Studies by cancer genome consortiums have identified frequent mutations in chromatin regulatory factors and histone proteins in human cancer, implicating them as major mediators in the pathogenesis of both hematological malignancies and solid tumors. Here, Morgan and Shilatifard review recent advances in our understanding of the role of chromatin in cancer, focusing on transcriptional regulatory complexes, enhancer-associated factors, histone point mutations, and alterations in heterochromatin-interacting factors.
Keywords: cancer, chromatin, histone proteins
Abstract
Changes in the pattern of gene expression play an important role in allowing cancer cells to acquire their hallmark characteristics, while genomic instability enables cells to acquire genetic alterations that promote oncogenesis. Chromatin plays central roles in both transcriptional regulation and the maintenance of genomic stability. Studies by cancer genome consortiums have identified frequent mutations in genes encoding chromatin regulatory factors and histone proteins in human cancer, implicating them as major mediators in the pathogenesis of both hematological malignancies and solid tumors. Here, we review recent advances in our understanding of the role of chromatin in cancer, focusing on transcriptional regulatory complexes, enhancer-associated factors, histone point mutations, and alterations in heterochromatin-interacting factors.
Fifteen years ago, in their paper “The Hallmarks of Cancer,” Hanahan and Weinberg (2000) laid out a conceptual framework for the properties of cancer cells. Cancer development is a complex process involving diverse tissue types of distinct developmental origins, cell–cell interactions, and a myriad of signaling pathways. Digesting decades worth of research, Hanahan and Weinberg (2000) extracted fundamental properties common to many cancer types. Some aspects of their six hallmarks of cancer (resisting apoptosis, self-sufficiency in growth signals, insensitivity to anti-growth signals, invasive metastasis, unlimited cellular proliferation, and sustained angiogenesis) can be viewed in the light of deregulated gene expression at the level of transcription. Indeed, many signal transduction pathways perturbed in cancer ultimately modulate the activity of transcriptional regulators (Hanahan and Weinberg 2000, 2011). Moreover, genomic instability is recognized as an enabling characteristic of cancer (Hanahan and Weinberg 2011). Thus, transcriptional control and structural maintenance of the genome at the level of chromatin are likely suspects in the hunt for culprits underlying cancer development.
The eukaryotic genome is packaged into a structure called chromatin that, at its most basic level, comprises the four core histones H2A, H2B, H3, and H4 wrapped inside ∼147 base pairs (bp) of DNA to form the nucleosome core particle (Kornberg 1974; Kornberg and Thomas 1974; Luger et al. 1997). Additionally, histone H1 functions as an internucleosome linker and is involved in the compaction of chromatin (Laybourn and Kadonaga 1991). The N-terminal tails of the core histones protrude out from the nucleosome and are subject to a diverse array of post-translational modifications that alter chromatin structure and dynamics (Campos and Reinberg 2009). Large families of proteins containing domains such as bromodomain, chromodomain, plant homeodomain (PHD) finger, Tudor domains, PWWP domains, and YEATS domains bind these modifications to effect diverse downstream chromatin-based processes (Taverna et al. 2007). Considering the importance of chromatin in regulating eukaryotic gene expression and maintaining genome stability, it is perhaps not wholly unexpected that recent genome-wide sequencing studies have uncovered cancer-associated mutations in genes encoding chromatin regulatory factors and enzymes (Fig. 1).
Figure 1.
Chromatin proteins mutated in cancer. A summary of cancer mutations that affect post-translational modifications of the histone H3 N-terminal tail. Protein classes are indicated by the fill color for the ovals ([red] methyltransferase; [green] demethylase; [orange] deacetylase; [blue] histone), whereas mutational status is indicated by the outline color ([gray] loss of function; [purple] overexpressed/hyperactive). Dashed lines indicate the residue of histone H3 that is expected to be modified due to the indicated cancer mutations.
The emerging picture of chromatin function in cancer is multifaceted and involves a complex interplay of chromatin-modifying enzymes. A recent review from Van Rechem and Whetstine (2014) highlights the diverse mutations in genes involved in histone lysine methylation pathways associated with human cancer. In some instances, alterations in chromatin itself, such as histone H3.3 Lys27-to-methionine mutations in pediatric glioma, are highly context-specific to a single cancer type (Schwartzentruber et al. 2012; Wu et al. 2012). In other cases, mutations of related pathway component genes such as MLL3, MLL4, and UTX within the COMPASS (complex of proteins associated with Set1) family occur in a range of cancers, suggesting a broader tumor suppressor role (Gui et al. 2011; Morin et al. 2011; Parsons et al. 2011; Pasqualucci et al. 2011; Jones et al. 2012; Lohr et al. 2012; Pugh et al. 2012; Herz et al. 2014a). As more cancer genomes are sequenced, perhaps one of the most stirring observations is co-occurrence as well as mutual exclusivity of mutations between related cancer types. These mutational signatures promise insight into not only cancer development but also the molecular signaling pathways underlying normal development. Paradoxically, whereas basic developmental biology research has supplied us with a rich understanding of the signal transduction pathways involved in cancer, the recent intense focus on cancer genomics may provide a better understanding of the interplay between cell signaling and chromatin during normal tissue development.
Mutations of Trithorax (Trx)/COMPASS and Polycomb-repressive complex 2 (PRC2) in cancer
The Trx and PCRC2 complexes were identified as factors controlling the developmentally regulated expression of the homeotic gene (Hox) clusters in Drosophila melanogaster (Ingham 1983; Jones and Gelbart 1990). Trx is essential for maintaining Hox gene activation, whereas PRC2 acts as a transcriptional repressor to prevent ectopic Hox expression. Genetically, these two complexes act in opposition to each other, suggesting that they converge on a common pathway (Ingham 1983; Hanson et al. 1999). Although the importance of Trx and PRC2 in developmental gene regulation has been established for some time (Ingham 1983; Jones and Gelbart 1990), the biochemical activity of these proteins remained elusive until ∼12 years ago. The first clues to the function of these complexes stemmed from the presence of a SET histone methyltransferase domain protein in both the Trx and PRC2 complexes. Studies in yeast revealed that Trx is a member of the COMPASS family of protein complexes that catalyzes methylation of histone H3 Lys4 (H3K4) (Miller et al. 2001; Krogan et al. 2002; Shilatifard 2012). Biochemical experiments and Drosophila genetics demonstrated that the Enhancer of Zeste [E(Z)] subunit of PRC2 is a histone methyltransferase specific for H3K27 (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). Consistent with Trx’s and PRC2’s respective roles as activators and repressors of transcription, histone H3K4 trimethylation (H3K4me3) is associated with active promoters, whereas histone H3K27me3 is associated with transcriptional silencing (Margueron and Reinberg 2011; Shilatifard 2012).
