Abstract
The peripheral nervous system (PNS) of embryonic and larval stage Drosophila consists of diverse types of sensory neurons positioned along the body wall. Sensory neurons, and associated end organs, show highly stereotyped locations and morphologies. The many powerful genetic tools for gene manipulation available in Drosophila make the PNS an advantageous system for elucidating basic principles of neural development. Studies of the Drosophila PNS have provided key insights into molecular mechanisms of cell fate specification, asymmetric cell division, and dendritic morphogenesis. A canonical lineage gives rise to sensory neurons and associated organs, and cells within this lineage are diversified through asymmetric cell divisions. Newly specified sensory neurons develop specific dendritic patterns, which are controlled by numerous factors including transcriptional regulators, interactions with neighboring neurons, and intracellular trafficking systems. In addition, sensory axons show modality specific terminations in the central nervous system, which are patterned by secreted ligands and their receptors expressed by sensory axons. Modality-specific axon projections are critical for coordinated larval behaviors. We review the molecular basis for PNS development and address some of the instances in which the mechanisms and molecules identified are conserved in vertebrate development.
Layout of the embryonic and larval peripheral nervous system
The insect sensory nervous system receives and transduces information from the outside world to the central nervous system (CNS), ultimately to promote appropriate larval and adult behavior. Inputs to the somatosensory system are diverse, and this is reflected in the many different morphologies of individual sensory organs, or sensilla. Somatosensory neurons reside along the basal (inner) surface of the epidermis, and spread sensory processes along the body wall or to specific sensory end organs 1, 2. The system of neurons is organized in a segmentally-repeated fashion, and the positioning of organs is stereotyped from animal to animal. This stereotypy allows investigators to focus their studies on individual identified sensory organs, a feature that has been instrumental in elucidating several important principles of neural development and sensory function.
In this review, we concentrate on body wall sensory elements of the peripheral nervous system (PNS), the organization of which is highly stereotyped. Sensory organs are organized loosely into dorsal (d), lateral (l), ventral’ (v’) and ventral (v) clusters (Figures 1A and 1B). These clusters consist of neurons that are classified as type I sensilla – neurons with single ciliated dendrites – and type II multidendritic (md) neurons. The md neurons spread complex, highly branched dendritic processes across the body wall or along internal scaffold structures, such as respiratory structures or connective strands 3. Within these broad categories, there are a large number of functionally specialized sensory organ subtypes. Type I neurons include the mechanosensory external sensory (es) organs, and internal chordotonal (ch) organs, which sense stretch or vibration (Figures 1C and 1D). Individual es organs are further distinguished by end organ morphology, including campanifom sensilla that extend a papilla, and trichoid sensilla that extend a long hair from a surrounding socket. The various chordotonal organs differ in their position, orientation, and the number of functional units, or scolopidia, that coalesce into a single organ (Figures 1C and 1D). Among the type II md neurons there are three broad subtypes, the bipolar dendrite (bd) neurons, the tracheal dendrite (td) neurons, and the dendritic arborization (da) neurons (Figure 1E). A major distinction between these groups of neurons is the substrates upon which dendrites grow.
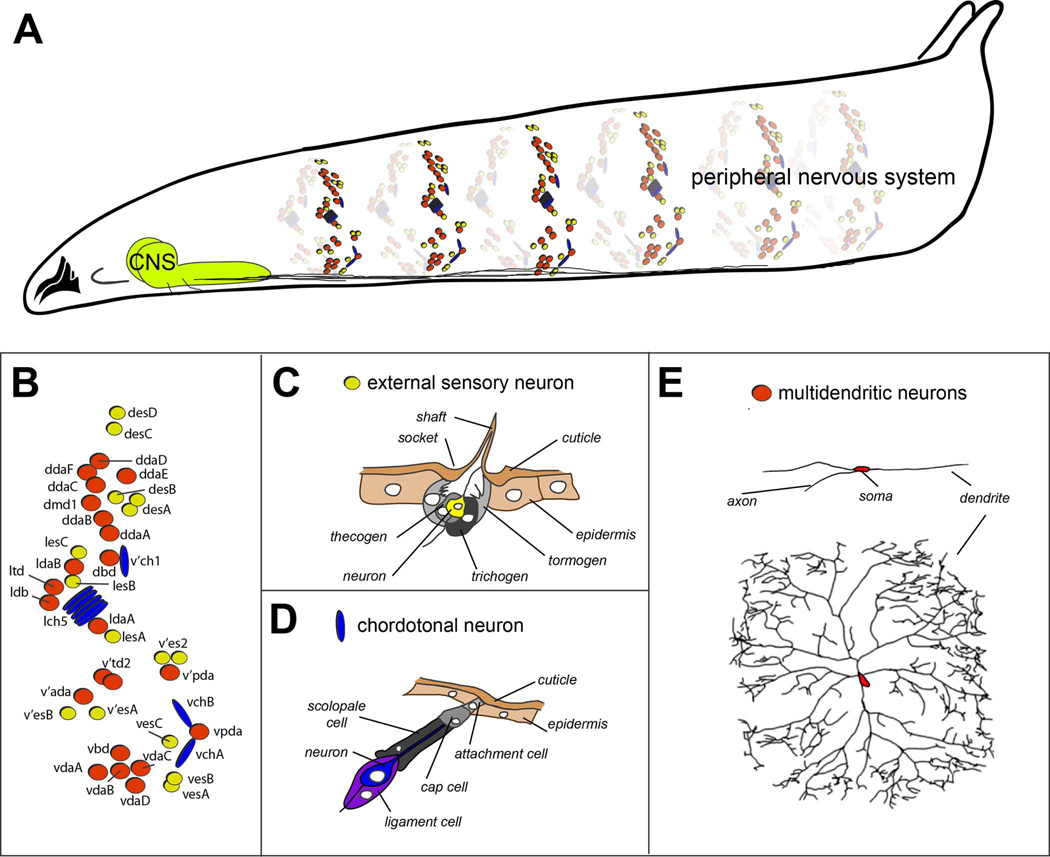
Figure 1. Organization of the embryonic and larval peripheral nervous system of Drosophila.
A. Drawing of a third instar Drosophila larva showing sensory elements that comprise the peripheral nervous system. For simplicity, only sensory neurons of a subset of abdominal segments are shown. Bundled sensory axons project to the central nervous system (CNS) that resides in the ventral and anterior part of the larva.
