Abstract
In differentiated cells, aging is associated with hypermethylation of DNA regions enriched in repressive histone post-translational modifications. However, the chromatin marks associated with changes in DNA methylation in adult stem cells during lifetime are still largely unknown. Here, DNA methylation profiling of mesenchymal stem cells (MSCs) obtained from individuals aged 2 to 92 yr identified 18,735 hypermethylated and 45,407 hypomethylated CpG sites associated with aging. As in differentiated cells, hypermethylated sequences were enriched in chromatin repressive marks. Most importantly, hypomethylated CpG sites were strongly enriched in the active chromatin mark H3K4me1 in stem and differentiated cells, suggesting this is a cell type–independent chromatin signature of DNA hypomethylation during aging. Analysis of scedasticity showed that interindividual variability of DNA methylation increased during aging in MSCs and differentiated cells, providing a new avenue for the identification of DNA methylation changes over time. DNA methylation profiling of genetically identical individuals showed that both the tendency of DNA methylation changes and scedasticity depended on nongenetic as well as genetic factors. Our results indicate that the dynamics of DNA methylation during aging depend on a complex mixture of factors that include the DNA sequence, cell type, and chromatin context involved and that, depending on the locus, the changes can be modulated by genetic and/or external factors.
Genomic DNA methylation is known to change during lifetime and aging (Jaenisch and Bird 2003). Some changes play important roles in development, but others occur stochastically without any apparent biological purpose (Fraga 2009; Feil and Fraga 2012). These molecular alterations, which are known as epigenetic drift, are currently being investigated as they have been proposed to account for many age-related diseases (Bjornsson et al. 2004; Heyn et al. 2013; Timp and Feinberg 2013). Various recent studies using 1.5K and 27K Illumina methylation arrays have identified a group of gene promoters in blood that becomes hypermethylated during aging (Christensen et al. 2009; Rakyan et al. 2010; Teschendorff et al. 2010; Bell et al. 2012; Fernandez et al. 2012). Interestingly, some of these studies have also shown that these DNA sequences are enriched in bivalent chromatin domains in embryonic stem cells (Rakyan et al. 2010; Fernandez et al. 2012; Heyn et al. 2012) and repressive histone marks such as H3K9me3 and H3K27me3 in differentiated cells (Rakyan et al. 2010), and that many of them are also frequently hypermethylated in cancer. However, drawing conclusions from some of these studies is limited by their low genome coverage (< 0.1%) and the location of the sequences analyzed (mainly at gene promoters). Further studies using Infinium HumanMethylation450 BeadChip and larger cohorts (Heyn et al. 2012; Hannum et al. 2013; Johansson et al. 2013) have, though, corroborated most of the previous observations with the 27K methylation arrays and have, in addition, identified new sets of genes that become hypermethylated and hypomethylated during aging in humans. Finally, a recent study that analyzed the genome-wide DNA methylation status of newborns, middle-aged individuals, and centenarians confirmed the results obtained with the methylation arrays and showed that aging is associated with overall hypomethylation, which primarily occurs at repetitive DNA sequences (Heyn et al. 2012). Most of the above studies were conducted with whole blood, and consequently, changes in cell heterogeneity during aging could have affected the results (Calvanese et al. 2012; Houseman et al. 2012). However, some genes presented consistent changes in different tissues, which indicates that in some cases, the changes truly are associated with aging (Rakyan et al. 2010; Horvath et al. 2012). Interestingly, Houseman et al. (2012) have recently reported an algorithm that, using the DNA methylation values of certain genes, estimates the relative proportion of the different blood cell types in a specific sample. This algorithm was successfully used by Liu et al. (2013) in a study to identify DNA methylation alterations associated with rheumatoid arthritis.
In addition to the studies using blood, other works have identified specific DNA methylation signatures of aging in differentiated cell types, including the brain (Hernandez et al. 2011; Numata et al. 2012; Guintivano et al. 2013; Lister et al. 2013), muscle (Zykovich et al. 2014), and saliva (Bocklandt et al. 2011). Two studies have analyzed DNA methylation during aging in human adult stem cells: Bork et al. (2010) used 27K methylation arrays to analyze the DNA methylation status of mesenchymal stem cells (MSCs) obtained from young (21–50 yr) and old donors (53–85 yr) and found similar DNA methylation changes over time during prolonged in vitro culture and in vivo aging. Using the same methylation arrays, Bocker et al. (2011) observed a bimodal pattern of methylation changes in older hematopoietic progenitor cells, with hypomethylation of differentiation-associated genes, as well as de novo methylation events resembling epigenetic mutations. Recent studies in mice have revealed a number of genome-wide alterations in DNA methylation (Taiwo et al. 2013) that might play an important role in the functional decline of hematopoietic stem cells (HSCs) during aging (Beerman et al. 2013). To study the role of DNA methylation in adult stem cell aging further, the present study used the HumanMethylation450 BeadChip to characterize the genome-wide DNA methylation status of bone marrow MSCs obtained from individuals aged between 2 and 92 yr. We then systematically compared our results with previously published data to identify the chromatin signatures associated with DNA methylation changes in adult stem cells and to determine whether these changes were also present in other tissues. Finally, we analyzed monozygotic (MZ) twins of different ages to determine the effect of genetic factors on the DNA methylation changes during aging identified in our study.
Results
Global DNA methylation profiling in adult MSCs
To identify DNA methylation changes during MSC aging, we compared the DNA methylation status of 429,789 CpG sites in 34 independently isolated primary MSCs, obtained from individuals from 2–92 yr old, using the HumanMethylation450 BeadChip (Illumina) (Supplemental Fig. S1; Supplemental Table 1).
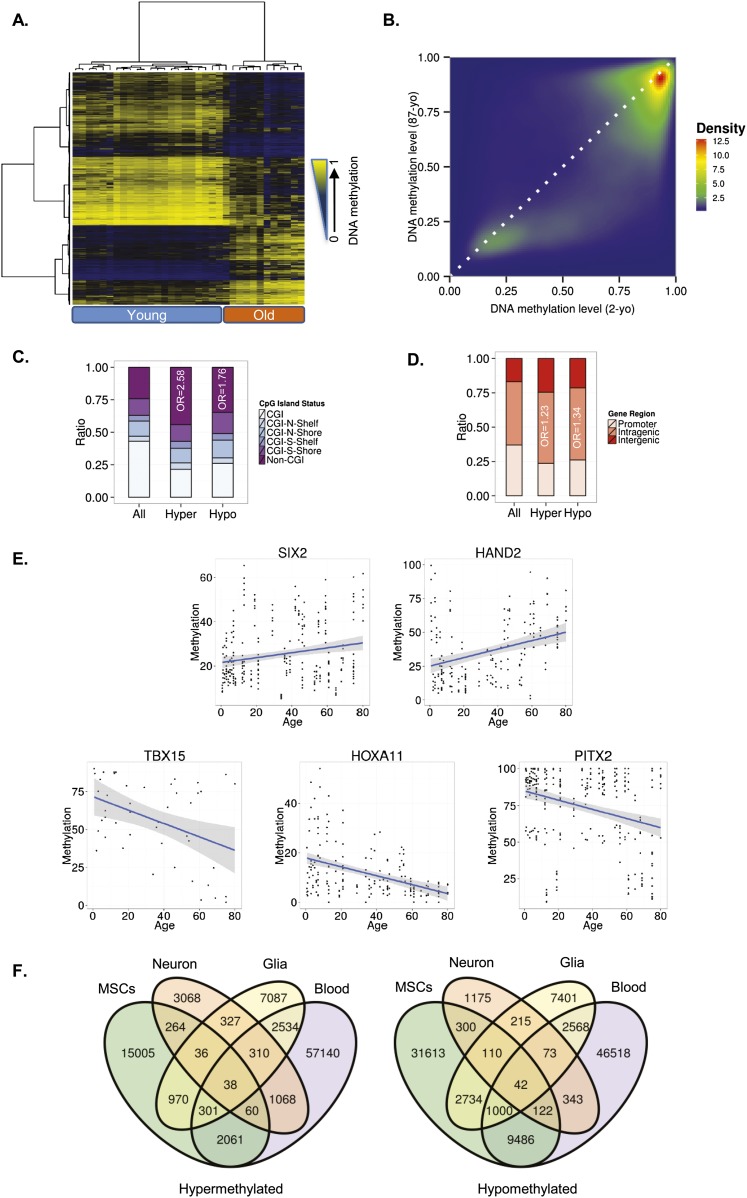
Using an empirical Bayes moderated t-test (see Methods), we first identified 64,142 autosomal CpG sites that were differentially methylated (dmCpGs; FDR < 0.05) between MSCs obtained from young (ages ranging from 2 to 22 yr) and elderly (aged between 61 and 91 yr) individuals. Hierarchical clustering of all samples using the dmCpGs alone enabled each sample to be correctly allocated to its corresponding age group (Fig. 1A). Of the dmCpG sites, 18,735 (29.20%) had become hypermethylated and 45,407 (70.80%) had become hypomethylated with aging (Fig. 1B; Supplemental Tables 2, 3).
