Abstract
Telomeres represent the repetitive sequences that cap chromosome ends and are essential for their protection. Telomere length is known to be highly heritable and is derived from a homeostatic balance between telomeric lengthening and shortening activities. Specific loci that form the genetic framework underlying telomere length homeostasis, however, are not well understood. To investigate the extent of natural variation of telomere length in Arabidopsis thaliana, we examined 229 worldwide accessions by terminal restriction fragment analysis. The results showed a wide range of telomere lengths that are specific to individual accessions. To identify loci that are responsible for this variation, we adopted a quantitative trait loci (QTL) mapping approach with multiple recombinant inbred line (RIL) populations. A doubled haploid RIL population was first produced using centromere-mediated genome elimination between accessions with long (Pro-0) and intermediate (Col-0) telomere lengths. Composite interval mapping analysis of this population along with two established RIL populations (Ler-2/Cvi-0 and Est-1/Col-0) revealed a number of shared and unique QTL. QTL detected in the Ler-2/Cvi-0 population were examined using near isogenic lines that confirmed causative regions on chromosomes 1 and 2. In conclusion, this work describes the extent of natural variation of telomere length in A. thaliana, identifies a network of QTL that influence telomere length homeostasis, examines telomere length dynamics in plants with hybrid backgrounds, and shows the effects of two identified regions on telomere length regulation.
Keywords: telomere, QTL, centromere-mediated genome elimination, haploid, Arabidopsis
TELOMERES are nucleoprotein structures that protect natural chromosome ends from DNA repair activities and nucleolytic resection. In the absence of a functional telomere, exposed chromosome termini trigger a strong DNA damage response resulting in chromosome end-to-end fusions and massive genomic instability. The canonical structure of nuclear telomeres comprises tandem repeats of TG-rich microsatellites that serve as a binding platform for a number of protein complexes. While the telomeric sequence and associated protein complexes are fairly conserved across eukaryotes, the length of telomeric arrays is known to vary extensively among different organisms; budding yeast telomeres average ∼300 bp (Gatbonton et al. 2006) and human telomeres ∼5–15 kb (De Lange 2010), whereas tobacco telomeres can reach 150 kb (Fajkus et al. 1995). A substantial variation in telomere length has also been shown among individuals of the same species. In one of the first studies examining intraspecies telomere length variation, Burr et al. found that telomeres from inbred lines of maize range from 1.8 to 40 kb (Burr et al. 1992). In humans, the length of telomeric repeats has long been implicated in the determination of replication capacity in somatic cells (Bodnar et al. 1998), and intra-individual variation in telomere size has been associated with cancer and age-associated pathologies (Blasco 2005).
Telomere length homeostasis is known to be determined by several antagonistic processes that regulate lengthening and shortening activities. Due to a process known as the end replication problem, telomeric repeats are subject to gradual attrition over numerous cell divisions and therefore contribute to replicative senescence (Watson 1972; Olovnikov 1973). Telomere shortening may also be further exacerbated by postreplicative nucleolytic processing or occasional rapid telomere deletions that are intratelomeric recombination events leading to stochastic excisions of larger telomeric fragments (Li and Lustig 1996). To counterbalance loss of telomeric DNA, the reverse transcriptase telomerase is able to synthesize de novo telomeric repeats at chromosome ends (Greider and Blackburn 1985). Telomeres can also be elongated under certain conditions by homologous recombination-based mechanisms collectively known as “alternative lengthening of telomeres” (Bryan et al. 1997). Telomere length homeostasis therefore can be seen as a carefully controlled balance between these opposing activities. While numerous genes affecting telomere length homeostasis have been identified by functional studies (Askree et al. 2004), little is known about the genetic framework underlying natural variation of telomere length. Large-scale linkage analysis of leukocyte telomere length in humans has previously suggested a number of genetic loci that contribute to telomere length variation (Vasa-Nicotera et al. 2005; Andrew et al. 2006; Zhu et al. 2013). Genome-wide association studies have also led to the identification of loci underlying natural variations including TERC, TERT, NAF1, OBFC1, and RTEL (Mangino et al. 2009; Codd et al. 2010, 2013; Levy et al. 2010; Mangino et al. 2012). While these studies have isolated a number of genetic determinants that contribute to telomere length variation, linkage mapping analysis in humans is classically hampered by genetic diversity and uncontrollable environments. This leads us to question whether species with more extensive mapping resources would be more suitable to identify new loci that influence telomere length.
Previous studies have indicated that Arabidopsis thaliana may present an ideal genetic model for analysis of intraspecies telomere length variation. A survey of 27 accessions has revealed a substantial natural variation with telomeres ranging from 2 to 9 kb (Shakirov and Shippen 2004; Maillet et al. 2006). Excellent genetic tools established in Arabidopsis include vast collections of natural accessions and a number of developed recombinant inbred line (RIL) mapping populations. In this study, we have taken advantage of these genetic resources and investigated the extent of telomere length variation in a large collection of natural accessions. To identify loci that contribute to telomere length variation, we decided to use quantitative trait loci (QTL) mapping approaches with established RILs to map causative loci. In addition to using published RIL populations, one population targeting elongated telomeres was created by centromere-mediated genome elimination, a new haploidization tool for Arabidopsis shown to facilitate the rapid generation of RILs (Ravi and Chan 2010; Seymour et al. 2012). Results from three separate RIL populations indicate that a number of shared and unique QTL underlie natural telomere length variation. Examination of Cvi-0/Ler-2 near isogenic lines (NILs) confirmed the lengthening and shortening effects of two QTL. Closer analysis of one NIL showed that telomere length can take a number of generations to establish homeostasis after segregation of a single causative region.
Materials and Methods
Plant materials
Seeds were obtained from the Nottingham Arabidopsis stock center (NASC); a list of all lines used can be found in Supporting Information, Table S1. Plants were grown at 22° under 16 hr light/8 hr dark conditions for 27 days unless stated otherwise. Five plants (1–5 g tissue) were used for pooled DNA extraction; extraction from single plants used three inflorescences.
DNA extraction
Plant tissue from pooled samples was ground in liquid nitrogen and transferred to 4 ml hexadecyltrimethylammonium bromide (CTAB) DNA extraction buffer (1.4 M NaCl, 20 g/liter CTAB; Sigma), 0.1 M Tris–HCl, pH 8), and 0.4 ml was used for inflorescences. After incubation at 65° for 1 hr, DNA was extracted with an equal volume of phenol:chloroform:isoamyl alcohol 25:24:1 (Sigma) and precipitated with isopropanol. The resulting pellet was washed with 70% ethanol and resuspended in 200 µl dH2O and subjected to RNAse treatment (2 µl of 10 mg/ml RNAse A at 37° for 1 hr).
