is correct. In any case, these aberrant SNPs need to be excluded from the analysis of the tumor samples to avoid the possibility of reporting false positives. In this regard, our experience suggests the necessity for having a set of normal controls, matched to the tissue type of interest, for determining a reliable SNP set for subsequent analyses.
The second limitation to this technique is shown in Supplementary Table S1 where the copy numbers obtained with the chips do not always match (numerically) to what is obtained using quantitative real-time PCR of the same SNPs. This is not surprising, because high-throughput assays are not designed to be quantitative but rather to give a wealth of qualitative information on a large number of probes. This last point introduces the third limitation to these assays, being that a large number of samples need to be assayed to make sure that the data being generated is correct. The lack of quantization in the microarray analyses makes conclusions derived from a limited number of samples suspect, and thus requires validation from an alternative method. By contrast, large sample numbers can overcome these statistical limitations such that if a relatively large number of tumors display the newly discovered areas of alteration, it is highly likely that these regions are relevant to the disease in question.
This last point is made even stronger if the results obtained from the SNP analysis can be correlated with RNA expression analysis via a microarray assay. Although this additional genomic method also is nonquantitative, the combination of these two methods in a large sample set greatly increases the probability of elucidating relevant findings. In our case, such analysis confirmed the vast majority of CNA regions with a very convincing statistical significance (P > 10−10). A sampling of the results obtained by such analysis is shown in Fig. 1, where A, B, and C show the distribution obtained for a homozygous deletion, a heterozygous deletion and one amplification, respectively. Clearly, higher copy numbers correlate with larger mRNA expression, whereas lower copy numbers display a lower level of mRNA. In the case of EGFR, some samples show a higher expression level without having an amplified gene, this being consistent with literature (31–33), because not all overexpression events are a result of genomic amplification. Similarly, in the case of CDKN2A, all the samples with low copy numbers show low expression, whereas some samples with normal copy numbers show low expression likely due to epigenetic-mediated down-regulation of the genes in this locus, as has been previously well described.
Interestingly, we found a gene on chromosome X that segregates in an almost perfect binary fashion according to gender (Fig. 4D). This gene, X inactivation-specific transcript (XIST; ref. 34), is known to have a principal role in the inactivation of the second X chromosome in females and formation of the Barr bodies. Thus, XIST expression levels should be high in females (X copy number 2) and low in males (X copy number 1), a finding clearly confirmed by our analysis and thereby further supporting the reliability of the technology.
To further validate the results obtained by the SNP arrays, we assayed a tissue microarray, containing all the samples used in this study, for total EGFR protein. The results obtained are concordant with what would be predicted from the CNA analysis (i.e., 100% of samples with predicted amplifications show very strong stain for the protein), indicating that the genomic alteration translates into a biological effect as previously reported (31–33). Consistent with the literature, we also found that some nonamplified samples showed strong staining, indicating possible translational regulation of the protein. In fact, all these samples showed higher mRNA levels for the EGFR transcripts than the tumors not demonstrating strong EGFR immunostaining (data not shown), supporting this explanation.
Our rationale for using thresholding in determining the areas of alteration comes from the need to use large numbers of samples in these analyses. If all the changes being observed were given equal value irrespective of the frequency at which they appear, the result would be very broad areas of CNAs/LOH due in part to the stochastic nature of the process producing them as well as the inherent noise in the microarray methodology. The transition from a continuous spectrum to one of discrete areas of amplification is shown in Fig. 3. This adjustment was necessary to avoid the formation of a quasi-continuum of alterations that would have obscured the regions of clear relevance. Interestingly, the results we observed with our thresholding is consistent with comments made by the original authors of the CCNT methodology who suggested that the algorithm performs better at amplifications larger than five copies (15).
Beyond the genomic surveying of specific diseased tissues, this technique could be used to elucidate the answers to certain basic genetic questions dealing with mechanisms of genomic alterations. As an example, all the areas we identified as homozygous deletions were also found as heterozygous deletions in a larger percentage of tumors, consistent with the notion that homozygous deletions start as heterozygous events. Another advantage of the 100K array analysis is that it allows one to highlight areas of the genome where gene conversion events have occurred as shown in Fig. 5A. To our knowledge, this is the only high-throughput technique that allows such determinations.