Figure 1.
DNA methylation changes during MSC aging. (A) Unsupervised hierarchical clustering and heatmap including the 15,000 most variable CpG sites with differential DNA methylation between young and old MSCs. Average methylation values are displayed from zero (blue) to one (yellow). (B) Density plot for differentially methylated CpG sites between representative young (2-yr-old [2-yo]) and old (87-yr-old [87-yo]) MSCs. (C) Distribution of differentially methylated CpGs relative to the CpG island. (D) Relative distribution of differentially methylated CpGs across different genomic regions. (E) Examples of aging-specific CpG methylation, in particular, genes further validated by pyrosequencing in an independent set of samples. For each of the genes of interest, a scatter plot of the percentage of methylation obtained for each sample and CpG of interest is shown. The two genes at the top show an age-dependent hypermethylation tendency, while the three genes at the bottom show hypomethylation with respect to age. Each point represents a single observation for a given sample and CpG of interest. The blue line represents a linear model fit. A 0.95 confidence interval of the fitted model is shown in gray. (F) Venn diagrams showing the number of CpG sites (hyper- and hypomethylated) shared by the different tissues
To study the characteristics of these dmCpG sites from a functional genomics point of view, we first determined their distribution within the different regions of the CpG islands (CGIs) (Wu et al. 2010). Interestingly, both hyper- and hypomethylated CpG sites were enriched in non-CGIs (χ2 test; P < 0.001, OR = 2.58 and P < 0.001, OR = 1.76, respectively) (Fig. 1C) and in intragenic DNA regions (χ2 test; P < 0.001, OR = 1.23 and P < 0.001, OR = 1.34, respectively) (Fig. 1D).
To validate the results obtained with the methylation arrays, we randomly selected five of the sequences previously identified, and we analyzed their methylation status by bisulfite pyrosequencing in an independent cohort of 46 MSCs obtained from individuals 7 mo to 80 yr old (Supplemental Table 1). In total, in the validation phase we obtained information on the DNA methylation status of 950 CpGs. The sequences selected corresponded to the genes HAND2 and SIX2, which become hypermethylated with aging, and to the genes TBX15, PITX2, and HOXA11, which become hypomethylated. Bisulfite pyrosequencing results showed that all the sequences selected for validation displayed the same DNA methylation dynamics during aging as in the study samples (Fig. 1E).
Tissue-specific DNA methylation changes during aging
Global DNA methylation patterns are tissue/cell type specific (Calvanese et al. 2012). To determine whether the CpG sites displaying DNA methylation changes during aging in adult stem cells are also affected in differentiated tissues, we used the same workflow described in the previous section to analyze the data obtained in previous aging studies which used the same methylation arrays with samples from blood (human whole blood from a mixed population of 426 Caucasian and 230 Hispanic individuals, with ages ranging from 19 to 101) and brain (neuronal and glial cells, from post-mortem frontal cortex of 29 healthy individuals [14 male, 15 female, aged 32.6 ± 16.1]) (Supplemental Fig. S1; Guintivano et al. 2013; Hannum et al. 2013). To reduce confounding factors in the blood data set, we first corrected for cellular heterogeneity with respect to the major cell subtypes (Houseman et al. 2012) to filter out only those associations that were the consequence of aging. Using this approach, we identified 63,512 hypermethylated and 60,155 hypomethylated sequences in blood (FDR < 0.05), 11,603 hypermethylated and 14,143 hypomethylated sequences in glial cells (FDR < 0.05), and 5171 hypermethylated and 2380 hypomethylated sequences in neural cells (FDR < 0.05) (Supplemental Fig. S2; Supplemental Tables 4, 5). As in MSCs, hypomethylated cytosines in the differentiated cells preferentially occurred at both non-CGI regions (χ2 test; blood, P < 0.001, OR = 2.35; neural, P < 0.001, OR = 1.74; glial, P < 0.001, OR = 3.03) and at intragenic regions (χ2 test; blood, P < 0.001, OR = 1.11; neural, P < 0.001, OR = 2; glial, P < 0.001, OR = 1.89) (Supplemental Fig. S2). However, in brain samples (neuronal and glial cells), hypermethylated cytosines occurred preferentially at both non-CGI regions (χ2 test; neural, P < 0.001, OR = 1.43; glial, P < 0.001, OR = 1.43) and at intragenic regions (χ2 test; neural, P < 0.001, OR = 1.1; glial, P < 0.001, OR = 1.1), while they occurred preferentially in both CGIs (χ2 test; P < 0.001, OR = 3.5) and at promoter regions (χ2 test; P < 0.001, OR = 1.49) in blood samples (Supplemental Fig. S2).
To identify possible cell type–independent DNA methylation signatures of aging, we created two additional data sets containing the hyper- and hypomethylated probes from selected subsets of the different tissues analyzed (Fig. 1F). This approach showed only a small overlap between MSCs and differentiated cells (42 hypomethylated and 38 hypermethylated), suggesting that systemic DNA methylation changes during aging are restricted to specific regions of the genome (Fig. 1F; Supplemental Tables 6, 7).
Hypermethylated CpG sites during aging are associated with repressive chromatin marks
In blood, DNA hypermethylation during aging has been shown to occur at gene promoters enriched in repressive histone marks such as H3K9me3 and H3K27me3 (Rakyan et al. 2010). To identify possible chromatin signatures associated with DNA hypermethylation during aging in adult MSCs, we compared our methylation data with previously published data on a range of histone modifications and chromatin modifiers in 10 different cell types obtained from healthy individuals (see Methods). In the present study, we found statistically significant associations with the repressive histone marks H3K9me3, H3K27me3, and EZH2 in most differentiated ENCODE cell lines (Fisher’s exact test; P < 0.001) (Fig.2), which is in line with previously published data (Rakyan et al. 2010). To determine whether these observations can be extrapolated to other cell types, we used the same approach to analyze the CpG sites that are hypermethylated during aging in blood, neural, and glial cells (Supplemental Table 4; Guintivano et al. 2013; Hannum et al. 2013). The results showed that hypermethylated CpG sites in blood and brain were enriched in the same chromatin marks identified in the adult MSCs (Fig. 2), suggesting that chromatin context is an important cell type–independent mark of DNA hypermethylation during aging. Analysis of the 38 commonly hypermethylated CpG sites in blood, MSCs, and neural and glial cells also showed statistically significant associations (FDR < 0.05) with the repressive histone marks H3K9me3, H3K27me3, and EZH2 found in some types of differentiated cells (Fig. 2).
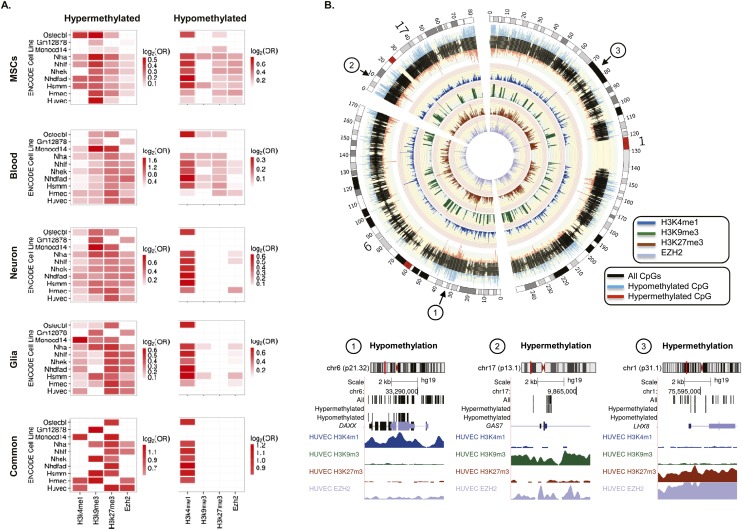
Figure 2.
Chromatin signatures associated with DNA methylation changes during aging. (A) Heatmaps showing significant enrichment of hyper- and hypomethylated CpG sites—identified in MSCs, blood, neurons, and glia—with different histone marks and chromatin modifiers contained in the UCSC Genome Browser Broad histone track from the ENCODE Project. Color code indicates the significant enrichment based on log2 odds ratio (OR). (B) Circular representation of three representative chromosomes (1, 6, and 17), indicating whether the CpGs were hypermethylated (red) or hypomethylated (blue) during MSC aging. Inner tracks display chromatin marks (H3K4me1, H3K9me3, H3K27me3, and EZH2) generated for HUVEC cells and associated with differentially methylated regions during aging. Broad histone peak information was averaged in 200-kbp genomic windows and represented as histogram tracks. Three examples of hypo- and hypermethylated DNA regions associated with specific chromatin signatures are displayed below.