Terminal restriction fragment analysis
Terminal restriction fragments (TRFs) were produced using Tru1I (Thermo Fischer Scientific) to digest 600 ng genomic DNA. Digests were separated on a 0.8% agarose gel and transferred to a membrane (Amersham Hybond–NX, GE Healthcare). Hybridizations were performed using 32P 5′-end-labeled T3AG3 oligonucleotide probes according to Fitzgerald et al. (1999). Signals were detected using a PharosFX plus phosphoimager (Bio-Rad), and telomere length was determined from 16-bit TIFF images using TeloTool (Gohring et al. 2014). The ladder was fitted with the recommended polynomial function of the third order. When telomeres displayed a mean corrected length <1.5 kb, mean length was recalculated with a nonlinear fit of the first order that gave more accurate measurements below detected ladder bands.
Flow cytometry
Inflorescences were prepared using the Partec Cystain UV precise P reagent kit and analyzed with the Partec Cyflow space flow cytometry system using FloMax software.
QTL analysis
Linkage maps were produced with R/qtl (Broman et al. 2003), and composite interval mapping (CIM) was performed using QTL cartographer V2.5 (Wang et al. 2010) with a walk speed of 1.0 cM. Significance threshold values were calculated using 1000 permutations.
384 SNP Array
Mapping populations were genotyped using a 384 SNP Illumina GoldenGate Genotyping Assay (http://www.illumina.com/Documents/products/technotes/technote_goldengate_design.pdf). Genome Studio software (http://www.illumina.com/applications/microarrays/microarray-software/genomestudio.html) and the R software environment (R Core Team 2013) were used for SNP calling.
Results
A large variation in telomere length is exhibited within natural Arabidopsis accessions
To examine the extent of natural variation, telomere length was measured in Arabidopsis worldwide and Sweden specific accessions (Table S1) (Long et al. 2013). A total of 229 accessions were subjected to TRF analysis (see Figure 1A for a sample blot) and analyzed for mean telomere length values. Because TRFs typically consist of heterogeneous smears that represent telomeric DNA extracted from an entire tissue sample, we questioned the best method to quantify mean telomere length. The recently published TeloTool software was shown to provide accurate telomere length measurement and therefore was used to extract mean length and range values from telomeric smears (Gohring et al. 2014). The overall distribution of mean telomere length over all natural accessions tested was found to range from ∼1 to 9 kb (Figure 1B and Table S1). Heritability was found to be high within this population (H2 = 0.95), which fits well with data obtained from previous studies (Vasa-Nicotera et al. 2005). Such extensive analysis shows the natural variation exhibited within numerous accessions and is shown to vary more than previously indicated (Shakirov and Shippen 2004; Maillet et al. 2006).
Figure 1.
Analysis of 229 natural accessions reveals extensive variation of telomere length. (A) Example TRF blot from 12 accessions. (B) Distribution of mean telomere length over 229 accessions. Range varies from ∼1 to 9.3 kb (see Table S1 for full list of accessions and telomere lengths).
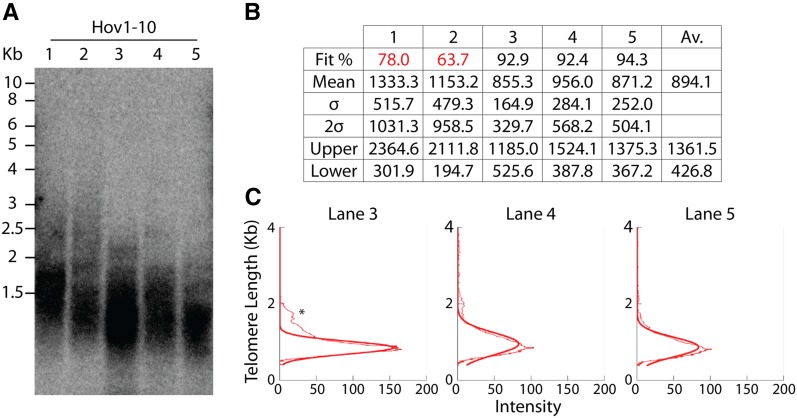
One Swedish accession, Hov1-10 (ID no. 6035), was found to contain telomeres with a mean length of 1065 bp (Figure 2). These data were surprising as studies in Arabidopsis have indicated that telomeres that shorten to <1 kb may become dysfunctional and that 300–400 bp is the minimal threshold required for telomere functionality (Heacock et al. 2004). To further examine this accession and confirm its telomere length, five individual plants were used to repeat TRF analysis. Hov1-10 plants were late flowering and started to bolt ∼3 months after sowing. DNA was extracted from 3-month-old plants that showed a similar length distribution over multiple individuals, indicating that telomere length homeostasis was stable within this accession (Figure 2A). To analyze these individuals further, TeloTool was used to measure mean and standard deviation from telomeric smears to ascertain minimal calculated telomere length values and signal profiles (Figure 2, B and C). To define upper and lower boundaries, two standard deviations were added or subtracted from the mean. Mean telomere length was found to be 894.1 bp when averaged from three samples (lanes 1 and 2 in Figure 2A were excluded due to poor fit qualities). The lower smear boundary was found to average only 426.8 bp, indicating that some telomeres within this accession operate at lengths close to the functional border of telomere length in Arabidopsis (Heacock et al. 2004).
Figure 2.
Hov1-10 displays extremely short telomeres. (A) TRF analysis of five individual Hov1-10 plants (ID no. 6035). (B) TeloTool analysis of telomeric smears showing fit quality percentage, mean, and standard deviation (σ). 2σ was used to define upper and lower boundaries, and averages were taken from lanes 3, 4, and 5. Fit quality was deemed too low in lanes 1 and 2 so these samples were excluded. (C) Initially, TeloTool data did not appear to match visually with the gel image from lane 3. Further analysis of intensity profiles showed that a small amount of higher-molecular-weight signal (marked with an asterisk) was present in this lane. This could be explained by incomplete digestion within this sample during TRF preparation. Because of the high fit quality in this lane, the extrapolated values were deemed to be correct and were used in the analysis.
