Abstract
Heritability is a central parameter in quantitative genetics, from both an evolutionary and a breeding perspective. For plant traits heritability is traditionally estimated by comparing within- and between-genotype variability. This approach estimates broad-sense heritability and does not account for different genetic relatedness. With the availability of high-density markers there is growing interest in marker-based estimates of narrow-sense heritability, using mixed models in which genetic relatedness is estimated from genetic markers. Such estimates have received much attention in human genetics but are rarely reported for plant traits. A major obstacle is that current methodology and software assume a single phenotypic value per genotype, hence requiring genotypic means. An alternative that we propose here is to use mixed models at the individual plant or plot level. Using statistical arguments, simulations, and real data we investigate the feasibility of both approaches and how these affect genomic prediction with the best linear unbiased predictor and genome-wide association studies. Heritability estimates obtained from genotypic means had very large standard errors and were sometimes biologically unrealistic. Mixed models at the individual plant or plot level produced more realistic estimates, and for simulated traits standard errors were up to 13 times smaller. Genomic prediction was also improved by using these mixed models, with up to a 49% increase in accuracy. For genome-wide association studies on simulated traits, the use of individual plant data gave almost no increase in power. The new methodology is applicable to any complex trait where multiple replicates of individual genotypes can be scored. This includes important agronomic crops, as well as bacteria and fungi.
Keywords: marker-based estimation of heritability, GWAS, genomic prediction, Arabidopsis thaliana, one- vs. two-stage approaches
NARROW-SENSE heritability is an important parameter in quantitative genetics, determining the response to selection and representing the proportion of phenotypic variance that is due to additive genetic effects (Jacquard 1983; Nyquist and Baker 1991; Ritland 1996; Visscher et al. 2006, 2008; Holland et al. 2010; Sillanpaa 2011). This definition of heritability goes back to Fisher (1918) and Wright (1920) almost a century ago. In plant species for which replicates of the same genotype are available (inbred lines, doubled haploids, clones), a different form of heritability, broad-sense heritability, is traditionally estimated by the intraclass correlation coefficient for genotypic effects, using estimates for within- and between-genotype variance. Broad-sense heritability is also referred to as repeatability and gives the proportion of phenotypic variance explained by heritable (additive) and nonheritable (dominance, epistasis) genetic variance.
With the arrival of high-density genotyping there is growing interest in marker-based estimation of narrow-sense heritability (WTCCC 2007; Yang et al. 2010, 2011; Speed et al. 2012; Vattikuti et al. 2012; Visscher and Goddard 2014). These estimates are obtained from mixed models containing random additive genetic effects, whose covariance structure is estimated from genetic markers. While marker-based heritability estimates have received much attention in human genetics, these are rarely reported for plant traits, despite their relevance in evolutionary genetics, in the dissection of complex traits, and in the ongoing debate on missing heritability (Manolio et al. 2009; Eichler et al. 2010; Brachi et al. 2011; Lee et al. 2011; Zuk et al. 2012). Heritability estimates are also of great relevance to plant breeders, as they give a measure for the breeding potential of a trait. In addition, state-of-the-art phenotyping platforms are making experiments more reproducible, increasing the relevance of marker-based estimation of heritability, as well as comparison of estimates from different experiments.
Although marker-based heritability estimation for plant traits can in principle be achieved within the same analytic framework that has been developed for human traits, there are important differences. First, heritability of human traits is usually estimated using panels of unrelated individuals, to avoid confounding with geographical or environmental effects (Browning and Browning 2011). For most plant species panels of unrelated individuals are not available, as plant genotypes will often share ancestry or adaptation, inducing dependence between genotypes. At the same time, however, plant genotypes can be evaluated under the same experimental conditions (e.g., common garden or growth chamber), and the usual assumption is that this eliminates genotype–environment correlations. Second, plant genotypes are often phenotyped in several genetically identical replicates. This is the case for all so-called immortal populations (Keurentjes et al. 2007; Wijnen and Keurentjes 2014). Mixed-model analysis can then be performed either on the individual plant (or plot) data or on genotypic means. In the literature on multi-environment trials (Smith et al. 2001, 2005; Oakey et al. 2006; Piepho and Williams 2006; Piepho et al. 2006, 2012; Boer et al. 2007; Verbyla et al. 2007; Stich et al. 2008; Möhring and Piepho 2009; Van Eeuwijk et al. 2010; Welham et al. 2010; Malosetti et al. 2013) these approaches are referred to as respectively one-stage and two-stage. These works consider mostly populations for which a pedigree is available, typically experimental populations. In the context of genomic prediction and genome-wide association studies (GWAS) for natural populations, mixed-model analysis is usually performed using a two-stage approach. The (usually tacit) assumption is that the genotypic means and kinship coefficients contain all the relevant information for estimating the genetic and residual variance. Here we investigate the feasibility of marker-based estimation of heritability with one- and two-stage approaches and look at how heritability estimates affect genomic prediction with the best linear unbiased predictor (G-BLUP) and GWAS. Although our analysis of observed phenotypes focuses on the model plant Arabidopsis thaliana, the asymptotic variances of different heritability estimators were also computed for diverse panels of Zea mays (Riedelsheimer et al. 2012; Van Heerwaarden et al. 2012) and Oryza sativa (Zhao et al. 2011).
In both published data (Atwell et al. 2010) and new experiments, we found very large standard errors and sometimes unrealistically high estimates of heritability, which could not be explained by varying linkage disequilibrium (Speed et al. 2012). Much better heritability estimates were obtained when mixed-model analysis was performed at the individual plant level. These estimates were based on kinship information as well as additional information on within-genotype variability, and in simulations they were found to be up to 13 times more accurate than heritability estimates based on genotypic means. In genomic prediction, correlation between simulated and predicted genetic effects increased in some cases by as much as 49%. This is a substantial improvement that shows the importance of accurate heritability estimates in plant breeding programs.
All reported heritability estimates can be obtained using our R-package heritability, which is freely available from CRAN (http://cran.r-project.org/web/packages/heritability/index.html). In contrast to existing packages such as emma (Kang et al. 2008), rrblup (Endelman 2011), and synbreed (Wimmer et al. 2012), it provides confidence intervals for heritability estimates. We also present software for GWAS: our program scan_GLS can efficiently perform GWAS directly on the individual plant or plot-level data, as well as on the means, incorporating a nondiagonal error covariance structure.
Materials and Methods
We assume a natural population of n genotypes, where for each genotype a quantitative trait is measured on a number of genetically identical replicates of immortal lines. These replicates can refer to either individual plants (e.g., Arabidopsis) or plots in a field trial (e.g., maize). For convenience we use the terms “replicate” and “individual plant data” as synonyms throughout. In either case, the observations on replicates should not be confused with having multiple observations on the same individual. We focus on inbred lines, but apart from a few different constants, all expressions are valid for any diploid species. We do not partition the environmental variance into different contributing factors (see, e.g., Visscher et al. 2008); hence the environmental variance just equals the error variance. For simplicity we first present marker-based estimation of heritability for a completely randomized design, in the absence of additional covariates. The expressions for more general designs are given in Appendix A, which also provides a brief overview of the existing methodology for marker-based heritability estimation. Details on G-BLUP, GWAS, and the phenotypic data are given in Appendixes C, D, and E.
Genotypic data
We analyze simulated and real traits for subpopulations of the RegMap, which contains 1307 worldwide accessions of A. thaliana that have been genotyped at 214,051 SNP markers (Horton et al. 2012). We considered four subpopulations: 298 Swedish accessions [Swedish RegMap (Horton et al. 2012; Long et al. 2013)], 204 French accessions [French RegMap (Horton et al. 2012; Brachi et al. 2013)], 350 accessions from the HapMap population (Li et al. 2010), and a subset of 250 accessions that we refer to as the structured RegMap (accession IDs are given in Supporting Information, Table S1). The structured RegMap accessions were chosen to have large differences in genetic relatedness (i.e., variation in kinship coefficients, across pairs of accessions). These differences were hence largest in the structured RegMap and smallest in the HapMap (Figure S1 and Figure S2). For the asymptotic distributions of heritability estimators given below, we also considered marker-based kinship matrices for three populations of crop plants: the panel described in Van Heerwaarden et al. (2012) (Z. mays, 400 accessions), the panel described in Riedelsheimer et al. (2012) (Z. mays, 280 accessions), and the panel from Zhao et al. (2011) (O. sativa, 413 accessions).
Phenotypic data
We estimated heritabilities for two traits from the literature and four traits from new experiments. Two flowering traits from Atwell et al. (2010) were analyzed: days to flowering time under long day and vernalization (LDV) and days to flowering time under long day (LD). In two new experiments (Appendix E), leaf area (LA) 13 days after sowing was measured for the Swedish RegMap and the HapMap, using the same automated phenotyping platform. In the final experiment, the HapMap was phenotyped for bolting time (BT) and leaf width (LW). Trait descriptions and abbreviations are given in Table 1. In all experiments the individual plants were grouped into complete blocks, but due to nongerminating seeds and dead plants, there is some unbalance for all of the traits.
Table 1. Abbreviations of the trait names.
| Abbreviation | Trait |
|---|---|
| LDV | Days to flowering time under long day and vernalization |
| LD | Days to flowering time under long day |
| LA(S) | Leaf area 13 days after sowing (Swedish RegMap) |
| LA(H) | Leaf area 13 days after sowing (HapMap) |
| BT | Bolting time (HapMap) |
| LW | Leaf width (HapMap) |
The first two traits were taken from Atwell et al. (2010).
Marker-based estimation of heritability
Let K denote the genetic relatedness or kinship matrix of the genotypes, with elements
| (1) |
(see, e.g., Patterson et al. 2006 and Goddard et al. 2010). The numbers xi,l ∈ {0, 2} denote the minor allele count [because we focus on inbred lines, there is a small difference with the standard expression for outbreeders under Hardy–Weinberg equilibrium, where xi,l ∈ {0, 1, 2}, and the constant 4 in (1) is replaced by 2] at marker l for genotype i, and fl ∈ [0, 1] is the minor allele frequency at marker l. The phenotypic response of replicate j of genotype i is modeled as
| (2) |
where G = (G1, … , Gn) has a distribution, and the errors Ei,j have independent normal distributions with variance . We aim to estimate the heritability
which has been referred to as the “chip” heritability (Speed et al. 2012) and under certain assumptions equals narrow-sense heritability (Appendix A). In contrast to the line heritability , the denominator contains the residual variance for a single individual . Given restricted maximum-likelihood (REML) estimates and , h2 can be estimated by
| (3) |
where the subscripts r stress the fact that and are estimates directly obtained from the replicates. Alternatively, h2 can be estimated using the genotypic means
| (4) |
Given REML estimates and for model (4), we have the heritability estimate
| (5) |
where the subscripts m indicate that the estimates are based on genotypic means. We omit the letters r and m either if these are clear from the context or when a statement holds for both models (2) and (4). In human association studies, r is usually 1, and and are identical.