DNA hypomethylation during aging preferentially occurs at H3K4me1-rich sites
To identify chromatin marks associated with CpG sites hypomethylated in aged MSCs, we aligned the DNA sequences identified in our study with the same database of histone modifications and chromatin modifiers described in the previous section. Of note is the fact that hypomethylation largely occurred at regions occupied by the active histone mark H3K4me1 in most of the ENCODE cell lines (FDR < 0.05) (Fig. 2).
To determine whether these associations occurred in differentiated cells, we used the same approach to analyze CpG hypomethylation during aging in blood, neural, and glial cells (Supplemental Table 5; Guintivano et al. 2013; Hannum et al. 2013). Blood and brain samples showed similar enrichment patterns to those of the MSCs in that hypomethylated CpG sites were preferentially located at regions enriched in H3K4me1 (Fig. 2). Interestingly, the analysis of the 42 commonly hypomethylated CpG sites in blood, MSCs, and neural and glial cells only showed statistically significant associations with H3K4me1 (FDR < 0.05) (Fig. 2A).
Dynamics of interindividual DNA methylation variability during aging
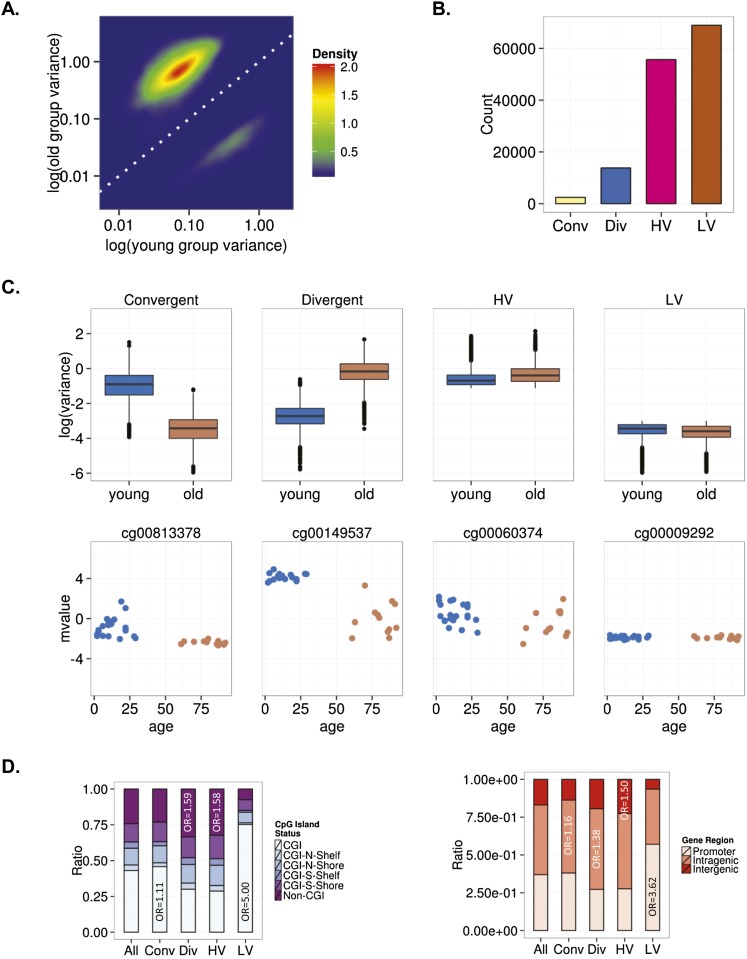
As in most previous studies on DNA methylation and aging, our analytical design allowed the identification of DNA sequences showing a specific tendency to change (hyper- or hypomethylation) during aging, but not other putative DNA regions exhibiting no change tendency (i.e., sequences that do not become hyper- or hypomethylated with aging but rather show an increase or a decrease in interindividual variability). To address this issue, we carried out an alternative data analysis on our MSCs based on the aging-dependent behavior of interindividual variability (i.e., DNA methylation scedasticity). Interindividual variability was higher in MSCs obtained from older individuals than in those obtained from younger individuals (Fig. 3A). Analysis of the scedasticity identified 16,243 heteroscedastic CpG sites, of which 2437 were convergent and 13,806 were divergent. We also identified 124,611 homoscedastic CpG sites, 68,927 showing low interindividual variability in both young and old individuals (LV) and 55,684 showing high interindividual variation in both populations (HV) (see Methods) (Fig. 3B,C; Supplemental Tables 8–11).
Figure 3.
Interindividual DNA methylation variability during MSC aging. (A) Density plot for CpG sites showing significant changes in variance in young and old MSCs. (B) Bar plot showing the number of age-dependent heteroscedastic (convergent and divergent) and homoscedastic (high [HV] and low [LV] variability) CpG sites in MSCs. (C) Box plots showing the classification of CpG sites into different groups based on the aging-dependent behavior of the interindividual variability. Representative examples of CpG sites for each group are shown below (mvalue: relative methylation values). (D) Distribution of homoscedastic and heteroscedastic CpGs relative to CpG island status and relative distribution across different genomic regions.
We studied these sequences from a functional genomics standpoint to identify factors associated with the behavior of DNA methylation changes during aging. We observed that divergent and HV CpG sites were preferentially enriched in non-CGIs (χ2 test; P < 0.001, OR = 1.59 and P < 0.001, OR = 1.58, respectively), and convergent and LV CpG sites in CGIs (χ2 test; P < 0.001, OR = 1.11 and P < 0.001, OR = 5.00, respectively) (Fig. 3D). Both divergent and convergent sequences were more abundant in intragenic regions (χ2 test; P < 0.001, OR = 1.38 and P < 0.001, OR = 1.16, respectively), with HV being more frequently found in intergenic regions (χ2 test; P < 0.001, OR = 1.50) and LV in promoter regions (χ2 test; P < 0.001, OR = 3.62) (Fig. 3D).
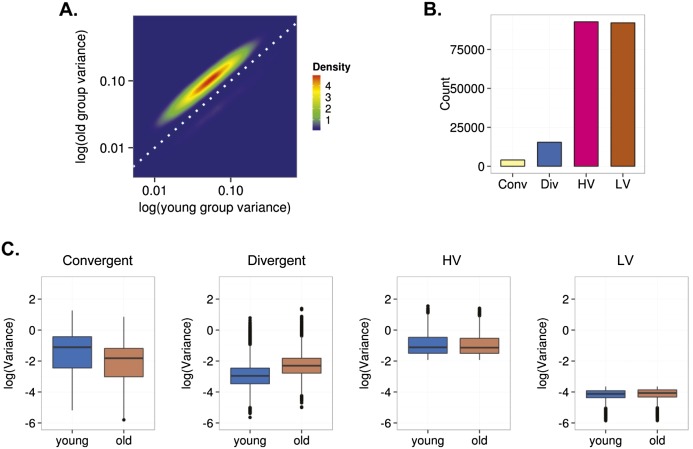
To determine whether scedasticity behavior can also identify DNA methylation changes during aging in differentiated cells, we repeated these same analyses on previously published blood DNA methylation data (Hannum et al. 2013). As the cohort of Hannum et al. (2013) contains DNA methylation data on more than 600 individuals, statistical analyses were carried out using a Brown-Forsythe test (see Methods). To discount a possible confounding effect of cell heterogeneity in the analysis of the scedasticity in blood, in addition to applying the algorithm described by Houseman et al. (2012), we carried out in silico functional analysis of the groups of genes established according to the behavior of the variance (see Methods). These analyses showed no significant associations between these groups of genes and any of the blood cell lineages examined (Supplemental Tables 12, 13). As in MSCs, interindividual variability was higher in blood obtained from older individuals than in blood obtained from younger individuals (Fig. 4A). Furthermore, in line with the findings for adult MSCs, in differentiated cells the analyses identified 19,454 heteroscedastic CpG sites, of which 4037 were convergent and 15,417 divergent. Of the homoscedastic CpG sites, 92,074 showed LV in both young and old individuals and 92,753 showed HV in both populations (Fig. 4B,C).
Figure 4.
Interindividual DNA methylation variability during aging of blood cells. (A) Density plot for CpG sites showing significant changes of variance in young and old individuals. (B) Bar plot showing the number of age-dependent heteroscedastic (convergent and divergent) and homoscedastic (high [HV] and low [LV] variability) CpG sites. (C) Box plots showing the classification of the CpG sites in different groups based on the aging-dependent behavior of the interindividual variability.