Generating a Pro-0/Col-0 mapping population using centromere-mediated genome elimination
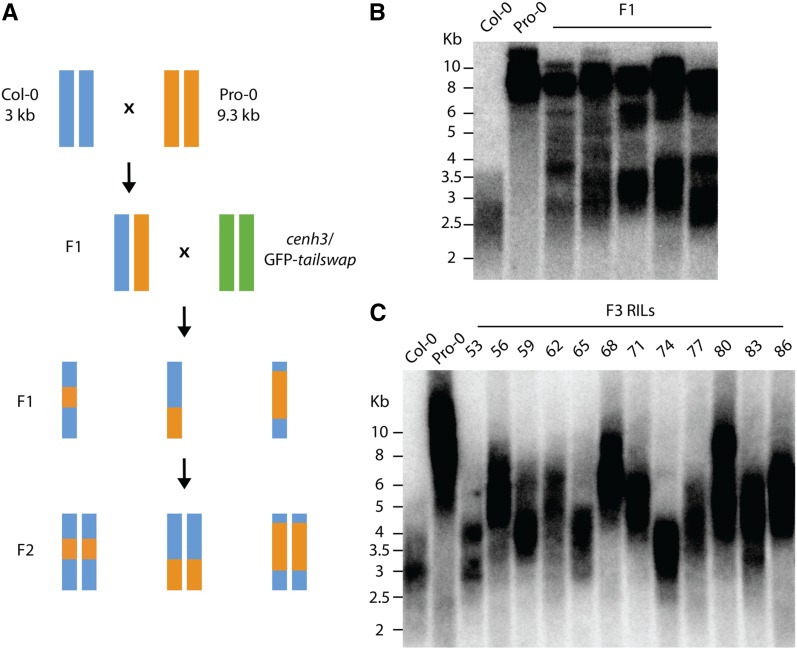
To uncover loci responsible for the large variation in telomere length among natural accessions, we first performed a genome-wide association mapping study (GWAS) that, unfortunately, did not highlight any significant loci (data not shown). It is possible that our data set was simply too small to identify significant loci or that GWAS cannot be used for certain complex traits and is limited in the detection of rare alleles (Korte and Farlow 2013). We therefore decided to adopt a QTL mapping population approach by constructing a RIL population using accessions with long (Pro-0: 9.3 kb, ID no. 8213) and average (Col-0: 3 kb, ID no. 6909) telomeres. Producing RIL populations is notoriously time-consuming, and selfing of recombinant lines to achieve homozygosity at all loci can take ∼2 years to complete. Centromere-mediated genome elimination has recently been shown to enable rapid generation of RIL populations through the production of doubled haploid lines (Ravi and Chan 2010; Seymour et al. 2012). Using this system, a RIL population derived from a cross between Col-0 and Pro-0 was produced (Figure 3A). TRFs performed on Pro-0/Col-0 F1 plants showed a bimodal telomere distribution that likely reflects long and short telomeres inherited from Pro-0 and Col-0 parents, respectively (Figure 3B). These data also are supported by previous observations suggesting that more than one generation is required to reach a new telomere length balance in a hybrid background (Maillet et al. 2006). Heterozygous Pro-0/Col-0 F1 plants were used to pollinate cenh3/GFP-tailswap haploid inducer plants resulting in ∼25–45% of viable, but infertile, haploid offspring (Ravi and Chan 2010). Haploid plants are expected to be recombinant for both F1 parents and therefore represent individual RILs. Infertile plants were screened by flow cytometry (Figure S1), and 246 haploid plants were subsequently identified. A small number of seeds (between ∼2 and 50 per plant) were recovered from haploid plants due to spontaneous chromosome doubling and diploidization in a subset of cells (Ravi and Chan 2010). Resulting plants were selfed by single-seed descent for three generations. TRF analysis in a subset of F3 lines revealed variation in telomere length within a subset of RILs (Figure 3C). Absence of the bimodal telomere length distribution typical for the F1 Pro-0/Col-0 lines (Figure 3B) demonstrated that telomeres had reached a new length homeostasis, presumably determined by a new genetic composition of the RIL lines.
Figure 3.
Construction of a new mapping population using centromere-mediated genome elimination. (A) Crossing scheme for Col-0/Pro-0 RIL population. F1’s derived from crosses between Col-0 and Pro-0 were used to pollinate cenh3/GFP-tailswap haploid inducer plants. Haploids segregated from these crosses are expected to be mosaic for both parental genomes. Diploids were recovered from haploid plants and selfed to the F5 generation. (B) TRF profiles of Col-0, Pro-0, and five individual F1 plants. (C) TRF analysis of a subset of F3 diploid RILs showing variation of telomere length. While F1 plants contained telomeres from both parents, analysis of F3 RILs shows that telomere length homeostasis adjusts to a newly defined balance when genomes from both parents are mixed.
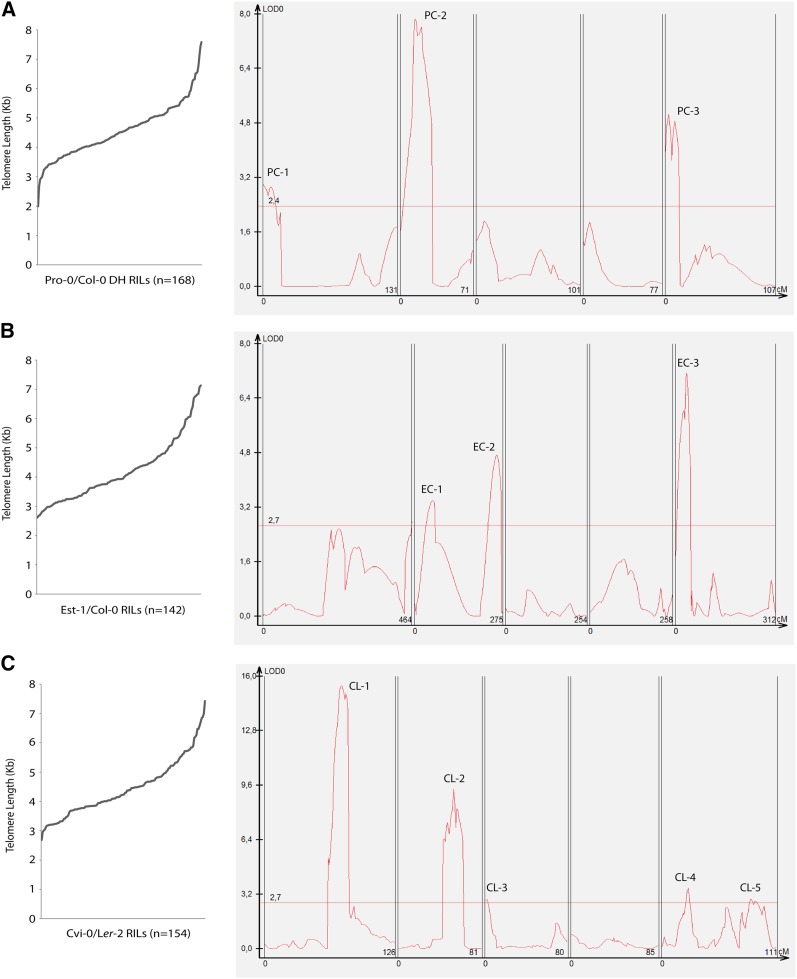
To determine the genotype of individual RIL lines, DNA extracted from five pooled F4 doubled haploid plants from each RIL line was subjected to a 384 SNP genotyping PCR array (Table S2) and phenotyped by TRF analysis (Table S1). From the original population of 246 haploids, 213 were used for genotyping, and 168 RILs were selected for mapping analysis based on the quality of genotyping data. After TRF analysis and quantification with TeloTool, RILs displayed a variation in mean telomere length between 1992.8 and 7581.7 bp (Figure 4A and Table S1). Composite interval mapping (CIM) highlighted three large QTL which were designated labels PC-1, PC-2, and PC-3 (Figure 4A). QTL PC-1 and PC-3 appear to be located near chromosome termini whereas PC-2 is located within chromosome 2. In summary, these results show that by constructing a new RIL population from a cross between Pro-0 and Col-0, it was possible to identify QTL that contribute to telomere length elongation.