Repeatability and broad-sense heritability
Given a completely randomized design with r replicates, repeatability, or intraclass correlation can be estimated by
| (6) |
where MS(G) and MS(Env) are the mean sums of squares for genotype and residual error, obtained from analysis of variance (ANOVA). In the case that MS(G) < MS(Env), is set to zero. See Singh et al. (1993) or (in the context of sib analysis) Lynch and Walsh (1998, p. 563). Here we stick to the widely used notation , although repeatability equals broad-sense heritability (H2) only under the assumption that all differences between genotypes are indeed genetic and not due to, e.g., genotype–environment correlation. In any case, no additional information on the genetic structure is used here, and hence the mean sums of squares for genotype contain all genetic effects, not only the additive ones. Consequently, is an estimator of (an upper bound on) broad-sense heritability.
It is known to have a small bias (Nyquist and Baker 1991; Singh et al. 1993), which tends to zero when the number of genotypes increases. Additional fixed effects can be included in the ANOVA, which may reduce MS(Env) and give a higher heritability estimate. In the case that the genotypes have differing numbers of replicates, genetic variance is estimated with , where
| (7) |
is the effective number of replicates (Lynch and Walsh 1998, p. 559). For a balanced design, .
Confidence intervals
Confidence intervals for broad-sense heritability H2 were obtained from classical theory (e.g., Lynch and Walsh 1998, p. 563) (for easy reference see File S1). Confidence intervals for h2 were constructed using and and the inverse average-information (AI) matrix, obtained from the AI-REML algorithm (Gilmour et al. 1995). This 2 × 2 matrix provides an estimate of the covariance matrix of . The delta method (see, e.g., Van der Vaart 2000) applied to the function then gives the asymptotic distribution of , from which a confidence interval for h2 is calculated. In the case that the heritability is low or high, confidence intervals may be partly outside [0, 1]. We set all negative values to zero and all values larger than one equal to one. An alternative is application of the delta method to the function . This gives confidence intervals for , which are then back transformed to confidence intervals for heritability. The latter intervals are referred to as “log-transformed” and the other intervals as “standard.”
When the likelihood as a function of h2 is monotonically increasing, the confidence intervals can become numerically unstable, alternating between [0, 1] and [1, 1]. However, we found that using the AI matrix obtained by setting h2 = 1 − ε, the interval was always very close to [0, 1], for any ε > 0. We therefore defined the interval to be [0, 1] in the case of a monotonically increasing likelihood.
Simulations
Each simulated trait consists of r = 3 replicates for a subset of 200 accessions randomly drawn from a subpopulation of the Arabidopsis RegMap (Horton et al. 2012). We simulate according to model (2), except that the genetic effects Gi are not purely polygenic, but a mixture of QTL effects at a small number of markers and a polygenic signal with genetic structure given by the kinship matrix defined in (1). More details are given in Appendix B. Different genetic architectures were considered, but all simulated genetic effects were additive. Hence, broad- and narrow-sense heritabilities were equal, and , , and were in this case different estimators of the (same) simulated heritability.
For the comparison of heritability estimates we simulated traits for the structured RegMap, the HapMap, and the Swedish and French RegMap. For comparing one- and two-stage association mapping and genomic prediction only the structured RegMap and the HapMap were considered. For genomic prediction and the comparison of heritability estimates we simulated three levels of heritability, for each subpopulation: low (0.2), medium (0.5), and high (0.8). For each heritability level, 5000 traits were simulated. For the comparison of one- and two-stage association mapping, 1000 traits were simulated with heritability 0.5. In all simulations, a completely randomized design was assumed.
Results
Asymptotic variance
Applying general mixed-model theory (Casella and McCulloch 2006, p. 387) to model (2), it follows that the REML estimators are asymptotically Gaussian with covariance
| (8) |
where for , X = 1nr, and Z is the nr × n incidence matrix assigning plants to genotypes. The number of replicates r is considered fixed here. The asymptotic arguments are all with respect to n, the number of genotypes. The behavior of as a function of n, h2, and K can also be derived from approximations recently proposed in Visscher and Goddard 2014, independently from the present work. Although this is not made explicit in the notation, , depends on the true (and usually unknown) values and through P and V. If are estimated based on genotypic means following model (4), has asymptotic covariance , which is obtained if we replace in (8) ZKZt by K and X = 1nr by X = 1n and substitute in the definition of P. The asymptotic variance of the heritability estimators and can now be obtained by application of the delta method (Van der Vaart 2000) to the function , with gradient
It then follows that given the true and ,
| (9) |
It is easily verified that both variances do not depend on the absolute values of and , but only on the ratio . To explore the potential gains in accuracy due to the use of individual plant data, we computed and the ratio of the two standard deviations, for several populations in A. thaliana, Z. mays, and O. sativa (Table 2) and three heritability levels (h2 = 0.2, 0.5, 0.8).
Table 2. Asymptotic distribution of marker-based heritability estimators.
| Population | r | h2 = 0.2 | h2 = 0.5 | h2 = 0.8 |
|---|---|---|---|---|
| Structured RegMap | 1 | 0.0702 (1.00) | 0.0762 (1.00) | 0.0497 (1.00) |
| Structured RegMap | 2 | 0.0428 (0.85) | 0.0406 (0.71) | 0.0187 (0.49) |
| Structured RegMap | 3 | 0.0337 (0.80) | 0.0323 (0.66) | 0.0153 (0.46) |
| Structured RegMap | 4 | 0.0289 (0.77) | 0.0284 (0.64) | 0.0139 (0.46) |
| HapMap | 1 | 0.1137 (1.00) | 0.1310 (1.00) | 0.1040 (1.00) |
| HapMap | 2 | 0.0356 (0.46) | 0.0292 (0.31) | 0.0139 (0.19) |
| HapMap | 3 | 0.0250 (0.40) | 0.0226 (0.29) | 0.0115 (0.18) |
| HapMap | 4 | 0.0204 (0.37) | 0.0199 (0.29) | 0.0106 (0.19) |
| Swedish RegMap | 1 | 0.0781 (1.00) | 0.0827 (1.00) | 0.0574 (1.00) |
| Swedish RegMap | 2 | 0.0391 (0.73) | 0.0344 (0.58) | 0.0160 (0.36) |
| Swedish RegMap | 3 | 0.0291 (0.67) | 0.0269 (0.54) | 0.0132 (0.34) |
| Swedish RegMap | 4 | 0.0244 (0.64) | 0.0237 (0.52) | 0.0121 (0.33) |
| French RegMap | 1 | 0.1141 (1.00) | 0.1135 (1.00) | 0.0768 (1.00) |
| French RegMap | 2 | 0.0497 (0.66) | 0.0416 (0.52) | 0.0192 (0.33) |
| French RegMap | 3 | 0.0360 (0.60) | 0.0322 (0.49) | 0.0158 (0.31) |
| French RegMap | 4 | 0.0297 (0.58) | 0.0283 (0.48) | 0.0145 (0.31) |
| VH et al. (2012) | 1 | 0.0680 (1.00) | 0.0725 (1.00) | 0.0502 (1.00) |
| VH et al. (2012) | 2 | 0.0329 (0.71) | 0.0290 (0.56) | 0.0136 (0.37) |
| VH et al. (2012) | 3 | 0.0244 (0.65) | 0.0227 (0.53) | 0.0113 (0.35) |
| VH et al. (2012) | 4 | 0.0204 (0.62) | 0.0200 (0.52) | 0.0103 (0.36) |
| RH et al. (2012) | 1 | 0.0698 (1.00) | 0.0679 (1.00) | 0.0399 (1.00) |
| RH et al. (2012) | 2 | 0.0374 (0.81) | 0.0346 (0.74) | 0.0168 (0.60) |
| RH et al. (2012) | 3 | 0.0286 (0.77) | 0.0280 (0.72) | 0.0141 (0.59) |
| RH et al. (2012) | 4 | 0.0244 (0.76) | 0.0249 (0.72) | 0.0129 (0.60) |
| Zhao et al. (2011) | 1 | 0.0642 (1.00) | 0.0625 (1.00) | 0.0329 (1.00) |
| Zhao et al. (2011) | 2 | 0.0399 (0.87) | 0.0351 (0.79) | 0.0156 (0.65) |
| Zhao et al. (2011) | 3 | 0.0311 (0.83) | 0.0280 (0.76) | 0.0128 (0.62) |
| Zhao et al. (2011) | 4 | 0.0264 (0.81) | 0.0246 (0.74) | 0.0116 (0.62) |
Shown are standard deviations of the estimators based on replicates and in parentheses the ratios of the standard deviations of the estimators based on replicates over those based on means. Asymptotic variances were obtained using the expressions in Equation 9. Four populations of A. thaliana (containing respectively 250, 350, 305, and 204 accessions), two populations of Z. mays [400 accessions (Van Heerwaarden et al. 2012) and 280 accessions (Riedelsheimer et al. 2012)] and one population of O. sativa [413 accessions (Zhao et al. 2011)] were considered. The second column gives the number of replicates (r). VH, Van Heerwaarden et al. (2012). RH, Riedelsheimer et al. (2012).
The use of individual plant data gave a substantial improvement in accuracy for all populations, which was largest for the Arabidopsis HapMap [81% reduction with respect to , for r = 4 and h2 = 0.8] and smallest for the structured RegMap, the rice population from Zhao et al. (2011), and the maize population from Riedelsheimer et al. (2012) (13–15% reduction when r = 2 and h2 = 0.2). For the maize population from Van Heerwaarden et al. (2012) and the other Arabidopsis populations the improvements were similar. The standard deviation of decreased with increasing heritability and increasing numbers of replicates. The ratio of the standard deviations of over those of also decreased substantially with increasing heritability, but remained similar when increasing the number of replicates beyond 2.
It should be emphasized that the ratios in Table 2 are asymptotic: although the expressions in (9) depend on the kinship matrix K of a finite population, they may be good approximations only for (very) large populations with population structure similar to that defined by K. Such populations could be modeled as a function of population genetic processes such as drift and selection (Rousset 2002; Fu et al. 2003). However, for real plant populations marker information is available for at most a few thousand genotypes and more often several hundred. To assess variance and bias of and in such populations we used simulated traits.
Heritability estimates for simulated data
We analyzed simulated traits for the four populations of A. thaliana, for different genetic architectures. We also used the simulations to compare these marker-based estimators with the broad-sense heritability estimator , which ignores genetic relatedness, and to investigate the quality of heritability confidence intervals.