B. Schematic of the arrangement of sensory neurons in a single abdominal hemisegment. External sensory organs are indicated by yellow circles, chordotonal organs by blue ovals, and multidendritic neurons by red circles.
C. Drawing of external sensory organ structure. Names of individual cellular elements are indicated. Drawing adapted, with permission, from Comprehensive Molecular Insect Science. Vol. 1: Hartenstein V. Development of Insect Sensilla. pp. 379–419, 2005.
D. Drawing of chordotonal organ structure. Names of individual cellular elements are indicated. Drawing adapted, with permission, from Comprehensive Molecular Insect Science. Vol. 1: Hartenstein V. Development of Insect Sensilla. pp. 379–419, 2005.
E. Tracings of multidendritic neurons. Two different neurons are shown, the dorsal bipolar dendrite neuron (top), and a class IV nociceptive neuron (bottom). Note the different degrees of dendritic branching shown by the two neurons. Tracing of class IV neuron reproduced, with permission from Grueber et al., 2003.
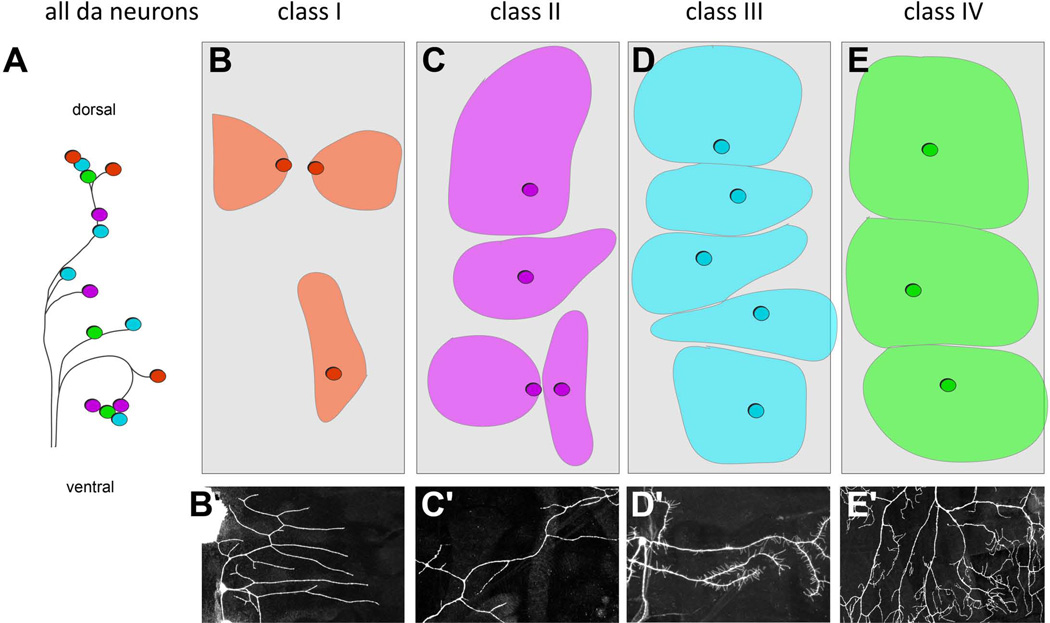
Among the da neurons, which extend dendrites across the epidermis, there is also considerable morphological diversity4, 5. The 15 da neurons per abdominal hemisegment are divided into four distinct classes (classes I-IV) based on their dendrite branching complexity and axon projection pattern 4, 6 (Figure 4). Class I and class II neurons have simple branching patterns, class III show numerous short actin-based protrusions extending from major branches, and class IV neurons innervate the entire epidermis with complex, space-filling arbors (Figures 4B’-4E’). Molecular and functional properties correlate with these morphological distinctions. Class I neurons likely function as proprioceptors, class III neurons as touch receptors, and class IV neurons as polymodal nociceptive (noxious sensing) neurons 7–12. The functions of class II neurons are not yet known, but there is some evidence that they also function as touch receptors 8. The molecular basis of the development of da neuron dendrite diversity will be discussed in more detail below. Thus, fitting with the many sensory requirements of larval stages, the elements of the sensory nervous system of insects are highly diversified.
Figure 4. Distinct dendritic morphologies and territories of different classes of dendritic arborization (da) neurons.
A. Schematic of arrangement of dendritic arborization neuron cell bodies. Different morphological classes of neurons are color coded red = class I, purple = class II, light blue = class III, and green = class IV.
B-E. Schematics of the territories occupied by the different classes of da neurons in each hemisegment. The approximate locations of sensory neuron cell bodies within each class are indicated by circles. The approximate dendritic coverage of the neuron is indicated by colored areas. Note tiling among class III neurons and among class IV neurons.
B'-E' Representative dendritic arbors of class I (B'), class II (C'), class III (D') and class IV (E') neurons. Neuron in C' adapted with permission from (Matthews et al., 2007).
The nomenclature for individual sensory neurons represents both their identity and their location along the epidermis (Figure 1B). In the naming scheme that is commonly used in recent literature, the names of individual neurons typically start with d, l, v’ or v to indicate the cluster to which the neuron belongs. This prefix is followed by a pair of letters to indicate the type of neuron; es for external sensory, ch for chordotonal, or da for dendritic arborization. This designation is followed by a letter or number that gives each neuron a unique identifier. Naming schemes have evolved over the years, an account of which is given by Orgogozo and Grueber, 2005 13.