The role of genetic factors on DNA methylation changes during aging
To study the role of genetic factors on DNA methylation changes during aging, we used the HumanMethylation450 BeadChip to analyze the DNA methylation status of 24 monozygotic twins from two age groups (young, 21–22 yr; and old, 58–66 yr). The effect of genotype was assessed comparing the Euclidean distance (ED) and the interindividual variability in methylation values between old and young monozygotic (MZ) pairs. To reduce possible bias due to cell heterogeneity, DNA methylation data were corrected with the algorithm described by Houseman et al. (2012). As in the larger cohort previously analyzed (Fig. 4), interindividual DNA methylation variability substantially increased during aging in the MZ twins (Fig. 5A). Interestingly, mean ED between MZ twins also increased (more than twofold) with age in 46,763 CpG sites (Fig. 5B; Supplemental Table 14), which indicates that at these CpG sites, the increase in interindividual methylation variability depends, at least in part, on nongenetic factors. In 24,782 of these sequences (Fig. 5B; Supplemental Table 15), the increase in ED (more than twofold) was higher than could be accounted for solely by interindividual variability, suggesting that in these CpG sites, genetic factors play a less important role in the regulation of DNA methylation changes during aging. However, in 21,908 of these sequences (Fig. 5B; Supplemental Table 16), the increase in ED (more than twofold) was less than could be accounted for solely by interindividual variability, which suggests that, in contrast, at these CpG sites genetic factors are more relevant for the regulation of DNA methylation during aging.
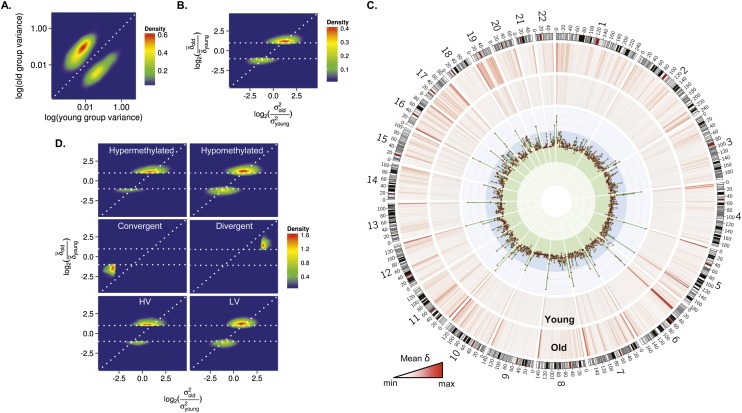
Figure 5.
Role of genetic factors in interindividual DNA methylation variability during aging. (A) Density plot for CpG sites showing significant changes of methylation variance in the blood cells of MZ twins during aging. (B) Density plot for comparison between the mean Euclidean distance (δ) and the interindividual variability (σ2) in methylation values between old and young MZ twins. The horizontal dotted lines represent a twofold change in the δ between MZ twins. (C) Circular representation of genome-wide CpG sites showing differences in the  between methylation values of young and old MZ twins. δ was averaged using a 2-Mbp window size. Inner tracks show genomic regions where the
between methylation values of young and old MZ twins. δ was averaged using a 2-Mbp window size. Inner tracks show genomic regions where the  was higher (blue region) or lower (green region) in old compared with young MZ twins. (D) Density plots for comparison between the
was higher (blue region) or lower (green region) in old compared with young MZ twins. (D) Density plots for comparison between the  and the σ2 in methylation values between old and young MZ twins, in hypermethylated, hypomethylated, heteroscedastic, and homoscedastic CpGs.
and the σ2 in methylation values between old and young MZ twins, in hypermethylated, hypomethylated, heteroscedastic, and homoscedastic CpGs.
Although the general trend was an increase in ED with age, ED between older MZ pairs decreased (more than twofold) for 22,542 CpG sites (Fig. 5B; Supplemental Table 17). As the EDs between older MZ individuals are greater than those between younger MZs in more than half the sequences identified, our results support the notion that, in general, DNA methylation patterns diverge with age, even in genetically identical individuals. In 11,624 sequences (Fig. 5B; Supplemental Table 18), the decrease in ED (more than twofold) was lower than could be accounted for solely by interindividual variability, which suggests that in these CpG sites, genetic factors play a more important role in the regulation of DNA methylation changes during aging. In 10,883 sequences (Fig. 5B; Supplemental Table 19), the decrease in ED was higher than could be accounted for solely by interindividual variability, indicating that in these CpG sites, genetic factors play a less important role in the regulation of DNA methylation during aging. As in the analysis of the previously published blood DNA methylation data, in silico functional analysis of the groups of genes identified in the monozygotic twins (Supplemental Tables 20, 21) suggested that, after correcting with the Houseman algorithm, cell heterogeneity had little impact on the Euclidean distances for changes in DNA methylation with age.
Comparative analysis of the interindividual variation and the EDs suggests that the effect of genotype on the regulation of DNA methylation changes during aging was locus-specific. Thus, to identify those DNA regions differentially affected by the genotype, we used Circos representations to study the genomic distribution of CpG sites that showed changes in ED with age (Fig. 5C). The results demonstrated that while CpG sites showing a decrease, or no difference, in ED between young and old MZs presented a random distribution, those showing an age-dependent increase in ED were strongly enriched in subtelomeric DNA regions. The greatest changes occurred at Chromosomes 11 and 19, and in general, clustering occurred at the same genomic regions in both young and old twins.
To study the effect of the genotype on DNA methylation and its interindividual variability during aging, we analyzed the twins’ data using similar strategies to those described in previous sections, identifying 41,987 hypermethylated, 56,923 hypomethylated, 1018 convergent, 1635 divergent, 58,680 HV, and 59,795 LV CpG sites (data not shown). The comparison of EDs between young and old MZ pairs for these groups of genes showed that the effect of genotype depended on the tendency and the scedasticity of the change (Fig. 5D).
ED increased (more than twofold) with age in 9.5% of the hypomethylated and in 14% of the hypermethylated CpGs, suggesting that genetic factors have a greater effect on the former during aging (Fig. 5D; Supplemental Tables 22, 23). ED increased more than twofold) in most (83.73%) of the divergent CpG sites and decreased more than twofold) in most (66.7%) of the convergent CpG sites (Fig. 5D; Supplemental Tables 24, 25). However, changes in interindividual variability were higher than the increase or decrease in ED (Fig. 5D), which indicates that genetic factors play a role in the regulation of DNA methylation of these DNA regions during aging. Interestingly, ED also increased in most of the HV and LV sequences more than twofold during aging (Fig. 5D). Furthermore, in most CpG sites, the increase in ED between the MZ twins was higher than the interindividual variability changes during aging (Fig. 5D), suggesting that genotype has little effect on epigenetic drift in homoscedastic DNA regions.
Discussion
Recent studies have shown that DNA methylation is altered during aging in a number of differentiated cell types (Rakyan et al. 2010; Teschendorff et al. 2010; Bell et al. 2012; Fernandez et al. 2012; Heyn et al. 2012; Numata et al. 2012; Guintivano et al. 2013; Hannum et al. 2013; Johansson et al. 2013). Here, we studied the dynamics and the context of DNA methylation changes during aging in human adult stem cells as they have been proposed to play an important role in aging (Sharpless and DePinho 2004). Indeed, a recent study in mice showed that epigenomic alterations of the DNA methylation landscape contribute to the functional decline of hematopoietic stem cells (HSCs) during aging (Beerman et al. 2013). To analyze our DNA methylation data, we first used an analytical strategy similar to that used in most of the previous studies on DNA methylation and aging (i.e., linear models). Using this approach, we identified 18,735 CpG sites that were hypermethylated and 45,407 that were hypomethylated during aging in MSCs, which provides support for the idea that, as in blood (Heyn et al. 2012), aging is associated with global DNA hypomethylation in MSCs. In addition, we validated five of the genes identified through the methylation arrays (HAND2, SIX2, TBX15, PITX2, and HOXA11) by bisulfite pyrosequencing, using an independent sample set of 46 MSCs obtained from individuals from 7 mo to 80 yr old. The results corroborated the data obtained from the methylation arrays and suggest that our genome-wide data can be extrapolated to independent sample sets of MSCs. HAND2 and SIX2 genes, which code for transcription factors, have also been found hypermethylated in several cancer types (Rauch et al. 2006; Tong et al. 2010; Jones et al. 2013). In contrast, the genes that are hypomethylated during MSC aging—TBX15, PITX2, and HOXA11—code for transcription factors involved in several differentiation and developmental processes (Singh et al. 2005; Gross et al. 2012; Gage et al. 2014).
Interestingly, 80 of the differentially methylated sequences identified in the MSCs were present in both blood and brain, which is in line with previous observations that suggest the existence of systemic DNA methylation changes during aging (Rakyan et al. 2010; Heyn et al. 2012). However, because many of the sequences were not common to different tissues, our data indicate that as has recently been proposed (Christensen et al. 2009; Day et al. 2013), systemic changes are restricted to specific loci, and cell type plays an important role in the regulation of DNA methylation changes over time.