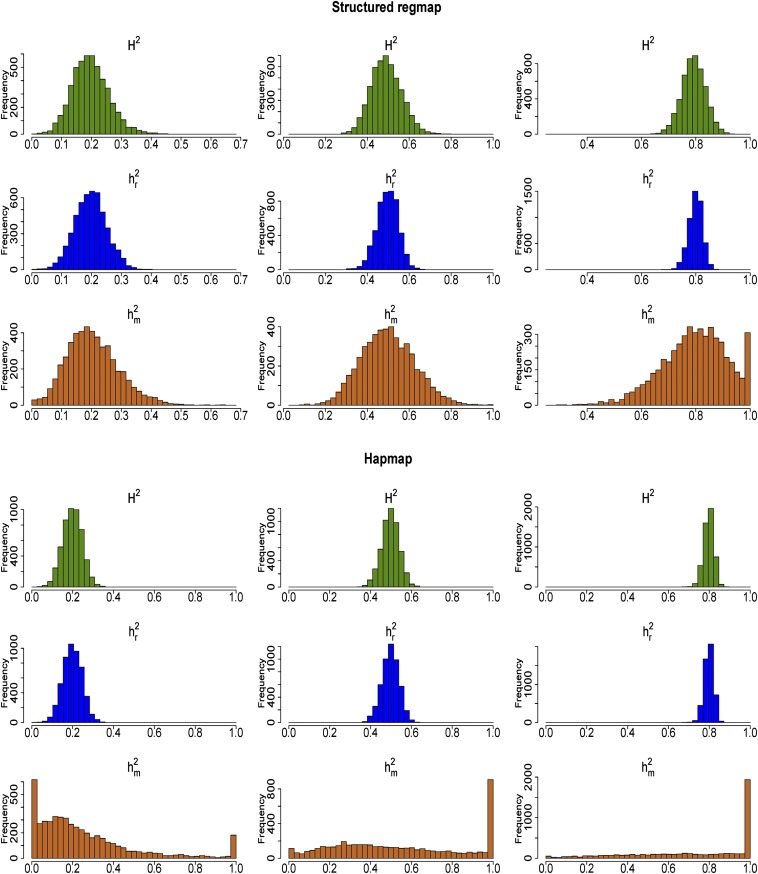
Heritability estimates for the structured RegMap and HapMap are given in Figure 1 and Table 3. Apart from in the HapMap, none of the estimators showed marked bias. In particular, had negligible bias, despite the fact that it does not account for genetic relatedness. As in the asymptotic results, had lower variance than , the difference being largest for the HapMap. The magnitude of the differences was larger than expected from the asymptotic variances in Table 2, and in the HapMap standard errors were up to 13 times larger. As in the asymptotic framework, differences were smallest for the structured RegMap and largest for the HapMap.
Figure 1.
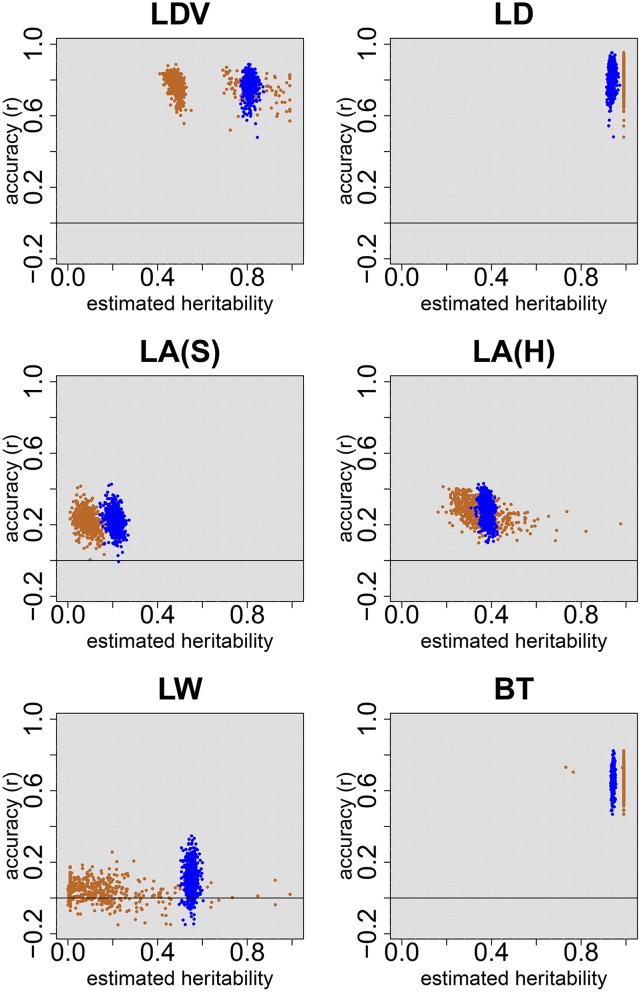
Heritability estimates for 5000 simulated traits for random samples of 200 accessions drawn from the structured RegMap (top) and the HapMap (bottom). Twenty QTL were simulated, which explained half of the genetic variance. The simulated heritability was 0.2 (left), 0.5 (center), and 0.8 (right). Within each panel, the first row shows the ANOVA-based estimates of broad-sense heritability, the second row the mixed-model-based estimates based on the individual plant data, and the third row the mixed-model-based estimates based on genotypic means.
Table 3. Comparison of the marker-based heritability estimators and for simulated data.
| Estimator | Bias | Standard error | Relative standard error |
|---|---|---|---|
| Structured RegMap | |||
| h2 = 0.2 | |||
| Broad sense (H2) | −0.00314 | 0.06079 | 0.70120 |
| Individual level () | −0.00268 | 0.05306 | 0.61204 |
| Means () | 0.00475 | 0.08670 | 1.00000 |
| h2 = 0.5 | |||
| Broad sense (H2) | −0.00938 | 0.07193 | 0.56365 |
| Individual level () | −0.00400 | 0.05093 | 0.39915 |
| Means () | −0.00164 | 0.12761 | 1.00000 |
| h2 = 0.8 | |||
| Broad sense (H2) | −0.00825 | 0.04309 | 0.36087 |
| Individual level () | −0.00138 | 0.02473 | 0.20712 |
| Means () | −0.00388 | 0.11940 | 1.00000 |
| HapMap | |||
| h2 = 0.2 | |||
| Broad sense (H2) | −0.00128 | 0.04663 | 0.19404 |
| Individual level () | −0.00154 | 0.04642 | 0.19317 |
| Means () | 0.05328 | 0.24033 | 1.00000 |
| h2 = 0.5 | |||
| Broad sense (H2) | −0.00288 | 0.04280 | 0.13754 |
| Individual level () | −0.00295 | 0.04120 | 0.13242 |
| Means () | 0.04400 | 0.31115 | 1.00000 |
| h2 = 0.8 | |||
| Broad sense (H2) | −0.00282 | 0.02385 | 0.08263 |
| Individual level () | −0.00231 | 0.02214 | 0.07672 |
| Means () | −0.06137 | 0.28863 | 1.00000 |
We simulated 5000 traits, for random samples of 200 accessions drawn from the structured RegMap and HapMap. Twenty unlinked QTL were simulated, which explained 50% of the genetic variance. The simulated heritability was 0.2, 0.5, and 0.8. Standard errors are given relative to those of .
Remarkably, also was considerably more accurate than . The additional marker information appeared to be less important here than the loss of information on within-genotype variability. The estimator , which includes both sources of information, outperformed . Differences were largest for the structured RegMap (12.7% reduction when h2 = 0.2, 42.6% reduction when h2 = 0.8), where the large differences in relatedness provide more information than in the case of the HapMap (almost no difference when h2 = 0.2 and 7% reduction when h2 = 0.8). Simulation results for the Swedish and French RegMap are given in Figure S3 and Table S2. In terms of standard deviations of , , and , these populations were somewhere in between the structured RegMap and HapMap.
For a substantial proportion of the simulated traits in both the HapMap and the structured RegMap, we observed a phenomenon that was not apparent from the asymptotic arguments above: was close to or equal to 1, in which case the likelihood as a function of h2 was monotonically increasing. This behavior is most likely to occur when the genetic relatedness matrix is close to compound symmetry (i.e., all genotypes being equally related), in which case the likelihood is constant in h2 (File S3). The small sample size can then lead to monotone likelihood profiles (Figure S4). Indeed = 1 occurred most often for the HapMap: even for a simulated heritability of 0.5, > 0.99 for 882 of the 5000 traits (Figure 1). For the HapMap and simulated h2 = 0.2, we also observed = 0.
The simulations were repeated for a genetic architecture where only 10% of the genetic variance consists of polygenic effects and the remaining 90% is determined by a single QTL (File S4). Even in this extreme scenario, still performed very well, having almost negligible bias. Also the improvement in standard error relative to remained similar.
Confidence intervals
For all simulated traits we calculated both standard and log-transformed confidence intervals for narrow-sense heritability (h2), using genotypic means as well as individual plant data. Also confidence intervals for broad-sense heritability (H2) were calculated. (We recall that only additive genetic effects were simulated, and hence h2 = H2.) Coverage and width of confidence intervals are given in Table 4 and Table S3 (Swedish and French RegMap). The standard 95% intervals obtained from the individual observations (i.e., those associated with ) had around 95% coverage in the simulations, while coverage was below 95% for the intervals based on genotypic means. The latter intervals also had larger width, which relates to the higher variance of in simulations. For the HapMap average width was larger than 0.5, even at a simulated heritability of 0.2. For the simulated traits with , the corresponding confidence interval was [0, 1].
