Abstract
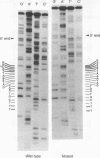
One of the eight endonuclease EcoRI fragments of yeast DNA that hybridize to yeast tRNATyr has been identified with the genetically defined nonsense-suppressor locus SUP4. This identification was achieved by analyzing the meiotic linkage between the genetic determinant for the SUP4 phenotype and that for an electrophoretic variant of the EcoRI fragment. The SUP4 gene was then cloned from an ochre-suppressing yeast strain and analyzed by DNA sequencing. A wild-type SUP4 gene and two other genetically unidentified tRNATyr genes were also sequenced. The sequence of the ochre suppressor differs from that of the wild-type genes by virtue of a G.C leads to T.A transversion in the base pair that codes for the wobble position base of the tRNATyr anticodon. All four genes contain, immediately to the 3' side of the anticodon triplet, a 14 base pair tract that is not present in mature tRNATyr. Although the four genes, which represent three unlinked chromosomal loci, all encode the same mature tRNA sequence, there is virtually no observable sequence homology between the three loci in the region preceding the 5' end of the mature tRNATyr sequences.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S. A modified uridine in the anticodon of E. coli tRNA I Tyr su + oc. Nucleic Acids Res. 1976 Feb;3(2):441–448. doi: 10.1093/nar/3.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J. S., Johnson P. F., Abelson J. Cloning of yeast transfer RNA genes in Escherichia coli. Science. 1977 Apr 8;196(4286):205–208. doi: 10.1126/science.322282. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Berget S. M., Moore C., Sharp P. A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. D., Brown D. D. The nucleotide sequence adjoining the 3' end of the genes coding for oocyte-type 5 S ribosomal RNA in Xenopus. J Mol Biol. 1976 Mar 25;102(1):1–14. doi: 10.1016/0022-2836(76)90070-x. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Panasenko S. M., Lehman I. R., Davis R. W. In vitro construction of bacteriophage lambda carrying segments of the Escherichia coli chromosome: selection of hybrids containing the gene for DNA ligase. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3416–3420. doi: 10.1073/pnas.72.9.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Gelinas R. E., Broker T. R., Roberts R. J. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977 Sep;12(1):1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Endow S. A., Roberts R. J. Two restriction-like enzymes from Xanthomonas malvacearum. J Mol Biol. 1977 May 25;112(3):521–529. doi: 10.1016/s0022-2836(77)80198-8. [DOI] [PubMed] [Google Scholar]
- Feinstein S. I., Altman S. Coding properties of an ochre-suppressing derivative of Escherichia coli tRNAITyr. J Mol Biol. 1977 May 25;112(3):453–470. doi: 10.1016/s0022-2836(77)80192-7. [DOI] [PubMed] [Google Scholar]
- Fink G. R., Styles C. A. Gene conversion of deletions in the his4 region of yeast. Genetics. 1974 Jun;77(2):231–244. doi: 10.1093/genetics/77.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne D. C., Leupold U. Suppressors in yeast. Curr Top Microbiol Immunol. 1974;64(0):1–47. doi: 10.1007/978-3-642-65848-8_1. [DOI] [PubMed] [Google Scholar]
- Hawthorne D. C. UGA mutations and UGA suppressors in yeast. Biochimie. 1976;58(1-2):179–182. doi: 10.1016/s0300-9084(76)80368-9. [DOI] [PubMed] [Google Scholar]
- Madison J. T., Kung H. K. Large oligonucleotides isolated from yeast tyrosine transfer ribonucleic acid after partial digestion with ribonuclease T1. J Biol Chem. 1967 Mar 25;242(6):1324–1330. [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Tizard R., Skryabin K. G., Gilbert W. Promotor region for yeast 5S ribosomal RNA. Nature. 1977 Jun 16;267(5612):643–645. doi: 10.1038/267643a0. [DOI] [PubMed] [Google Scholar]
- Olson M. V., Montgomery D. L., Hopper A. K., Page G. S., Horodyski F., Hall B. D. Molecular characterisation of the tyrosine tRNA genes of yeast. Nature. 1977 Jun 16;267(5612):639–641. doi: 10.1038/267639a0. [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Manning J., Davidson N. Sequence arrangement of the rDNA of Drosophila melanogaster. Cell. 1977 Feb;10(2):213–214. doi: 10.1016/0092-8674(77)90215-x. [DOI] [PubMed] [Google Scholar]
- Piper P. W., Wasserstein M., Engbaek F., Kaltoft K., Celis J. E., Zeuthen J., Liebman S., Sherman F. Nonsense suppressors of Saccharomyces cerevisiae can be generated by mutation of the tyrosine tRNA anticodon. Nature. 1976 Aug 26;262(5571):757–761. doi: 10.1038/262757a0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J., Esposito R. E., Esposito M. S. The effect of ochre suppression on meiosis and ascospore formation in Saccharomyces. Genetics. 1977 Jan;85(1):35–54. doi: 10.1093/genetics/85.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Liebman S. W., Stewart J. W., Jackson M. Tyrosine substitutions resulting from suppression of amber mutants of iso-1-cytochrome c in yeast. J Mol Biol. 1973 Jun 25;78(1):157–168. doi: 10.1016/0022-2836(73)90435-x. [DOI] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Jackson M., Gilmore R. A., Parker J. H. Mutants of yeast defective in iso-1-cytochrome c. Genetics. 1974 Jun;77(2):255–284. doi: 10.1093/genetics/77.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. P. Unequal crossover and the evolution of multigene families. Cold Spring Harb Symp Quant Biol. 1974;38:507–513. doi: 10.1101/sqb.1974.038.01.055. [DOI] [PubMed] [Google Scholar]
- Smith J. D. Transcription and processing of transfer RNA precursors. Prog Nucleic Acid Res Mol Biol. 1976;16:25–73. doi: 10.1016/s0079-6603(08)60755-2. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas M., Cameron J. R., Davis R. W. Viable molecular hybrids of bacteriophage lambda and eukaryotic DNA. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4579–4583. doi: 10.1073/pnas.71.11.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Valenzuela P., Bell G. I., Masiarz F. R., DeGennaro L. J., Rutter W. J. Nucleotide sequence of the yeast 5S ribosomal RNA gene and adjacent putative control regions. Nature. 1977 Jun 16;267(5612):641–643. doi: 10.1038/267641a0. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977 Feb;10(2):193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]