Sensillum specification: variations on a basic plan
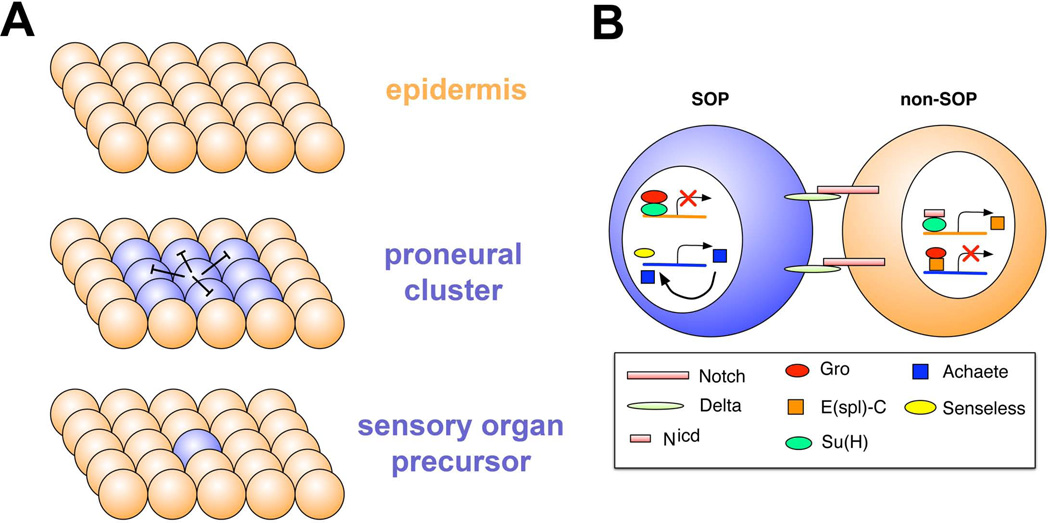
Despite the diversity of insect sensillum structures, most, if not all, arise via a highly conserved sequence of cell-cell interactions and cell divisions. An initial step is the generation of proneural clusters, groups of ectodermal cells with the capacity to become neurons (Figure 2). Neural capacity is subsequently restricted to a single cell within this cluster, the neural precursor (Figure 2). At this point the neural precursor undergoes a series of stereotyped cell divisions to give rise to neurons and the support cells that become associated with the sensory organ (Figure 2).
Figure 2. Sensory organ precursor specification.
A. Proneural clusters (blue spheres) typified by expression of basic helix-loop-helix transcription factors, arise from a field of equipotential epidermal cells (orange). From the proneural cluster a single sensory organ precursor emerges, which inhibits proneural gene expression in surrounding cells, a process termed lateral inhibition.
B. Molecular basis for lateral inhibition. Delta ligand and Notch receptor are expressed in future SOP and non-SOP cells. Delta levels become higher in the SOP, which promotes higher Notch signaling in non-SOP cells. The Notch intracellular domain collaborates with the Suppressor of Hairless protein to promote expression of Enhancer of split in non-SOP cells. Enhancer of split acts together with the Groucho transcriptional repressor to shut off expression of the proneural gene achaete. In the future SOP, Groucho and Suppressor of hairless repress expression of Enhancer of split, so that proneural gene expression can persist. High-level expression of proneural genes is promoted by Senseless and also by proneural factors themselves in a positive feedback loop. Drawing adapted with permission from (Castro et al. Development, 2005)121.
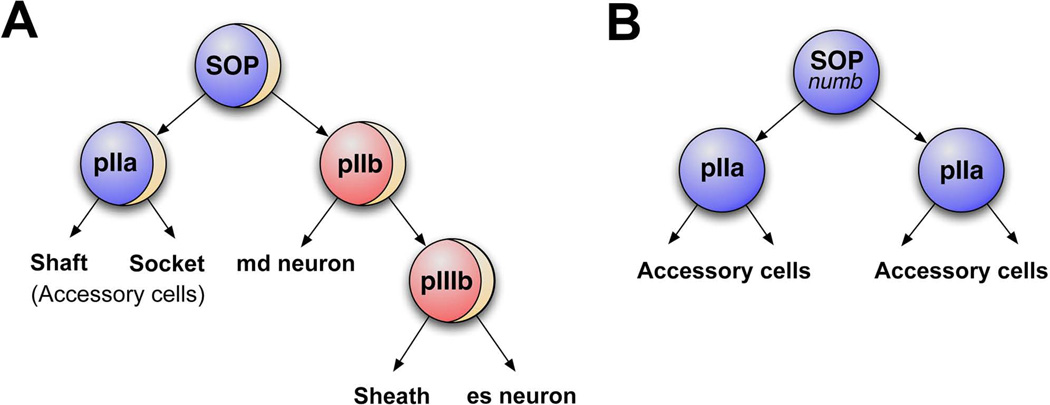
Given that the series of divisions that give rise to different sensilla is conserved, even though the type of organ that is produced may differ, a conserved ground plan for sensillum development was proposed by Lai and Orgogozo (2004) 14. The proposed canonical sensillum lineage consists of a sensory organ precursor (SOP) cell, pI, that undergoes four cell divisions to generate a total of five cells: two “outer cells” generated by the pIIa division, and three “inner cells” generated by the pIIb lineage 14 (Figure 3A). Md and es neurons are in some cases born from the same SOP, and in these lineages, neurons derive from progeny of the pIIb cell – the first division directly gives rise to an md neuron and a pIIIb cell, and subsequent division of pIIIb generates the es neuron along with a sheath cell (Figure 3A). Outer and inner terminal cell fates can differ in a lineage-dependent manner to generate organ diversity. For example ch lineages generate attachment cells and cap cells from the pIIa division, whereas in es organ lineages the analogous division generates the external shaft of the bristle and the socket that anchors this shaft. Specific lineages also generate specialized sensilla by adjustments in lineage-specific cell death or proliferation 15, 16. For example, some lineages produce only md neurons, known as the “solo-md” lineages, due to programmed cell death of pIIa and pIIIb cells 16. Thus, from a canonical cell lineage, many different types of organs can be produced via changes in terminal cell fate.
Figure 3. Asymmetric cell division in the SOP lineage diversifies cell fates.
A. Wildtype external sensory organ precursor lineage (ESOP). Tan arc in the SOP, pIIb, and pIIIb represents a crescent of asymmetrically localized Numb protein. The SOP divides to produce pIIa (blue) and pIIb (red) cells. pIIb inherits Numb and undergoes another round of numb-dependent asymmetric cell division which gives rise to an md neuron and pIIIb (red). pIIIb in turn generates a sheath cell and external sensory (es) neuron. The pIIa cell generates accessory cells, the shaft cell and socket cell.
B. In the absence of numb the SOP divides into two identical pIIa cells, which divide to give rise to accessory cells. No neurons are produced by SOPs mutant for numb.
Genetic specification of the sensillum lineages
Given the highly stereotyped sequence of cell divisions described above, which leads to a largely invariant number of sensilla per animal, the sensillum lineage has been an important model for understanding principles of neuronal specification. The process of neuronal specification begins with expression of proneural genes in proneural clusters. Proneural factors include members of the Achaete-Scute complex (ASC; consisting of the genes achaete, scute, lethal of scute, and asense), atonal (ato) and absent md neurons and olfactory sensilla (amos) 17–19,20. The ASC complex is required in proneural clusters that produce es organs and md neurons derived from the esSOP lineage. Loss of function mutations of individual members of the ASC genes indicate that they act redundantly 21. ato controls the production of ch organs and a subset of md neurons 18, and amos the production of the remaining md neurons 19.