Table 4. Marker-based estimation of heritability.
| Interval | Coverage | Interval width |
|---|---|---|
| Structured RegMap | ||
| h2 = 0.2 | ||
| Broad sense | 0.864 | 0.178 |
| Individual level (standard) | 0.926 | 0.201 |
| Individual level (log-transformed) | 0.957 | 0.203 |
| Means (standard) | 0.912 | 0.314 |
| Means (log-transformed) | 0.965 | 0.322 |
| h2 = 0.5 | ||
| Broad sense | 0.735 | 0.160 |
| Individual level (standard) | 0.948 | 0.196 |
| Individual level (log-transformed) | 0.956 | 0.194 |
| Means (standard) | 0.909 | 0.460 |
| Means (log-transformed) | 0.961 | 0.433 |
| h2 = 0.8 | ||
| Broad sense | 0.671 | 0.086 |
| Individual level (standard) | 0.941 | 0.094 |
| Individual level (log-transformed) | 0.941 | 0.094 |
| Means (standard) | 0.923 | 0.422 |
| Means (log-transformed) | 0.946 | 0.492 |
| HapMap | ||
| h2 = 0.2 | ||
| Broad sense | 0.948 | 0.178 |
| Individual level (standard) | 0.945 | 0.181 |
| Individual level (log-transformed) | 0.969 | 0.182 |
| Means (standard) | 0.835 | 0.538 |
| Means (log-transformed) | 0.916 | 0.678 |
| h2 = 0.5 | ||
| Broad sense | 0.935 | 0.160 |
| Individual level (standard) | 0.948 | 0.164 |
| Individual level (log-transformed) | 0.952 | 0.163 |
| Means (standard) | 0.861 | 0.806 |
| Means (log-transformed) | 0.984 | 0.832 |
| h2 = 0.8 | ||
| Broad sense | 0.924 | 0.084 |
| Individual level (standard) | 0.948 | 0.085 |
| Individual level (log-transformed) | 0.950 | 0.085 |
| Means (standard) | 0.892 | 0.882 |
| Means (log-transformed) | 0.914 | 0.915 |
Shown are width and coverage confidence intervals obtained from the individual plant data and the genotypic means. Results for broad-sense heritability intervals are reported for comparison. We simulated 5000 traits, for random samples of 200 accessions drawn from the structured RegMap (top) and HapMap (bottom). Twenty unlinked QTL were simulated, which explained 50% of the genetic variance. The simulated heritability was 0.2, 0.5, and 0.8.
Differences between the log-transformed and the standard intervals were small for . For the use of the log-transformed intervals gave bigger improvements, although coverage was still below 95%. Also the confidence intervals for broad-sense heritability had insufficient coverage. This is due to model misspecification: the analysis of variance does not incorporate the genetic structure, and uncertainty in the estimates is therefore underestimated. Coverage was still around 93% for the HapMap, whereas for the structured RegMap it decreased to 67.6% for h2 = 0.8.
Heritability estimates for real data
The heritability estimates , , and were calculated for the six traits. Genotypic means were calculated as the best linear unbiased estimators (BLUEs) for the genetic effects, in a linear model containing fixed effects for both genotype and additional design effects (Appendix A). In the calculation of the nondiagonal covariance structure of the BLUEs was taken into account. In the case of and , design effects were directly included in the mixed model.
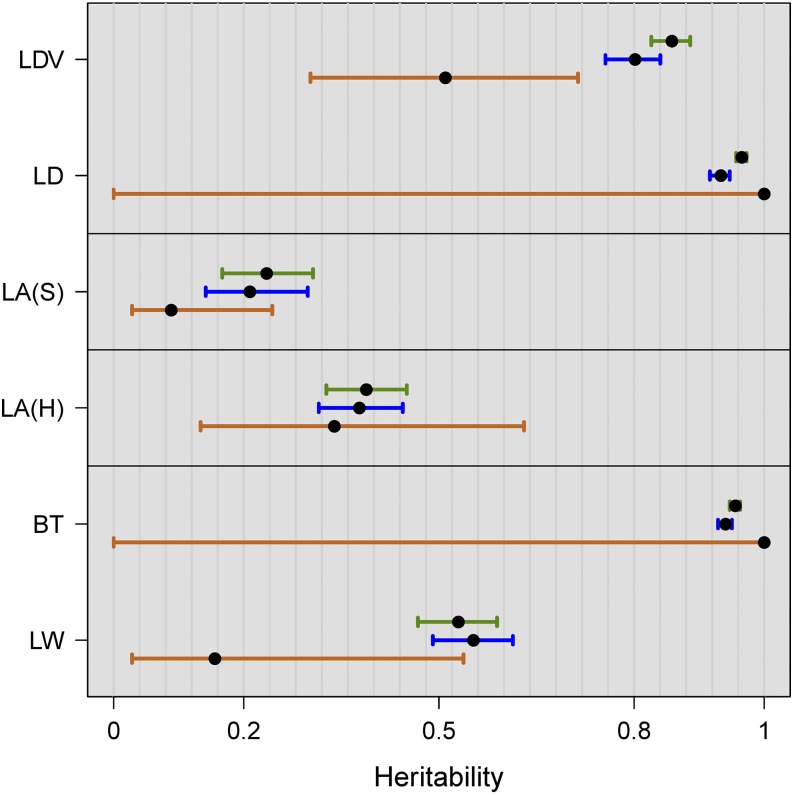
For the flowering traits from Atwell et al. (2010), our estimates of broad-sense heritability were 0.858 for LDV and 0.966 for LD (Table 5). For several reasons (File S2), these values were lower than those reported in supplementary table 7 of Atwell et al. (2010): 0.94 for LDV and 0.99 for LD. For both traits, marker-based heritability estimates based on individual plant data () were substantially lower, but still high (0.80 for LDV and 0.93 for LD). Marker-based heritability estimates obtained from genotypic means () were very different: much lower in case of LDV (0.51) and 1 for LD. (Table 5 and Figure 2). In the latter case, the likelihood as a function of h2 was monotonically increasing, just as we observed for some of the simulated traits. The estimate of residual variance (not reported) was virtually zero in this case, and the confidence interval was equal to the whole unit interval. Also in the leaf area experiments there were substantial differences between and , especially for the Swedish RegMap ( vs. ). Heritability estimates were larger for the HapMap ( vs. ), which has greater genetic diversity. Again, confidence intervals associated with were wider than those associated with . In the final experiment, heritability estimates for bolting time (BT) were very similar to those for LD: = 1, and and were close to 1. For LW we found , much lower than . The latter was slightly larger than the broad-sense heritability estimate , but the confidence intervals largely overlap.
Table 5. Heritability estimates and confidence intervals for two flowering traits from Atwell et al. (2010) (LDV and LD) and four traits from new experiments.
| Trait | |||
|---|---|---|---|
| LDV | 0.801 (0.756,0.840) | 0.510 (0.303,0.714) | 0.858 (0.826,0.886) |
| LD | 0.933 (0.917,0.947) | 1.000 (0.000,1.000) | 0.966 (0.957,0.973) |
| LA(S) | 0.209 (0.141,0.298) | 0.088 (0.028,0.244) | 0.235 (0.167,0.306) |
| LA(H) | 0.378 (0.315,0.444) | 0.339 (0.134,0.631) | 0.388 (0.327,0.451) |
| BT | 0.941 (0.929,0.950) | 1.000 (0.000,1.000) | 0.956 (0.947,0.963) |
| LW | 0.553 (0.491,0.614) | 0.155 (0.028,0.538) | 0.530 (0.468,0.589) |
Three estimators were used: the marker-based estimator using individual plant data (), the marker-based estimator using genotypic means (), and the ANOVA-based estimator of broad-sense heritability (). Horizontal lines separate traits measured in different experiments. Trait abbreviations are given in Table 1.
Figure 2.
Heritability estimates and confidence intervals for two flowering traits from Atwell et al. (2010) (LDV and LD) and four traits from new experiments. Three estimators were used: the ANOVA-based estimator of broad-sense heritability (, green), the marker-based estimator using individual plant data (, blue) and the marker-based estimator using genotypic means (, brown). Traits from different experiments are separated by the black horizontal lines. Trait abbreviations are given in Table 1. Confidence intervals associated with the marker-based estimates are constructed using the logarithmic transformation described in Materials and Methods.
Genomic prediction with G-BLUP
The G-BLUP depends on the estimated genetic and residual variance and hence on the estimated heritability (Henderson 1975; Robinson 1991). Therefore genomic prediction with G-BLUP could potentially be improved when individual plant data are used, instead of genotypic means. We compared G-BLUP based on genotypic means and individual plant data using simulated traits, each including a training set of n = 200 genotypes and a validation set of m = 50 genotypes, for which only marker information was available. For both sets of genotypes, we obtained the G-BLUP () predicting the true (simulated) genetic effects G and calculated the prediction accuracy in terms of the correlation (r) between and G.
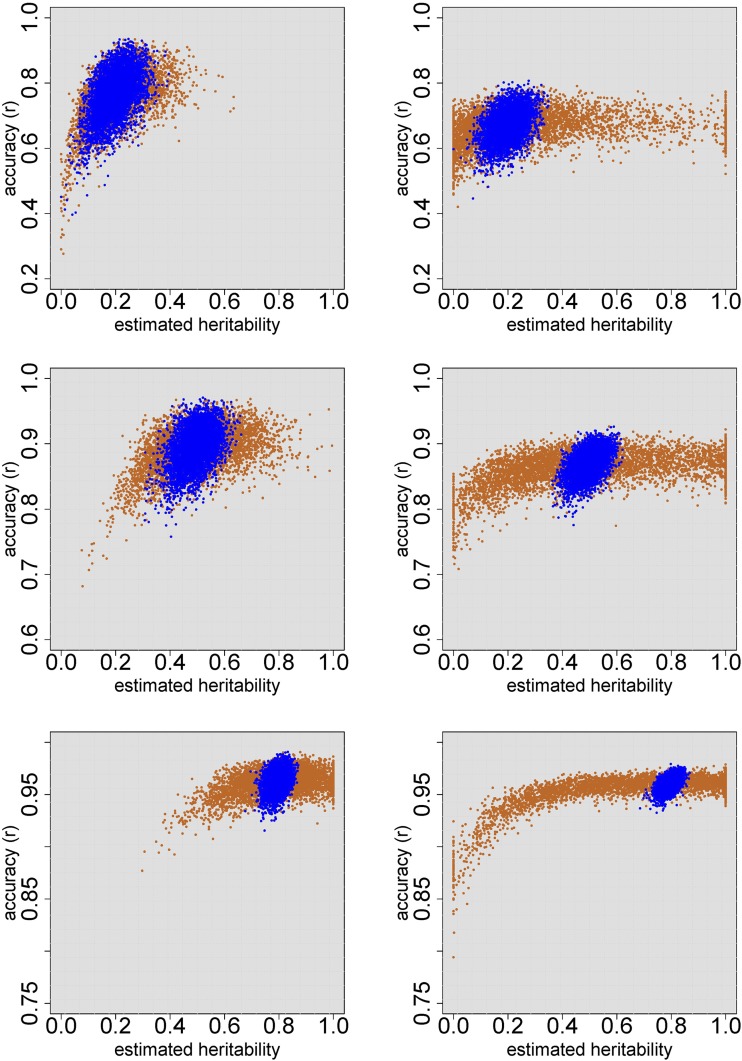
For the training and validation sets, accuracy decreased when the estimated heritability was too far from the simulated heritability (Figure 3, Figure 4, and Table S5). For the validation sets there were larger differences in accuracy across simulations, because of the additional randomness in the selection of the validation genotypes, creating varying degrees of connectedness with the training sets. When using individual plant data, heritability estimates were never far from the simulated heritability, and prediction accuracy was close to a constant, depending on the simulated heritability. Using only genotypic means, heritability was often severely over- or underestimated, in which case accuracy decreased substantially. This decrease was largest when heritability was underestimated, which follows from the mathematical expressions for G-BLUP (Materials and Methods and File S5).