The first link between Trx function and cancer was made when it was observed that childhood mixed-lineage leukemias (MLLs) contain a translocation occurring at chromosome 11q23 involving the MLL1 gene, one of the two mammalian Trx homologs (Ziemin-van der Poel et al. 1991; Rowley 1993; Mohan et al. 2010, 2011). These translocations remove the C-terminal portion of MLL1, containing its catalytic SET histone methyltransferase domain, and create an in-frame fusion to generate gain-of-function chimeric proteins (Mohan et al. 2010, 2011). Recent work has elucidated the molecular mechanism underlying the oncogenic activity of these MLL1 fusions. A number of the most common MLL1 gene translocation partners, including AF4, AF9, ENL, and ELL, are components of the macromolecular complex called the super elongation complex (SEC) (Lin et al. 2010; Luo et al. 2012). SEC associates with positive transcriptional elongation factor B (PTEF-b), a cyclin-dependent kinase (CDK) that promotes RNA polymerase II elongation by phosphorylating its C-terminal domain and other basal factors within the preinitiation complexes (Smith et al. 2011). Thus, MLL1-SEC fusion proteins cause aberrant activation of MLL1 targets through misregulation of transcription elongation. MLL1 is required for normal hematopoietic stem cell function (Hess et al. 1997; Ernst et al. 2004), and MLL1 fusions likely result in altered stem cell properties that promote tumor formation.
Whereas MLL1 gene mutations involve a characteristic chromosomal translocation in a specific tumor type, PRC2 appears to a have a more complex role in cancer. Frequent point mutations of the EZH2 gene are observed in non-Hodgkin lymphoma (follicular and diffuse large B-cell lymphoma) (Morin et al. 2010). These affect the EZH2 catalytic site and convert Tyr641 (Y641) to a variety of other amino acids, with asparagine being the most common substitution. In vitro, these mutants are unable to methylate an unmodified histone peptide (Morin et al. 2010). However, subsequent studies revealed that these mutations are not inactive but rather possess an altered activity. Remarkably, EZH2 Y641 mutants show increased activity toward the di- and trimethylated states (Sneeringer et al. 2010). Thus, tumor cells with Y641 mutations in the EZH2 gene contain increased H3K27me3. This finding is intriguing because H3K27 monomethylation (H3K27me1), H3K27 dimethylation (H3K27me2), and H3K27me3 were recently shown to have distinct enrichment patterns across the genome, with H3K27me2 being implicated in the suppression of enhancer function (Ferrari et al. 2014). In addition to Y641, the A677G EZH2 mutant exhibits a similar increase in H3K27me3 accompanied by a decrease in H3K27me2 (McCabe et al. 2012a). In contrast, a recently characterized A687V mutant displayed both increased H3K27me3 and H3K27me2 (Ott et al. 2014). Remarkably, a Drosophila mutation of E(Z) that mimics the Trx loss-of-function phenotype has also been shown to possess hyperactive methyltransferase activity (Bajusz et al. 2001; Stepanik and Harte 2012). The E(Z) Trx mimic mutation [E(z)(Trm)] converts Arg741 (Arg727 in human EZH2) to lysine (R741K), suggesting that this position may also be important for regulating PRC2 catalytic activity (Bajusz et al. 2001; Stepanik and Harte 2012). The activating nature of these mutations makes PRC2 an attractive target for therapeutic intervention. Recently, a small molecule inhibitor of EZH2, GSK126, was shown to specifically inhibit the growth of B-cell lymphomas containing activating EZH2 mutations, whereas tumor lines with wild-type EZH2 were largely unaffected (McCabe et al. 2012b).
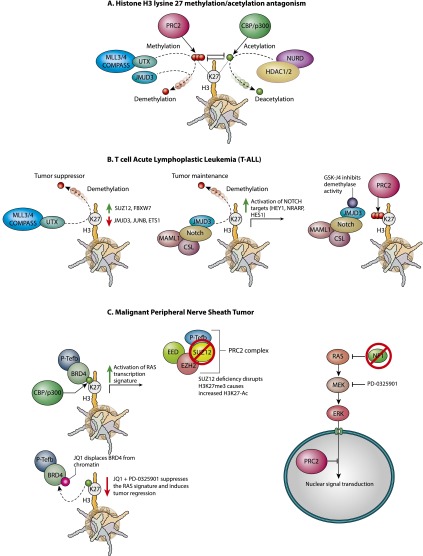
While EZH2 activating mutations are common in non-Hodgkin lymphoma, loss of PRC2 activity is associated with cancer development in other contexts. Inactivating mutations of the PRC2 components EZH2, SUZ12, and EED are detected in T-cell acute lymphoblastic leukemia (T-ALL) (Fig. 2; Ntziachristos et al. 2012; Simon et al. 2012; Zhang et al. 2012). Removal of the H3K27 methyl mark is catalyzed by the Jumonji domain containing demethylases UTX/KDM6A and JMJD3/KDM6B (Kooistra and Helin 2012). A recent study explored whether disruption of UTX and JMJD3 activity might provide a therapeutic benefit for T-ALL by increasing H3K27me3 levels (Ntziachristos et al. 2014). Surprisingly, UTX and JMJD3 have strikingly distinct roles in T-ALL. UTX acts as a tumor suppressor, as mice with a NOTCH1-driven model of T-ALL succumb to disease more rapidly on a UTX mutant genetic background (Ntziachristos et al. 2014). In contrast, JMJD3 is highly expressed in T-ALL versus normal T cells and is required for leukemogenesis, as mice with JMJD3 mutant T-ALL show improved survival rates. GSK-J4, an inhibitor of KDM6-type demethylases (Kruidenier et al. 2012), causes cell cycle arrest and apoptosis in T-ALL cells but not myeloid leukemia or normal hematopoietic progenitors (Ntziachristos et al. 2014). Remarkably, GSK-J4 treatment results in gene expression changes that resemble knockdown of JMJD3 but are inversely correlated with the changes observed for UTX knockdown. Chromatin immunoprecipitation (ChIP) combined with sequencing (ChIP-seq) revealed a significant overlap between JMJD3 and NOTCH1 targets, including genes with known oncogenic function such as HEY1, NRARP, and HES1. Strikingly, these genes gain H3K27me3 and are repressed upon JMJD3 depletion or GSK-J4 treatment. It is unclear why GSK-J4 appears to inhibit JMJD3 but not UTX functional activity in T-ALL, but perhaps the molecule has a higher affinity for JMJD3 in vivo. Recent studies have also suggested that GSK-J4 may also target KDM5-type demethylases but with an affinity five to 10 times lower than JMJD3 and UTX (Heinemann et al. 2014). Despite these caveats, GSK-J4 appears to be a promising drug for modulating chromatin modifications and perhaps a chemotherapeutic agent.
