Abstract
Transcriptional profile based subtypes of cancer are often viewed as identifying different diseases from the same tissue origin. Understanding the mechanisms driving the subtypes may be key in development of novel therapeutics but is challenged by lineage-specific expression signals. Using a t-test statistics approach we compared gene expression subtypes across twelve tumor types, which identified eight transcriptional superclusters characterized by commonly activated disease pathways and similarities in gene expression. One of the largest superclusters was determined by the upregulation of a proliferation signature, significant enrichment in TP53 mutations, genomic loss of CDKN2A (p16ARF), evidence of increased numbers of DNA double strand breaks and high expression of cyclin B1 protein. These correlations suggested that abrogation of the P53 mediated apoptosis response to DNA damage results in activation of cell cycle pathways and represents a common theme in cancer. A second consistent pattern, observed in nine of eleven solid tumor types, was a subtype related to an activated tumor-associated stroma. The similarity in transcriptional footprints across cancers suggested that tumor subtypes are commonly unified by a limited number of molecular themes.
INTRODUCTION
Cancer is a genetic disease in which genomic abnormalities alter the transcriptome, thereby directly or indirectly deregulating the pathways that control proliferation and survival. Large scale efforts to systematically catalogue the landscape of somatic alterations that contributes to tumorigenesis, such as The Cancer Genome Atlas (TCGA), have shown that extensive genomic heterogeneity within and across tumor types exists, but that alterations in pathways such as the p53 pathway or the receptor tyrosine kinase pathway represent common themes1–7. The transcriptomic diversity in cancer has been captured by robust expression subtypes that are characterized by similarity to gene signatures related to developmental lineages and cellular differentiation8–10. Furthermore, molecular subtypes are frequently found to associate with somatic alterations, such as EGFR abnormalities in the classical subtype of glioblastoma10, or the enrichment of NF1 deletions and mutations in the primitive group of lung squamous carcinoma4. Classifying patients into subgroups on the basis of their expression profiles may have clinical relevance including correlations with clinical parameters such as drug response, tumor stage, or survival outcome11, 12.
The associations between transcriptional profile and genomic abnormalities suggest that regulatory networks could be uncovered through integrated analysis of RNA expression, DNA copy number, mutation and other genomic data types. However, this analysis may be hindered by the dominant effect of cellular differentiation on transcription levels, which is unrelated to tumorigenesis. One example is the above mentioned glioblastoma subtypes, which not only associate with genomic abnormalities but also show preferential activation of different neural cell signatures10 and may represent different cells of origin or differentiation down alternative neural cell pathways. Similarly, unsupervised clustering of expression profiles from acute myeloid leukemia identified associations with the French-American-British-classification which is based on cellular morphology and resemblance to various stages of normal hematopoietic development13. By comparing transcriptional signatures across different tumor types, the effects of cellular lineage may be minimized allowing commonalities related to the tumorigenic process to be recognized. For example, TCGA recently reported that the breast carcinoma basal subtype shares genomic as well as transcriptomic features with high-grade serous ovarian cancer, leading to the speculation that therapeutic strategies that are successful in the treatment of ovarian carcinoma may have similar efficacy in the poor prognosis basal breast cancers3.
We hypothesized that common tumorigenic processes exist across cancer types and that oncogenic pathways can be exposed through pan-cancer comparison of expression subtypes from different tissue origins. To validate our hypothesis, we analyzed the expression profiles of 3,444 samples from twelve tumor types, available through The Cancer Genome Atlas consortium. Our analysis identified common components in the expression subtype gene signatures across different tumor types, thereby eliminating the contribution of lineage and exposed the presence of Pan-cancer superclusters. Finally, we provided further insights into the molecular basis of these superclusters through annotation with genomic abnormalities, pathway activation scoring and clinical annotation.
RESULTS
Transcriptome based pan-cancer clustering is primarily driven by tumor lineage and histology
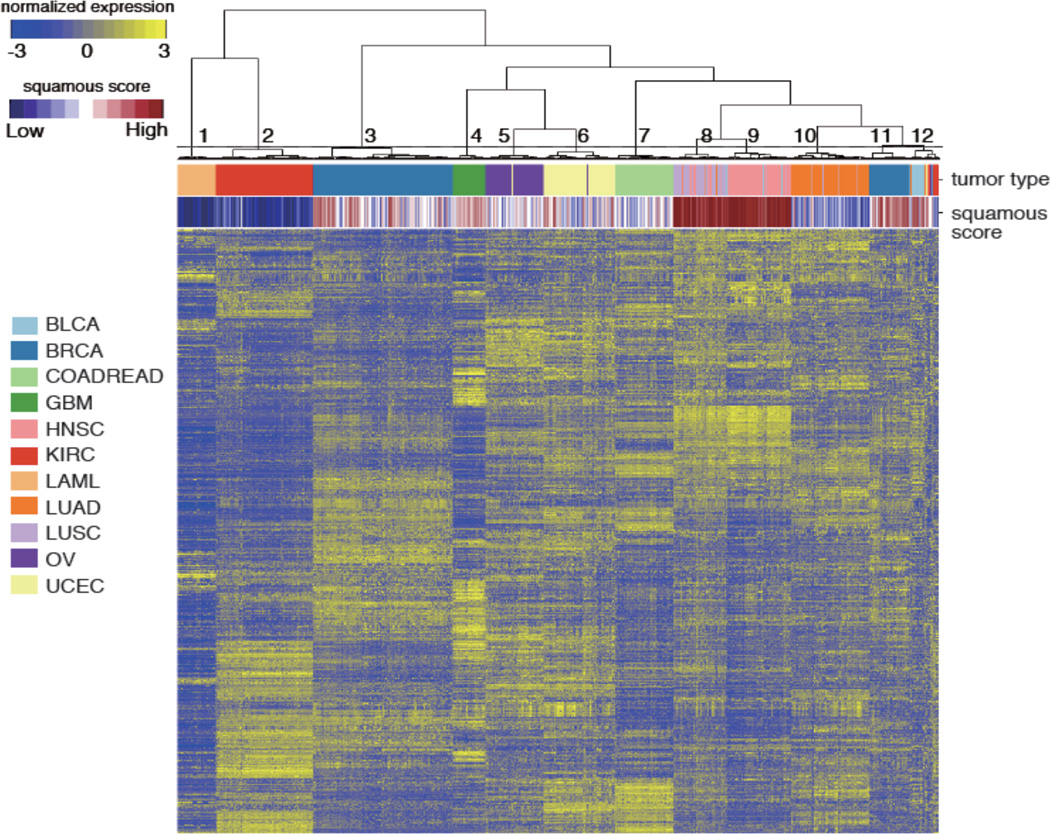
To identify pan-cancer gene expression subtypes, we performed unsupervised hierarchical clustering of 3,444 expression profiles from twelve different tumor types data sets: acute myeloid leukemia (LAML, n = 173), bladder urothelial carcinoma (BLCA, n = 96), breast cancer (n = 817), colon adenocarcinoma (COAD, n = 192), rectal adenocarcinoma (READ, n = 71), glioblastoma (GBM, n= 154), head and neck squamous cell carcinoma (HNSC, n = 303), clear cell renal cell carcinoma (KIRC, n = 470), lung adenocarcinoma (LUAD, n = 353), lung squamous cell carcinoma (LUSC, n = 220), ovarian cancer (OV, n = 262), and uterine corpus endometrial carcinoma (UCEC, n = 333) using the top 1,500 genes with the largest variance across all samples. Expression data was downloaded from TCGA. Visual inspection of the dendrogram and gene expression heatmap strongly suggested twelve clusters, ten of which were highly enriched for a specific tumor type (Figure 1, Supplementary Table 1). Squamous cell carcinoma samples of lung, head and neck and a subset of bladder urothelial carcinoma were found in two clusters, suggesting that the squamous histology is a driving force behind these subsets. We obtained comparing gene expression profiles from lung, cervical and esophageal cancers and cell lines from public resources and derived a squamous cell signature by comparing squamous cancers to adenocarcinomas. We used the squamous cell signature to annotate all samples according to their level of “squamous-ness” and found that cluster eight and nine, containing 87.7% of lung squamous and 99.7% of head and neck squamous cancers, showed high levels of squamous marker expression. Clusters eleven, made up by 22.5% of breast cancers and cluster twelve, which included 70.8% of the bladder cancer samples, complemented by 6.8% of renal cancers, 4.5% of lung squamous cancers and 25 tumor samples from all tumor types except LAML, additionally correlated with squamous cell phenotype. In summary, tumor lineage and squamous histology were the major drivers behind the unsupervised clustering across tumor types.