Figure 3.
Prediction accuracy (r) of G-BLUP on training sets of 200 accessions, for 5000 simulated traits, for the structured RegMap (left) and HapMap (right). Within both populations, each trait was simulated for a randomly drawn training set of 200 accessions. Genetic effects were predicted using G-BLUP, based on a mixed model either for the individual plants (blue) or for the genotypic means (orange). Twenty QTL were simulated, which explained 50% of the genetic variance. The simulated heritability was 0.2 (top), 0.5 (middle), and 0.8 (bottom).
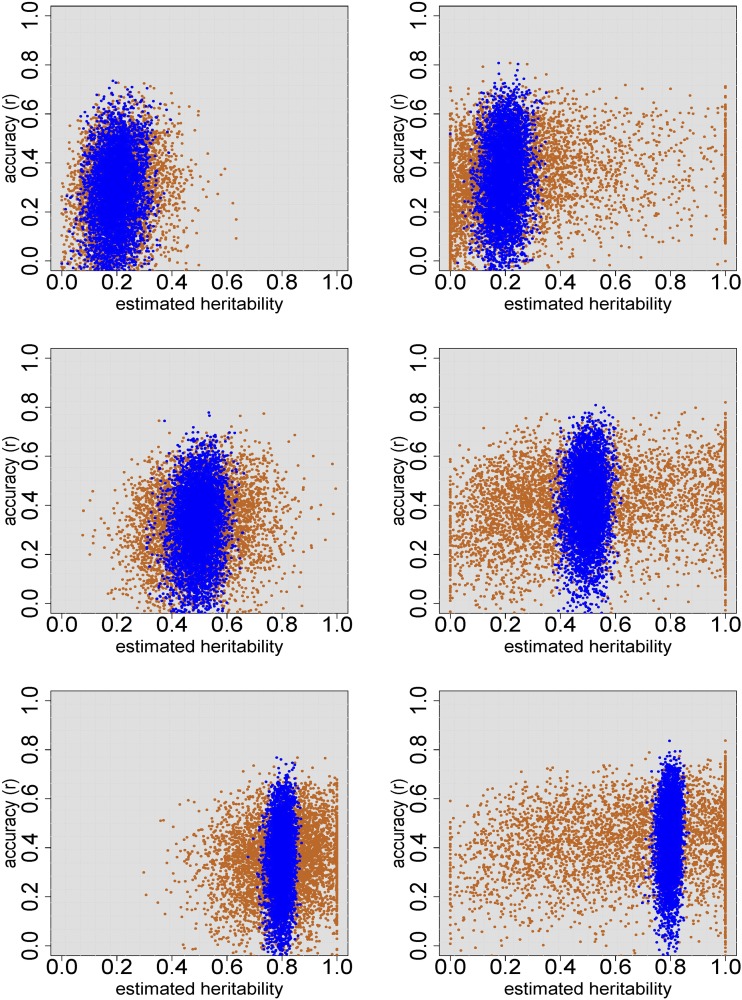
Figure 4.
Prediction accuracy (r) of G-BLUP on validation sets of 50 accessions, for 5000 simulated traits, for the structured RegMap (left) and HapMap (right). Within both populations, each trait was simulated for a randomly drawn training set of 200 accessions. Genetic effects for a randomly drawn validation set of 50 accessions were predicted using G-BLUP, based on a mixed model either for the individual plants (blue) or for the genotypic means (orange). Twenty QTL were simulated, which explained 50% of the genetic variance. The simulated heritability was 0.2 (top), 0.5 (middle), and 0.8 (bottom).
In the HapMap population with simulated heritability 0.8, the estimate was between 0.7 and 0.9 for 4998 of the 5000 simulated traits, and accuracy (r) averaged over these simulations was 0.431 (Table 6). The heritability estimate based on means () was, however, between 0.1 and 0.3 for 8.4% of the simulated traits, and for 2.6% of them it was even below 0.1. Averaged over the latter group of traits, accuracy was only 0.289. Consequently, for these traits an improvement in accuracy of 49% could be realized by genomic prediction based on individual plant data instead of genotypic means. Averaged over all simulated traits, the prediction accuracy obtained from genotypic means was not much lower, because of the large “safe zone” where the approaches performed similarly: accuracy was at least 0.4 once the estimated heritability was above 0.5. It is impossible, however, to determine whether a given trait is within this zone, using genotypic means only. When individual plant data are available, the heritability estimates and could be compared, and the G-BLUP based on genotypic means could be used if and were similar.
Table 6. Prediction accuracy (r) of G-BLUP for 5000 simulated traits, for the HapMap population.
| Interval | (%) | (%) | r (replicates) training set | r (means) training set | r (replicates) validation set | r (means) validation set |
|---|---|---|---|---|---|---|
| [0, 0.1) | 0.00 | 2.58 | 0.890 | 0.289 | ||
| [0.1, 0.3) | 0.00 | 8.34 | 0.937 | 0.373 | ||
| [0.3, 0.5) | 0.00 | 12.34 | 0.954 | 0.409 | ||
| [0.5, 0.7) | 0.04 | 15.90 | 0.942 | 0.959 | 0.208 | 0.423 |
| [0.7, 0.9) | 99.96 | 15.62 | 0.961 | 0.961 | 0.431 | 0.443 |
| [0.9, 1] | 0.00 | 45.22 | 0.961 | 0.448 | ||
| [0, 1] | 100 | 100 | 0.961 | 0.956 | 0.431 | 0.428 |
Each trait was simulated for a randomly drawn training set (200 accessions) and validation set (50 accessions). Genetic effects were predicted using G-BLUP, based on a mixed model either for the individual plants (replicates) or for the genotypic means. The second and third columns contain the percentage of the 5000 traits for which the corresponding heritability estimates ( and ) were contained in the intervals in the first column. The remaining columns show the correlation (r) between simulated and predicted genetic effects, averaged over these traits. The simulated heritability was 0.8; 20 QTL were simulated, which explained 50% of the genetic variance.
Similar results were obtained for prediction accuracy observed in cross-validations on the six observed traits (Figure 5). Averaged over 500 validation sets, prediction accuracies obtained with individual plant data and means were almost the same for LDV, LD, and BT (respectively 0.77, 0.82, and 0.67). For leaf area 13 days after sowing (Swedish RegMap) [LA(S)], leaf area 13 days after sowing (HapMap) [LA(H)], and LW, average accuracies were respectively 0.22, 0.27, and 0.04 when using genotypic means and 0.23, 0.28, and 0.11 when using individual plant data. For LDV, the heritability estimates obtained using genotypic means followed a bimodal pattern, which was largely due to accession 8233, for which only a single observation was available and genotypically is an outlier (Figure S6). Despite the fact that the standard errors and correlations among the genotypic means were taken into account, training sets including this accession produced much lower heritability estimates than in cases where it was assigned to the validation set. This, however, did not lead to lower prediction accuracy. More generally, the relation between low prediction accuracy and underestimating heritability was less clear than in Figure 4, which may be due to the additional uncertainty in the estimation of fixed effects and the fact that this concerns a subsampling of observed traits, rather than simulated traits. Nonetheless, genomic prediction based on individual plant data again performed at least as well as the standard approach based on means and much better in the case of LW.
Figure 5.
Prediction accuracy (r) of G-BLUP in 500 cross-validations, for six traits observed on A. thaliana. In each cross-validation, the accessions are randomly partitioned into a training set of 80% and a validation set of 20% of the accessions. Predictions for the individual plant data for the accessions in the validation set were obtained from Equation C6 in Appendix C. The genetic effects in (C6) were estimated either using individual plant data (blue) or using genotypic means (orange).
GWAS
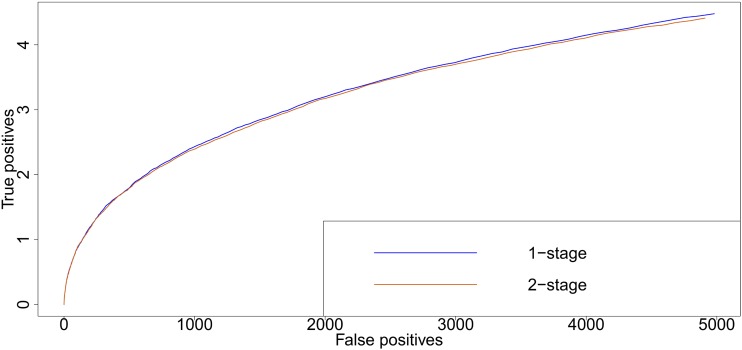
Many state-of-the-art methods for GWAS (Kang et al. 2010; Lippert et al. 2011; Lipka et al. 2012; Zhou and Stephens 2012) use the same mixed model as in marker-based estimation of heritability, apart from the additional marker effect (Appendix D). When testing the significance of this marker effect, the estimated genetic and residual variances (and hence the heritability) determine the correction for population structure or genetic effects elsewhere in the genome. This suggests that poor estimation of the variance components and may also affect association mapping, especially when these are estimated in a model without marker effects and then kept fixed when calculating generalized least-squares (GLS) estimates of marker effects (Kang et al. 2010). The GLS estimate of the fixed effects (including the marker effect) is unbiased since the expectation of Y is Xβ. Hence , even conditionally on a poor estimate of . However, P-values for the significance of marker effects will be more sensitive to poor estimates of than the effect estimates themselves. We investigated this with GWAS on simulated traits for the HapMap and RegMap populations, which were performed on genotypic means (two-stage) and individual plant data (one-stage). Rank correlations between one- and two-stage −log10 P-values were almost 1 when was close to the simulated heritability and decreased when under- or overestimated heritability (Figure S7). However, even then correlations were still high, and the resulting loss in power appeared to be limited (Figure 6). For the RegMap (results not shown) differences between receiver operating characteristic curves were even smaller. In contrast to the accuracy of genomic prediction, these differences remained small when restricting the set of simulated traits to those for which underestimated the simulated heritability (curves not shown). This may be explained by the fact that the correlation between and tends to zero for large numbers of genotypes (Casella and McCulloch 2006).
Figure 6.