The factors determining the behavior of DNA methylation during aging have received much attention during the last few years. Recent works have shown that genes that are hypermethylated in blood during aging are associated with the presence of bivalent chromatin domains in embryonic stem cells (Rakyan et al. 2010; Teschendorff et al. 2010; Fernandez et al. 2012; Heyn et al. 2012) and with repressive histone marks (H3K27me3/H3K9me3) in differentiated cells (Rakyan et al. 2010). Our data indicate that the same repressive histone marks in differentiated cells are also present in sequences in those MSCs that are hypermethylated during aging, implying that, independent of morphogenic potential and/or cell type, these repressive histone marks are associated with DNA methylation gain during aging. Of note, our data provide new evidence that sequences that are hypomethylated in MSCs and differentiated cells during aging are strongly enriched in the active chromatin mark H3K4me1, which suggests that this histone modification is a cell type–independent chromatin signature of DNA hypomethylation during aging. Interestingly, H3K4me1 has recently been associated with enhancers (Rada-Iglesias et al. 2010), genomic regions that play a fundamental role in cis-regulation of gene function. In addition, a recent study has shown that DNA hypomethylation within specific transposable elements is associated with tissue-specific enhancer marks, including H3K4me1, suggesting that these sequences might play an important role in tissue-specific epigenetic gene regulation (Xie et al. 2013), which implies that H3K4me1-associated DNA hypomethylation could play a role in the deregulation of gene expression during aging (Bahar et al. 2006). Further parallel studies analyzing DNA hypomethylation in enhancers and gene expression during aging should shed light on this matter. Collectively, our data indicate that although there are few altered DNA sequences which are common to different cell types, the chromatin signatures associated with DNA hyper- and hypomethylation during aging were similar for different tissues, supporting the notion that chromatin context is associated with the dynamics of systemic DNA methylation changes during aging. The reasons why the repressive histone marks H3K27me3/H3K9me3 favor hypermethylation and the active histone mark H3K4me1 promotes hypomethylation during aging are not known. A simple explanation could be the preferential location of DNA methyltransferases (DNMTs) at repressive chromatin regions (Jeong et al. 2009). Repressive chromatin regions could be predisposed to becoming hypermethylated due to the abundance of DNMTs. In contrast, active chromatin regions would be more susceptible to losing methylation because the low levels of DNMTs at these regions make it more difficult to maintain DNA methylation patterns after mitosis. This possibility is supported by the fact that post-mitotic tissues such as brain (Numata et al. 2012; Guintivano et al. 2013) and muscle (Zykovich et al. 2014) present far fewer hypomethylated sequences during aging than do highly mitotic cells such as blood and MSCs. Further studies analyzing the genome-wide distribution of DNMTs during aging are needed to support this possibility.
One possible limitation of our study is the purification and the in vitro culture of MSCs (Calvanese et al. 2008; Choi et al. 2012), although this should have no great impact when comparing young and old MSCs as both sets of samples were cultured under exactly the same conditions. Moreover, cell heterogeneity, which is a major issue in DNA methylation studies (Houseman et al. 2012; Guintivano et al. 2013), has less impact in relation to MSCs because they are more homogeneous than blood cell populations. However, to minimize the impact of cell heterogeneity in our analysis of blood, we corrected DNA methylation data with a recently published algorithm (Houseman et al. 2012), which yielded slightly different sequences to those previously proposed, suggesting that some of the DNA changes previously identified might be cell-type dependent. However, as this algorithm considers only the major cell subtypes, possible variations driven by minor subtypes would not be detected. Another limitation of our study is that the differences in the number of individuals analyzed and different data analyses undertaken make the interpretation of the comparison of age-dependent DNA methylation changes in different cell types difficult. However, the conserved pattern of chromatin signatures in stem and differentiated cells suggests that H3K9me3/H3K27me3 and H3K4me1 are truly tissue-independent histone marks of DNA hyper- and hypomethylation, respectively, during aging.
As in most previous studies on DNA methylation and aging, CpG sites showing DNA methylation changes during lifetime associated with a specific tendency (i.e., hyper- or hypomethylation) were identified. However, using this analytical approach means that other possible changes occurring at CpG sites displaying high interindividual variability in both young and old individuals and/or age-dependent interindividual variability are overlooked. To address this issue, we reanalyzed the DNA methylation data to characterize the age-dependent interindividual variability (i.e., scedasticity).
Using this approach, we identified 16,243 heteroscedastic (2437 convergent and 13,806 divergent) and 55,684 homoscedastic CpG sites with high (HV) and 68,927 with low (LV) interindividual variability. Most of these CpG sites were not identified through linear model analysis, leading us to suggest that DNA methylation changes during aging might be more frequent than has previously been thought. Interestingly, although there were some CpG sites that converged during aging, most of the heteroscedastic changes were divergent, providing support for the notion that interindividual DNA methylation variability increases during lifetime (Gemma et al. 2013; Ong and Holbrook 2013). Although the behavior of adult stem cell populations during aging is still poorly understood (Pollina and Brunet 2011), the clonal expansion or decline of specific cell populations could affect the interpretation of changes of interindividual DNA methylation variability with aging. As it has been proposed that the number of MSCs declines with age (Stolzing et al. 2008), it is possible that the increase in interindividual variability might in fact be even larger than was observed in our study.
Functional genomics analyses of the groups of CpG sites established according to the behavior of the variance revealed that low variable CpG sites were enriched in CGIs and gene promoters. As DNA methylation occurring at CGI promoters has been proposed to play an important role in gene regulation (Bird 1986; Bird and Wolffe 1999; Calvanese et al. 2012), our results could indicate that the DNA methylation involved in gene regulation is protected against the stochastic epigenetic changes that occur during lifetime (Feil and Fraga 2012). Interestingly, analysis of the interindividual variability of DNA methylation during aging in blood, showed that, as in adult stem cells, the DNA methylation patterns of differentiated cells also diverge with age, thereby supporting the notion that a systemic epigenetic drift occurs during the lifetime of higher organisms (Feil and Fraga 2012; Issa 2014). To confirm that the sequences identified in blood after correcting with the Houseman algorithm were not affected by cell heterogeneity, we carried out in silico functional analysis to discard a possible blood cell lineage–dependent regulation. The analyses showed no meaningful associations, which further supports our contention that after correcting with the Houseman algorithm, cell heterogeneity had a minor impact on our blood DNA methylation data.
Previous reports have demonstrated that genetic factors play an important role in the regulation of DNA methylation during aging (Heijmans et al. 2007; Coolen et al. 2011; Gertz et al. 2011; Bell et al. 2012). To determine whether the effect of genotype is different depending on the intrinsic behavior of the DNA changes during aging at each specific CpG site, we analyzed the DNA methylation status of MZ twins of different ages. The results showed that interindividual variability increased with aging, in agreement with the notion that epigenetic drift during lifetime occurs even in genetically identical individuals (Fraga et al. 2005; Wong et al. 2010; Pirazzini et al. 2012; Talens et al. 2012; van Dongen et al. 2012). However, our results also showed that the DNA methylation status of some CpG sites may converge during lifetime. Specifically, the analysis of genetically identical individuals revealed that the effect of genotype depended on the intrinsic behavior of the DNA methylation changes during aging. For example, although the mechanisms underlying methylation convergence are still largely unknown, our MZ data indicate that genetic factors must be involved, at least in part, as the intrapair changes were similar to, or even less than, the interindividual variations. In addition, in contrast to the convergent and divergent CpG sites, genotype seems to play a less important role in whether the CpG sites display high or low interindividual variability, as evidenced by the fact that the increase in ED in the homoscedastic sequences for MZ twin pairs during aging was higher than the differences explained by interindividual variability. Of particular note is the finding that genotype had the lowest effect on the CpG sites, displaying high interindividual variability in young and old individuals, evidenced by the increase in ED in MZ twins during aging being similar to or even higher than the increase in interindividual variability. Our results indicate that these CpG sites, which have received little attention until now, might be important targets of environmental and/or stochastic epigenetic variation during development and aging. Although we have reduced the effect of cell heterogeneity and immune status over time (Allegretta et al. 1990) by using the Houseman algorithm (Houseman et al. 2012) and performing several functional in silico analyses of the groups of the genes showing age-related changes in ED, we cannot completely discount a partial effect of these in our results.