Figure 4.
Composite interval mapping of RIL populations. Distribution of telomere length in RIL populations and results from CIM analysis (A) Pro-0/Col-0. (B) Cvi-0/Ler-2. (C) Est-1/Col-0.
QTL mapping of multiple RIL populations reveals a number of shared and unique QTL
Next we examined whether telomere length variation is associated with the same set of QTL in different Arabidopsis accessions. In addition to QTL mapping performed with Pro-0/Col-0 RILs, two established RIL populations were also used for telomere length analysis. Col-0/Est-1 (Balasubramanian et al. 2009) and Cvi-0/Ler-2 (Alonso-Blanco et al. 1998a,b) populations have been used in a number of studies to identify QTL contributing to variation in traits such as flowering time and root skewing (Alonso-Blanco et al. 1998a; Vaughn and Masson 2011). Telomere length was first measured in 142 Est-1/Col-0 RILs and found to range between 2618.7 and 7126.8 bp (Table S1). Telomere length of parental lines averaged over 10 samples (corrected mean) was 3242.6 bp for Col-0 and 6817.3 bp for Est-1, showing a variance of RILs between the two parental values. CIM analysis conducted using these data revealed the presence of three QTL, which were labeled EC-1, EC-2, and EC-3 (Figure 4B). Interestingly, two of these loci seemed to align roughly with two QTL regions identified in the Pro-0/Col-0 population (EC-1 and PC-2, PC-3 and EC-3). Next, we examined telomere length in the Cvi-0/Ler-2 population that indicated transgressive variation between RILs with values much larger than shown in parental lines. Ler-2 and Cvi-0 were shown to measure 4299.8 and 5004.7 bp, respectively (average of 10 control samples, corrected mean), which contrasts to a total Cvi-0/Ler-2 RIL population range of 2684.7–7423.7 bp. This suggests antagonistic epistasis among different loci within each parent and that separation of the causative alleles by crosses with other accessions can confer lengthening and shortening effects that surpass parental allele combinations. Five QTL (CL-1–CL-5, Figure 4C) that seemed to feature five completely novel regions in comparison to the other two populations were identified in this population. In summary, analysis of three populations has revealed novel and shared QTL that can be tested for their contributions to telomere length homeostasis.
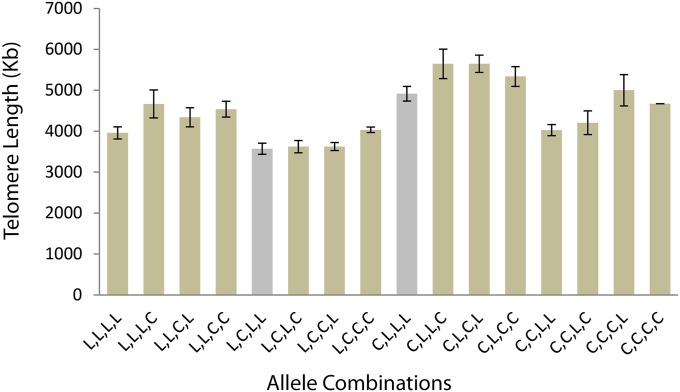
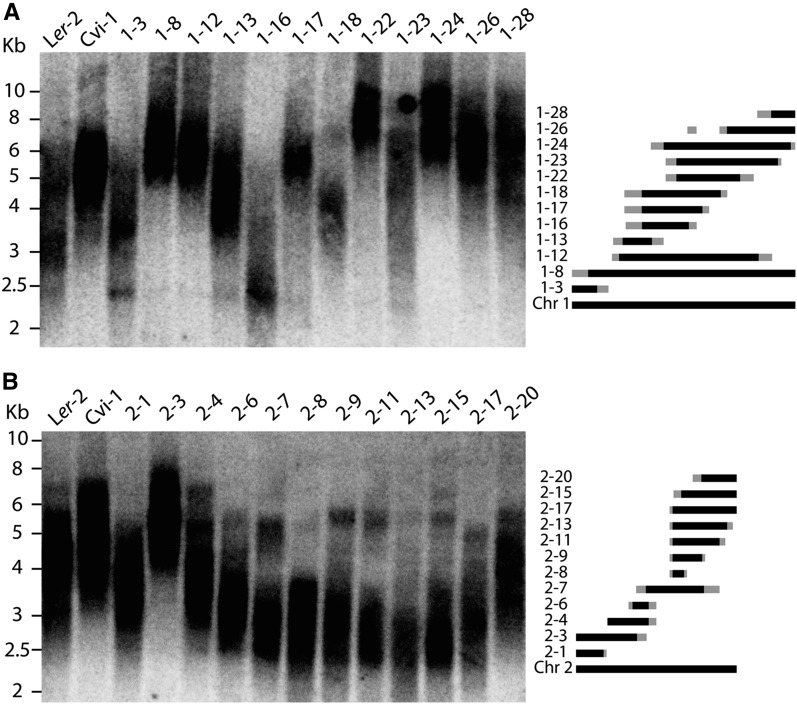
To examine QTL effects, NILs were used to analyze changes in telomere length when respective QTL regions were segregated as Ler-2 or Cvi-0. A large catalog of NILs corresponding to the Cvi-0/Ler-2 population was previously established and comprises insertions of Cvi-0 into the Ler-2 background that span the entire genome (Keurentjes et al. 2007). Because of this established resource, we primarily centered our analysis on the Cvi-0/Ler-2 population. To begin dissecting Cvi-0/Ler-2 QTL, we first wanted to predict the effects of single regions on telomere length regulation. RILs were sorted based on the genotype at four QTL positions, which were signified by the most significant marker (CL-1:EG.198C/200L-Col, CL-2:BH.195L-Col, CL-4:CC.400L-Col, CL-5:CC.262C). In this analysis, CL-3 was excluded to simplify data as this QTL exhibited only a small effect. The mean telomere length of RILs comprising the relevant genotype combinations is displayed in Figure 5. Cvi-0 insertions into CL-1 and CL-2 at respective QTL positions generally conferred a lengthening (CL-1) and shortening (CL-2) effect (Figure 5, gray bars). Next, we wanted to confirm experimentally the effect of single loci on telomere length using the established NIL population. NILs spanning chromosome 1 and 2 were subsequently selected and tested by TRF analysis. Data from Figure 6A show that insertions of Cvi-0 into the QTL region on chromosome 1 results in telomere elongation (Figure 6A). NILs 1–8 and 1–12 represent Cvi-0 insertions covering large portions of chromosome 1, and both exhibited elongated telomeres in comparison to the Ler-2 wild-type line. Lines 1–22 and 1–24 appeared to display drastically elongated telomeres and correspond to the region defined by the QTL. Surprisingly, 1–16 showed drastic shortening, which could mean that this region corresponds to two causative alleles that, when separated, confer considerable elongation or shortening effects. This phenomenon is described in other systems where large QTL are found to contain a number of linked causative alleles (Liti and Louis 2012). In contrast, NILs spanning chromosome 2 suggested that this region confers a shortening effect as demonstrated by the “smile” pattern in the region corresponding to the QTL (Figure 6B). The lengthening (CL-1) and shortening (CL-2) effects of relevant QTL are therefore confirmed experimentally with NILs, indicating that both contain causative loci that contribute to telomere regulation.