The potentially significant genomic alterations observed in our glioma samples will need to be further characterized by both in vitro and in vivo techniques to ascertain their biological relevance. Nevertheless, it is encouraging to see that the methodology we have used successfully identified all of the commonly established genomic alterations in gliomas described to date and at frequencies similar to those reported in the literature using standard genetic approaches (i.e., PCR, Southern blot analysis; refs. 22, 23). For example, homozygous and heterozygous deletions of CDKN2A were found in 21% and 34% of glioblastoma samples, respectively, whereas the 7p11.2 amplicon (EGFR) was found altered in 43% of GBMs and 15% of astrocytomas. The similarity of these data to those established in the literature adds validity to both our technique and the data we derived from our sample set. Additionally, the similarity in the pattern of genomic alterations, as determined by LOH or CNA analyses, shows the robustness of our analyses methodology in view of the different data elements used for each analysis and the fact that LOH is calculated as an indirect statistical quantity (due to our lack of germ line DNA to perform a direct comparison).
Beyond the discovery of novel genomic alterations that could lead to new targets for therapy, this genomic survey technique may also allow for a more rational, biology-based classification of gliomas. It is clear from the examination of Figs. 2 to 4 that the different WHO-recognized histopathologic subtypes have substantially different patterns of genomic alterations (oligodendrogliomas are the majority of tumors showing 1p/19, EGFR amplification are most common in GBMs, etc.). There are, however, some elements that seem common to the majority of the tumors (such as the amplifications at 1p36, 4q16, and 7q22) that may point to primary changes essential in the molecular pathogenesis of all glioma subtypes.
Relative to the potential use of SNP-based genomic surveys for tumor subtype classification, it is of interest that a class of tumors whose exact classification has been clinically troublesome for years (oligoastrocytomas or mixed gliomas) do not seem to conform to a single group but instead can be subdivided into two distinct entities: one being very similar genotypically to GBMs (chromo-somes 7, 19, and 20 amplification; chromosomes 10 and 9 deletions), whereas the other group contain many alterations typical of oligodendrogliomas (1p/19q deletions). This can be seen in Figs. 2 to 4 (and in more detail in Fig. 5B), where some of the typical genomic alterations for GBMs and oligodendrogliomas are recapitulated in one of the two subgroups of mixed gliomas. Although this observation is intriguing, data from a significantly larger number of mixed gliomas will be required before we can conclude with any certainly that mixed gliomas are in fact two different tumor types rather than one.
In summary, the use of high-resolution techniques has allowed us to identify a wealth of new genomic abnormalities in gliomas, confirm the existence of previously reported ones, and obtain a preliminary picture of a system for the creation of a biologically based classification of such tumors. Further investigation will be required to confirm whether the areas of genomic alteration identified in this analysis encode genes responsible for the pathogenesis of gliomas and/or offer potentially new targets for therapeutic intervention.
Supplementary Material
Acknowledgments
Grant support: Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and National Cancer Institute grant CA095809 (T. Mikkelsen).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Supplementary information was provided as Tables S1 to S4 and Figs. S1 and S2, describing all the regions found to be altered in the present study. All raw files for the Genotyping Microarrays can be found at the caArray depository and our Rembrandt (Repository of Molecular Brain Neoplasia Data) publicly accessible database (http://Rembrandt.nci.nih.gov).
Unpublished results.
References
- 1.Hartmann C, Mueller W, von Deimling A. Pathology and molecular genetics of oligodendroglial tumors. J Mol Med. 2004;82:638–655. doi: 10.1007/s00109-004-0565-9. [DOI] [PubMed] [Google Scholar]
- 2.Reifenberger G, Collins VP. Pathology and molecular genetics of astrocytic gliomas. J Mol Med. 2004;82:656–670. doi: 10.1007/s00109-004-0564-x. [DOI] [PubMed] [Google Scholar]
- 3.Wang SI, Puc J, Li J, et al. Somatic mutations of PTEN in glioblastoma multiforme. Cancer Res. 1997;57:4183–4186. [PubMed] [Google Scholar]
- 4.Raizer JJ. HER1/EGFR tyrosine kinase inhibitors for the treatment of glioblastoma multiforme. J Neurooncol. 2005;74:77–86. doi: 10.1007/s11060-005-0603-7. [DOI] [PubMed] [Google Scholar]
- 5.Haas-Kogan DA, Prados MD, Tihan T, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005;97:880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 6.Hirose Y, Aldape KD, Chang S, Lamborn K, Berger MS, Feuerstein BG. Grade II astrocytomas are subgrouped by chromosome aberrations. Cancer Genet Cytogenet. 2003;142:1–7. doi: 10.1016/s0165-4608(02)00791-4. [DOI] [PubMed] [Google Scholar]