Proneural genes all belong to the basic-helix-loop-helix (bHLH) family of transcription factors that bind to specific regions of DNA and coordinate the expression of multiple other genes. Class II bHLH proteins, such as the ASC genes, are expressed in tissue-specific patterns and act as heterodimers with more broadly expressed bHLH proteins, the class I bHLH factors 22. Interactions between class I and class II bHLH proteins lead to strong DNA binding activity, whereas homodimers of class I proteins and homo and heterodimers of different class II proteins show little or no DNA binding activity 23, 24. In Drosophila, the class I protein is encoded by the daughterless gene. Loss of Daughterless leads to loss of all sensory neurons 25. Daughterless is not required for neuronal precursor formation, but is required for the expression of multiple neuronal precursor genes, including prospero, deadpan, and scratch 26. Thus, bHLH-dependent transcriptional regulation plays a key role in neuronal specification.
Much of our understanding of SOP specification derives from studies of embryonic development and from studies of macrochaetae on the notum of the adult fly. The expression of proneural genes is very dynamic in embryonic stages. Studies of the achaete (ac) gene showed that it is first expressed in clusters of cells, the “competent” stage, at embryonic stage 10 (approximately 4.5 hours of development at 25°C)27. One cell within each cluster soon begins to express higher levels of achaete and this cell becomes the SOP 27 (Figure 2). Selection of a single SOP depends critically on the membrane receptor Notch. Notch plays no direct role in emergence of proneural clusters, but is central to a process termed lateral inhibition, which limits the number of precursor cells that differentiate within each cluster (Figure 2B) 27, 28. The SOP cells present high levels of the transmembrane Delta protein, which acts as a ligand and binds to the Notch receptor, activating the Notch pathway in surrounding cells. How the one cell is selected to adopt the precursor identity is not yet known, however the resulting bias in Delta-Notch signaling sends all but one cell within the cluster into an epidermal fate. This effect is mediated by the Notch intracellular domain (Nicd) which translocates to the nucleus, and together with the Suppressor of Hairless [Su(H)] protein, activates expression of Enhancer of split complex [E(spl)-C] genes 29, 30. E(spl)-C proteins cooperate with the transcriptional co-repressor Groucho (Gro) to repress neuronal fate by down regulating the expression of proneural genes and the zinc finger transcription factor Senseless (Sens)31. High levels of Sens promote further expression of proneural genes, whereas low levels of Sens in other cells in the proneural field appear to repress proneural genes 31. High expression of proneural genes is maintained through a positive feedback loop (Figure 2B) 32.
Different proneural genes are expressed in different subsets of proneural clusters and are sufficient for the specification of neuronal precursors. Some insight into how bHLH activity is linked to specific sensory organ differentiation has come from studies of the role of Ato. Ato is expressed only transiently in SOPs so likely initiates expression of a cascade of differentiation genes that act directly to specify ch fate 33. Expression profiling of ato-expressing cells revealed hundreds of genes that are regulated by the activity of this transcription factor. Highly represented among this group are genes involved in neurogenesis and ciliogenesis, such as the transcriptional regulator Regulatory factor X (Rfx), and dilatory, which encodes a predicted coiled-coil protein that localizes to the ciliary base33, 34. The finding that Ato controls pathways involved in ciliogeneses fits well with ciliated dendrites being a distinguishing feature of ch organ neurons. This study provides an example of how proneural genes could define specific features of the sensilla types that they promote.
Asymmetric cell division in the SOP lineage diversifies cell fates
One fundamental question in neuronal development is how cells with distinct fates are generated from a common precursor cell. Divisions of precursor cells that produce unequal daughter cells are termed asymmetric divisions, and are a key mechanism of cellular diversification during development. Different scenarios could be envisioned to give rise to asymmetric fate decisions, including specification by environmental factors, or by intrinsic factors that are localized to one cell but not the other. The first example of such an asymmetrically localized cell fate determinant in fly neural precursors was the Numb protein (Figure 3A). Mutations in the numb gene cause loss of sensory neurons in Drosophila embryos. Recall that sensory organs consist not only of neurons, but also external cells, such as bristles, that arise from the same lineage. Loss of neurons in numb mutants was coupled to gain of external cells 35, suggesting that a fate switch had occurred in the SOP lineage (Figure 3B). Studies of protein localization using antibodies directed against Numb revealed that, remarkably, Numb protein was segregated to only one of the two daughter cells during division of the SOP 36 (Figure 3A). Furthermore, employing a genetic approach to misexpress Numb so that it was present in both daughter cells caused them to take equivalent fates, indicating that the differential segregation of Numb normally confers distinct fates to cells in this lineage 36.
These experiments raised several major questions that have provided a basis for numerous ensuing studies. The volume of work is too large to cover in depth here, so we address these major points in abbreviated form and refer to readers other reviews on the subject 37–40. One key problem relevant to the generation of sensory organ diversity has been to understand how Numb exerts its effect on cell fate. Studies of this problem have focused on interactions with the Notch pathway, already introduced above as a key factor in lateral inhibition. Notch is involved not only in SOP specification, but also in subsequent cell fate decisions in the SOP lineage. Loss of either Notch or Delta causes the opposite phenotype as loss of Numb, namely conversion of external cells into internal ones. Notch is expressed in both daughter cells, as is the Delta ligand. However, in the one daughter cell that inherits Numb, Notch signaling is antagonized 41, 42.
Numb is able to associate with endocytic proteins, such as EPS15 and alpha-adaptin 43–45 so has been hypothesized to control membrane localization of proteins important for asymmetric fate specification. Work focused on cell division in the central nervous system revealed that a four-pass transmembrane protein called Sanpodo (Spdo) acts upstream of Notch signaling in asymmetric cell division 46. Numb was shown to inhibit the plasma membrane localization of Spdo, thereby inhibiting the activity of Notch. Recent studies of the SOP lineage in the fly notum have clarified how this happens 47–49. It appears that normal steady state membrane and endosomal localization of Spdo and Notch is independent of Numb. Rather, Numb interacts with the AP-1 complex to regulate endocytic recycling of Spdo and Notch. In the absence of Numb, recycling to the membrane is permitted. By contrast, in the presence of Numb, Spdo becomes localized to endosomes. In this way, the asymmetric segregation of Numb results in down regulation of Notch signaling in the cell that inherits Numb and diversification of sister cell fates.