Receiver operating characteristics for the GWAS simulations. One thousand traits were simulated, for random samples of 200 accessions drawn from the HapMap. Ten QTL were simulated, at randomly chosen SNP positions (see Simulations in Materials and Methods for more details). The QTL explained 75% of the genetic variance. The simulated heritability was 0.5. The curves were obtained by moving from a small (conservative) significance threshold to a larger one. Simulated QTL were counted as true positive if their P-value was below the threshold; all other SNPs below the threshold were counted as false positive.
Software
In most of our analyses and simulations we used the commercial R package asreml (Butler et al. 2009), which contains a fast implementation of the AI algorithm. We also made our own implementation of the AI algorithm, together with functions for estimating heritability. These are contained in the R package heritability, which is freely available online. In contrast to other packages such as emma and synbreed, it provides confidence intervals for heritability.
For GWAS on simulated traits we developed the command-line program scan_GLS, which is available on request. scan_GLS performs generalized least-squares calculations conditional on variance components estimated in a model without markers, as proposed by Kang et al. (2010), and can efficiently handle genetically identical individuals. State-of-the-art association mapping software can perform association mapping on genetically identical individuals only when these are given different identifiers, i.e., as if they were different genotypes. This leads to large genotypic data files and increases computation time. The GLS calculations in scan_GLS are more efficient since they use the fact that ZKZt and K have the same rank. This has been proposed in Lippert et al. (2011) (supplement), but to our knowledge this has not been implemented in the Fast-LMM software. Although we did not find one-stage GWAS to be more powerful in the present study, the ability to perform fast association mapping for genetically identical individuals is useful in the context of a compressed kinship matrix (Bradbury et al. 2007; Zhang et al. 2010; Lipka et al. 2012). scan_GLS also includes a function to perform GLS calculations with nondiagonal residual variance structure, allowing association mapping with extra (possibly nongenetic) random effects.
Discussion
We have presented new methodology for marker-based estimation of heritability for plant traits, which accounts for genetic relatedness and includes information on within-genotype variability, available from replicates. Our approach offers an alternative to mixed-model analysis based on genotypic means, which is the current practice in GWAS and genomic prediction with G-BLUP. Although mixed models can indeed estimate heritability from only kinship coefficients and genotypic means, we observed very large standard errors and sometimes unrealistic estimates of heritability, in both published data and new experiments. Using simulations and statistical arguments we showed that marker-based estimation of heritability based on genotypic means has indeed severe limitations when applied to commonly used association panels in A. thaliana. The main reason for this is the lack of information on within-genotype variability: heritability estimates are exclusively based on the (usually small) differences between genotypes. This is feasible in human cohorts with many thousands of individuals, but gives insufficient information in plant populations with only several hundred different genotypes, even if standard errors of genotypic means are taken into account. Much more accurate heritability estimates were obtained with mixed-model analysis at the individual plant or plot level. The resulting heritability estimates had accuracies similar to those reported for human diseases (see e.g., Speed et al. 2012). In our simulations with 200 genotypes, the difference in accuracy relative to that of estimates obtained from genotypic means was larger than what was expected from the asymptotic variances. The reason for this larger difference appeared to be the monotone likelihood profile occurring in a substantial part of the simulations, giving heritability estimates of zero or one. However, even without this phenomenon, heritability estimations using individual plant data are more precise, and the asymptotic approximations appear to provide a lower bound on the gain in accuracy.
Mixed-model analysis at the individual plant level can also improve accuracy of G-BLUP, in particular when the actual heritability is underestimated. Too much shrinkage then leads to lower prediction accuracy. In GWAS, where the interest is in the estimated marker effects rather than the variance components, inclusion of the individual plant gave almost no increase in power. However, the possibility to include covariates observed at the individual plant level may be an important practical advantage. While two-stage procedures are usually considered preferable in complex multiexperiment settings (Welham et al. 2010; Piepho et al. 2012), a one-stage approach may give a more convenient and less error-prone analysis of a single experiment with a simple design.
State-of-the-art phenotyping platforms can measure plant traits with increasing accuracy and throughput. Compared to human traits, the key advantages are that phenotyping is performed under experimental conditions and can include different individuals of the same genotype. Our findings suggest that to fully exploit these advantages, statistical analysis at the individual plant level is necessary. Obviously, this requires the availability of the individual plant data, as well as covariates. For many studies in the literature this information is, however, not available, since most online resources store only genotypic means. More specifically, our results are relevant in the light of the missing heritability debate. Although the aim here was not to propose specific explanations for missing heritability, any such explanation clearly requires an accurate estimate of heritability in the first place. Standard errors for heritability are commonly reported for human traits, but usually absent in the A. thaliana literature. We demonstrated that these standard errors can be extremely large when using mixed models at a genotypic means level. Explaining missing heritability based on such estimates then becomes an unreasonable goal.
For two flowering traits, from both a published and a new experiment, = 1. In these cases, also and the broad-sense heritability estimates () were very high. This can partly be explained by the discrete scale (in whole days) on which the traits were measured. Some genotypes therefore had exactly equal phenotypic values in all or many of the replicates. The heritability estimates for LD were also affected by the fact that in the Atwell et al. (2010) data, all nonflowering plants were given a phenotypic value of 200. However, the estimates = 1 also occurred for some of our simulated (Gaussian) traits and have a more fundamental reason: the small number of genotypes (compared to human studies) leads to monotone likelihood profiles and very large standard errors, making it hard to make any statement about heritability.
Recently, Speed et al. (2012) showed that heritability estimates may become biased when linkage disequilibrium (LD) is not constant over the genome and proposed LD-adjusted kinship (LDAK) matrices to correct for this. To assess the effect on and we recalculated these heritability estimates, using an LD-adjusted kinship matrix (Figure S5 and Table S4). For all traits the estimates were very close to the values obtained using the unadjusted kinship matrix. The estimates on the other hand were quite different for several traits. Nevertheless, the same problems occurred: very large confidence intervals and estimates that appear biologically unrealistic. We conclude that is more sensitive to the choice of kinship matrix than and that the different behavior of and cannot be explained by LD being different over the genome.
The mixed-models analysis at the individual plant level proposed here can be extended in several ways, for example by partitioning the genetic variance into different chromosomal contributions, as in Yang et al. (2011), or by including epistatic effects. The latter has been proposed for experimental populations with known pedigrees (Oakey et al. 2007), but marker-based estimation of epistatic effects for natural populations appears possible only with more advanced statistical methodology, for example semiparametric mixed models and reproducing kernel Hilbert spaces (Gianola and Van Kaam 2008; Howard et al. 2014). Another direction for future research is the estimation of heritability in the presence of additional random effects, which would increase the applicability to agricultural field trials (where the raw data are usually at plot rather than individual plant level). Although our approach allows for missing values, it does assume that all design effects can be modeled as fixed. Field trials are often laid out in incomplete blocks, containing only a small number of the genotypes under study. Differences between such blocks are usually best modeled using random block effects. The definition of heritability in such contexts is, however, far from obvious: Oakey et al. (2007) proposed generalized heritability, but for natural populations this definition is not equivalent to the classical definition of heritability (Oakey et al. 2007, p. 813). In this case the ratio of the estimated genetic variance over the total phenotypic variance could be used as a lower bound on heritability. Nevertheless, we have demonstrated that mixed-model analysis at the individual plant or plot level offers important advantages over mixed models for genotypic means.
Supplementary Material
Acknowledgments
We are grateful to Bjarni Vilhjálmsson, Arthur Korte, Susanna Atwell, and Magnus Nordborg for valuable discussions on heritability. We thank Doug Speed for his comments on the use of his LDAK software. Jian Yang is acknowledged for his comments on the calculation of confidence intervals in his GCTA software and Brian Cullis for feedback on the use of asreml-R. Christian Riedelsheimer, Tobias Schrag, and Albrecht Melchinger are acknowledged for providing the SNP data from Riedelsheimer et al. (2012). Finally, we thank two referees for helpful suggestions regarding the terminology.
W.K. was partially funded by the Learning from Nature project of the Dutch Technology Foundation (STW), which is part of the Netherlands Organisation for Scientific Research (NWO). The work was also funded by the Generation Challenge Program–Integrated Breeding Platform project 2.2.5 (M.M. and F.v.E.), the Technological Top Institute Green Genetics (P.J.F.), NWO Earth and Life Sciences (ALW) (P.J.F.), and the Centre for BioSystems Genomics (R.K. and J.K.).
Author contributions:W.K. and F.v.E. designed the research. W.K. performed the research. W.K. wrote the article, with input from M.M., B.E., F.v.E., P.J.F., M.B., J.K., and R.K. Software was written by W.K. (R package heritability) and M.B. (scan_GLS). Experimental data were obtained by P.J.F. [traits LA(S) and LA (H)] and R.K. (traits BT and LW).
Appendix A
Marker-Based Estimation of Heritability
We start with a brief summary of existing methodology and the required assumptions and then define marker-based heritability estimates given genetically identical replicates measured in an unbalanced design.
Marker-based estimation of heritability given a single observation per genotype
Phenotypic variance can be partitioned as
where the genetic variance is decomposed into additive, dominance, and interaction effects (Falconer and Mackay 1996), the dominance effect being absent for inbred lines. Heritability in the broad sense is defined as , while the narrow-sense heritability takes into account only the additive genetic effects, which determine breeding values and the selection response. Defining the residual variance to be the sum of the environmental and nonadditive genetic variance terms (), we can write
| (A1) |
Marker-based estimates of narrow-sense heritability can be obtained using mixed models with random genetic effects. The covariances between these effects are modeled by a genetic relatedness matrix (GRM) estimated from markers, with elements given by (1). Given a single observation per genotype, the standard infinitesimal model is
| (A2) |
where μ is the intercept, G = (G1, …, Gn) has a distribution, and the errors Ei have independent normal distributions with variance . The optional term models the effect of k additional covariates. Under model (A2), heritability can be estimated by
| (A3) |
where and are REML estimates of additive genetic and residual variance (Yang et al. 2011; Speed et al. 2012).
It has been noted (e.g., Hayes et al. 2009) that using model (A2) with genetic relatedness matrix (1) is equivalent to assuming that the effects of the standardized marker scores are drawn independently from Gaussian distributions with variance . Consequently, the model can account only for additive genetic effects, and nonadditive effects will get into the residual variance. This is why is an estimate of narrow-sense heritability. The preceding argument also highlights the fact that is a marker-based estimate, which requires the (frequently made) assumptions that every causal locus is tagged by one of the markers, and that LD is constant across the genome (Speed et al. 2012). For the case that LD is not constant over the genome, Speed et al. (2012) proposed LD-adjusted kinship matrices.