- 7.Roerig P, Nessling M, Radlwimmer B, et al. Molecular classification of human gliomas using matrix-based comparative genomic hybridization. Int J Cancer. 2005;117:95–103. doi: 10.1002/ijc.21121. [DOI] [PubMed] [Google Scholar]
- 8.Hui AB, Lo KW, Yin XL, Poon WS, Ng HK. Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab Invest. 2001;81:717–723. doi: 10.1038/labinvest.3780280. [DOI] [PubMed] [Google Scholar]
- 9.Bourdon V, Plessis G, Chapon F, Guarnieri J, Derlon JM, Jonveaux P. Chromosome imbalances in oligodendroglial tumors detected by comparative genomic hybridization. Ann Genet. 2004;47:105–111. doi: 10.1016/j.anngen.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Matsuzaki H, Dong S, Loi H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 11.Di X, Matsuzaki H, Webster TA, et al. Dynamic model based algorithms for screening and genotyping over 100K SNPs on oligonucleotide microarrays. Bioinformatics. 2005;21:1958–1963. doi: 10.1093/bioinformatics/bti275. [DOI] [PubMed] [Google Scholar]
- 12.Miller CL, Diglisic S, Leister F, Webster M, Yolken RH. Evaluating RNA status for RT-PCR in extracts of post-mortem human brain tissue. Biotechniques. 2004;36:628–633. doi: 10.2144/04364ST03. [DOI] [PubMed] [Google Scholar]
- 13.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 14.Seo J, Bakay M, Chen YW, Hilmer S, Shneiderman B, Hoffman EP. Interactively optimizing signal-to-noise ratios in expression profiling: project-specific algorithm selection and detection P-value weighting in Affymetrix microarrays. Bioinformatics. 2004;20:2534–2544. doi: 10.1093/bioinformatics/bth280. [DOI] [PubMed] [Google Scholar]
- 15.Huang J, Wei W, Zhang J, et al. Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum Genomics. 2004;1:287–299. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fridlyand J, Snijders A, Pinkel D, Albertson DG, Jain A. Application of Hidden Markov Models to the analysis of the array CGH data. J Multivariate Anal. 2004;90:132–153. [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aldape KD, Ballman K, Furth A, et al. Immunohistochemical detection of EGFRvIII in high malignancy grade astrocytomas and evaluation of prognostic significance. J Neuropathol Exp Neurol. 2004;63:700–707. doi: 10.1093/jnen/63.7.700. [DOI] [PubMed] [Google Scholar]
- 19.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 20.Nigro JM, Takahashi MA, Ginzinger DG, et al. Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quanti-tative polymerase chain reaction assay. Am J Pathol. 2001;158:1253–1262. doi: 10.1016/S0002-9440(10)64076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array CGH data for down-stream analyses. Bioinformatics. 2005;21:4084–4091. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- 22.Hartmann C, Kluwe L, Lucke M, Westphal M. The rate of homozygous CDKN2A/p16 deletions in glioma cell lines and in primary tumors. Int J Oncol. 1999;15:975–982. doi: 10.3892/ijo.15.5.975. [DOI] [PubMed] [Google Scholar]
- 23.Jen J, Harper JW, Bigner SH, et al. Deletion of p16 and p15 genes in brain tumors. Cancer Res. 1994;54:6353–6358. [PubMed] [Google Scholar]
- 24.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, James CD, Frederick L, Alderete BE, Jenkins RB. PTEN/MMAC1 mutations and EGFR amplification in glioblastomas. Cancer Res. 1997;57:5254–5257. [PubMed] [Google Scholar]
- 26.Smith JS, Tachibana I, Passe SM, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–1256. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- 27.Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994;145:1175–1190. [PMC free article] [PubMed] [Google Scholar]
- 28.Nishikawa R, Ji XD, Harmon RC, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci U S A. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coriell LL, Greene AE, Mulivor RA. Origin of cell-culture lines. N Engl J Med. 1983;308:1296–1297. doi: 10.1056/NEJM198305263082119. [DOI] [PubMed] [Google Scholar]
- 31.Tabori U, Rienstein S, Dromi Y, et al. Epidermal growth factor receptor gene amplification and expression in disseminated pediatric low-grade gliomas. J Neurosurg. 2005;103:357–361. doi: 10.3171/ped.2005.103.4.0357. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki S, Dobashi Y, Sakurai H, Nishikawa K, Hanawa M, Ooi A. Protein overexpression and gene amplification of epidermal growth factor receptor in nonsmall cell lung carcinomas. An immunohistochemical and fluorescence in situ hybridization study. Cancer. 2005;103:1265–1273. doi: 10.1002/cncr.20909. [DOI] [PubMed] [Google Scholar]
- 33.Schober R, Bilzer T, Waha A, et al. The epidermal growth factor receptor in glioblastoma: genomic amplification, protein expression, and patient survival data in a therapeutic trial. Clin Neuropathol. 1995;14:169–174. [PubMed] [Google Scholar]
- 34.Brown CJ, Hendrich BD, Rupert JL, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.