The role of Numb and its functional homolog Numblike in mammals has likewise been the subject of intensive investigation. Numb/Numblike control neurogenesis and help maintain neuroepithelial integrity 50, 51. Studies also indicate that Numb plays a role in cell cycle progression and terminal asymmetric cell division in the developing mouse retina 52. The mechanism involved appears similar to that observed in Drosophila, namely asymmetric inheritance of Numb and differential Notch signaling in sibling cells 52.
Execution of SOP identity
As described above, different SOPs give rise to a wide diversity of sensilla types. Proteins such as Numb and Notch are important for diversifying fates within a lineage, but how is a lineage fated to generate different types of organs, for example chordotonal versus external sensory organs? Several studies have identified transcription factors that are expressed in SOPs and progeny and execute fate decisions within a lineage, including the homeodomain transcription factors (TFs) Cut and Poxn. In the PNS, Cut is expressed in a complex pattern in both md neurons and es organs, but not ch organs 53. The role of Cut in md neuron specification will be discussed below. Expression of Cut in developing es organs is crucial for their specification, and in the absence of Cut, es organs are transformed into a ch identity 54. Loss of another factor, Poxn causes a transformation of multiply-innervated chemosensory organs to mechanoreceptors 55. The choice between two different types of external sensory organs, campanifom sensilla and trichoid sensilla, is specified by a pair of redundant homeobox genes, BarH1 and BarH2 2, 56. In the absence of both Bar genes, campaniform sensilla are transformed to hair structures characteristic of trichoid sensilla. Notably, Bar proteins are found in internal cells, but phenotypes manifest in the external cells, suggesting non-autonomous control 2, 56. Hamlet, a zinc finger transcription factor, regulates the decision between type I and type II neurons 57. Furthermore, loss of Hamlet also transforms glial cells to hair cells (trichogen) 58, showing that the function of cell fate determinants in these lineages can be context-dependent. Thus, SOPs are specified to form distinct cell types by the TFs they express in conjunction with the series of asymmetric cell divisions that cause divergence in cell fate.
Control of sensory dendrite morphogenesis
Dendritic shapes and territories impact neuronal connectivity and function, so understanding how dendrites develop is an important goal. Sensory neuron dendrite development involves sequential stages, including outgrowth, branching, targeting, and stopping of growth 59. The stereotypy of dendritic arbor development of md neurons has provided a useful system for understanding how dendritic morphology is specified, and each of the above steps has been dissected in some detail in this system 60, 61.
One powerful approach for decoding the genetic control of neural development in Drosophila is the forward genetic screen, in which the genome is mutagenized, for example using the mutagen ethylmethanesulfonate (EMS), and lines carrying mutations are examined for defective development of a feature of interest (such as axon guidance or dendrite patterning). A transgenic approach was developed for labeling developing md neuron dendrites in vivo and screening for EMS-induced mutations that disrupt the normal embryonic dendrite pattern 59. From this screen several mutations that affect dendrite outgrowth, routing, and branching were identified, including membrane proteins such as Flamingo, cytoskeletal regulators such as Enabled, and transcription factors including Sequoia and Hamlet 59, 62, 63. These studies also showed that the establishment of axon and dendrite morphology was under at least partially distinct control in sensory neurons, prompting further investigation of pathways that specifically control dendritic morphogenesis (see below). These results provided a basis for studying the genetic control of dendritic morphogenesis in the Drosophila PNS.
Transcriptional control of dendritic diversity
Dendrites show remarkably diverse morphologies, fitting with distinct functions and connectivity of neurons. How do diverse and stereotyped morphologies of neurons arise during development? The varied morphologies and functions of md neurons have made this a powerful system for studies of dendritic diversity. Several studies have identified TFs that are expressed in a class-specific manner and control the distinctive dendrite morphology of the cells in which they are expressed. The homeodomain transcription factor Cut, introduced above in the context of es organ development, regulates the diversification of dendrites in a protein level-dependent manner 64. Cut is not expressed in class I neurons, and is expressed in increasing levels in class II, IV, and III neurons. Class III neurons show the highest level of Cut and loss of Cut simplifies the dendrites of these neurons to resemble the morphologies of non-expressing cells. Conversely, over-expression of Cut is sufficient to produce class III-like dendritic extensions. The actin binding protein Singed has been shown to function downstream of Cut in promoting the numerous actin-based protrusions of class III arborizations 65. Expression of the Ig superfamily member Turtle was shown to be positively regulated by Cut, and to mediate effects of Cut on dendrite morphogenesis 66. Moreover, Cut promotion of dendritic terminal branching requires activity of the RhoGEF Trio, suggesting that they act in a common pathway 67.
Cux1/2, mouse homologs of Cut, are important for determining the dendritic branching pattern and spine development of upper layer cortical neurons, and mutations in Cux2 affect working memory 68. Comparisons of gene expression profiles indicate that the role of vertebrate Cux genes in spine maturation is mediated through regulation of X-linked lymphocyte regulated (Xlr) 3b and Xlr4b, which may function as chromatin remodelers 68. Thus, Cut/Cux factors have a conserved role in dendritic maturation.
Expression of the Broad/Tramtrack/Bric a brac (BTB) zinc finger transcription factor Abrupt (Ab) is restricted to the class I neurons, which have the least complex branching pattern 69, 70. Loss of Abrupt in class I neurons leads to more complex branching patterns, whereas ectopic expression in neuronal classes with more complex branching pattern causes dendritic simplification 69, 70. Thus, Cut and Abrupt represent two transcription factors with essentially opposite effects on dendritic complexity, with one factor (Cut) promoting dendrite arborization, and the other (Abrupt) restricting dendritic growth, to produce neurons with distinctive morphologies. Class IV neuron morphogenesis is orchestrated by the transcription factor Collier/Knot, which is selectively expressed in complex class IV neurons, and is sufficient to promote excessive branching in neurons that otherwise have a simpler arborization 71–73. Knot promotes expression of the microtubule severing protein Spastin 71, which is hypothesized to create extra sites for branch initiation and promote arbor complexity. Notably another microtubule severing protein, katanin p60-like 1 (Kat-60L1) was also shown to regulate class IV dendrite branch number and length, but in a manner distinct from Spastin 74. Whereas Spastin depletes stable microtubules, Kat-60L1 appears to increase polymerizing microtubules and support terminal branch stabilization 74. A comparison of class I and class IV neuron transcriptional profiles revealed several hundred target genes for Knot and Abrupt, including both overlapping and distinct regulatory targets 75. Despite the differential control of dendritic arborization by Abrupt and Knot, all common targets showed similar direction of regulation (up or down) by the two factors. One of the common target genes encodes a homophilic cell adhesion molecule, Teneurin-m (Ten-m). Ten-m was expressed highly in class I neurons and promotes directional growth of dendrites and the characteristic comb-like arbor of these cells 75. By contrast, low expression in class IV neurons promotes their radial shape. The directional preference is likely endowed by interactions between dendrites and overlying epidermal cells. These results argue that differential quantitative control of regulators of morphogenesis that are shared in common between different neurons underlies dendritic diversification 75.