Figure 5.
QTL combinations of Cvi-0/Ler-2 population. To examine the effects of QTL, the most significant markers were taken from each of the four major trans QTL positions: CL-1:EG.198C/200L-Col, CL-2:BH.195L-Col, CL-4:CC.400L-Col, and CL-5:CC.262C. Average telomere length was taken from RILs in each combination. Gray bars represent lines where a Cvi-0 insertion at a single locus conferred telomere lengthening or shortening. Error bars represent ±SE.
Figure 6.
Analysis of NILs confirms effects of two loci on telomere length. Cvi-0 insertions into the causative region encompassing the QTL showed telomere lengthening in chromosome 1 (A) and shortening in chromosome 2 (B). Figure illustrating known positions of Cvi-0 insertions is adapted from Keurentjes et al. (2007). Black bars represent known positions of Cvi-0 introgressions, and gray bars indicate crossover areas between markers selected for genotyping.
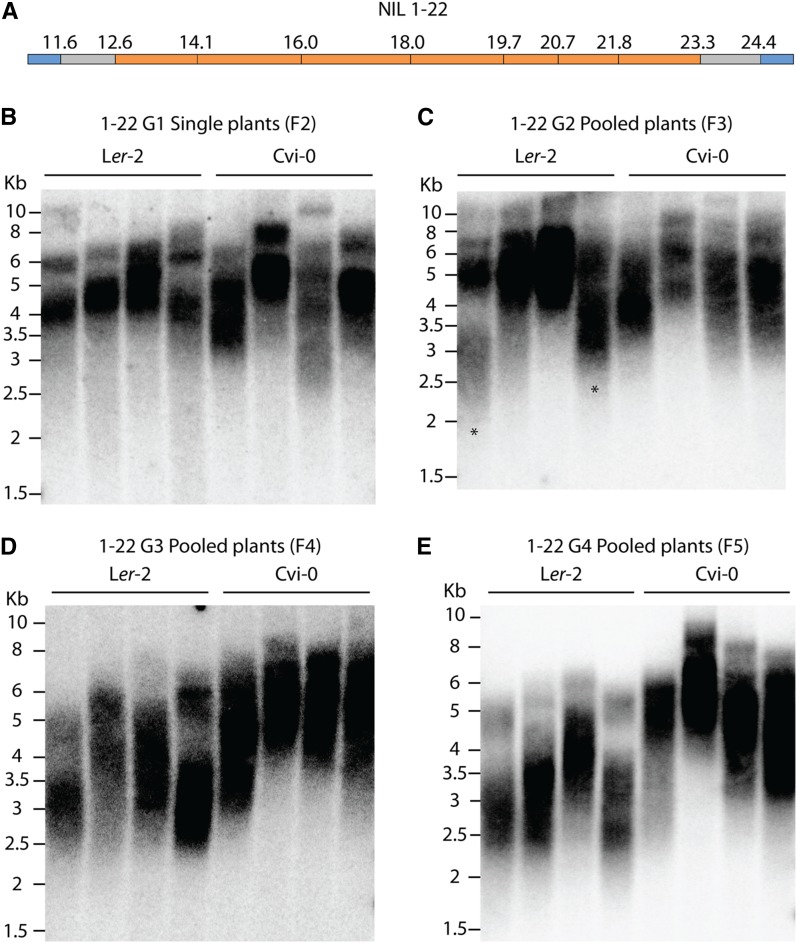
To study the rate of telomere lengthening or shortening when changing a single causative allele, NIL 1–22 (corresponding to QTL CL-1 leading to a elongation effect) was backcrossed to the Ler-2 parent, and plants homozygous for Ler-2 or Cvi-0 in this region were established in the F2 generation. First, the position of the Cvi-0 insertion in NIL 1–22 was genotyped with cleaved amplified polymorphism (CAPS) markers designed to span chromosome 1 (Table S3); this confirmed the insertion at ∼12–24 Mb (Figure 7A). TRFs were performed on five individual F2 plants that segregated as Cvi-0 or Ler-2. F2 lines showed no difference in telomere length, which appeared to be uniform over all samples (Figure 7B). This suggests that telomere length homeostasis is not quickly established after exchanging one causative region. Each individual was then selfed, five plants from the next generation were pooled, and TRFs were performed. Gradual shortening was seen in two samples, suggesting that telomeres were gradually adjusting to a new homeostatic balance (Figure 7C). Selfing for four generations in total revealed a distinct change in lines that segregated as Cvi-0 or Ler-2 (Figure 7D). This result confirms the presence of a causative locus within this 10-mb region. It is evident that telomere length homeostasis is not quickly established within this line and requires a number of generations before the effect of the insertion can be seen.
Figure 7.
Segregation of Cvi-0 region from NIL 1–22 confirms its effect on telomere elongation. (A) NIL 1–22 was genotyped with CAPS markers and found to comprise a Cvi-0 insertion in chromosome 1 between ∼12 and 24 Mb. (B) Homozygous NIL 1–22 was backcrossed to the parental Ler-2 line, and single plants homozygous for Ler-2 or Cvi-0 at this region were identified in the F2. Analysis of these plants showed no apparent difference in telomere length. (C) Pooled plants in the second generation were analyzed, and telomere shortening was observed in two Ler-2 samples (marked by asterisks). (D) Third generation and (E) fourth generation show clear change in telomere length, indicating that this region contains the causative locus.
Discussion
Variation of telomere length among natural populations has been shown within a number of species. Studies from budding yeast, humans, and mice have used this variation to map novel loci that contribute to telomere length variation (Li and Lustig 1996; Zhu et al. 1998; Vasa-Nicotera et al. 2005; Liti et al. 2009). One important example was first shown in mice interspecies crosses when a region on chromosome 2 was first identified to play a role in telomere length regulation through mapping with an F1 backcross population (Zhu et al. 1998). Later work revealed that the helicase Rtel was found to underlie this locus, and mutants displayed drastic telomere-related phenotypes (Ding et al. 2004). More recently, it was shown that Rtel is important in the disassembly of T-loops during replication (Vannier et al. 2012). Mapping approaches in humans have been much more extensive, and a number of loci were initially identified to contribute to leukocyte telomere length variation (Vasa-Nicotera et al. 2005; Andrew et al. 2006; Mangino et al. 2009). The causative gene in one locus from chromosome 12, identified by Vasa-Nicotera et al. (2005), was eventually shown to encode for Bicaudal-D homolog 1 (BICD1) (Mangino et al. 2008). GWAS have also identified a number of loci of known to influence telomere length (Mangino et al. 2009; Codd et al. 2010, 2013; Levy et al. 2010; Mangino et al. 2012). Data from QTL mapping studies were not replicated in independent studies, although variants of TERC and OBFC1 were discovered in a number of GWAS (Codd et al. 2010, 2013; Mangino et al. 2012). In addition, work in budding yeast has also used natural variation of telomere length for mapping approaches identifying loci containing YKu80 and TLC1 (Gatbonton et al. 2006; Liti et al. 2009). These studies therefore highlight the power of mapping-based approaches in identifying new loci that contribute to telomere length homeostasis.