Figure 1. Unsupervised hierarchical clustering of 3,444 samples across twelve different tumor types.
Unsupervised hierarchical clustering of the expression profiles from 3,444 tumor samples was performed by using the top 1,500 most variable genes across all samples according to median absolute deviation (MAD). Sidebars indicate tumor type and the squamous score for each sample. Cluster number is labeled on the dendrogram.
Comparison of expression subtypes across cancer
To be able to extract transcription-based tumorigenic processes without being limited by the dominating influence of tumor lineage, we performed a pan-cancer comparison on the basis of gene expression subtypes. Subtype classification annotation was obtained from the OV, COADREAD, UCEC, LUSC, BLCA, LUAD, GBM, LAML, HNSC, and KIRC TCGA disease working group2, 4–7, 10, 14–17. To identify expression subtypes of BRCA, we clustered samples for each tumor type using the non-negative matrix factorization (NMF) algorithm on the 1,500 variably expressed genes18. This resulted in five subtypes for BRCA. In total, 45 expression subtypes were included for further analyses (Supplementary Table 2).
To decrease the contributions from tumor lineage and to identify patterns that supersede tumor subtypes, we calculated t-test scores by comparing gene expressions levels of each tumor subtype to other clusters of the same cancer. This resulted in a t-test score matrix of 45 columns (subtypes) and 11,186 rows, representing genes that were common to the four gene expression platforms. To show similarity between expression subtypes, we calculated pairwise Pearson’s correlation coefficient for all subtype combinations using the top 1,500 genes amongst t-scores, ranked by the median absolute deviation (MAD) and performed hierarchical cluster analysis of the 45 expression subtypes. We observed the highest correlation density, silhouette scores when cutting the dendrogram tree at eight clusters (Supplementary Figure 1–3). The superclusters consisted of at least two subtypes and named them “supercluster 1” to “supercluster 8” (Figure 2). We did not observe correlations of tissue source site with subtype or supercluster, nor did we find batch effects related to gene expression platforms (data not shown).
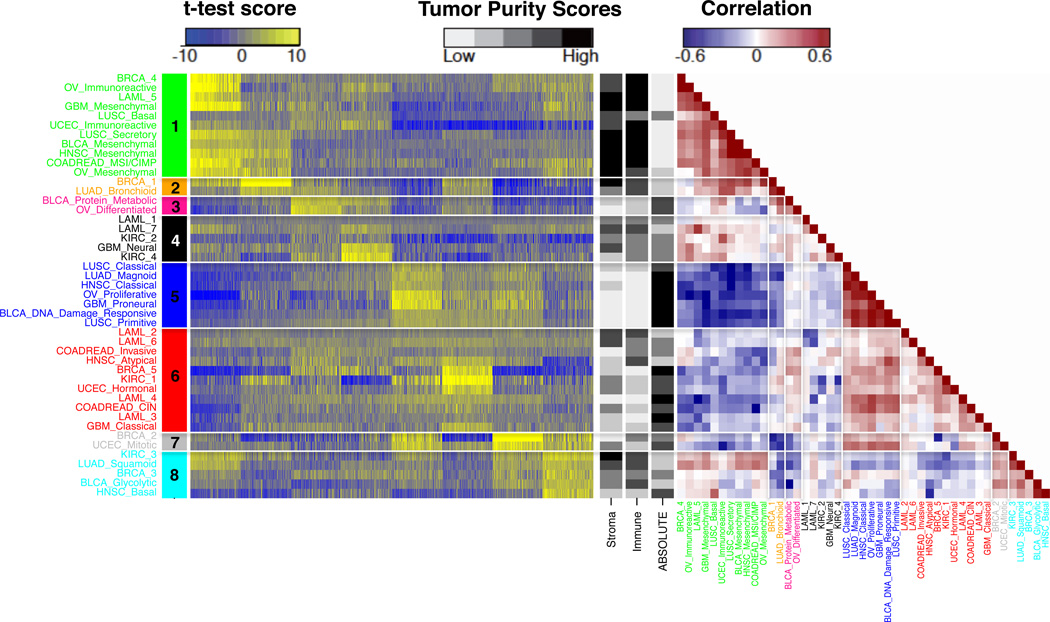
Figure 2. Identification of superclusters.
Left panel: Heatmap of differences in expression levels between superclusters. Displayed are the t-test scores of the top 200 ranked genes per supercluster (mean), computed per subtype versus the other subtypes of the same cancer. Row and column represent 44 subtypes and 1,600 top ranked genes, respectively. Right: Correlation heatmap of fourty-four subtypes, divided into eight superclusters by hierarchical clustering (See Supplementary Figure 1). Black and white scaled sidebars indicate the stromal and immune scores (16) and tumor purity inferred using ABSOLUTE24 for each subtype.
To validate the stability and robustness of eight superclusters, we adopted two methods. First, we randomly selected different percentages of samples (50–90%) and used the reduced data sets to evaluate whether superclusters were retained. The validation rate of supercluster formation removing 50, 60, 70, 80 or and 90% of samples was consistently higher than 90%, suggesting that the superclusters we identified were robust and reproducible. Next, we compared superclusters detected in the TCGA data set with those identified in an independent validation data set. We used subtype gene signatures to classify 2,550 expression profiles from publicly available resources (Supplementary Table 3). We found that 41 of 45 subtypes were detected in the validation data set and clustering these subtypes showed that five superclusters (supercluster 1, 2, 3, 5, and 8) were retained in their entirety (Supplementary Figure 9). Further details are described in the Supplementary Text.