Our data indicate that the differences in the effect of genotype on DNA changes during lifetime depend largely on the genomic region involved, which is in agreement with previously published data (Wong et al. 2010). In line with this, the greatest DNA methylation changes for MZs were clustered at subtelomeric DNA regions, which suggests that the regulation of DNA methylation at these sequences is largely independent of genetic factors. Interestingly, subtelomeric DNA methylation has been shown to be affected by environmental factors (SM Tajuddin, AF Amaral, AF Fernández, S Chanock, DT Silverman, A Tardón, A Carrato, M García-Closas, BP Jackson, EG Toraño, et al., unpubl.). It is worth noting that although for most CpG sites the ED in young twins was lower than for older twins, they still clustered in the same subtelomeric regions, providing support for the previous proposal that epigenetic drift starts early in life (Kaminsky et al. 2009; Ollikainen et al. 2010; Wong et al. 2010; Martino et al. 2013) and accumulates during lifetime at particular CpG sites that, for still unknown reasons, evade the control of genetic factors (Fraga 2009).
Collectively, our results indicate that the dynamics of DNA methylation during lifetime in humans is associated with a complex mixture of factors. These include the DNA sequence itself, tissue type, and, in particular, the chromatin context, where repressive histone modifications such us H3K9me3 and H3K27me3 are related to DNA hypermethylation and, most notably, the active histone mark H3K4me1 is related to DNA hypomethylation. Finally, depending on the locus, the changes appear to be modulated by genetic and/or external factors.
Methods
Isolation and culture of MSCs
MSCs were purchased from Lonza (Verviers), Millipore (Billerica), and Inbiobank or directly obtained from young and elderly donors. After informed consent, bone marrow aspirates were obtained from one group of patients, and from a second group, bone scrapings were obtained following hip replacement surgery. Mononuclear cells were isolated by Ficoll density centrifugation (400 g, 25 min, 20°C) and washed twice by sedimentation with phosphate buffer (300 g, 5 min), and the cells were resuspended in MSC medium (DMEM plus 10% FBS) and seeded into culture flasks (Nunc) at 1.5 × 10−5 cells/cm2 and allowed to adhere for 24 h. MSCs were then cultured (37C, 5% CO2) in MSC medium. DNA methylation analyses were carried out at cell passages 4–6 (Supplemental Table 1).
MZ twins’ samples
Genomic DNA from 24 samples from the Italian Twin Registry, corresponding to 12 pairs of MZ twins, was extracted from buffy coats following standard procedures. Two different age groups were included for array-based DNA methylation profiling; one included individuals between 21 and 22 yr old (“young” MZ twins), and the other individuals between 58 and 66 yr (“old” MZ twins). The sample distribution by gender was the same in both groups.
Genome-wide DNA methylation analysis with high-density arrays
Microarray-based DNA methylation profiling was performed with Illumina's Infinium HumanMethylation450 BeadChip (Bibikova et al. 2011). Bisulfite conversion of DNA was performed using the EZ DNA methylation kit (Zymo Research) following the manufacturer’s procedures, with the modifications described in the Infinium assay methylation protocol guide. Processed DNA samples were then hybridized to the BeadChip, following the Illumina Infinium HD methylation protocol. Genotyping services were provided by the Centro Nacional de Genotipado (CEGEN-ISCIII) (www.cegen.org).
Data sets of blood and brain samples
DNA methylation data of blood (Hannum et al. 2013) and brain (neuron and glia) (Guintivano et al. 2013) samples produced with the HumanMethylation450 BeadChip were used to compare with the results obtained in MSCs. DNA methylation β-value data were downloaded from GEO accession numbers GSE40279 and GSE41826. The data analysis workflow is outlined in Supplemental Figure S1.
HumanMethylation450 BeadChip data preprocessing
IDAT files from the HumanMethylation450 BeadChip were processed further using the R/Bioconductor package minfi (R package version 1.7.15). In order to adjust for the different probe design types present in the HumanMethylation450 BeadChip architecture, red and green signals from the IDAT files were corrected using the SWAN algorithm (Makismovic et al. 2012). No background correction or control probe normalization was applied. Probes where at least two samples had detection P-values > 0.01 were filtered out. In accordance with the method of Du et al. (2010), both beta-values and M-values were computed and employed across the analysis pipeline. M-values were used for all the statistical analyses, assuming homoscedasticity (with the exception of the blood heterogeneity adjustment), while beta-values were mostly used for the intuitive interpretation and visualization of results.
Filtering confounding probes
Probes located in the X/Y chromosomes were removed from the data set when differential methylation profiles were analyzed. Probes that had been found to cohybridate with probes in the sex chromosomes (Lemire et al. 2013) were also removed. We used the information from the SNP137Common track from the UCSC Genome Browser (Sherry et al. 2001) in order to remove those probes with an SNP located inside their 2-bp central region.
Batch effect correction
Multidimensional scaling (MDS) was employed to detect whether there was any significant batch effect depending on the different HumanMethylation450 BeadChip plates that comprised the experiments. When there was, the ComBat method implemented in the R/Bioconductor package sva (R package version 3.7.0) was used to adjust the data sets accordingly, employing the variable “age” as the outcome of interest and the sample plate as a batch covariate.
White blood cell heterogeneity adjustment
Methylation data for the blood and twins’ data sets were adjusted for blood cell heterogeneity, with respect to the major cell subtypes, using the method described in Houseman et al. (2012). In order to feed this method, we used the original 27K database of purified white blood cell subtypes included in the original implementation of the algorithm. The correction was performed on the beta-values due to the fact that the 27K database was expressed using those units. M-values were obtained from the corrected beta-values for subsequent downstream analyses.
Detection of differentially methylated probes
For the MSC data set, the 34 samples were divided into two groups: young (ages ranging from 2 to 22 yr) and old (ages between 61 and 92 yr). Similarly, samples in the twins’ data set were divided into young (ages ranging from 21–22 yr) and old (age between 58 and 66 yr). For the neuron and glia data sets, the two groups were defined by taking those individuals whose age was below the 33rd percentile (young) and above the 66th percentile (old). Blood samples were not divided into groups, and the age predictor was used as a quantitative covariate. For the MSCs’, twins’, and neuron and glia data sets, significant methylation of a probe was determined by the moderated t-test implemented in the R/Bioconductor package limma (Smyth 2005). Probes in the blood data set were tested with a linear regression. A linear model, with methylation level as response and age as the only predictor, was used on all the data sets. P-values were corrected for multiple testing using the Benjamini-Hochberg method for controlling the false-discovery rate (FDR). A significance level of 0.05 was employed to determine differentially methylated probes. An additional threshold of effect size was applied, meaning that only those probes with the strongest differences between groups (the top 70%) were selected. The application of this threshold is essential to remove those differences prone to coming from technical artifacts, and consequently, ensure a more biologically sound statistical data analysis (Pan et al. 2005). Our threshold was adjusted according to the differences in M-values between groups in the brain and MSC data sets and the slope coefficients extracted from the blood data set linear regression model.
Analysis of variability trends
To analyze aging-dependent behavior of DNA interindividual variability (i.e., DNA methylation scedasticity), two groups, corresponding to young (samples where age is below the 33rd percentile) and old (those where age is above the 66th percentile) individuals, were selected for all the data sets. This separation allows the method to focus on the global tendency of the variability and to be less dependent on a fixed, underlying model. A Brown-Forsythe test for the equality of variances was used to determine which probes in the blood data set had significantly different variability in methylation between the two groups. For the remaining data sets, and due to the small number of available samples and low statistical power for conducting a variance test, a simple descriptive approach was used, labeling a probe as having a significant difference in methylation variability when the absolute value of the base-2 logarithm of the change of the variances for the two groups was greater than threefold. We did not apply any threshold of effect size for any of the data sets. For the blood data set, P-values were corrected for multiple testing using FDR (Benjamini-Hochberg method), and a significance level of 0.05 was used to determine which probes had a significant trend in variability. Two special subsets of probes with no significant trends in variability were generated: (1) HV (high variance), for those probes with variance values above the 75th percentile of the whole set of variances for both the young and old sample groups, and (2) LV (low variance), generated with those probes where both young and old variances were below the 25th percentile.
The in silico functional analysis and interpretation of the groups of genes established according to the behavior of the variance in blood was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) and the “Gene ontology” and “UP_TISSUE” categories (Dennis et al. 2003; Huang et al. 2009).
Measuring intra- and interindividual distance
EDs between twins were computed for every probe in the original twins’ data set using beta-values. In a simple scenario, the ED accounts for the absolute difference between the beta-values of the two siblings. Differences in distances were computed as the base-2 logarithm of the fold change between the average ED for the young and old sample groups.