Mapping efforts in plants, however, have not yielded any clearly defined candidates so far. Burr et al. (1992) initially showed variation of telomere length within maize RIL populations (Burr et al. 1992). QTL mapping in maize later identified a number of contributing loci and suggested a rad51-like protein as a candidate (Brown et al. 2011). QTL from this study, however, are not confirmed experimentally with NILs, and candidates are selected from large QTL regions that are not fine-mapped. Because only one population was examined in this study, only contributing alleles from the parental lines were shown in QTL plots. Approaches in Saccharomyces cerevisiae have previously used multiple cross combinations for linkage analysis, revealing large numbers of QTL in comparison to conventional single-cross populations (Cubillos et al. 2011). Analysis of the same phenotypic trait over a number of different cross combinations showed strain-dependent and context-dependent QTL combinations. The use of multiple populations therefore can allow mapping of QTL that contribute over a variety of strains. The extensive genetic resources that have been established in Arabidopsis, including numerous recombinant inbred line populations, would provide the ideal basis for a similar approach in plants. Taking this into account, a study encompassing three RIL populations was undertaken and has revealed many new loci that contribute to telomere length. Using multiple populations, a map of genetic loci has been constructed showing many associated regions (Figure 8). Shared QTL regions (e.g., EC-1/PC-2 and EC-3/PC-3; see Figure 8) indicate the presence of strain-dependent QTL that are shared between populations containing Col-0. In addition, context-dependent QTL were also identified showing causative regions that are unique to each population (Figure 8). Although it is not possible at this stage to discern the causative genes, overlaying putative and known telomere-associated proteins onto the physical map of the genome with displayed physical QTL regions indicates some similarities with genes contained in regions from each population (Figure 8). This is, for example, illustrated in the Cvi-0/Ler-2 population where QTL are positioned over each member of the MRN complex. The MRN complex is known in Arabidopsis to play roles in the regulation of telomere length (Gallego and White 2001; Bundock and Hooykaas 2002, and NBS1 is known to play a synergistic role with telomerase at telomeres (Najdekrova and Siroky 2012). Fine-mapping and further analysis of regions identified in this study could begin to elucidate the network that controls telomere length homeostasis in natural accessions.
Figure 8.
Summary of QTL positions. Approximate QTL positions are indicated by known positions of significant markers. Horizontal marks present on each chromosome represent 1-Mb intervals. Results indicate that there are shared and unique QTL between Cvi-0/Ler-2 (blue), Est-1/Col-0 (orange), and Pro-0/Col-0 (green) RIL populations. Positions of genes with putative or known effects on telomere biology are highlighted (STN1, Song et al. 2008; CTC1, Surovtseva et al. 2009; TEN1, Leehy et al. 2013; KU70, Riha et al. 2002; KU80, Gallego et al. 2003; TRB1/2/3, Kuchar and Fajkus 2004; TER1 and TER2, Cifuentes-Rojas et al. 2012; TERT, Fitzgerald et al. 1999; POT1a/b/c, Shakirov et al. 2005 and Rossignol et al. 2007; RPA70a, Takashi et al. 2009; RAD50, Gallego and White 2001; NBS1, Najdekrova and Siroky 2012; MRE11, Bundock and Hooykaas 2002; NAP57, Kannan et al. 2008; STEP1, Kwon and Chung 2004; WHY1, Yoo et al. 2007; and TAC1, Ren et al. 2004).
Natural variation of telomere length in Arabidopsis was initially shown by Shakirov and Shippen where telomere length was measured in 14 accessions and found to range between 2 and 9 kb (Shakirov and Shippen 2004). In another study, telomeres were found to range between 2 and 12 kb over 18 accessions (Maillet et al. 2006). These values, however, represent boundaries of telomeric smears and do not represent the statistical mean as calculated in this study by TeloTool. Analysis of 229 worldwide and Sweden specific accessions with TeloTool, a new program that is able to accurately quantify mean telomere length and range from TRF blots, allowed a fully comprehensive study of natural variation. When measuring telomeric smears in Hov1-10, telomeres were found to average 1065 bp and decrease to an average of 426.8 bp (Figure 2); these lengths are much shorter than indicated previously within a natural accession. Studies on telomere shortening in the Arabidopsis telomerase mutant have given great insights into the rate of telomere shortening over numerous generations (Fitzgerald et al. 1999; Riha et al. 2001). Telomeres were found to shorten by 250–500 bp per generation, and developmental defects were observed after G6 and G10 plants were deemed completely terminal (Riha et al. 2001). Also in this study, the shortest telomeric bands of the G8 mutants were found to be ∼500 bp. Analysis of G9 tert telomeres by primer extension telomere repeat amplification later showed that telomeres ranged between 300 and 900 bp (Heacock et al. 2004). This study also showed that telomere–subtelomere fusions in G9 tert mutants have a mean of 265 bp, defining this as the average length where telomere fusions occur at a dysfunctional telomere. The shortest telomeres of Hov1-10, therefore, seem to be very close to the minimal functional size limit as described in these studies. Additional experiments examining telomere structure and genome stability could clarify whether telomere function is compromised in this accession.
In addition to accessions that display drastically shortened telomere phenotypes, telomeres of the Pro-0 accession were shown to average ∼9.3 kb. Such extreme elongation was previously observed in Arabidopsis ku70 mutants and was shown to be a result of uninhibited telomerase activation at chromosome ends (Riha et al. 2002; Riha and Shippen 2003). To begin elucidating factors that lead to telomere lengthening in a natural background, a new mapping population was constructed using the cenh3 haploid inducer system (Ravi and Chan 2010). The major advantage of this system includes the fast generation time of RIL populations from doubled haploids (Seymour et al. 2012). Results from F1 crosses showed bimodal telomere length, suggesting that telomeres inherited from both parents are not immediately adjusted to the new genetically set homeostasis. Nevertheless, the bimodal distribution is not apparent in subsequent generations. Processes that mediate rapid changes in telomere length such as telomere rapid deletion and ALT have previously been described in Arabidopsis and could contribute to the establishment of new length homeostasis in early generations (Watson and Shippen 2007). Mapping of QTL by CIM analysis revealed two major loci that contribute to elongated telomeres; while none of the Pro-0/Col-0 QTL include KU70 or KU80, they do involve POT1 genes that encode proteins known to bind the TER1 component of telomerase (Surovtseva et al. 2007; Cifuentes-Rojas et al. 2011). Backcrosses of relevant RILs and further characterization of these loci would provide valuable information on important factors influencing telomere elongation.