While tumor lineage played a dominant role when clustering individual samples, comparing cancer on the basis of expression subtypes showed extensive intermingling of clusters from different tumor types. Each of the eight superclusters included subtypes from at least two different tumor types. The largest “supercluster 1” combined subtypes from nine different cancer origins (LAML, BLCA, BRCA, COADREAD, GBM, HNSC, LUSC, OV, and UCEC). Correlations between subtypes, which were based on t-test vectors of subtype specific differences in gene expression, ranged between 0.3 and 0.8 in the strongest groups, such as supercluster 1 and supercluster 5 (Figure 2).
A tumor-associated normal cell supercluster
To link superclusters to disease pathways, we used single sample gene set enrichment analysis (ssGSEA) and PAthway Recognition Algorithm using Data Integration on Genomic Models (PARADIGM), respectively19, 20. ssGSEA quantifies the activation level of a gene set in a particular sample by comparing the expression based ranking of genes within the gene set relative to all genes. Gene sets (n = 8,513) were obtained from the Molecular Signatures Database (MSigDB version 3.1)21. ssGSEA scores were calculated for each gene set and each sample. To extract gene sets associated to superclusters, we computed a t-statistic of the ssGSEA scores for each subtype by comparing one subtype versus others, within tumor type. Next, gene sets were ranked in descending order according to the lower quartile of t-test scores amongst the subtype members of each supercluster (Supplementary Table 4). PARADIGM infers integrated pathway activities (IPAs) by integrating transcriptional levels and DNA copy number profiles20. We generated IPAs from >1,200 curated signal transduction, transcriptional, and metabolic pathways22, and estimated a score for each subtype and each IPA based on a t-statistic by comparison of one subtype with others per tumor type (Supplementary Table 5).
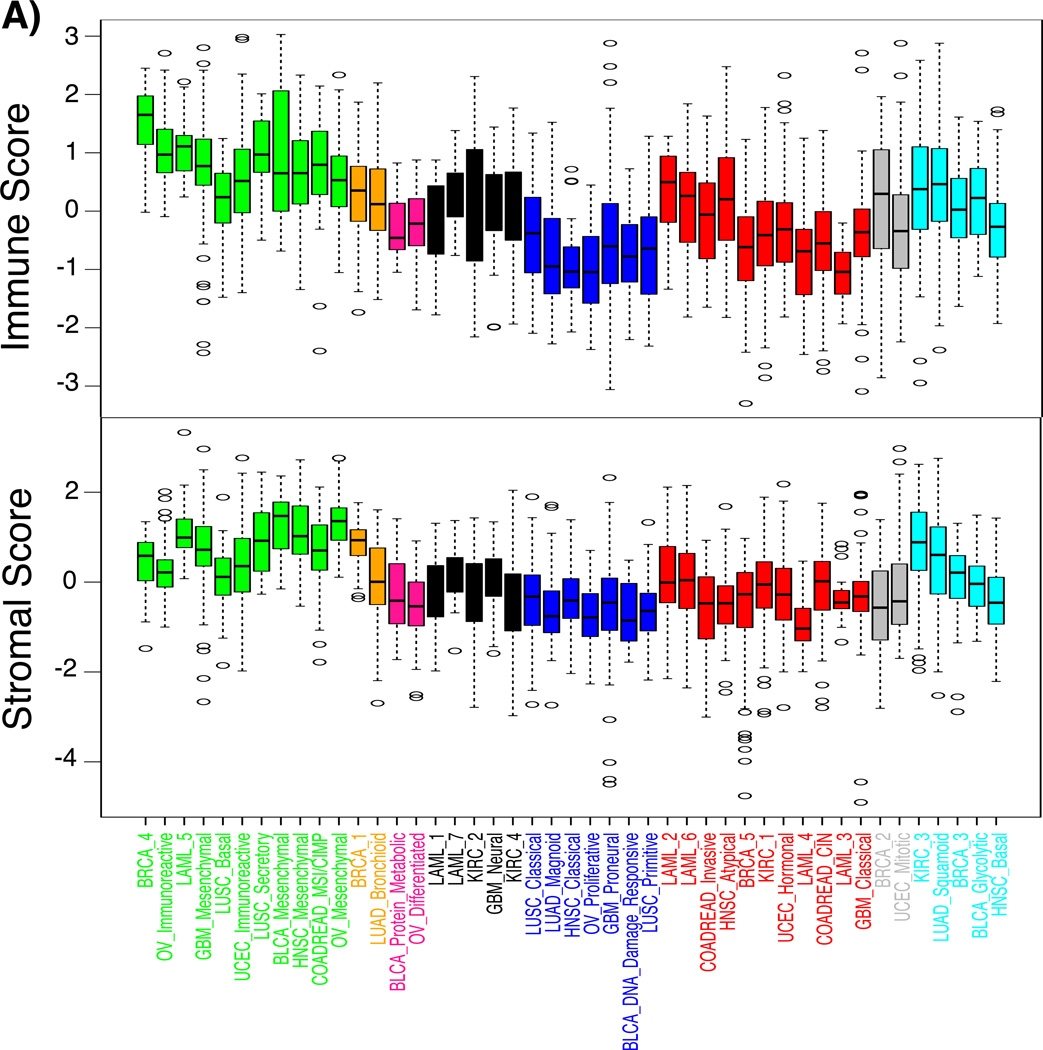
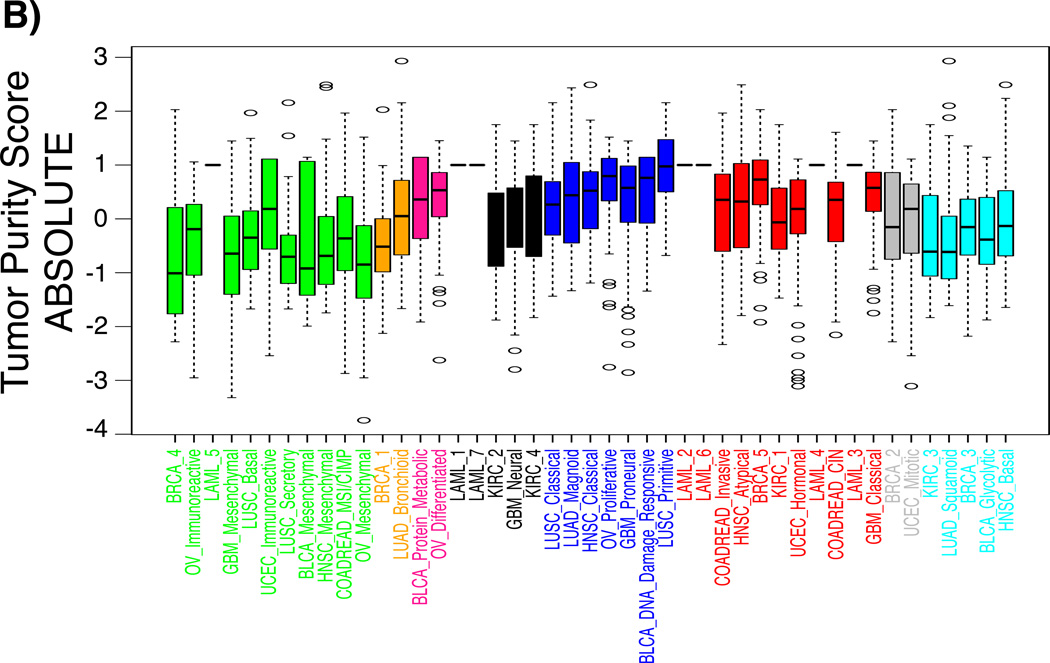
Both ssGSEA and PARADIGM found overrepresented pathways related to “immune system” or “cell adhesion” in supercluster 1, suggesting that this supercluster was linked to tumor microenvironment and tumor-associated normal cells (Figure 3). We used our ESTIMATE method to determine the level of infiltrating stromal or immune cells in each sample23. As expected, both stromal and immune scores were significantly higher in supercluster 1 compared to other superclusters (stromal score, posthoc maximum P = 0.00065; immune score, posthoc maximum P = 1.3E-15) (Figure 4). The increased level of immune cell scores pertained to the BRCA_4, OV_Immunoreactive, LAML_5, and LUSC_Secretory subtypes, whereas stromal scores were increased in the BLCA_Mesenchymal, HNSC_Mesenchymal, LUSC_secretory, COADREAD_MSI/CIMP, OV_Mesenchymal, GBM_Mesenchymal and LAML_5 classes. The association of supercluster 1 and increased volumes of tumor-associated normal cells was confirmed by relatively lower tumor purity (posthoc maximum P = 0.00867; Figure 4B), with tumor purity being estimated by applying the ABSOLUTE method on DNA copy number profiles24.