Figure 2.
Drugging the histone H3K27 methyl/acetyl switch in cancer. (A) Antagonism between H3K27 methylation and acetylation machinery. H3K27 methylation and acetylation are mutually exclusive, and the PRC2 and CBP/p300 complexes act in opposition to one another. In addition, deacetylation of H3K27ac by the HDAC1/2–NURD complex promotes PRC2-mediated repression, whereas demethylation of H3K27me3 by UTX within COMPASS or JMJD3 is required for acetylation to occur. (B) In NOTCH-driven T-ALL, the histone H3K27 demethylases UTX and JMJD3 have distinct functions. UTX acts as a tumor suppressor by activating genes such as FBXW7 that negatively regulate the NOTCH pathway. In contrast, JMJD3 exists in a complex with NOTCH and is responsible for activation of oncogenic NOTCH targets. Inhibition of JMJD3 with the small molecule GSK-J4 promotes PRC2-mediated H3K27me3 at NOTCH target genes, resulting in their silencing. (C) MPNSTs often carry mutations in the genes encoding the components of both the RAS pathway inhibitor NF1 and the PRC2 component SUZ12. In this cell type, PRC2 functions to suppress RAS target genes. Reduced H3K27 methylation by PRC2 results in increased H3K27ac, increased recruitment of BRD4, and amplification of the RAS transcriptional signature. Inhibition of BRD4 with JQ1 in combination with dampening of the RAS pathway with the MEK inhibitor PD-0325901 suppresses RAS targets, resulting in tumor regression.
Loss of PRC2 components EED and SUZ12 is often detected in combination with mutation of NF1 and CDKN2A genes in malignant peripheral nerve sheath tumors (MPNSTs) (Fig. 2; Lee et al. 2014; Zhang et al. 2014). In addition to loss of H3K27me3, PRC2 mutant MPNSTs also display increased H3K27 acetylation (H3K27ac) levels, an effect observed for loss of PRC2 in multiple contexts (Tie et al. 2009; Pasini et al. 2010; Herz et al. 2014b). Histone H3K27 methylation and acetylation are mutually exclusive modifications that correlate with gene silencing and activation, respectively. Strong evidence suggests that complexes responsible for implementing these modifications act in opposition to one another (Tie et al. 2009; Pasini et al. 2010; Reynolds et al. 2012; Herz et al. 2014b). A recent study examined whether these increased acetylation levels in MPNSTs could serve as a therapeutic target. Bromodomain-containing protein 4 (BRD4) is a member of the bromodomain and extraterminal (BET) family of chromatin-associated proteins that bind to acetylated histone H3 and H4 via tandem bromodomains (Belkina and Denis 2012; Shi and Vakoc 2014). The small molecule JQ1 binds to the BRD4 bromodomains and evicts the protein from chromatin (Filippakopoulos et al. 2010). JQ1 has shown promise as a potential chemotherapeutic agent in a number of cancers in part due to regulation of the c-Myc oncogene by BRD4 (Filippakopoulos et al. 2010; Delmore et al. 2011; Chapuy et al. 2013; Knoechel et al. 2014). BRD4 localizes to enhancers containing H3K27ac, and the effects of JQ1 may involve disruption of enhancer activity (Chapuy et al. 2013; Loven et al. 2013). As PRC2 mutant MPNSTs display increased H3K27ac levels, BRD4 is an attractive target in this context. Indeed, loss of SUZ12 and NF1 (a negative regulator of the oncogenic RAS pathway) in MPNSTs renders them sensitive to treatment with JQ1 in combination with inhibition of the RAS pathway by the MEK inhibitor PD-0325901 (De Raedt et al. 2014). Interestingly, the RAS pathway and PRC2 appear to synergize in MPNSTs, as SUZ12 loss promotes cell proliferation in NF1 mutant cells but not wild-type cells, and, moreover, SUZ12 loss enhances the RAS transcriptional signature (De Raedt et al. 2014). MEK inhibition has been shown to inhibit PRC2 activity in embryonic stem cells, suggesting a potential negative feedback loop (Marks et al. 2012; Tee et al. 2014). It will be important to determine the molecular details of the connection between RAS/MEK signaling and PRC2 activity and examine whether this link is conserved in multiple types of cancer.
PRC2 gene mutations in cancer highlight both the biochemical complexity of chromatin-modifying pathways and the rich potential for therapeutic intervention. In PRC2 loss-of-function cancer models, inhibition of BRD4, which binds to increased acetylated histones in PRC2 mutant cells, as well as inhibition of H3K27me3 demethylases show therapeutic effects. In contrast, drugs inhibiting EZH2 activity are more appropriate for non-Hodgkin lymphomas carrying hyperactive EZH2 mutations. Thus, the search for therapeutic targets should take into consideration cross-regulation between histone modification pathways (i.e., methylation and acetylation) as well as effectors of those pathways, such as bromodomain and chromodomain proteins that bind to modified histones. These studies also highlight the importance of determining the precise mutational status of individuals to determine what pathways should be targeted for treatment.