The parameter can be interpreted as additive genetic variance only if K is scaled appropriately. For the kinship matrix used here this is the case, but for different types of kinship matrix (e.g., identity by state), it is necessary to divide each coefficient by tr(PKP)/(n − 1), for P = In − 11′/n. Under the condition that tr(PKP) = (n − 1),
| (A4) |
Marker-based estimation of heritability given genetically identical replicates, for unbalanced designs
When for each genotype a number of genetically identical individuals are observed, (A2) generalizes to
| (A5) |
where Yi,j is the phenotypic response of replicate j of genotype i, xi,j is a vector of fixed effects, G = (G1, … , Gn) has a distribution, and the errors Ei,j have independent normal distributions with variance . Equation A5 is similar to Equation 2 apart from the additional covariates and the generally unbalanced design. Analogous to the case of balanced designs, we use the estimator defined in (3) for REML estimates and obtained for model (A5).
Heritability estimates based on genotypic means can be constructed as in Equations 4 and 5, except that these genotypic means are no longer equal to the arithmetic averages . We fit the linear model
| (A6) |
where is the vector of phenotypic observations, ri is the number of replicates of genotype i, XC is the design matrix of the extra fixed effects, and is the vector of independent Gaussian errors with variance . This model is identical to (A5), apart from the factor genotype now being fixed instead of random. The intercept is included in the incidence matrix XG, which has dimension N × n, for .
We obtain least-squares estimates for the genotypic effects in model (A6), which form the basis for the subsequent analysis. In mixed-model terminology, this is the BLUE for g. In (A6) the only random term is E, i.e., we consider an ordinary linear model, as the additional covariates are all assumed to be fixed. The least-squares estimator is given by
| (A7) |
where X = [XG XC]. This vector has covariance matrix
| (A8) |
Consequently, the covariance matrix of is
The mixed model (4), used for the estimation of heritability, now naturally generalizes to
| (A9) |
where, to avoid having twice the letter g in the same equation, we abused the notation by writing instead of . Hence the matrix r−1In in (4) is replaced by R. Given REML estimates and for this model, we use in (5) as heritability estimate. As in the case of a completely randomized design, is a so-called two-stage estimator. The fact that genotype is taken as a fixed effect in the first stage (A6) and as random in the subsequent mixed-model analysis may appear inelegant, but is common in two-stage analyses and necessary to avoid shrinking twice.
In the case of a completely randomized design with ri replicates without further covariates, and
Note that in contrast to the estimator of broad-sense heritability described below, it is not necessary to define an average number of replicates. In the case of a randomized complete block design with r replicates, XC is the nr × (r − 1) design matrix for the factor block, and is the n × n matrix with elements (r − 1)/n. Hence, R is not diagonal, even though the design is balanced. The off-diagonal elements are equal in this case and small (provided n is sufficiently large). With n = 200 and r = 3 for example, the diagonal elements of R are 0.336667 and the off-diagonal elements are 0.003333. For unbalanced designs, some of the off-diagonal elements may be larger and more influential.
Appendix B
Simulations
Data for genotypes i = 1, … , n with replicates j = 1, … , r were simulated following the model
| (B1) |
where K was scaled such that tr(PKP) = n − 1, for P = In − 11′/n (see Equation A4).
We assumed q QTL located at marker positions, with effect sizes αm (m = 1, … , q) and minor allele frequencies fm (m = 1, … , q); i.e., genotypes AA and aa occur with frequencies fm and (1 − fm), αm being the effect of a single allele. Let xi,m ∈ {0, 1, 2} denote the marker score at QTL m for genotype i. In the case of A. thaliana inbred lines, xi,m ∈ {0, 2} and
| (B2) |
where
is the total (additive) genetic variance. For outbreeding species under Hardy–Weinberg equilibrium, the constant 4 is replaced by a 2. In (B2) we assume linkage equilibrium between the QTL.
For each simulated trait, QTL locations were sampled randomly from all available markers whose minor allele frequency >10%. To enforce linkage equilibrium between the sampled QTL, the sampled locations were discarded and drawn again if the difference between and v2 = 4(α1, … , αq)S(α1, … , αq)′ was too large, S being the sample covariance matrix of the marker scores at QTL positions. It was required that min(v1, v2)/max(v1, v2) > 0.97.
Let
| (B3) |
denote the proportion of the genetic variance explained by the QTL. In our main set of simulations we choose , γ = 0.5, q = 20 QTL. For the comparison of one- and two-stage association mapping, we choose γ = 0.75, q = 10. To achieve the desired level of heritability (for given γ and ), we set and , and α1, … , αq were chosen equally large, such that for each m. The QTL effects were given random signs. In File S4 we repeat the simulations for γ = 0.1 and q = 1.
For the purpose of genomic prediction, 50 additional genotypes were drawn randomly from the same subpopulation of the RegMap. These were assigned only genotypic values, defined as the sum of QTL effects and polygenic effects gi (i = 201, … , 250). The polygenic effects were simulated such that , Ktotal being the kinship matrix of the training and validation sets combined.
Appendix C
Genomic Prediction with G-BLUP
Genomic prediction with G-BLUP relies on the same models used for marker-based estimation of heritability and can be based on individual plant data (one-stage) or genotypic means (two-stage). We now provide the expressions for one- and two-stage G-BLUP, which are based on models (A5) and (A9) in Appendix A.
One- and two-stage G-BLUP for the training and validation sets
First we consider the G-BLUP for the training set, for which phenotypic observations are available. Including the intercept in the design matrix, model (A5) can be rewritten as
| (C1) |
where N is the total number of individuals and Z is the N × n incidence matrix assigning individuals to genotypes. The BLUP of G = (G1, … , Gn)t and the BLUE of β are given by
| (C2) |
where N is the total number of individuals, Z is the N × n incidence matrix assigning individuals to genotypes, and . Numerically and can be more conveniently obtained by solving the mixed-model equations (see, e.g., Henderson 1975 or Robinson 1991). Similar expressions hold for predictions based on the genotypic means following model (A9),
| (C3) |
where, as in (A9), denotes the vector of genotypic means obtained from preliminary linear model analysis (not necessarily the arithmetic averages). Since we assume that all covariates have been accounted for already in this preliminary analysis, the only fixed effect in (C3) is an intercept, with design matrix X = 1n.
Assuming the one-stage model (C1) and given by (C2), the genetic effects Gpred = (Gn+1, … , Gn+m)t of m unobserved (but genotyped) genotypes can be predicted by the conditional means
| (C4) |
where Kpred.obs is the m× n matrix of kinship coefficients for the unobserved vs. observed genotypes. Assuming the two-stage model (A9) and given by (C3), the predictor is given by
| (C5) |
Prediction of new observations and cross-validation
For new observations Ypred at the individual plant level, we have the one-stage predictor
| (C6) |
where Xpred and Zpred are the corresponding design matrices. For predictions with genotypic means, we replace by and β by the estimate obtained within the linear model (A6) in the preliminary stage.
Appendix D
Genome-Wide Association Studies
In mixed-model-based GWAS (Kang et al. 2010; Lippert et al. 2011; Lipka et al. 2012; Zhou and Stephens 2012) the phenotype of genotype i is modeled as
| (D1) |
where xi is the marker score, γ is the marker effect, and the genotypic effects G = (G1, … , Gn) follow a distribution. This model assumes a single observation per genotype. If observations on genetically identical individuals are available, the Yi’s can be replaced by genotypic means , as in (A7)–(A9). Model (D1) then generalizes to
| (D2) |
This amounts to a two-stage approach: first the genotypic means are calculated using (A7), and next association mapping is performed following (D2). In practice a GWAS is often performed on the arithmetic averages , which implicitly assumes a balanced completely randomized design, without any missing values or replicate effects. Here we use the more general model (D2). Apart from the additional marker effect, this is the same model we used in (A9) to construct the heritability estimate .
Alternatively, association mapping can be based on the one-stage model
| (D3) |
where the term (XC)i,jβ models additional covariates, as in (A7). In the two-stage GWAS, this information is accounted for in the genotypic means . In models (D2) and (D3) we test the hypothesis γ = 0 using the F-test, conditional on estimates of the variance components obtained from a model without markers (Kang et al. 2010). When marker effects are small, these estimates are a good approximation of the exact estimates, obtained when genetic and residual variances are reestimated for each marker.
Appendix E
Phenotypic Data
Collection of phenotypic data
For the two flowering traits from Atwell et al. (2010) (LDV and LD), details can be found in the original publication. In the case of LD, all plants that had not flowered by the end of the experiment were given a phenotypic value of 200 days. The leaf area trait [LA(S)/LA(H)] was measured in two separate experiments on the Swedish RegMap and the HapMap, using the same phenotyping platform. The plants were imaged top down for projected leaf area every day. The images were taken using near-infrared light (790 nm) to not influence the photoperiod during the night measurements. In our final experiment, BT and LW were measured for the HapMap. BT was noted as the number of days after vernalization on which the plant started to bolt. LW was measured on photographs taken from the longest leaf 2 weeks after flowering.
Plant growth conditions
In the leaf area experiments, seeds from the Arabidopsis Swedish RegMap (298 accessions) and the Arabidopsis HapMap (350 accessions) were stratified at 4° for 4 days and sown on Rockwool blocks that had been covered with a black foamed PVC sheet to prevent algal growth and provide a uniform background for automated image analysis. The growth conditions were a light intensity of 200 μmol⋅m−2⋅sec−1, 10-hr short day, 20°/18° day/night cycle, and 70% relative humidity (RH). In the experiment measuring BT and LW on the HapMap, seeds were sown on filter paper with demi water and stratified at 4° in dark conditions for 5 days. Following stratification, seeds were transferred to a culture room (16-hr LD, 24°) to induce seed germination for 42 hr. Germinating seeds were then transplanted to wet Rockwool blocks of 4 × 4 cm in a climate chamber with a light intensity of 125 μmol⋅m−2⋅sec−1 16-hr LD, 20°/18° day/ night cycle, and 70% RH. All plants were watered every morning for 5 min at 9 am with 1/1000 Hyponex solution (Hyponex, Osaka, Japan). Nineteen days after germination, all plants were vernalized for 6 weeks in a cold room (12 hr light, 4°). After the 6-week vernalization period plants were transferred back to the same climate chamber in the same order, but given more space to grow.
Genotypic means
All experiments were laid out as randomized complete block designs, apart from the final HapMap experiment (BT and LW), where the same randomization was used within each replicate. In all experiments we included a replicate (complete block) effect. The numbers of replicates in the different experiments were six (LD and LDV), four [LA(S)/LA(H)], and three (BT and LW). Due to nongerminating seeds and dead plants, some accessions had lower numbers of replicates. For the leaf area experiments [LA(S)/LA(H)] we additionally included a row and column effect to model the within-image position of each plant (x = 1, 2, 3 and y = 1, 2, 3, 4). These factors correct for technical artifacts, which are known to be consistent across replicates.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167916/-/DC1.