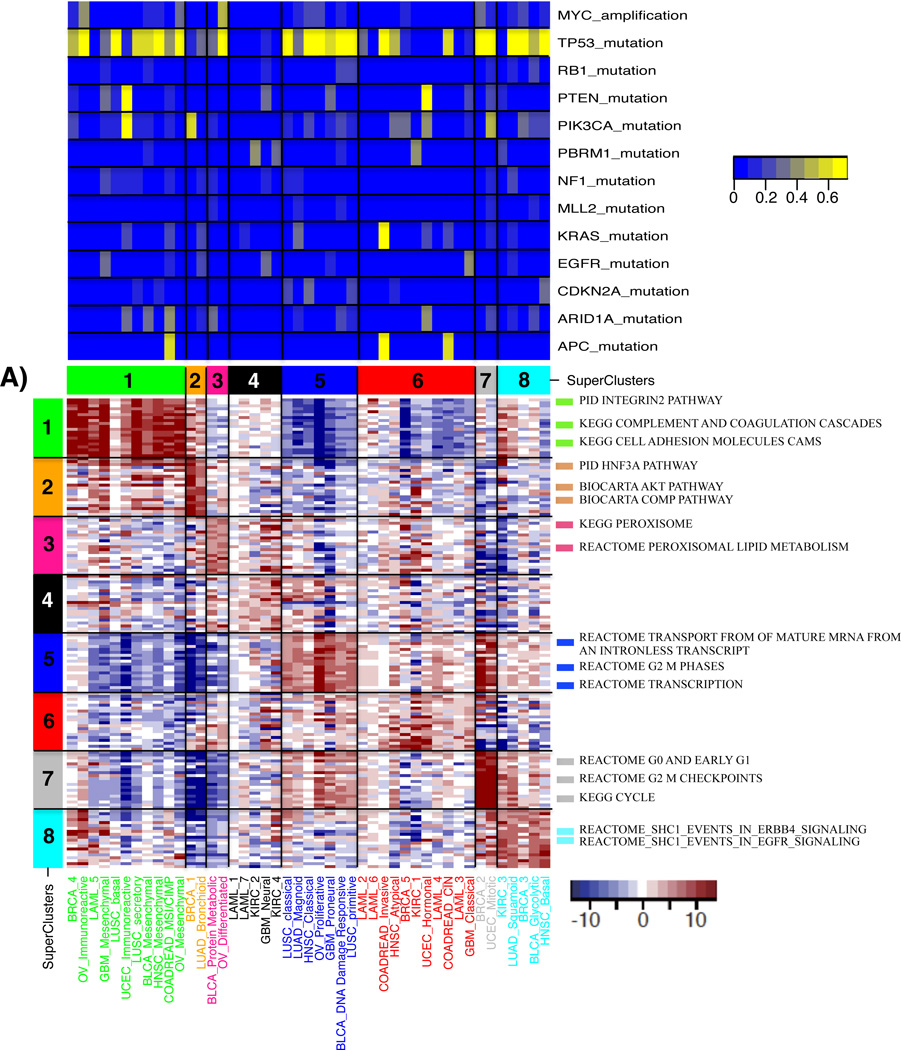
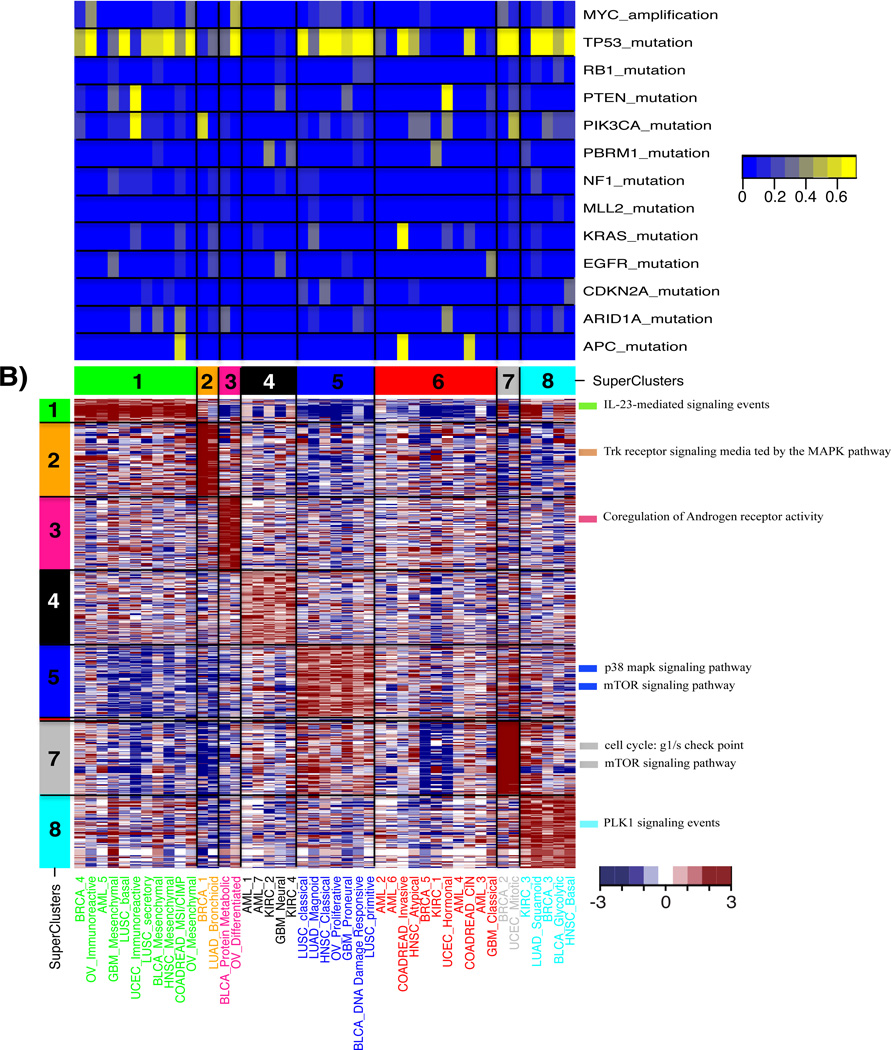
Figure 3. Molecular characteristics of superclusters by pathway analyses.