Misregulation of enhancer chromatin in cancer
Enhancers are noncoding DNA elements that play an essential role in transcriptional regulation by conferring tissue-specific gene expression patterns (Smith and Shilatifard 2014). Although enhancers have been intensely studied for several decades, their precise mode of action is not fully understood. Enhancers can act across very long ranges of intervening DNA to activate a specific promoter (Blackwood and Kadonaga 1998; Bulger and Groudine 2011). Enhancer–promoter communication involves the formation of chromatin loops mediated by cohesin complexes and other trans-acting factors (Cuylen and Haering 2010). However, the mechanisms that restrict enhancer activity to a single promoter in the presence of multiple promoter choices are unclear.
Enhancers carry a unique chromatin structure characterized by the presence of 3K4me1 (Heintzman et al. 2009). In addition, histone H3K27ac distinguishes active enhancers from poised enhancers (Creyghton et al. 2010). Promoters typically contain H3K4me3 implemented by the Set1A/B and MLL1/2 COMPASS-like complexes, whereas MLL3/4 COMPASS catalyzes H3K4me1 at enhancers (Herz et al. 2012; Smith and Shilatifard 2014). Acetylation of H3K27 is mediated by the acetyltransferases CREBBP (CBP) and EP300 (p300) (Pasini et al. 2010). Besides H3K4me1 methylase activity, MLL3/4 complexes also contain the H3K27 demethylase KDM6A (UTX), raising the possibility that removal of H3K27 methyl marks by the MLL3/4 complex facilitates acetylation by CBP and p300 (Cho et al. 2007; Issaeva et al. 2007; Herz et al. 2012; Hu et al. 2013).
Recent genome-wide studies have identified mutations in genes for the regulators of enhancer chromatin in cancer (Herz et al. 2014a). Mutations of the H3K4 monomethylases MLL3 and MLL4 as well as their cofactor, UTX, within the COMPASS family have been identified in a range of malignancies, including the pediatric brain cancer medulloblastoma (Parsons et al. 2011; Jones et al. 2012; Pugh et al. 2012), non-Hodgkin lymphoma (Morin et al. 2011; Pasqualucci et al. 2011; Lohr et al. 2012), and bladder cancer (Gui et al. 2011). MLL4 is particularly frequently mutated in non-Hodgkin lymphomas and often co-occurs with mutations in the histone acetyltransferase gene CREBBP and activating mutations of EZH2 (Morin et al. 2011; Okosun et al. 2014). Mutations of EP300 and CREBBP have been found to co-occur with UTX in bladder cancer (Gui et al. 2011). MEF2B, a transcription factor involved in recruiting CREBBP and EP300 to target sites in chromatin, is frequently mutated in non-Hodgkin lymphoma (Morin et al. 2011). Intriguingly, the majority of these mutations result in single amino acid changes at one of four positions (K4, Y69, N81, and D83) (Morin et al. 2011), and a subset of these mutations results in increased MEF2B transcriptional activation activity by loss of binding to the corepressor CABIN1 (Ying et al. 2013) Another enhancer-associated factor, LIM domain-binding protein 1 (LDB1), is mutated in medulloblastoma (Pugh et al. 2012). LDB1 is involved in the formation of chromatin loops in both Drosophila and mammalian cells and participates in enhancer–promoter communication (Morcillo et al. 1997; Deng et al. 2012; Krivega et al. 2014).
A large body of evidence implicates enhancer malfunction in cancer, and much remains to be learned about the molecular mechanisms of this process (Herz et al. 2014a). For instance, how does mutation in genes for factors such as EP300 and CREBBP that are thought to function globally at most enhancers play a role in cancer development? Inappropriate enhancer–promoter communication is known to play a role in the pathogenesis of some cancers. For instance, the classical chromosomal translocation found in Burkitt’s lymphoma places the c-Myc gene under the regulation of the immunoglobulin heavy chain enhancer, thus boosting its expression in B cells, resulting in lymphomagenesis. Recent studies of acute myeloid leukemia with a chromosomal translocation near the GATA2 and EVI1 genes revealed that this inversion allows a GATA2 enhancer to inappropriately activate EVI1 expression (Groschel et al. 2014; Yamazaki et al. 2014). This raises the possibility that mutation in genes for enhancer-associated factors may lead to defective enhancer–promoter restriction, perhaps allowing for promiscuous activation of oncogenic gene products (Herz et al. 2014a).
Histone gene mutations in cancer
Mutations and translocations in genes encoding chromatin regulatory proteins such as the MLL family within COMPASS have been linked with oncogenesis for many years (Shilatifard 2012); however, cancer-associated mutations of histone genes themselves were only recently identified. Genome sequencing studies of aggressive pediatric brainstem glioma uncovered point mutations in histone H3 (Schwartzentruber et al. 2012; Wu et al. 2012). These mutations convert Lys27 to methionine (H3K27M) or Gly34 to arginine or valine (H3G34R/V), occurring primarily in the replication-independent histone H3.3 (H3F3A) and, to lesser extent, the replication-dependent histone H3.1 (HIST1H3B) (Schwartzentruber et al. 2012; Wu et al. 2012). Strikingly, these mutations occur in only a single copy of the multiple histone H3 genes, suggesting that they have gain-of-function activity. Mutations of H3K27M and H3G34R/V define distinct subtypes of glioma, as they occur in distinct regions of the brain and display unique molecular characteristics (Sturm et al. 2012).
Histological examination of tumors harboring the H3K27M mutation revealed a dramatic reduction in the levels of H3K27me3 (Venneti et al. 2013). Further molecular studies revealed that H3K27M as well as other histone lysine-to-methionine mutants act as dominant inhibitors of histone lysine methylation pathways in tissue culture (Chan et al. 2013; Lewis et al. 2013). Histone H3K27M expression in Drosophila recapitulates the phenotype observed for depletion of the PRC2 component E(Z) and mirrors the phenotype of replacing all histone H3s with a H3K27R mutant (Pengelly et al. 2013; Herz et al. 2014b). In contrast, the H3K34R/V mutations do not dominantly inhibit bulk H3K27me3 or H3K36me3 in trans but do block methylation of H3K36 in cis (Lewis et al. 2013).