Histone enrichment analysis
In order to analyze the enrichment of a histone mark on a given subset of probes, we used the information contained in the UCSC Genome Browser Broad histone track from the ENCODE Project (Supplemental Table 26; Rosenbloom et al. 2010, 2012). Histone peak data for every histone modification and chromatin modifier in hESCs and 10 different cell types obtained from healthy individuals were downloaded from the UCSC Genome Browser. Small peaks were discarded when they were completely contained within wider peaks. Following the ENCODE Broad histone methods description, discrete intervals of ChIP-seq fragment enrichment were identified using Scripture, a scan statistics approach, under the assumption of uniform background signal (http://genome.ucsc.edu//cgi-bin/hgTrackUi?db=hg19&g=wgEncodeBroadHistone).
For each combination of cell line and mark, a 2 × 2 contingency table was built to determine its association with the input subset of probes. Probes in the array were classified according to whether they belonged to the subset or not, and whether they intersected with a significant Broad peak for the given combination of cell line and mark. A Fisher’s exact test was used to determine if the given subset of probes was significantly enriched for each combination of cell line and mark. P-values were corrected for multiple testing using FDR (Benjamini-Hochberg method), and a significance level of 0.05 was used to determine which probes had significant enrichment. The base-2 logarithm of the odds ratio was used as a measure of effect size.
Genomic region analysis
The probes in the microarray were assigned a genomic region according to their position relative to the transcript information extracted from the R/Bioconductor package TxDb.Hsapiens.UCSC.hg19.knownGene (R package version 2.9.2). A probe was said to be in a “promoter” region if it was located inside the first exon, the 5′ UTR, or a region up to 2 kbp upstream of the transcription start site (TSS) of any given transcript. Similarly, a probe was labeled as “intragenic” if it was inside any intron or any exon other than the first. Intergenic probes were determined as those not falling into either of the two previous categories. According to this definition, a probe could be in both a promoter and an intragenic region at the same time for different transcripts. A contingency table was built for each selected subset of probes and a given genomic region, with one variable indicating whether a given probe belonged or not to the subset, and the other indicating whether a given probe was labeled with the selected region. Significance of the association was determined by a Pearson’s χ2 test with Yates’ continuity correction. A significance level of 0.05 was used to determine whether a subset was dependent with respect to a given genomic region. An odds ratio was used as a measure of effect size.
CGI status analysis
The CGI locations used in the analyses were obtained from the R/Bioconductor package FDb.InfiniumMethylation.hg19 (R package version 1.0.1). The generation procedure of these CGIs is described by Wu et al. (2010). “CpG shores” were defined as the 2-kbp regions flanking a CGI. “CpG shelves” were defined as the 2-kbp regions either upstream of or downstream from each CpG shore. Probes not belonging to any of the regions thus far mentioned were assigned to the special category “non-CGI.” Each probe was assigned to only one of the categories. A 4 × 2 contingency table was constructed for every subset of probes in order to study the association between the given subset and the different CGI categories. A χ2 test was used to determine if any of the categories had a significant association with the given subset. For each of the CGI status levels, a 2 × 2 contingency table was defined and another χ2 test was used to independently evaluate the association of the given subset with each status level, a significance level of 0.05 being employed for all tests. Effect size was reported as the odds ratio for each of the individual tests.
Microarray background correction
Although it is sometimes referred to as a genome-wide solution, the HumanMethylation450 BeadChip only covers a fraction of the entire genome. In its 27K predecessor, the probes were mainly located at gene promoter regions, while in addition to the promoter probes, the HumanMethylation450 BeadChip includes probes located inside genes and in intergenic regions (Dedeurwaerder et al. 2011).
The irregular distribution of probes can lead to unwanted biases when studying whether a selected subset of probes is enriched with respect to any functional or clinical mark. A reference to the background distribution of features was included in every type of statistical test performed in order to prevent our conclusions from being driven by the irregular distribution of probes. In qualitative tests (CGI status, genomic region, or histone mark enrichment), the contingency matrix was built to represent the background distribution of the microarray. Thus, any significant result would indicate a departure from the fixed background distribution and ignore any manufacturer bias.
Circos data track smoothing
In order to plot the CpG information on the Circos genome-wide graphs (Krzywinski et al. 2009), smoothing was applied to our data. Broad histone peak information from the UCSC Genome Browser was averaged by partitioning the genome into intervals of 200 kbp and assigning to each a score corresponding to the average of the Broad peak scores found within it. CpG locations were not smoothed. Distances in the twins’ data set were averaged using a 2-Mbp window size.
Bisulfite pyrosequencing
DNA methylation patterns of representative dmCpGs during aging were analyzed by bisulfite pyrosequencing in an independent sample set of 46 MSCs obtained from individuals of different ages (Supplemental Table 1). Bisulfite modification of DNA was performed with the EZ DNA methylation-gold kit (Zymo Research) following the manufacturer’s instructions. The set of primers for PCR amplification and sequencing were designed using the specific software PyroMark assay design (version 2.0.01.15). Primer sequences were designed to hybridize with CpG free sites to ensure methylation-independent amplification (Supplemental Table 27). After PCR amplification of the region of interest with the specific primers, pyrosequencing was performed using PyroMark Q24 reagents and a vacuum prep workstation, equipment, and software (Qiagen). A linear regression model was fitted to the pyrosequencing methylation data using age as a predictor.
Data analysis workflow
All the necessary steps for upstream and downstream analyses were defined and implemented using the Snakemake tool (Köster and Rahmann 2012). This tool helps data scientists to generate a reproducible and inherently parallel processing pipeline. The source code of the workflow is included as Supplemental Material.
Data access
The HumanMethylation450 BeadChip data sets from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE52114 (subseries GSE52112 and GSE52113).
Supplementary Material
Acknowledgments
We thank Ronnie Lendrum for manuscript preparation and Tim Triche Jr. for his invaluable advice. This work has been financially supported by the Plan Nacional de I+D+I 2008-2011/2013-2016/FEDER (PI11/01728 to A.F.F., PI 12/0615 to J.A.R., PI10/0449 to P.M., and PI11/0119 to C.B.); the ISCIII-Subdirección General de Evaluación y Fomento de la Investigación (Miguel Servet contracts CP11/00131 to A.F.F. and CP07/0059 to C.B.); the Spanish Ministry of Health (PS09/02454 and PI12/01080 to M.F.F.); the Spanish National Research Council (CSIC; 200820I172 to M.F.F.); IUOPA (to C.F. and G.F.B.); Fundacion Cientifica de la AECC (to R.G.U. and P.M.); Fundación Ramón Areces (to M.F.F.); and FICYT (to E.G.T.). J.G.-C. receives funding from the Fondo de Investigaciones Sanitarias (FIS; PI05/2217 and PI08/0029) and the Madrid Regional Government (S-BIO-0204-2006 and S2010/BMD-2420). J.A.R. receives funding from the Fondo de Investigaciones Sanitarias (ISCIII-FIS PI 12/0615). P.M. is also supported by MINECO (SAF2013/43065), ERANET E-Rare (PI112/03112), and Fundación Sandra Ibarra. P.M. also acknowledges support from Obra Social “La Caixa/Fundacio Josep Carreras.” The IUOPA is supported by the Obra Social Cajastur, Spain.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.169011.113.