Top: percentage of altered samples per subtype. a) A heatmap of the t-test statistics per subtype calculated by using the ssGSEA scores for the canonical pathways gene sets from MsigDB. Top 20 pathways ranked by t-test statistic scores per supercluster are shown in the heatmap. Representative gene sets with high scores are shown on the right side of heatmap. T-test statistic scores for all pathways and subtypes are summarized in Supplementary Table 4. (b) A heatmap of the t-test statistics per subtype calculated by the PARADIGM integrated pathway activities (IPAs) for the entities. The top 200 entities, ranked by t-test statistic scores per supercluster, are shown. Representative pathways are displayed on the right side of heatmap.
Figure 4. The presence level of stromal and immune cells in tumor tissues per each subtype.
Boxplots of a) stromal and immune scores, c) tumor purity scores per subtype revealed the different distribution of each score among superclusters. The x-axis denotes subtypes and the y-axis represents stromal, immune, and tumor purity scores that were z-transformed per each tumor type.
Cell cycle activation is observed across cancer lineages
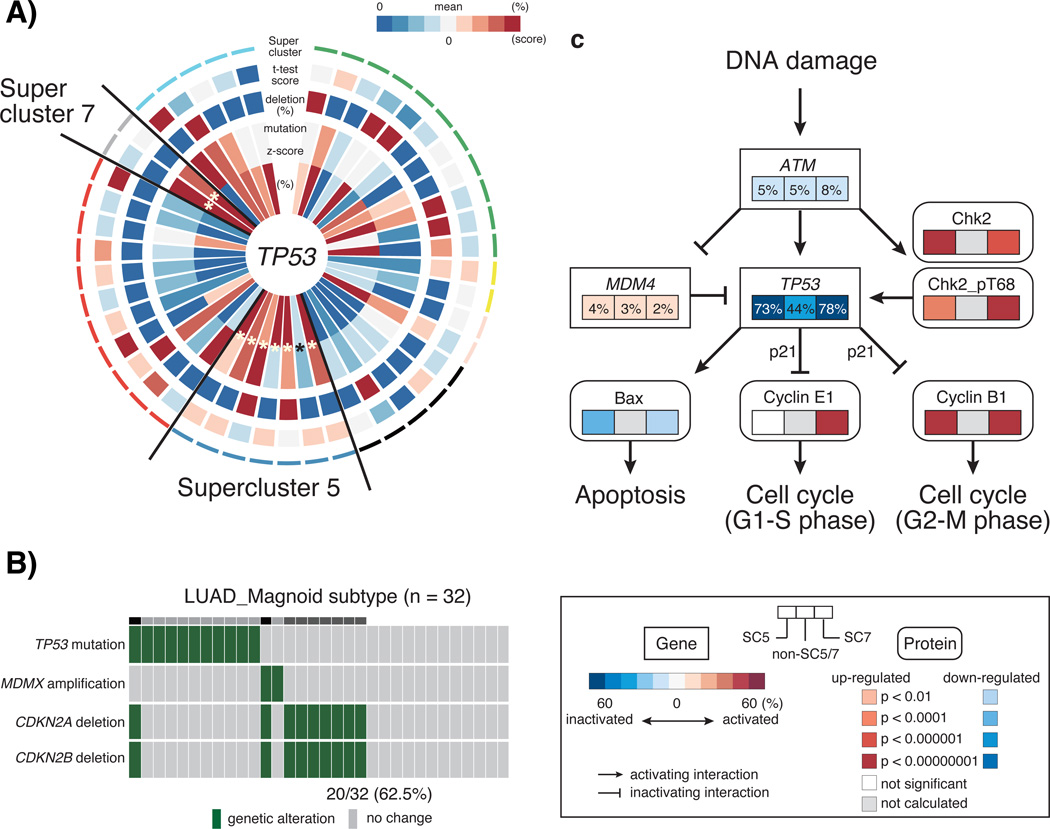
Supercluster 5 contained subtypes from six of the twelve tumor types and ssGSEA and PARADIGM analysis highlighted an association between this set of expression subtypes and increased cell cycle activity (Figure 3, Supplementary Table 4 and 5). We used reverse phase protein array (RPPA) profiles available for a subset of samples and found that the protein expression levels of mitotic cycle marker cyclin B1 (CCNB1) was significantly up-regulated in supercluster 5 relative to other superclusters (P = 1.56E-8) (Supplementary Figure 4). CCNB1, in complex with cyclin-dependent kinase 1, regulates the G2 phase of the cell cycle during which DNA is checked for chromosome replication errors25. We compared the number of DNA copy number segments across subtypes as a proxy for the number of double strand DNA breaks and found a statistically significant increase in the number of copy number segments in association with supercluster 5 (P = 4.78E-7; Supplementary Figure 5). In addition to CCNB1, high protein levels of DNA repair gene CHEK2 suggested the presence of DNA damage (P = 4.29E-10), which under normal conditions would direct cells to enter apoptosis. Interestingly, a higher number of mutations in the apoptosis and cell cycle regulator TP53 were observed in supercluster 5 (77% of samples; Figure 5a), which may explain why cells are not entering apoptosis despite a high level of DNA damage. In addition, all subtypes except OV_Proliferative in supercluster 5 harbored deletions of cyclin-dependent kinase inhibitor 2A (CDKN2A) in more than 10% of samples (Supplementary Figure 6, Supplementary Table 6 and 7). Supercluster 5 member LUAD_Magnoid, did not harbor many TP53 mutations, but instead correlated to a high frequency of CDKN2A/CDKN2B homozygous deletions (62.5% of samples; Figure 5b), which control the G1 cell cycle phase (Figure 5c). The association between TP53 mutation and CDKN2A deletion showed a trend towards mutually exclusivity (Fisher’s exact test, P = 0.10). A similar set of significant observations was made for supercluster 7. This supercluster consisted of a BRCA and a UCEC subtype, two tumor types that were not found amongst supercluster 5 members. In addition to protein expression of CCNB1, supercluster 7 showed higher expression of CCNE1 (P = 2.08E-22 and 4.6E-17) and lower expression of CCND1 compared to the other superclusters (P = 9.58E-10) (Supplementary Figure 4). Furthermore, a significantly increased frequency of MYC amplification was observed in supercluster 7 (BRCA_2, P = 0.000018; UCEC_Mitotic, P = 0.0027; (Supplementary Figure 6, Supplementary Table 8), which leads to cell cycle progression and potentially distinguished supercluster 7 from supercluster 5. In summary, these results suggested that supercluster 5 and supercluster 7 are dominated by somatic alterations that affect the G2-M phase and S-phase of the cell cycle, respectively (Figure 5c).