The precise mechanism of H3K27M action is still unclear. In vitro methyltransferase assays and immunoprecipitation followed by Western blotting suggest that H3K27M interacts strongly with EZH2 (Chan et al. 2013; Lewis et al. 2013). However, using an unbiased proteomic approach, we failed to detect increased enrichment of PRC2 subunits relative to wild-type H3.3 control (Herz et al. 2014b). In contrast, we found increased association of the bromodomain protein BRD4, which is consistent with increased histone acetylation levels observed in H3K27M mutant cells (Herz et al. 2014b). It is also intriguing that mutations in genes encoding for the PRC2 components do not appear to be prevalent in these pediatric gliomas.
Recent studies of chondroblastoma and giant cell tumors of bone revealed additional histone H3.3 gene mutations associated with distinct disease phenotypes (Behjati et al. 2013). Remarkably, 95% of chondroblastoma samples analyzed carried a mutation of the H3.3 gene at Lys36 to methionine (H3.3K36M) in the H3F3B gene, whereas 92% of giant cell tumors of bone harbored mutations of H3.3 Gly34 to tryptophan or leucine (Behjati et al. 2013). Similar to H3.3K27M, H3.3K36M can dominantly inhibit methylation of H3K36 (Lewis et al. 2013).
Histone gene mutations in cancer are not restricted to histone H3. Recent work in follicular lymphoma identified mutations in a number of histone H1 genes (Lohr et al. 2012; Morin et al. 2013; Li et al. 2014; Okosun et al. 2014). Whereas histone H2A, H2B, H3, and H4 constitute the nucleosome core, histone H1 acts as a linker histone and is involved in chromatin compaction. Like H3 gene mutations, H1 gene mutations are primarily single amino acid substitutions; however, instead of occurring at a few specific positions, the H1 gene mutations are scattered throughout the H1 globular domain (Lohr et al. 2012; Morin et al. 2013; Li et al. 2014; Okosun et al. 2014). Molecular analysis of one of these mutants, H1S102F, revealed that it has a reduced capacity to associate with chromatin (Morin et al. 2013) and binding to DNA methyltransferase 3B (DNMT3B) (Li et al. 2014). This suggests that histone H1 may lead to defective chromatin compaction and cause transcriptional misregulation or result in genomic instability. It will be important to examine the molecular function of histone H1 gene mutations in B-cell lymphoma in more detail.
Analysis of mutations that co-occur or are mutually exclusive to histone H3 gene mutations have been insightful. For instance, in pediatric glioblastoma, mutations of histone H3K27M, H3G34R/V, and isocitrate dehydrogenase 1 (IDH1) are mutually exclusive and occur in tumors with different molecular signatures, neuroanatomic locations, and prognostic outcome (Sturm et al. 2012). H3K27M mutants lose H3K27 methylation, whereas H3G34V/R mutants display DNA CpG hypomethylation, and IDH1 mutants have a CpG hypermethylation phenotype (Sturm et al. 2012; Turcan et al. 2012). Mutations of the IDH1 gene are particularly prevalent in glioma but are also detected in leukemias (Parsons et al. 2008; Mardis et al. 2009). IDH1 alterations occur in the substrate-binding site at position Arg132, and most mutations convert this residue to histidine (IDH1 R132H), although other substitutions have also been detected (Parsons et al. 2008; Mardis et al. 2009). Under normal circumstances, IDH1 converts isocitrate to α-ketoglutarate and nicotinamide adenine dinucleotide phosphate (NADP+) to NADPH. However, the mutant IDH1 R132H enzyme generates the 2-hydroxyglutarate in place of α-ketoglutarate (Dang et al. 2009). This metabolite inhibits α-ketoglutarate-dependent enzymes (including Jumonji-containing histone demethylases) as well as TET family methylcytosine dioxygenases thought to be involved in the process of DNA demethylation by converting 5-methylcytosine to 5-hydroxy-methylcytosine (Figueroa et al. 2010; Chowdhury et al. 2011; Xu et al. 2011; Lu et al. 2012). Thus, cells with the mutant IDH1 gene display CpG hypermethylation as well as increased histone lysine methylation. Interestingly, histone H3G34R/V gene mutations show an opposite effect on CpG methylation (Sturm et al. 2012). It is also notable that H3G34R/V gene mutations tend to co-occur with mutations of the histone H3.3 chaperone genes ATRX and DAXX, suggesting that altered histone incorporation into chromatin may play a role in these cancers (Schwartzentruber et al. 2012).
Recently, mutations in the gene for BMP receptor ACVR1/ALK2 were detected in pediatric glioma with H3K27M mutations (Buczkowicz et al. 2014; Fontebasso et al. 2014; Taylor et al. 2014; Wu et al. 2014). These point mutations convert ACVR1 into a constitutively active form, and several cancer-associated mutations are identical to those found in the rare but devastating bone formation disorder fibrodysplasia ossificans progressiva (FOP) (Shore et al. 2006). Interestingly, ACVR1 mutations tend to overlap with H3.1K27M mutations. Mutations of H3.3K27M are more prevalent than H3.1K27M in pediatric gliomas (Schwartzentruber et al. 2012; Wu et al. 2012), and these mutational types have distinct properties, as patients with H3.1K27M show an early disease onset with tumors located in the pons, whereas H3.3K27M tumors are located at multiple brain regions along the midline (Buczkowicz et al. 2014; Fontebasso et al. 2014; Taylor et al. 2014; Wu et al. 2014). Determining the biological significance of these mutational signatures will be important to understanding pediatric glioma and may shed light onto other developmental disorders, such as FOP.
Maintenance of genome stability through heterochromatin
In eukaryotic cells, chromosomal structures such as pericentromeric regions and telomeres are associated with blocks of condensed heterochromatin (Maison and Almouzni 2004). Heterochromatin is characterized by histone hypoacetylation and methylation of histone H3 at Lys9, which serves as a binding substrate for the chromodomain protein heterochromatin protein-1 (HP-1) (Fig. 3; Maison and Almouzni 2004). These features are essential for normal chromosome function and establish a transcriptionally repressed state (James and Elgin 1986; Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001; O’Sullivan and Karlseder 2010). Maintenance of heterochromatic silencing is dependent on both histone H3K9 methyltransferases and HP-1 proteins (Fig. 3; Eissenberg et al. 1992; Bannister et al. 2001; Nakayama et al. 2001). Moreover, heterochromatin is epigenetically stable through a self-reinforcing circuit by which HP-1 associates with the DNA replication machinery and recruits H3K9 histone methyltransferase complexes (Aagaard et al. 1999; Murzina et al. 1999; Maison and Almouzni 2004; Quivy et al. 2008).