References
- Allegretta M, Nicklas JA, Sriram S, Albertini RJ. 1990. T cells responsive to myelin basic protein in patients with multiple sclerosis. Science 247: 718–721. [DOI] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. . 2006. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 441: 1011–1014. [DOI] [PubMed] [Google Scholar]
- Beerman I, Bock C, Garrison BS, Smith ZD, Gu H, Meissner A, Rossi DJ. 2013. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell 12: 413–425. [DOI] [PubMed] [Google Scholar]
- Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, et al. . 2012. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet 8: e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. . 2011. High density DNA methylation array with single CpG site resolution. Genomics 98: 288–295. [DOI] [PubMed] [Google Scholar]
- Bird AP. 1986. CpG-rich islands and the function of DNA methylation. Nature 321: 209–213. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. 1999. Methylation-induced repression: belts, braces, and chromatin. Cell 99: 451–454. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Cui H, Gius D, Fallin MD, Feinberg AP. 2004. The new field of epigenomics: implications for cancer and other common disease research. Cold Spring Harb Symp Quant Biol 69: 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. 2011. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood 117: e182–e189. [DOI] [PubMed] [Google Scholar]
- Bocklandt S, Lin W, Sehl ME, Sanchez FJ, Sinsheimer JS, Horvath S, Vilain E. 2011. Epigenetic predictor of age. PLoS ONE 6: e14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork S, Pfister S, Witt H, Horn P, Korn B, Ho AD, Wagner W. 2010. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell 9: 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V, Horrillo A, Hmadcha A, Suarez-Alvarez B, Fernandez AF, Lara E, Casado S, Menendez P, Bueno C, Garcia-Castro J, et al. . 2008. Cancer genes hypermethylated in human embryonic stem cells. PLoS ONE 3: e3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V, Fernandez AF, Urdinguio RG, Suarez-Alvarez B, Mangas C, Perez-Garcia V, Bueno C, Montes R, Ramos-Mejia V, Martinez-Camblor P, et al. . 2012. A promoter DNA demethylation landscape of human hematopoietic differentiation. Nucleic Acids Res 40: 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen MW, Statham AL, Qu W, Campbell MJ, Henders AK, Montgomery GW, Martin NG, Clark SJ. 2011. Impact of the genome on the epigenome is manifested in DNA methylation patterns of imprinted regions in monozygotic and dizygotic twins. PLoS ONE 6: e25590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MR, In YH, Park J, Park T, Jung KH, Chai JC, Chung MK, Lee YS, Chai YG. 2012. Genome-scale DNA methylation pattern profiling of human bone marrow mesenchymal stem cells in long-term culture. Exp Mol Med 44: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, et al. . 2009. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 5: e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day K, Waite LL, Thalacker-Mercer A, West A, Bamman MM, Brooks JD, Myers RM, Absher D. 2013. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol 14: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. 2011. Evaluation of the Infinium Methylation 450K technology. Epigenomics 3: 771–784. [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3. [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. 2010. Comparison of β-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. 2012. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 13: 97–109. [DOI] [PubMed] [Google Scholar]
- Fernandez AF, Assenov Y, Martin-Subero JI, Balint B, Siebert R, Taniguchi H, Yamamoto H, Hidalgo M, Tan AC, Galm O, et al. . 2012. A DNA methylation fingerprint of 1628 human samples. Genome Res 22: 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF. 2009. Genetic and epigenetic regulation of aging. Curr Opin Immunol 21: 446–453. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. . 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci 102: 10604–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage PJ, Kuang C, Zacharias AL. 2014. The homeodomain transcription factor PITX2 is required for specifying correct cell fates and establishing angiogenic privilege in the developing cornea. Dev Dyn 243: 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemma C, Ramagopalan SV, Down TA, Beyan H, Hawa MI, Holland ML, Hurd PJ, Giovannoni G, David Leslie R, Ebers GC, et al. . 2013. Inactive or moderately active human promoters are enriched for inter-individual epialleles. Genome Biol 14: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Varley KE, Reddy TE, Bowling KM, Pauli F, Parker SL, Kucera KS, Willard HF, Myers RM. 2011. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet 7: e1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Krause Y, Wuelling M, Vortkamp A. 2012. Hoxa11 and Hoxd11 regulate chondrocyte differentiation upstream of Runx2 and Shox2 in mice. PLoS ONE 7: e43553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA. 2013. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics 8: 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, et al. . 2013. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 49: 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. 2007. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet 16: 547–554. [DOI] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, et al. . 2011. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet 20: 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ, et al. . 2012. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci 109: 10522–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Moran S, Esteller M. 2013. Aberrant DNA methylation profiles in the premature aging disorders Hutchinson-Gilford Progeria and Werner syndrome. Epigenetics 8: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MP, van Eijk K, van den Berg LH, Ophoff RA. 2012. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol 13: R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Issa JP. 2014. Aging and epigenetic drift: a vicious cycle. J Clin Invest 124: 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet (Suppl) 33: 245–254. [DOI] [PubMed] [Google Scholar]
- Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA. 2009. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol 29: 5366–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A, Enroth S, Gyllensten U. 2013. Continuous aging of the human DNA methylome throughout the human lifespan. PLoS ONE 8: e67378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A, Teschendorff AE, Li Q, Hayward JD, Kannan A, Mould T, West J, Zikan M, Cibula D, Fiegl H, et al. . 2013. Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. PLoS Med 10: e1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, et al. . 2009. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 41: 240–245. [DOI] [PubMed] [Google Scholar]
- Köster J, Rahmann S. 2012. Snakemake: a scalable bioinformatics workflow engine. Bioinformatics 28: 2520–2522. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire M, Butcher DT, Grafodatskaya D, Zanke BW, Weksberg R. 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. . 2013. Global epigenomic reconfiguration during mammalian brain development. Science 341: 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Reinius L, Acevedo N, Taub M, Ronninger M, et al. . 2013. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol 31: 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makismovic J, Gordon L, Oshlack A. 2012. SWAN: Subset-quantile within array normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol 13: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino D, Loke YJ, Gordon L, Ollikainen M, Cruickshank MN, Saffery R, Craig JM. 2013. Longitudinal, genome-scale analysis of DNA methylation in twins from birth to 18 months of age reveals rapid epigenetic change in early life and pair-specific effects of discordance. Genome Biol 14: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, Colantuoni C, Weinberger DR, Kleinman JE, Lipska BK. 2012. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet 90: 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollikainen M, Smith KR, Joo EJ, Ng HK, Andronikos R, Novakovic B, Abdul Aziz NK, Carlin JB, Morley R, Saffery R, et al. . 2010. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet 19: 4176–4188. [DOI] [PubMed] [Google Scholar]
- Ong ML, Holbrook JD. 2013. Novel region discovery method for Infinium 450K DNA methylation data reveals changes associated with ageing in muscle and neuronal pathways. Aging Cell 13: 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KH, Lih CJ, Cohen SN. 2005. Effects of threshold choice on biological conclusions reached during analysis of gene expression by DNA microarrays. Proc Natl Acad Sci 102: 8961–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirazzini C, Giuliani C, Bacalini MG, Boattini A, Capri M, Fontanesi E, Marasco E, Mantovani V, Pierini M, Pini E, et al. . 2012. Space/population and time/age in DNA methylation variability in humans: a study on IGF2/H19 locus in different Italian populations and in mono- and di-zygotic twins of different age. Aging 4: 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollina EA, Brunet A. 2011. Epigenetic regulation of aging stem cells. Oncogene 30: 3105–3126. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2010. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, et al. . 2010. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res 20: 434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch T, Li H, Wu X, Pfeifer GP. 2006. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res 66: 7939–7947. [DOI] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. . 2010. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res 38: D620–D625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, et al. . 2012. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res 40: D912–D917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. 2004. Telomeres, stem cells, senescence, and cancer. J Clin Invest 113: 160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Petry M, Haenig B, Lescher B, Leitges M, Kispert A. 2005. The T-box transcription factor Tbx15 is required for skeletal development. Mech Dev 122: 131–144. [DOI] [PubMed] [Google Scholar]
- Smyth GK. 2005. limma: linear models for microarray data. In Bioinformatics and computational biology solutions using R and Bioconductor (ed. Gentleman VCR, et al.), pp. 397–420. Springer, New York. [Google Scholar]
- Stolzing A, Jones E, McGonagle D, Scutt A. 2008. Age-related changes in human bone marrow-derived mesenchymal stem cells: consequences for cell therapies. Mech Ageing Dev 129: 163–173. [DOI] [PubMed] [Google Scholar]
- Taiwo O, Wilson GA, Emmett W, Morris T, Bonnet D, Schuster E, Adejumo T, Beck S, Pearce DJ. 2013. DNA methylation analysis of murine hematopoietic side population cells during aging. Epigenetics 8: 1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talens RP, Christensen K, Putter H, Willemsen G, Christiansen L, Kremer D, Suchiman HE, Slagboom PE, Boomsma DI, Heijmans BT. 2012. Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell 11: 694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. . 2010. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res 20: 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timp W, Feinberg AP. 2013. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer 13: 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong WG, Wierda WG, Lin E, Kuang SQ, Bekele BN, Estrov Z, Wei Y, Yang H, Keating MJ, Garcia-Manero G. 2010. Genome-wide DNA methylation profiling of chronic lymphocytic leukemia allows identification of epigenetically repressed molecular pathways with clinical impact. Epigenetics 5: 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J, Slagboom PE, Draisma HH, Martin NG, Boomsma DI. 2012. The continuing value of twin studies in the omics era. Nat Rev Genet 13: 640–653. [DOI] [PubMed] [Google Scholar]
- Wong CC, Caspi A, Williams B, Craig IW, Houts R, Ambler A, Moffitt TE, Mill J. 2010. A longitudinal study of epigenetic variation in twins. Epigenetics 5: 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Caffo B, Jaffee HA, Irizarry RA, Feinberg AP. 2010. Redefining CpG islands using hidden Markov models. Biostatistics 11: 499–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Hong C, Zhang B, Lowdon RF, Xing X, Li D, Zhou X, Lee HJ, Maire CL, Ligon KL, et al. . 2013. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat Genet 45: 836–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykovich A, Hubbard A, Flynn JM, Tarnopolsky M, Fraga MF, Kerksick C, Ogborn D, MacNeil L, Mooney SD, Melov S. 2014. Genome-wide DNA methylation changes with age in disease-free human skeletal muscle. Aging Cell 13: 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.