Figure 5. Association of activating cell cycle with TP53 mutation in supercluster.
a) A CIRCOS plot of TP53 alterations per subtypes. Starting from the center, the plot displays the following variables: 1. Proportional mutation frequency per subtype (percentage of samples mutated versus total number of samples), 2. Relative mutation frequency per subtype (z-score transformation of the percentage of samples mutated versus total number of samples, across all subtypes within a tumor type), 3. Relative deletion frequency per subtype (percentage of samples homozygously deleted versus total number of samples), 4. t-test score of TP53 gene expression levels per subtype versus other subtypes, within each tumor type, and 5. supercluster annotation. b) An alteration of cell cycle signaling pathway in LUAD_Magnoid subtype. Genetic alterations per samples shows green bar. c) A schema of altered cell cycle signaling per supercluster 5 (left box), supercluster 7 (right box), and the others (middle box). b) Mutual exclusivity between TP53 mutation and CDKN2A/B deletion in LUAD_Magnoid subtype. Genetic alterations are represented in green, wildtype in grey. c) Cell cycle pathway alterations in supercluster 5 (left box), supercluster 7 (right box), and others (middle box). Rectangles reflect the frequency of gene alteration for a specific gene. Rounded rectangles represent differentially expressed proteins among three groups. Colors indicate the association of gene alteration with altered gene activity (red, activation; blue, inactivation). Significance of protein up- and down-regulation was defined by p-value after comparing one supercluster to the others.
Common molecular characteristics of superclusters were associated with clinical outcome
To examine whether superclusters were consistently associated with survival, we compared clinical outcome between superclusters (Cox proportional hazards survival regression analysis, hazard ratios and 95% confidence intervals, Supplementary Figure 7A). An important aspect of this analysis is that we accounted for differences in survival between tumor types by normalizing outcome measures within each tumor type, and evaluated whether multiple subtype members of the same supercluster were showing a similar direction in outcome. Two significant correlations were observed. The BRCA_1 and LUAD_Bronchoid subtypes, which combined made up supercluster 2, associated with better prognosis (BRCA_1, P = 0.1; LUAD_Bronchioid, P = 0.02) and corresponding to the relatively favorably outcome we noted that 79.2 % of these samples were diagnosed as early stage (stage I/II) (Supplementary Table 9). In contrast, three of five subtypes in supercluster 8 correlated with adverse outcome (KIRC_3, P = 0.0012; LUAD_Squamoid, P = 0.06; BRCA_3, P = 0.00017). These subtypes had in common pathways related to SHC1 events in EGFR and ERBB4 signaling, which are related to drug resistance as well as pro-mitotic and survival pathways26, 27. We did not find consistent associations between superclusters and demographics such as age and gender (Supplementary Figure 7B, and Supplementary Table 9).
DISCUSSION
Here, we presented a comprehensive pan-cancer comparison of transcriptional patterns across twelve different tumor types. Unsupervised analysis of cancer expression profiles showed the dominating effects of tumor lineage and histology on cluster formation. However, comparing cancers using molecular subtype profiles revealed the presence of recurrent disease related expression patterns. These findings suggest that grouping tumor samples on the basis of their transcriptional profile, which has been described for many tumor types8, 9, 13, 28, generally follows a limited number of themes, some of which we have identified as occurring across tumor lineage.
We observed a supercluster characterized by presence of tumor-associated normal cells, which included subtypes from nine of eleven solid tumors. The role of the tumor microenvironment is increasingly being appreciated, most prominently as immunotherapeutics such as anti-PD1 and anti-CTL4A that have shown efficacy in advanced melanoma and other tumor types29–32. The persistent presence of subsets of stroma and immune cell associated tumors across many cancers may suggest a role for microenvironment produced growth factors. Alternatively, these tumors may produce chemotactic factors that attract tumor-associated normal cells. Whether the presence of increased number of immune cells will signal responsiveness to immunomodulatory therapeutics will need to be assessed clinically for this subset of tumors. A second large supercluster, plus a third smaller supercluster, unified the cell cycle and apoptosis pathways, through combined presence of an increased level of DNA double strand breaks, mutations in TP53, loss of CDKN2A, and cell cycle protein levels. While TP53 mutations are frequent across cancer lineages and may generally serve an anti-apoptotic role, we speculate that in the context of the proliferation transcriptomic signature, these mutations were selected to negate the apoptotic signals resulting from high levels of DNA damage. The resulting protein and gene expression signature reflect the consequences of an altered cell cycle pathway. However, our observations do not necessarily indicate that targeting of the cell cycle pathway by means of CDK4/CDK6 inhibitors will be more effective in supercluster 5 / supercluster 7 tumors than in other cancers as it is unclear that the observed pathway changes are functional or a consequence of changes in proliferation and viability. Further comparative experiments of cell cycle performance metrics, such as cell proliferation assays, are needed to show that increased cell cycle pathway activity is associated with increased proliferation.
Although we were able to identify some correlations with patient outcomes in superclusters, we were unable to identify a consistent pattern of clinical outcome in relation to superclusters. It is important to note that the survival data in several TCGA data sets is limited by the length of follow up3, 6.
In summary, comparison of large numbers of gene expression profiles across many tumor types highlights the relevance of a number of transcriptional footprints that unify a decade of cancer subtype research.
MATERIAL AND METHODS
Data preparation
The TCGA level 3 for gene mutation, copy number, and expression data were downloaded from TCGA Data portal (https://tcga-data.nci.nih.gov/tcga/). Twelve tumor types (acute myeloid lymphoma, bladder urothelial carcinoma, breast carcinoma, colon adenocarcinoma, rectal adenocarcinoma, glioblastoma multiforme, head and neck squamous cell carcinoma, clear cell renal cell carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, ovarian serous cystadenocarcinoma, and uterine corpus endometrial carcinoma) from four platforms (Agilent G4502A; Affymetrix HG-U133Plus2.0; Affymetrix HT-HG-U133A; Illumina HiSeq) were used in this study (Table 1).
Table 1.