Communicating editor: A. H. Paterson
Literature Cited
- Atwell S., Huang Y. S., Vilhjalmsson B. J., Willems G., Horton M., et al. , 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer M. P., Wright D., Feng L., Podlich D. W., Luo L., et al. , 2007. A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177: 1801–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B., Morris G., Borevitz J., 2011. Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol. 12: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B., Villoutreix R., Faure N., Hautekeete N., Piquot Y., et al. , 2013. Investigation of the geographical scale of adaptive phenological variation and its underlying genetics in Arabidopsis thaliana. Mol. Ecol. 22: 4222–4240. [DOI] [PubMed] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., et al. , 2007. Tassel: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- Browning S. R., Browning B. L., 2011. Population structure can inflate SNP-based heritability estimates. Am. J. Hum. Genet. 89: 191–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, D. G., B. R. Cullis, A. R. Gilmour, and B. J. Gogel, 2009 ASReml-R Reference Manual. Queensland Department of Primary Industries and Fisheries, Brisbane, Australia.
- Casella S. R. S. G., McCulloch C. E., 2006. Variance Components. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- Eichler E. E., Flint J., Gibson G., Kong A., Leal S. M., et al. , 2010. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11: 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J. B., 2011. Ridge regression and other kernels for genomic selection with r package rrblup. Plant Genome 4: 250–255. [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996 Introduction to Quantitative Genetics, Ed. 4. Prentice-Hall, Englewood Cliffs, NJ. [Google Scholar]
- Fisher R. A., 1918. The correlation between relatives on the supposition of mendelian inheritance. Trans. R. Soc. Edinb. 52: 399–433. [Google Scholar]
- Fu R., Gelfand A. E., Holsinger K. E., 2003. Exact moment calculations for genetic models with migration, mutation, and drift. Theor. Popul. Biol. 63: 231–243. [DOI] [PubMed] [Google Scholar]
- Gianola D., van Kaam J. B. C. H. M., 2008. Reproducing kernel Hilbert spaces regression methods for genomic assisted prediction of quantitative traits. Genetics 178: 2289–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour A. R., Thompson R., Cullis B. R., 1995. Average information REML: an efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 51: 1440–1450. [Google Scholar]
- Goddard M. E., Wray N. R., Verbyla K., Visscher P. M., 2010. Estimating effects and making predictions from genome-wide marker data. Stat. Sci. 24: 517–529. [Google Scholar]
- Hayes B. J., Visscher P. M., Goddard M. E., 2009. Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47–60. [DOI] [PubMed] [Google Scholar]
- Henderson C. R., 1975. Best linear unbiased estimation and prediction under a selection model. Biometrics 31: 423–447. [PubMed] [Google Scholar]
- Holland, J. B., W. E. Nyquist, and C. T. Cervantes-Martinez, 2010 Estimating and Interpreting Heritability for Plant Breeding: An Update, pp. 9–112. John Wiley & Sons, New York/Hoboken, NJ. [Google Scholar]
- Horton M. W., Hancock A. M., Huang Y. S., Toomajian C., Atwell S., et al. , 2012. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, R., A. L. Carriquiry, and W. D. Beavis, 2014 Parametric and nonparametric statistical methods for genomic selection of traits with additive and epistatic genetic architectures. G3 (Bethesda): 4: 1027–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquard A., 1983. Heritability: one word, three concepts. Biometrics 39: 465–477. [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S.-y., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes J. J. B., Bentsink L., Alonso-Blanco C., Hanhart C. J., Blankestijn-De Vries H., et al. , 2007. Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175: 891–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H. H., Wray N. R., Goddard M. E., Visscher P. M., 2011. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 88: 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Y. Huang, J. Bergelson, M. Nordborg, and J. O. Borevitz, 2010 Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21199–21204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka A. E., Tian F., Wang Q., Peiffer J., Li M., et al. , 2012. Gapit: genome association and prediction integrated tool. Bioinformatics 28: 2397–2399. [DOI] [PubMed] [Google Scholar]
- Lippert C., Listgarten J., Liu Y., Kadie C. M., Davidson R. I., et al. , 2011. FaST linear mixed models for genome-wide association studies. Nat. Methods 8: 833–835. [DOI] [PubMed] [Google Scholar]
- Long Q., Rabanal F. A., Meng D., Huber C. D., Farlow A., et al. , 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45: 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998 Genetics and Analysis of Quantitative Traits, Ed. 1. Sinauer Associates, Sunderland, MA.
- Malosetti M., Ribaut J.-M., van Eeuwijk F. A., 2013. The statistical analysis of multi-environment data: modeling genotype-by-environment interaction and its genetic basis. Front. Physiol. 4: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhring J., Piepho H. P., 2009. Comparison of weighting in two-stage analyses of series of experiments. Crop Sci. 49: 1977–1988. [Google Scholar]
- Nyquist W. E., Baker R., 1991. Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 10: 235–322. [Google Scholar]
- Oakey H., Verbyla A., Pitchford W., Cullis B., Kuchel H., 2006. Joint modeling of additive and non-additive genetic line effects in single field trials. Theor. Appl. Genet. 113: 809–819. [DOI] [PubMed] [Google Scholar]
- Oakey H., Verbyla A., Cullis B., Wei X., Pitchford W., 2007. Joint modeling of additive and non-additive (genetic line) effects in multi-environment trials. Theor. Appl. Genet. 114: 1319–1332. [DOI] [PubMed] [Google Scholar]
- Patterson N., Price A. L., Reich D., 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepho H., Williams E., 2006. A comparison of experimental designs for selection in breeding trials with nested treatment structure. Theor. Appl. Genet. 113: 1505–1513. [DOI] [PubMed] [Google Scholar]
- Piepho H. P., Büchse A., Truberg B., 2006. On the use of multiple lattice designs and alpha-designs in plant breeding trials. Plant Breed. 125: 523–528. [Google Scholar]
- Piepho H.-P., Möhring J., Schulz-Streeck T., Ogutu J. O., 2012. A stage-wise approach for the analysis of multi-environment trials. Biom. J. 54: 844–860. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C., Czedik-Eysenberg A., Grieder C., Lisec J., Technow F., et al. , 2012. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat. Genet. 44: 217–220. [DOI] [PubMed] [Google Scholar]
- Ritland K., 1996. A marker-based method for inferences about quantitative inheritance in natural populations. Evolution 50: 1062–1073. [DOI] [PubMed] [Google Scholar]
- Robinson G. K., 1991. That blup is a good thing: the estimation of random effects. Stat. Sci. 6: 15–32. [Google Scholar]
- Rousset F., 2002. Inbreeding and relatedness coefficients: What do they measure? Heredity 88: 371–380. [DOI] [PubMed] [Google Scholar]
- Sillanpaa, M. J., 2011. On statistical methods for estimating heritability in wild populations. Mol. Ecol. 20: 1324–1332. [DOI] [PubMed] [Google Scholar]
- Singh M., Ceccarelli S., Hamblin J., 1993. Estimation of heritability from varietal trials data. Theor. Appl. Genet. 86: 437–441. [DOI] [PubMed] [Google Scholar]
- Smith A., Cullis B., Thompson R., 2001. Analyzing variety by environment data using multiplicative mixed models and adjustments for spatial field trend. Biometrics 57: 1138–1147. [DOI] [PubMed] [Google Scholar]
- Smith A. B., Cullis B. R., Thompson R., 2005. The analysis of crop cultivar breeding and evaluation trials: an overview of current mixed model approaches. J. Agric. Sci. 143: 449–462. [Google Scholar]
- Speed D., Hemani G., Johnson M. R., Balding D. J., 2012. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet. 91: 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stich B., Mohring J., Piepho H.-P., Heckenberger M., Buckler E. S., et al. , 2008. Comparison of mixed-model approaches for association mapping. Genetics 178: 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart, A. W., 2000 Asymptotic Statistics (Cambridge Series in Statistical and Probabilistic Mathematics). Cambridge University Press, Cambridge/London/New York. [Google Scholar]
- van Eeuwijk F. A., Bink M. C. A. M., Chenu K., Chapman S. C., 2010. Detection and use of qtl for complex traits in multiple environments. Curr. Opin. Plant Biol. 13: 193–205. [DOI] [PubMed] [Google Scholar]
- van Heerwaarden, J., M. B. Hufford, and J. Ross-Ibarra, 2012 Historical genomics of North American maize. Proc. Natl. Acad. Sci. USA 109: 12420–12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattikuti S., Guo J., Chow C. C., 2012. Heritability and genetic correlations explained by common snps for metabolic syndrome traits. PLoS Genet. 8: e1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbyla A., Cullis B., Thompson R., 2007. The analysis of qtl by simultaneous use of the full linkage map. Theor. Appl. Genet. 116: 95–111. [DOI] [PubMed] [Google Scholar]
- Visscher P. M., Goddard M. E., 2014. A general unified framework to assess the sampling variance of heritability estimates using pedigree or marker-based relationships. Genetics 199: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Medland S. E., Ferreira M. A. R., Morley K. I., Zhu G., et al. , 2006. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Hill W. G., Wray N. R., 2008. Heritability in the genomics era–concepts and misconceptions. Nat. Rev. Genet. 9: 255–266. [DOI] [PubMed] [Google Scholar]
- Welham S. J., Gogel B. J., Smith A. B., Thompson R., Cullis B. R., 2010. A comparison of analysis methods for late-stage variety evaluation trials. Aust. N. Z. J. Stat. 52: 125–149. [Google Scholar]
- Wijnen C. L., Keurentjes J. J., 2014. Genetic resources for quantitative trait analysis: novelty and efficiency in design from an Arabidopsis perspective. Curr. Opin. Plant Biol. 18: 103–109. [DOI] [PubMed] [Google Scholar]
- Wimmer V., Albrecht T., Auinger H.-J., Schoen C.-C., 2012. synbreed: a framework for the analysis of genomic prediction data using r. Bioinformatics 28: 2086–2087. [DOI] [PubMed] [Google Scholar]
- Wright S., 1920. The relative importance of heredity and environment in determining the piebald pattern of guinea-pigs. Proc. Natl. Acad. Sci. USA 6: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WTCCC , 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., Visscher P. M., 2011. Gcta: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Ersoz E., Lai C.-Q. Q., Todhunter R. J., Tiwari H. K., et al. , 2010. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Tung C.-W. W., Eizenga G. C., Wright M. H., Ali M. L., et al. , 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44: 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O., Hechter E., Sunyaev S. R., Lander E. S., 2012. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA 109: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.