Some other transcriptional regulators are broadly expressed and mediate specific aspects of dendritic growth and branching. The spineless gene is expressed in all da neurons and in the absence of Spineless, normally diverse arbors converge on a similar dendritic complexity 76. The dar1 gene encodes a novel Kruppel-like factor expressed in all multidendritic neurons, but not in es neurons or chordotonal neurons 77. In dar1 mutants dendrites of all classes of da neurons are simplified, whereas overexpression of dar1 strongly promotes branching 77. Dar1 appears to restrict expression of Spastin to maintain proper levels of this factor and promote dendrite growth 77.
These studies together indicate that cell-type specific expression of transcriptional regulators is key to generating diverse dendritic architectures, however the suite of transcriptional regulators so far studied in detail is a small proportion of those shown to regulate dendritic patterning. For example, over 70 candidate transcriptional regulators were identified in an RNAi-based screen 78. Further exploration of these genes should provide important new insights into combinatorial transcriptional control of dendrite patterning. Moreover, many other regulators are likely awaiting identification. Recent advances in expression profiling of all, or subsets, of da neurons 79 should begin to shed further light on this important problem.
Specifying the locations of dendritic branches along an arbor
Sensory neurons are characterized not only by the complexity (i.e. number) of dendrites, but also by the positioning of dendrites throughout the arbor. For example, dendritic branches are not distributed uniformly along class IV dendrites, rather a greater density of branches is seen distally, nearer the margins of the field (Figures 4E’ and 5A). The locations of branches could have important consequences for how neurons receive information.
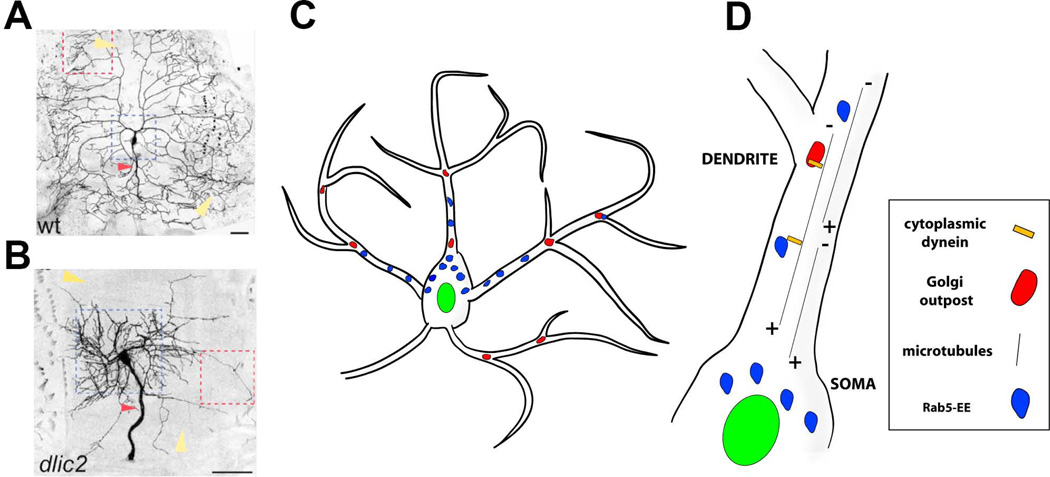
Figure 5. Molecular mechanisms that specify the location of dendritic branches along an arbor.
A, B. Images of class IV ddaC neurons that are wildtype (A) and mutant for dlic2 (B). The field size of dlic2 mutant neurons and dendritic arbors are abnormal (yellow arrowheads). The dlic2 mutant neurons show a shift in dendritic branches from distal to proximal (indicated by dotted blue squares). The red arrowheads indicate sensory axons, which are also thickened in dlic2 mutants. Reproduced with permission from Zheng et al., 2008.
C. Schematic representing Golgi outposts (red) and Rab5 early endosomes (Rab5-EE, blue).
D. Close up view of a dendritic branch indicating dynein-dependent microtubule minus (−) end directed movement of organelles to distal dendritic branches.
A genetic screen approach identified components of the dynein complex as important regulators of dendrite branch positioning along class IV dendrites (Figure 5B). Dynein is a microtubule motor complex that traffics molecules and organelles along microtubules (Figure 5C-5D). Dendrites and axons have distinct microtubule polarity, with dendritic microtubules primarily (−)-end distal (dendritic microtubules have mixed polarity in rodents) and axonal microtubules oriented with (+)-end distal 80, 81(Figure 5D). Dynein is the motor that moves cargo along the (−) end of microtubules, whereas kinesin is the major motor for trafficking toward the (+) end of microtubules. Thus, outbound trafficking in dendrites depends primarily on movements of dynein and dynein-dependent cargo. Removal of dynein light intermediate chain (dlic) from individual class IV neurons resulted in a dramatic shift of branches from distal to more proximal regions of the field (Figures 5A-5B) 82, 83. The number of dendritic branches within these crowded dendritic arbors was also reduced. Similar phenotypes were caused by mutations in other components of the dynein complex such as dynein heavy chain (dhc) and dynein intermediate chain (dic) 82, 83 Thus, dynein-dependent trafficking of cargo along dendritic microtubules is important for the development of complex sensory neuron dendrites, in particular, the characteristic distribution of branches along dendritic arbors.
What are the cargoes that are moved by dynein for proper branching morphology? Dynein appears to influence dendritic branching at least in part by regulating the distribution of a satellite secretory system, the Golgi outposts, in distal dendrites (Figure 5C-5D) 83. The Golgi apparatus is a major component of the cellular secretory pathway that resides primarily in the cell soma. Neuronal dendrites, but not axons, contain Golgi outposts at numerous branch points. Studies in vertebrate neurons showed that Golgi outposts are important for dendritic branching 84, and they have also been shown to operate in an evolutionary conserved manner in fly sensory neuron dendrites 85. Golgi outposts promote branching, and branch maintenance, within dendritic arbors of Drosophila class IV da neurons by supporting acentrosomal microtubule nucleation 86. In dlic mutants Golgi outposts remain localized to the proximal dendrites. Similarly, disruption of the Golgi adaptor protein Lava Lamp, which attaches Golgi to dynein, results in the accumulation of Golgi outposts at proximal dendrites 83. Altogether then, it seems that dynein-dependent trafficking of Golgi outposts is important for defining the spatial distribution of dendritic branches along an arbor.