Figure 3.
Maintenance of genome stability through the heterochromatin pathway. Centromeric heterochromain is essential for normal segregation of chromosomes during mitosis, and defects in this pathway result in aneuploidy. H3K9me3 and binding of HP-1 are hallmarks of heterochromatin. At pericentromeric heterochromatin, SETDB1 monomethylates H3K9, whereas Suv39 converts H3K9me1 to H3K8me2/3. Disruption of Suv39 function results in aneuploidy and lymphoma development in mice. Active deacetylation is also important for centromeric heterochromatin. Treatment of cells with the class I and II histone deacetylase inhibitor TSA results in abnormal mitosis. Similarly, Suv39 and SETDB1 are essential for telomeric heterochromatin. At telomeres, disruption of Suv39 results in loss of H3K9me2/3 and a depletion of HP-1 recruitment. However, Suv39 mutant telomeres contain increased H3K9me1 mediated by SETDB1 and exhibit abnormal telomere lengthening. Overexpression of SETDB1 has been reported in some cancers. Whereas centromeres depend on type I and II HDACs, the sirtuin deacetylase SIRT6 is essential at telomeres. Lack of SIRT6 in mice results in telomere fusions and premature senescence. In other contexts, SIRT6 functions as a tumor suppressor.
Heterochromatin plays an essential role in genomic stability at multiple levels. Mice doubly mutant for the H3K9 methyltransferases Suv39h1 and Suv39h2 lack H3K9 methylation at pericentric heterochromatin, exhibit aneuploidy and male germline meiosis defects, and develop B-cell lymphomas (Peters et al. 2001). Mutations in the gene for the H3K9 methyl-binding protein HP1 also disrupt genomic stability through both aberrant centromere and telomere function. HP1 mutant flies display defective chromosome segregation as well as telomere fusions (Kellum and Alberts 1995; Fanti et al. 1998). Interestingly, cells mutant for Suv39h1 and Suv39h2 exhibit abnormally elongated telomeres (Garcia-Cao et al. 2004). These mutant telomeres have reduced H3K9me2/3 and loss of HP-1 binding but display increased H3K9me1 (Garcia-Cao et al. 2004). This is consistent with the function of SETDB1 as a H3K9 monomethyltransferase, whereas Suv39h1/2 act as H3K9 di- and trimethylases (Loyola et al. 2009). Intriguingly, studies suggest that amplification of SETDB1 may play a role in development of human cancer as well as in a zebrafish model of melanoma (Ceol et al. 2011; Rodriguez-Paredes et al. 2014). It remains to be examined whether these oncogenic effects may be mediated through abnormal telomere lengthening.
Maintenance of histone hypoacetylation is also important for heterochromatin function. Treatment of cells with the class I and II histone deacetylase inhibitor trichostatin A (TSA) results in loss of HP1 binding to pericentric regions and relocalization of these domains to the nuclear periphery (Taddei et al. 2001). TSA also causes abnormal mitotic structures consistent with a defect in centromere function (Taddei et al. 2001). SIRT6, a histone deacetylase specific for histone H3K9, is essential for maintenance of telomeric heterochromatin (Michishita et al. 2008). SIRT6 mutant mice display genomic instability and exhibit a premature aging phenotype (Mostoslavsky et al. 2006). Human cells depleted for SIRT6 display telomere hyperacetylation, chromosome end-to-end fusions, and premature senescence that can be rescued by overexpression of telomerase (Michishita et al. 2008). While Sirt6-null mice exhibit premature aging and early death, they do not develop spontaneous tumors (Mostoslavsky et al. 2006). However, immortalized Sirt6-null mouse embryonic fibroblasts (MEFs) are able to form tumors in immunocompromised mice even in the absence of transformation with an activated oncogene (Sebastian et al. 2012). Moreover, SIRT6 is frequently deleted in human cancer, and a conditional mutant mouse model revealed it to act as a tumor suppressor in an intestinal cancer model in vivo (Sebastian et al. 2012).
Recent work has linked heterochromatin function to the regulation of DNA replication. Methylation of histone H3K9 and K36 have been linked to DNA replication in fission yeast (Kim et al. 2008). The mammalian Jumonji domain protein KDM4A/JMJD2A is a histone lysine demethylase specific for methylated H3K9 and K36 (Klose et al. 2006; Whetstine et al. 2006). Studies in Caenorhabditis elegans and mammalian cells revealed that KDM4A overexpression positively regulates S-phase progression, whereas depletion slowed DNA replication and induced cell death (Black et al. 2010). Moreover these effects were dependent on HP1 levels, implying that KDM4A influences cell cycle in part by removing H3K9me3 and evicting HP1. A follow-up study revealed that KDM4A is amplified in human cancers, and overexpression in tissue culture results in focal copy number gains during DNA replication (Black et al. 2013). However, these copy number gains are transient and become resolved during entry into G2/M through an as-yet-undetermined mechanism (Black et al. 2013). These copy gains are suppressed by increasing the cellular concentration of the H3K9 methyltransferase Suv39h1 or HP1-γ. Interestingly, expression of the mutant histones H3.3K9M or H3.3K36M, which inhibit bulk methylation of H3K9 and H3K36, respectively, also results in copy number gains. Recent work has shown that H3.3K9M disrupts heterochromatic transcriptional silencing in D. melanogaster (Herz et al. 2014b). Whereas the function of H3K9 methylation in heterochromatin is well established, studies by Whetstine and colleagues (Black et al. 2010, 2013) implicate H3K36 methylation in a pathway involving heterochromatin machinery that controls mammalian DNA replication. In yeast, H3K36 methylation restricts nucleosome dynamics over transcribed regions and prevents “cryptic” transcription (Smolle et al. 2013). Perhaps a similar mechanism is involved in restricting access of DNA replication machinery.