A list of the Cancer Genome Atlas data sets.
| Tumor type* | Platform | No. of samples |
No. of subtypes |
Subtyping | No. of samples with mutation data |
No. of samples with copy number data |
|---|---|---|---|---|---|---|
| BLCA | RNASeq | 122 | 4 | TCGA | 95 | 86 |
| BRCA | Agilent | 530 | 5 | Self | 510 | 501 |
| COADREAD | Agilent | 220 | 3 | TCGA | 94 | 209 |
| GBM | Affymetrix | 515 | 4 | TCGA | 230 | 471 |
| HNSC | RNASeq | 279 | 4 | TCGA | 299 | 280 |
| KIRC | RNASeq | 417 | 4 | TCGA | 390 | 408 |
| LAML | Affymetrix | 169 | 7 | TCGA | 178 | 172 |
| LUAD | RNASeq | 230 | 3 | TCGA | 124 | 230 |
| LUSC | RNASeq | 177 | 4 | TCGA | 177 | 177 |
| OV | Affymetrix | 488 | 4 | TCGA | 315 | 488 |
| UCEC | RNASeq | 265 | 3 | TCGA | 219 | 216 |
BLCA, bladder urothelial carcinoma; BRCA, breast cancer; COAD, colon adenocarcinoma; READ, rectal adenocarcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, clear cell renal cell carcinoma; LAML, acute myeloid leukemia; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; UCEC, uterine corpus endometrial carcinoma.
Cluster Assignments and Consensus clustering using Non-negative Matrix Factorization (NMF)
We used the molecular classification provided by the TCGA disease analysis working group of eleven out of twelve tumor types (Table 1). Since consensus clustering was originally used for cancer subtype discovery studies in TCGA, we applied consensus clustering using NMF18 to the breast cancer Agilent expression data. The NMF algorithm is a linear algebra matrix factorization algorithm that depends on the initialization and a parameter k representing the number of clusters. Thus, the consensus clustering algorithm was ran 100 times reporting highly stable clusters. We used the top ~1,500 genes (as used in previous TCGA papers) ranked according to the median absolute deviation to run NMF over each of the three tissues that did not have TCGA subtype information. The number k of clusters (subtypes) was chosen using the cophenetic correlation coefficient; approximated ties were resolved by maximizing the number of clusters and samples per cluster. We did not use PAM50 signature to obtain breast cancer subtypes because PAM50 signature is not based on consensus clustering and is applied to real-time PCR based expression33. However, supplementary table 10 shows some overlap between PAM50 subtypes and our NMF subtypes.
Scoring the presence level of tumor-associated normal cells and the extent of squamous cell phenotype using gene expression data
Stromal and immune scores were defined by ESTIMATE algorithm23. Briefly, through comparison of tumor samples with high presence of tumor-associated stroma, and infiltrating leukocytes, as well as cell sorting of tumor samples, we generated a signature predictive of presence of stroma and a signature predictive of the presence of immune cells, using single sample gene set enrichment analysis (ssGSEA)19. We calculated ssGSEA scores for our stromal and immune signatures that predicted the presence level of stromal and immune cells in the tumor tissue. We also used the purity predictions generated by the ABSOLUTE algorithm24 which uses DNA copy number data to infer tumor purity. The data was obtained from The Cancer Genome Atlas Pan-Cancer data set34.
To identify a squamous signature specifically related to the presence of squamous cell carcinoma, four microarray data sets were obtained from the Gene Expression Omnibus (GSE10245, lung cancer; GSE28571, lung cancer; GSE26886, esophageal cancer; and GSE27388, cervical cancer). Next, squamous cell carcinoma samples with adenocarcinoma samples using the significance analysis of microarray (SAM)35 method to detect significantly up-regulated expressed genes (> 2 fold & q < 0.0001) in the squamous cell carcinoma group compared to adenocarcinoma group for each microarray data set. For those three respective data sets, we extracted 226, 419, 189, and 99 up-regulated genes in squamous cell carcinomas. In total, 254 squamous-related genes were identified in at least two data sets. Next, to consider the influence of tumor purity, we extracted 97 up-regulated genes in squamous cell carcinoma cell lines (n=26) compared to adenocarcinoma cell lines (n=49) from the CCLE expression data set36. Of these, we selected 25 overlapping genes involved in squamous cell carcinoma. Finally, one gene was excluded as it was previously found to be associated with “normal hematopoietic-cell related genes”23, resulting in a final set of 24 squamous cell carcinoma related genes (squamous signature). We confirmed that ssGSEA scores for this signature (named as “squamous score”) could detect squamous cell phenotype in a validation data set (GSE2109), consisted of 66 squamous cell carcinomas, 9 adenosquamous cell carcinomas, 2 adenocarcinomas with squamous differentiation, and 1,311 non-squamous cell carcinomas.
Supercluster identification and validation
To identify superclusters, we first generated t-statistics for all genes, by comparing gene expression levels of each subtype and the other subtypes of the same tumor type. This resulted in t-statistic vectors for each of the 45 gene expression subtypes from twelve tumor types. We then calculated Pearson’s correlation between all tissue subtype, using the t-statistic vectors. Then, we applied hierarchical clustering using squared Euclidean distance37. To maximize the tightness within clusters we used complete agglomeration. To generate the clusters, we cut the tree using the following criteria: a) maximize the average or minimize the negative silhouette scores among clusters, b) maximize the correlation among clusters, c) include clusters composed of more than one tissue minimizing the number of subtypes from the same tissue, and d) obtain the least number of clusters.
To validate superclusters we used two methods: 1) removing different percentages of samples and identifying superclusters using the reduced data sets, and 2) identifying superclusters on other public data sets.
The first method is similar to one used to validate oncogenic signatures for multiple cancers38. For each tissue, we removed 10 to 50% of the samples 100 times each. We calculated the frequency of times two subtypes coincided in the same cluster and then identified supeclusters using this frequency (as a correlation measure) and cutting the hierarchical tree to obtain the same number of clusters as for the original TCGA data. Finally, we compared if the superclusters identified from the reduced data sets were similar to the original superclusters.
We also validated superclusters using public data. Supplementary table 11 shows the data sets used for this experiment. For each tumor type, we selected these data sets from Affymetrix HG-U133 Plus 2.0 based gene expression data with maximum sample size in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/). To avoid batch effect, we did not merge some public data sets in the same tumor type. The public data sets have a total of 2,550 samples. To identify superclusters on public data sets, we first assigned a subtype to each respective tissue sample and then identify superclusters cutting the hierarchical tree similar as for original data. As for the reduced data sets, then we matched the superclusters acquired using public data sets to the original superclusters.
TCGA subtype definition for public data
In order to use public data, we needed to classify samples according to TCGA subtypes. Therefore, we first obtained subtype signatures for each tissue. If available, subtype signature were extracted from the TCGA respective tissue publications. Otherwise, we ran SAM35 comparing one subtype versus the others and selected the top 200 up-regulated genes. For BLCA, BRCA, COADREAD, LUAD, LUSC, and UCEC, subtypes signatures derived from the top 200 SAM genes. For GBM, HNSC, KIRC, LAML, and OV, we used the subtypes signatures from the respective publication7, 10, 12, 14, 15. We tested two classification algorithms, PAMR39 and SVM40, on the TCGA data and selected the most accurate per tissue using a 10-fold cross validation scheme. Supplementary table 12 shows SVM was selected for all tissues except LUSC.