In addition to the novel Pro-0/Col-0 population, established Est-1/Col-0 and Cvi-0/Ler-2 RILs indicated a number of shared and unique QTL (Figure 4, Figure 5, and Figure 8). Two of these loci were confirmed through analysis with a population of NILs; the QTL CL-1 indicated a lengthening effect whereas CL-2 appeared to contribute to shortening (Figure 6). When focusing on chromosome 1, analysis of a number of NILs spanning this chromosome showed contradicting effects (Figure 6A). NIL 1–22 was shown to exhibit a lengthening effect whereas another line, NIL 1–16, actually showed a telomere-shortening phenotype. It is possible that two closely linked causative genes are present within this single large-QTL region that, when separated, confer opposite phenotypic effects. The presence of multiple linked causative loci underlying a single QTL has also been recorded in S. cerevisiae. Four genes controlling sporulation efficiency were found within a single QTL, each of which also produced opposing effects (Ben-Ari et al. 2006). When NIL 1–22 containing the causative region on chromosome 1 was backcrossed, plants segregated as Cvi-0 and Ler-2 in this region displayed a clear difference in telomere length after four generations (Figure 7). TRFs indicate that the rate of change is subtle and occurs gradually over generations. This also mimics previous work in yeast where the return to telomere homeostasis was examined over many generations (Marcand et al. 1999). This study shows that, after induction of a shortened telomere, telomerase is able to contribute to its elongation over many generations; the elongation rate, however, slows down when the telomere reaches its homeostatic balance. This is also displayed at elongated telomeres where, in the presence of telomerase, telomeres shorten at a constant rate until they reach the point of equilibrium where the rate of degradation decreases. It can also be in Arabidopsis that, when shuffling multiple alleles that control telomere length homeostasis, drastic changes in telomere length are seen as a consequence of telomerase elongation or telomere rapid deletion. This would serve to establish a rough equilibrium of telomere length quickly, whereas more subtle fine-tuning of telomere length would occur more slowly. In the case of NIL 1–22, because only one allele is changed within this line, reestablishment of telomere length homeostasis progresses at a slower rate, taking a larger number of generations to reach equilibrium. Modulation of single alleles therefore may not have such a drastic effect initially, but will change length by a large amount over numerous generations. This could also depend on the nature of investigated polymorphism as others may have immediate effects. Rates of telomere elongation and shortening are seen to be more drastic in studies where key genes involved in telomere metabolism are inactivated (Riha et al. 2002; Surovtseva et al. 2007; Cifuentes-Rojas et al. 2011). Comparisons between published mutants and natural polymorphisms, however, are not straightforward as such polymorphisms will not necessarily knock out gene function, but are rather expected to modulate gene expression or protein functions and interactions.
Acknowledgments
This work was supported by the Austrian Science Fund (FWF; Y418-B03), by a European Molecular Biology Organization Installation Grant, and by the European Research Council (ERC-2010-AdG #268962–MAXMAP).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.172163/-/DC1.
Communicating editor: A. Houben
Literature Cited
- Alonso-Blanco C., El-Assal S. E. D., Coupland G., Koornneef M., 1998a Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C., Peeters A. J., Koornneef M., Lister C., Dean C., et al. , 1998b Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14: 259–271. [DOI] [PubMed] [Google Scholar]
- Andrew T., Aviv A., Falchi M., Surdulescu G. L., Gardner J. P., et al. , 2006. Mapping genetic loci that determine leukocyte telomere length in a large sample of unselected female sibling pairs. Am. J. Hum. Genet. 78: 480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree S. H., Yehuda T., Smolikov S., Gurevich R., Hawk J., et al. , 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 101: 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Schwartz C., Singh A., Warthmann N., Kim M. C., et al. , 2009. QTL mapping in new Arabidopsis thaliana advanced intercross-recombinant inbred lines. PLoS ONE 4: e4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari G., Zenvirth D., Sherman A., David L., Klutstein M., et al. , 2006. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2: e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M. A., 2005. Telomeres and human disease: ageing, cancer and beyond. Nat. Rev. Genet. 6: 611–622. [DOI] [PubMed] [Google Scholar]
- Bodnar A. G., Ouellette M., Frolkis M., Holt S. E., Chiu C. P., et al. , 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279: 349–352. [DOI] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Brown, A. N., N. Lauter, D. L. Vera, K. A. McLaughlin-Large, T. M. Steele et al., 2011 QTL mapping and candidate gene analysis of telomere length control factors in maize (Zea mays L.). G3 (Bethesda) 1: 437–450. [DOI] [PMC free article] [PubMed]
- Bryan T. M., Englezou A., Dalla-Pozza L., Dunham M. A., Reddel R. R., 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3: 1271–1274. [DOI] [PubMed] [Google Scholar]
- Bundock P., Hooykaas P., 2002. Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14: 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr B., Burr F. A., Matz E. C., Romeroseverson J., 1992. Pinning down loose ends: mapping telomeres and factors affecting their length. Plant Cell 4: 953–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C., Kannan K., Tseng L., Shippen D. E., 2011. Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc. Natl. Acad. Sci. USA 108: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cifuentes-Rojas C., Nelson A. D., Boltz K. A., Kannan K., She X., et al. , 2012. An alternative telomerase RNA in Arabidopsis modulates enzyme activity in response to DNA damage. Genes Dev. 26: 2512–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd V., Mangino M., van der Harst P., Braund P. S., Kaiser M., et al. , 2010. Common variants near TERC are associated with mean telomere length. Nat. Genet. 42: 197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd, V., C. P. Nelson, E. Albrecht, M. Mangino, J. Deelen et al., 2013 Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 45: 422–427, 427e1–2. [DOI] [PMC free article] [PubMed]
- Cubillos F. A., Billi E., Zorgo E., Parts L., Fargier P., et al. , 2011. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol. Ecol. 20: 1401–1413. [DOI] [PubMed] [Google Scholar]
- de Lange T., 2010. How shelterin solves the telomere end-protection problem. Cold Spring Harb. Symp. Quant. Biol. 75: 167–177. [DOI] [PubMed] [Google Scholar]
- Ding H., Schertzer M., Wu X., Gertsenstein M., Selig S., et al. , 2004. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell 117: 873–886. [DOI] [PubMed] [Google Scholar]
- Fajkus J., Kovarik A., Kralovics R., Bezdek M., 1995. Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol. Gen. Genet. 247: 633–638. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M. S., Riha K., Gao F., Ren S., McKnight T. D., et al. , 1999. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 96: 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M. E., White C. I., 2001. RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M. E., Jalut N., White C. I., 2003. Telomerase dependence of telomere lengthening in Ku80 mutant Arabidopsis. Plant Cell 15: 782–789.12615949 [Google Scholar]