Concluding remarks
The role of chromatin proteins in cancer is complex and highly context-specific. Relatively few chromatin modifiers seem capable of independently causing cancer development; they are typically mutated in combination with essential tumor suppressors and cell cycle regulators such as p53 and CDKN2A. Although some chromatin regulators, such as the MLL3/4-UTX of the COMPASS family, may play a broad tumor suppressor role in various cancers, many mutant chromatin proteins are highly tissue-specific. Moreover, in the case of PRC2, both hyperactivating and loss-of-function mutations are found in cancers of distinct origins. Whereas B-cell lymphomas tend to acquire hyperactivating mutations of the EZH2 gene in combination with loss of MLL4, other cancer types, such as T-ALL and MPNST, harbor inactivating mutations in genes encoding for PRC2 components EZH2, EED, and SUZ12. These differences likely reflect tissue-specific functions for PRC2. Thus, it is important to determine the precise molecular consequence of altered chromatin proteins, particularly in the case of point mutations that may cause either loss of function or gain of function. As the majority of cancer-associated mutations in chromatin protein-encoding genes have yet to be functionally characterized, biochemical analysis of these mutants may lead to exciting new avenues of research.
The rapid proliferation of next-generation genome sequencing promises to reveal not only novel mutations involved in cancer but also co-occurring and mutually exclusive mutations. These will likely connect developmental signaling pathways to their downstream chromatin effector proteins. In the instance of the NF1 mutant MPNST, PRC2 appears to dampen the RAS signaling pathway. Similarly, in T-ALL, PRC2 antagonizes the NOTCH1 signal transduction pathway, whereas the H3K27 demethylase JMJD3 directly associates with NOTCH1 to remove PRC2-deposited H3K27me3. Interestingly, recent studies have identified activating mutations in the gene for the BMP receptor ACVR1/ALK2 in combination with histone H3.1K27M gene mutations, suggesting a potential connection between BMP–SMAD1/5/8 signaling and PRC2-mediated repression. Thus, future collaborative efforts between clinicians, geneticists, biochemists, and developmental biologists may shed light onto both the mechanisms underlying cancer development and the connection between cell signaling pathways and the chromatin signatures of cancer.
Acknowledgments
We are grateful to the members of the Shilatifard laboratory for conversation and discussion during the writing of this review. Studies in A.S.’s laboratory are supported by grants R01CA150265, R01GM069905, and R01CA89455 to A.S.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.255182.114.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. . 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J 18: 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajusz I, Sipos L, Gyorgypal Z, Carrington EA, Jones RS, Gausz J, Gyurkovics H. 2001. The Trithorax-mimic allele of Enhancer of zeste renders active domains of target genes accessible to polycomb-group-dependent silencing in Drosophila melanogaster. Genetics 159: 1135–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. [DOI] [PubMed] [Google Scholar]
- Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, et al. . 2013. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 45: 1479–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkina AC, Denis GV. 2012. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer 12: 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Allen A, Van Rechem C, Forbes E, Longworth M, Tschop K, Rinehart C, Quiton J, Walsh R, Smallwood A, et al. . 2010. Conserved antagonism between JMJD2A/KDM4A and HP1γ during cell cycle progression. Mol Cell 40: 736–748. [DOI] [PubMed] [Google Scholar]
- Black JC, Manning AL, Van Rechem C, Kim J, Ladd B, Cho J, Pineda CM, Murphy N, Daniels DL, Montagna C, et al. . 2013. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell 154: 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT. 1998. Going the distance: a current view of enhancer action. Science 281: 60–63. [DOI] [PubMed] [Google Scholar]
- Buczkowicz P, Hoeman C, Rakopoulos P, Pajovic S, Letourneau L, Dzamba M, Morrison A, Lewis P, Bouffet E, Bartels U, et al. . 2014. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet 46: 451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. 2009. Histones: annotating chromatin. Annu Rev Genet 43: 559–599. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, et al. . 2011. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471: 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. . 2013. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. . 2013. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24: 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. . 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. . 2011. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 12: 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. . 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Haering CH. 2010. A new cohesive team to mediate DNA looping. Cell Stem Cell 7: 424–426. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. . 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. . 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149: 1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, Helin K, Hornick JL, Mautner V, Kehrer-Sawatzki H, et al. . 2014. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514: 247–251. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Morris GD, Reuter G, Hartnett T. 1992. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. 2004. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell 6: 437–443. [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell 2: 527–538. [DOI] [PubMed] [Google Scholar]
- Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stutzer A, Fischle W, Bonaldi T, Pasini D. 2014. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 53: 49–62. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. . 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. . 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, Bechet D, Faury D, De Jay N, Ramkissoon LA, et al. . 2014. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet 46: 462–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. 2004. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36: 94–99. [DOI] [PubMed] [Google Scholar]
- Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, van der Velden VH, Havermans M, Avellino R, van Lom K, et al. . 2014. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 157: 369–381. [DOI] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. . 2011. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 43: 875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2000. The hallmarks of cancer. Cell 100: 57–70. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hanson RD, Hess JL, Yu BD, Ernst P, van Lohuizen M, Berns A, van der Lugt NM, Shashikant CS, Ruddle FH, Seto M, et al. . 1999. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci 96: 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann B, Nielsen JM, Hudlebusch HR, Lees MJ, Larsen DV, Boesen T, Labelle M, Gerlach LO, Birk P, Helin K. 2014. Inhibition of demethylases by GSK-J1/J4. Nature 514: E1–E2. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. . 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. 2012. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev 26: 2604–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Hu D, Shilatifard A. 2014a. Enhancer malfunction in cancer. Mol Cell 53: 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Morgan M, Gao X, Jackson J, Rickels R, Swanson SK, Florens L, Washburn MP, Eissenberg JC, Shilatifard A. 2014b. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345: 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. 1997. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood 90: 1799–1806. [PubMed] [Google Scholar]
- Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. 2013. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol 33: 4745–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW. 1983. Differential expression of bithorax complex genes in the absence of the extra sex combs and trithorax genes. Nature 306: 591–593. [DOI] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. 2007. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 27: 1889–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Elgin SC. 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol 6: 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RS, Gelbart WM. 1990. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 126: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, et al. . 2012. Dissecting the genomic complexity underlying medulloblastoma. Nature 488: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Alberts BM. 1995. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci 108: 1419–1431. [DOI] [PubMed] [Google Scholar]
- Kim HS, Rhee DK, Jang YK. 2008. Methylations of histone H3 lysine 9 and lysine 36 are functionally linked to DNA replication checkpoint control in fission yeast. Biochem Biophys Res Commun 368: 419–425. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442: 312–316. [DOI] [PubMed] [Google Scholar]
- Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, Gillespie SM, Fernandez D, Ku M, Wang H, et al. . 2014. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet 46: 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. 2012. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13: 297–311. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184: 868–871. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Thomas JO. 1974. Chromatin structure; oligomers of the histones. Science 184: 865–868. [DOI] [PubMed] [Google Scholar]