Identification of statistically significant differences in genetic alterations per subtype
To associate subtypes within superclusters to DNA alterations (somatic mutations, focal amplifications, and homozygous deletions), we counted alterations per subtype and used a chi square test to test for overrepresentation relative to the remaining samples of that tumor type. Since we were looking for over-representations, we avoided cases where counts were lower than expected setting its p-value to 1. Only DNA alterations that involve genes and samples included in gene expression data were considered.
For mutation data, we ignored “Silent”, “Non-stop mutation”, or “RNA” mutations. For copy number variation data, we used TCGA discretized (values −2, −1, 0, 1, and −2) and transformed the data to −1, 0, +1 representing deletions, no alteration, and focal amplifications respectively, which were independently evaluated.
To select gene alterations associated to a supercluster, we ranked the genes according to a conservative p-value, which was estimated by the first quartile of included subtypes p-values. This will highlight genes having alterations in more than one subtype.
Genomic relationships in supercluster
To identify the similarity of genetic alterations (mutations, amplification, and homozygous deletions) in supercluster 5 and 7, we first extracted genes genetically altered in at least 10% of each subtype composed of supercluster 5 and 7. Of extracted genes, we selected overlapped genes among more than three fourth subtypes within supercluster 5 or 7, respectively. By comparing the frequency of altered samples per gene between supercluster 5, 7, and the others, we found the similarity of genetic alterations in supercluster 5 and 7.
Gene set enrichment analysis using ssGSEA
To find gene sets related to superclusters, we ran single sample GSEA (ssGSEA). We transformed the ssGSEA sample scores to estimate a subtype t-statistic by comparing each subtype to the other subtypes of the same tissue. We used 9,707 gene sets obtained from MSigDB21 (n = 8,513) plus subsets generated by combining up-regulated and down-regulated genes having the same subset name. To avoid intrinsic similarity between gene sets, we filtered out gene sets whose jaccard coefficient index was higher than 0.7 resulting in 8,907 gene sets. The results from this filtering are indicated as “jaccard” in supplementary table 4. Also, we reported the results from using the complete list of gene sets (identified as “complete” in Supplementary Table 4). To achieve clearer biological insights, we focused on canonical pathways belonging to Biocarta, Reactome41, KEGG42, and PID43 (“canonical” in Supplementary Table 4).
To select interesting gene sets related to a supercluster, gene sets were ranked by a representative t-statistic, which was estimated by first quartile from the subtypes t-statistics belonging to the supercluster. This would emphasize gene sets having consistently high t-statistics (and thus higher ssGSEA scores) along the supercluster.
Pathway activity analysis using PARADIGM
PARADIGM algorithm20 estimates pathway activities levels per sample from curated biological entities modeling the process of transcription, translation, and protein activation considering data as evidence under probabilistic inference. We used expression and copy number alteration data to run PARADIGM. We initially used 56,418 entities covering ~1,250 pathways. Similar to the ssGSEA analysis, pathways whose jaccard coefficient index were higher than 0.7 were filtered. We kept those pathways with a higher number of entities, which provide better description of the pathway. We also filtered PARADIGM output entities that were repeated in the same pathway. We finally used 42,214 entities corresponding to ~1,000 pathways.
Similarly to ssGSEA analysis, PARADIGM sample scores were transformed to subtype t-statistics. To focus on activated PARADIGM entities, entities were selected according to a summary supercluster t-statistic, which was estimated from the first quartile from the t-statistics of the subtypes belonging to the supercluster. Only entities having positive t-statistic in all supercluster subtypes were considered. This will select entities that are consistently positive values in all subtypes of the supercluster. To select PARADIGM pathways related to superclusters, we used a hypergeometric test to evaluate the likelihood of observing a high number of selected entities from a pathway. Supplementary Table 5 show the entities and pathways resulted from this analysis.
Validation of activated cell cycle using protein expression
We downloaded reverse-phase protein array (RPPA) expression data (syn1710429), which composed of 205 total and phosphorylated proteins, from Synapse BETA (https://www.synapse.org/). Of 2,704 samples, 2,125 common samples between mRNA expression data and RPPA data (BLCA, n=51; BRC, n=407; COADREAD, n = 124; GBM, n=184; HNSC, n=206; KIRC, n=384; LUAD, n=181; LUSC, n=111; OV, n=288; UCEC, n=189) were used in the subsequent analysis. To compare cell-cycle related proteins across different tumor types, we performed z-transformation per each tumor type.
Evaluation of genomic instability
We used TCGA level 3 copy number alteration data based on Affymetrix SNP 6 to calculate the number of segments per each sample. To compare the number of segments across different tumor types, we excluded samples with extreme number of segments that were detected by using the generalized extreme studentized deviate (GESD) test44 and performed z-transformation per each tumor type.
Statistical analysis
We conduced all computations with R 2.13.245, and used standard statistical tests as appropriate, including Pearson’s correlation analysis, unpaired t-test, 1-way ANOVA, Fisher’s exact test, and Cox proportional hazards univariate analysis. We downloaded TCGA clinical information on March 2013 from TCGA data portal [https://tcga-data.nci.nih.gov/tcga/].
Supplementary Material
ACKNOWLEDGEMENTS
The results published here are in whole or part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at “http://cancergenome.nih.gov”.
This work was supported in part by the U.S. National Cancer Institute (grant number CA143883 to The University of Texas MD Anderson Genome Data Analysis Center). HK is supported in part by the Odyssey Program and the Theodore N, Law Endowment for Scientific Achievement at The University of Texas MD Anderson Cancer Center.
REFERENCES
- 1.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas Research Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 9.Verhaak RG, Wouters BJ, Erpelinck CA, Abbas S, Beverloo HB, Lugthart S, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 12.Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123:517–525. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter V, Yin X, Wilkerson MD, Cabanski CR, Zhao N, Du Y, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013;8:e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Cancer Genome Atlas Research Network. Diversity of Lung Adenocarcinoma Revealed by in Depth Molecular Characterization. 2013 Submitted. [Google Scholar]
- 17.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014 doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinsson-Ahlzén HS, Liberal V, Grünenfelder B, Chaves SR, Spruck CH, Reed SI. Cyclin-dependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol Cell Biol. 2008;28:5698–5709. doi: 10.1128/MCB.01833-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Bennett K, Stukalov A, Fang B, Zhang G, Yoshida T, et al. Perturbation of the mutated EGFR interactome identifies vulnerabilities and resistance mechanisms. Mol Syst Biol. 2013;9:705. doi: 10.1038/msb.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, Zhang C, Croucher DR, Soliman MA, St-Denis N, Pasculescu A, et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013;499:166–171. doi: 10.1038/nature12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Everitt BS, Landau S, Leese M, Stahl D, Shewhart WA, Wilks SS. Cluster Analysis. Wiley; 2011. [Google Scholar]
- 38.Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noble WS. What is a support vector machine? Nat Biotechnol. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 41.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosner B. Percentage Points for a Generalized ESD Many Outlier Procedure. Technometrics. 1983;25:165–172. [Google Scholar]
- 45.Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.