Disruption of the endosomal pathway and related components also affects dendritic growth and branching. Early endosomes are the first stop during sorting of endocytosed vesicles for delivery into different pathways, such as recycling or degradative pathways. The small GTPase Rab5 is an important regulator of the early endocytic pathway and associates with Dlic in Drosophila neurons (Figure 5D) 82. The mobility of endosomes in dendritic branches depends on dynein for their transport to the distal dendrites 82. In the absence of Rab5, dendrites become highly simplified throughout the arbor. The branching pattern is not identical to dlic mutants in that there is not a distal to proximal shift in dendrites. In neurons mutant for both dlic and rab5, the proximal dendritic hyperbranching seen in dlic mutants is suppressed. Thus, dynein traffics branching machinery to distal dendrites whereas Rab5 is responsible generally for addition of terminal branches 82.
In summary, the dynein complex is an important regulator of dendrite development, controlling the transport of Golgi and endosomal components essential for spatial patterns of dendritic branching and growth along an arbor.
Dendritic field patterning
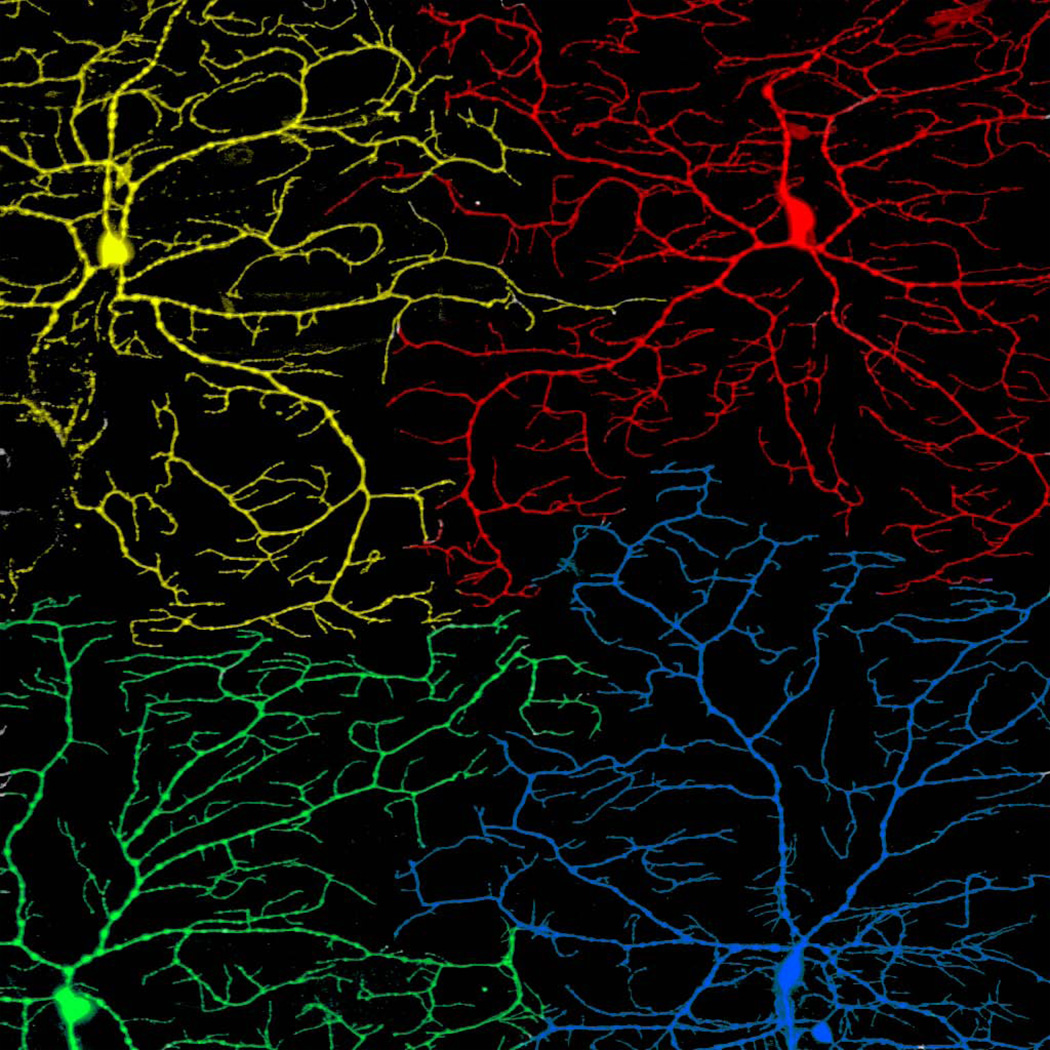
Two key mechanisms that control the spatial patterning of dendritic branches in the PNS are self-avoidance and tiling. Self-avoidance refers to the tendency of dendrites from the same cell to avoid crossing, resulting in evenly-spaced dendrites and minimal redundancy in coverage (Figure 6A). Tiling is an analogous phenomenon that occurs between the dendrites of different, but functionally and morphologically similar, sensory neurons. The lack of crossing between these heteroneuronal dendrites results in contiguous, but non-overlapping fields, similar to how tiles cover a floor (Figure 7). Tiling therefore ensures that all of sensory space is covered once, and only once, by neurons of the same type.