- Krivega I, Dale RK, Dean A. 2014. Role of LDB1 in the transition from chromatin looping to transcription activation. Genes Dev 28: 1278–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. 2002. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277: 10753–10755. [DOI] [PubMed] [Google Scholar]
- Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, et al. . 2012. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488: 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120. [DOI] [PubMed] [Google Scholar]
- Laybourn PJ, Kadonaga JT. 1991. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science 254: 238–245. [DOI] [PubMed] [Google Scholar]
- Lee W, Teckie S, Wiesner T, Ran L, Prieto Granada CN, Lin M, Zhu S, Cao Z, Liang Y, Sboner A, et al. . 2014. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet 46: 1227–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S, Fox H, Jacobi K, Saiya-Cork K, Bixby D, et al. . 2014. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood 123: 1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37: 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. . 2012. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci 109: 3879–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, Nakatani Y, Dent SY, Almouzni G. 2009. The HP1α-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep 10: 769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. . 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Luo Z, Lin C, Shilatifard A. 2012. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13: 543–547. [DOI] [PubMed] [Google Scholar]
- Maison C, Almouzni G. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5: 296–304. [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. . 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. . 2012. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149: 590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al. . 2012a. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci 109: 2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A 3rd, Diaz E, et al. . 2012b. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492: 108–112. [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. . 2008. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci 98: 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A. 2010. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer 10: 721–728. [DOI] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. 2011. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol 31: 4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Baylies MK, Dorsett D. 1997. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev 11: 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. . 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. . 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff RD, Scott DW, Ding J, Roth A, Chiu R, et al. . 2013. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 122: 1256–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. . 2006. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124: 315–329. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208. [DOI] [PubMed] [Google Scholar]
- Murzina N, Verreault A, Laue E, Stillman B. 1999. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell 4: 529–540. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. [DOI] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. . 2012. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 18: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, Loizou E, Holmfeldt L, Strikoudis A, King B, et al. . 2014. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 514: 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al. . 2014. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 46: 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Karlseder J. 2010. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 11: 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott HM, Graves A, Pappalardi MB, Huddleston M, Halsey WS, Hughes A, Groy A, Dul E, Jiang Y, Bai Y, et al. . 2014. A687V EZH2 is a driver of histone H3 lysine 27 (H3K27) hypertrimethylation. Mol Cancer Ther 13: 3062–3073. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. . 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al. . 2011. The genetic landscape of the childhood cancer medulloblastoma. Science 331: 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, et al. . 2010. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res 38: 4958–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O, et al. . 2011. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet 43: 830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. 2013. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339: 698–699. [DOI] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. . 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, et al. . 2012. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488: 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy JP, Gerard A, Cook AJ, Roche D, Almouzni G. 2008. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol 15: 972–979. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B. 2012. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J 31: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Martinez de Paz A, Simo-Riudalbas L, Sayols S, Moutinho C, Moran S, Villanueva A, Vazquez-Cedeira M, Lazo PA, Carneiro F, et al. . 2014. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene 33: 2807–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. 1993. Rearrangements involving chromosome band 11Q23 in acute leukaemia. Semin Cancer Biol 4: 377–385. [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. . 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231. [DOI] [PubMed] [Google Scholar]
- Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. . 2012. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151: 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Vakoc CR. 2014. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 54: 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore EM, Xu M, Feldman GJ, Fenstermacher DA, Cho TJ, Choi IH, Connor JM, Delai P, Glaser DL, LeMerrer M, et al. . 2006. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet 38: 525–527. [DOI] [PubMed] [Google Scholar]
- Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, Boucher G, Chagnon P, Drouin S, Lambert R, et al. . 2012. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev 26: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. 2014. Enhancer biology and enhanceropathies. Nat Struct Mol Biol 21: 210–219. [DOI] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 25: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle M, Workman JL, Venkatesh S. 2013. reSETting chromatin during transcription elongation. Epigenetics 8: 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. 2010. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci 107: 20980–20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanik VA, Harte PJ. 2012. A mutation in the E(Z) methyltransferase that increases trimethylation of histone H3 lysine 27 and causes inappropriate silencing of active Polycomb target genes. Dev Biol 364: 249–258. [DOI] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. . 2012. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22: 425–437. [DOI] [PubMed] [Google Scholar]
- Taddei A, Maison C, Roche D, Almouzni G. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol 3: 114–120. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14: 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KR, Mackay A, Truffaux N, Butterfield YS, Morozova O, Philippe C, Castel D, Grasso CS, Vinci M, Carvalho D, et al. . 2014. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet 46: 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee WW, Shen SS, Oksuz O, Narendra V, Reinberg D. 2014. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell 156: 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. . 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rechem C, Whetstine JR. 2014. Examining the impact of gene variants on histone lysine methylation. Biochim Biophys Acta 1839: 1463–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. 2013. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 23: 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. . 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481. [DOI] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. . 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, et al. . 2014. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 46: 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. . 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Suzuki M, Otsuki A, Shimizu R, Bresnick EH, Engel JD, Yamamoto M. 2014. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell 25: 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying CY, Dominguez-Sola D, Fabi M, Lorenz IC, Hussein S, Bansal M, Califano A, Pasqualucci L, Basso K, Dalla-Favera R. 2013. MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat Immunol 14: 1084–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. . 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang Y, Jones S, Sausen M, McMahon K, Sharma R, Wang Q, Belzberg AJ, Chaichana K, Gallia GL, et al. . 2014. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet 46: 1170–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemin-van der Poel S, McCabe NR, Gill HJ, Espinosa R 3rd, Patel Y, Harden A, Rubinelli P, Smith SD, LeBeau MM, Rowley JD, et al. . 1991. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci 88: 10735–10739. [DOI] [PMC free article] [PubMed] [Google Scholar]