- Gatbonton T., Imbesi M., Nelson M., Akey J. M., Ruderfer D. M., et al. , 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2: 907–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohring J., Fulcher N., Jacak J., Riha K., 2014. TeloTool: a new tool for telomere length measurement from terminal restriction fragment analysis with improved probe intensity correction. Nucleic Acids Res. 42.: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H., 1985. Identification of a specific telomere terminal transferase-activity in Tetrahymena extracts. Cell 43: 405–413. [DOI] [PubMed] [Google Scholar]
- Heacock M., Spangler E., Riha K., Puizina J., Shippen D. E., 2004. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 23: 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K., Nelson A. D., Shippen D. E., 2008. Dyskerin is a component of the Arabidopsis telomerase RNP required for telomere maintenance. Mol. Cell. Biol. 28: 2332–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes J. J. B., Bentsink L., Alonso-Blanco C., Hanhart C. J., Vries H. B. D., et al. , 2007. Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175: 891–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A., Farlow A., 2013. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchar M., Fajkus J., 2004. Interactions of putative telomere-binding proteins in Arabidopsis thaliana: identification of functional TRF2 homolog in plants. FEBS Lett. 578: 311–315. [DOI] [PubMed] [Google Scholar]
- Kwon C., Chung I. K., 2004. Interaction of an Arabidopsis RNA-binding protein with plant single-stranded telomeric DNA modulates telomerase activity. J. Biol. Chem. 279: 12812–12818. [DOI] [PubMed] [Google Scholar]
- Leehy K. A., Lee J. R., Song X., Renfrew K. B., Shippen D. E., 2013. MERISTEM DISORGANIZATION1 encodes TEN1, an essential telomere protein that modulates telomerase processivity in Arabidopsis. Plant Cell 25: 1343–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D., Neuhausen S. L., Hunt S. C., Kimura M., Hwang S. J., et al. , 2010. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc. Natl. Acad. Sci. USA 107: 9293–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Lustig A. J., 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 10: 1310–1326. [DOI] [PubMed] [Google Scholar]
- Liti G., Louis E. J., 2012. Advances in quantitative trait analysis in yeast. PLoS Genet. 8: e1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Haricharan S., Cubillos F. A., Tierney A. L., Sharp S., et al. , 2009. Segregating YKU80 and TLC1 alleles underlying natural variation in telomere properties in wild yeast. PLoS Genet. 5: e1000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q., Rabanal F. A., Meng D., Huber C. D., Farlow A., et al. , 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45: 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet G., White C. I., Gallego M. E., 2006. Telomere-length regulation in inter-ecotype crosses of Arabidopsis. Plant Mol. Biol. 62: 859–866. [DOI] [PubMed] [Google Scholar]
- Mangino M., Brouilette S., Braund P., Tirmizi N., Vasa-Nicotera M., et al. , 2008. A regulatory SNP of the BICD1 gene contributes to telomere length variation in humans. Hum. Mol. Genet. 17: 2518–2523. [DOI] [PubMed] [Google Scholar]
- Mangino M., Richards J. B., Soranzo N., Zhai G., Aviv A., et al. , 2009. A genome-wide association study identifies a novel locus on chromosome 18q12.2 influencing white cell telomere length. J. Med. Genet. 46: 451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangino M., Hwang S. J., Spector T. D., Hunt S. C., Kimura M., et al. , 2012. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet. 21: 5385–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Brevet V., Gilson E., 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18: 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najdekrova L., Siroky J., 2012. NBS1 plays a synergistic role with telomerase in the maintenance of telomeres in Arabidopsis thaliana. BMC Plant Biol. 12: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov A. M., 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41: 181–190. [DOI] [PubMed] [Google Scholar]
- R Core Team 2013 R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: http://www.R-project.org/. [Google Scholar]
- Ravi M., Chan S. W., 2010. Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618. [DOI] [PubMed] [Google Scholar]
- Ren S., Johnston J. S., Shippen D. E., McKnight T. D., 2004. TELOMERASE ACTIVATOR1 induces telomerase activity and potentiates responses to auxin in Arabidopsis. Plant Cell 16: 2910–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K., Shippen D. E., 2003. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K., McKnight T. D., Griffing L. R., Shippen D. E., 2001. Living with genome instability: plant responses to telomere dysfunction. Science 291: 1797–1800. [DOI] [PubMed] [Google Scholar]
- Riha K., Watson J. M., Parkey J., Shippen D. E., 2002. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21: 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P., Collier S., Bush M., Shaw P., Doonan J. H., 2007. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 120: 3678–3687. [DOI] [PubMed] [Google Scholar]
- Seymour D. K., Filiault D. L., Henry I. M., Monson-Miller J., Ravi M., et al. , 2012. Rapid creation of Arabidopsis doubled haploid lines for quantitative trait locus mapping. Proc. Natl. Acad. Sci. USA 109: 4227–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov E. V., Shippen D. E., 2004. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16: 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov E. V., Surovtseva Y. V., Osbun N., Shippen D. E., 2005. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 25: 7725–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Leehy K., Warrington R. T., Lamb J. C., Surovtseva Y. V., et al. , 2008. STN1 protects chromosome ends in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 19815–19820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y. V., Shakirov E. V., Vespa L., Osbun N., Song X., et al. , 2007. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 26: 3653–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y. V., Churikov D., Boltz K. A., Song X., Lamb J. C., et al. , 2009. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashi Y., Kobayashi Y., Tanaka K., Tamura K., 2009. Arabidopsis replication protein A 70a is required for DNA damage response and telomere length homeostasis. Plant Cell Physiol. 50: 1965–1976. [DOI] [PubMed] [Google Scholar]
- Vannier J. B., Pavicic-Kaltenbrunner V., Petalcorin M. I., Ding H., Boulton S. J., 2012. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806. [DOI] [PubMed] [Google Scholar]
- Vasa-Nicotera M., Brouilette S., Mangino M., Thompson J. R., Braund P., et al. , 2005. Mapping of a major locus that determines telomere length in humans. Am. J. Hum. Genet. 76: 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn, L. M., and P. H. Masson, 2011 A QTL study for regions contributing to Arabidopsis thaliana root skewing on tilted surfaces. G3 (Bethesda) 1: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., C. J. Basten, Z. B. Zeng, 2010 Windows QTL Cartographer 2.5 Department of Statistics, North Carolina State University, Raleigh, NC.
- Watson J. D., 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239: 197–201. [DOI] [PubMed] [Google Scholar]
- Watson J. M., Shippen D. E., 2007. Telomere rapid deletion regulates telomere length in Arabidopsis thaliana. Mol. Cell. Biol. 27: 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. H., Kwon C., Lee M. M., Chung I. K., 2007. Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 49: 442–451. [DOI] [PubMed] [Google Scholar]
- Zhu L. X., Hathcock K. S., Hande P., Lansdorp P. M., Seldin M. F., et al. , 1998. Telomere length regulation in mice is linked to a novel chromosome locus. Proc. Natl. Acad. Sci. USA 95: 8648–8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Voruganti V. S., Lin J., Matsuguchi T., Blackburn E., et al. , 2013. QTL mapping of leukocyte telomere length in American Indians: the Strong Heart Family Study. Aging (Albany, N.Y. Online) 5: 704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]