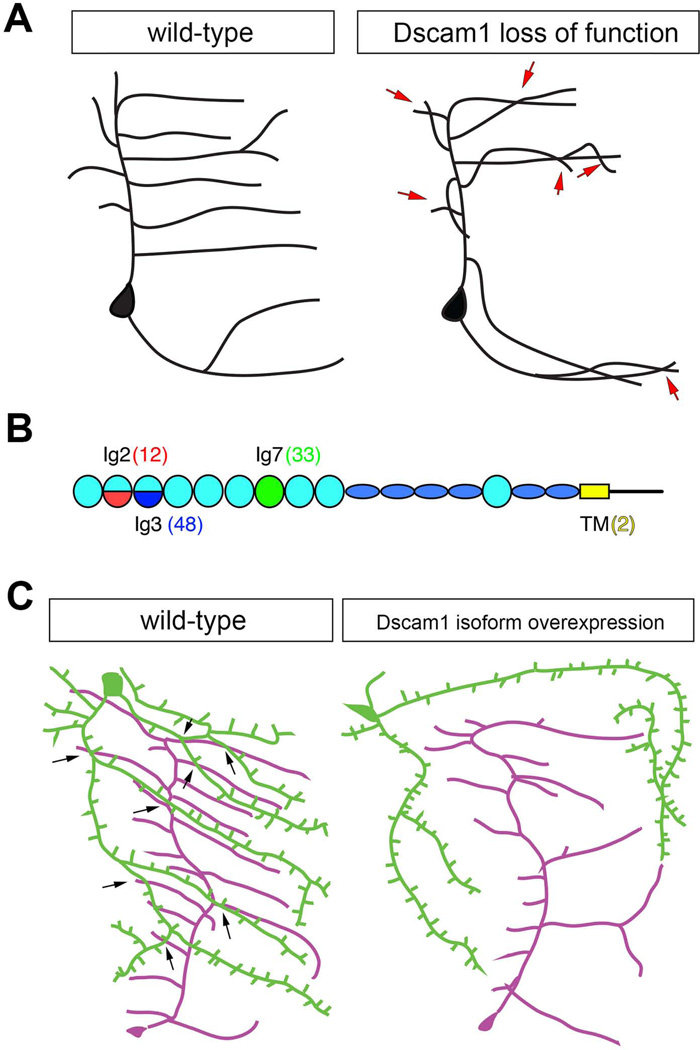
Figure 6. Self-avoidance control by Dscam1.
A. Schematic drawing of sensory dendrites in wild-type and Dscam1 loss of function mutants. Overlaps between sister branches are indicated by red arrows.
B. The Dscam1 locus consists of a series of exon cassettes (exon 4, 6, 9, and 17) that via alternative splicing can together generate over 38,000 distinct isoforms. Exons 4, 6, and 9 code for subregions of extracellular Ig domains 2 (red) and 3 (blue), or the entire Ig7 (green), respectively. Exon 17 generates either of two alternative transmembrane domains (yellow). Numbers in parentheses indicate the number of potential forms of each alternatively spliced domain. Flattened blue ovals indicate FN3 domains.
C. In wild-type larvae overlap is observed between dendritic branches of two different dendritic arborization sensory neurons, schematized as green and magenta neurons. Note the extensive crossing between branches of different neurons (arrows), but no overlap between branches of the same cell. Overexpression of a single Dscam1 isoform in cells that normally overlap leads to repulsion and separation of their fields such that few crossings among branches are observed.
Figure 7. Tiling of da neuron dendritic arbors.
Tiling between adjacent dendrites leads to non-overlapping dendritic fields and complete territory coverage. Shown is a confocal micrograph of class IV da neurons in a second instar larva. The cell bodies, axons, and dendrites of four different neurons are painted different colors to illustrate the tiling between them.
Control of dendritic self-avoidance
Self-avoidance was first studied experimentally in leech somatosensory axons, which extend across the body wall in very characteristic patterns. Lesions of specific axon branches in the leech leads to invasion of the vacated space by remaining axons, indicating that a self-recognition system underlies non-overlapping axon terminal arborization 87. The phenomenon seen in Drosophila sensory neuron dendrites is highly analogous to the leech system. Single branches avoid crossing each other and nearby branches invade the territory vacated by ablated branches (Figure 6A)4, 88, 89. By contrast, dendrites from different cells can overlap freely with one another. These studies point to a mechanism of self-avoidance in which branches are able to specifically recognize and avoid other branches from the same neuron. Dendrites from different cells overlap and thus must not share the same recognition code.
What is the molecular basis for discrimination of self and non-self branches, and how might the majority, or perhaps all, sensory neurons have their own unique surface identity? Studies of self-avoidance in Drosophila sensory neurons have focused on the Dscam1 gene. Dscam1 is a cell surface receptor of the immunoglobulin superfamily. Remarkably, the Dscam1 locus generates over 38,000 isoforms by extensive alternative splicing, with the majority of splicing occurring in the extracellular region within three of the ten different Ig domains 90. Different neurons express multiple different Dscam1 isoforms 91, 92. Biochemical studies indicate that Dscams bind homophilically in an isoform specific manner 93, 94. These studies raised the possibility that Dscam1 could provide a recognition code unique to each neuron. Sensory neurons lacking Dscam1 show excessive overlap of dendritic branches, consistent with a defect in self-recognition and repulsion (Figure 6B) 95–97. Single Dscam1 isoforms expressed ectopically in neurons are able to rescue self-avoidance defects, indicating that isoform diversity is not needed for self-recognition and repulsion. By contrast, if single isoforms are expressed in cells that normally have overlapping arbors, these arbors will now avoid crossing, as if they begin to recognize each other as “self” (Figures 6C). Analysis of Dscam1 splicing reporters in vivo indicated that exon choice is probabilistic, and did not reveal evidence for cell type-specific or spatially-specific alternative splicing of exon 4 98. Thus, the model for self-avoidance controlled by Dscam1 invokes the use of multiple Dscam1 isoforms, expressed in a probabilistic fashion in neurons, to provide different identification tags on the cell surface. Dendrites belonging to the same neuron will have the same set of Dscams, but given the large number of isoforms to choose from, neighboring neurons will be unlikely to share the same recognition code and thus will co-exist. It has recently been shown that, in rodents, starburst amacrine cells (SACs) use a very similar mechanism of molecular recognition during dendritic self-avoidance, albeit with a completely different group of cell surface receptors. In mammals, the clustered protocadherins, a class of highly diverse transmembrane proteins, allow discrimination of self and non-self 99. Knockout of gamma-protocadherins leads to excessive dendrite crossing, and a single isoform is sufficient for self-recognition and repulsion. Misexpression of a single isoform in neighboring SACs disrupts co-existence that normally is seen among amacrine cell dendrites. Thus, although the molecules mediating self-avoidance in fly and mammalian systems are distinct, they operate via remarkably similar principles.
In addition to the recognition specificity provided by the Dscam1 code, self-avoidance requires that sensory dendrites are restricted to a 2D plane so that processes are assured of coming into contact. This restriction is controlled by integrin receptors for the extracellular matrix (ECM). In the absence of integrins, dendritic segments become enclosed within invaginations of overlying epidermal cells, which prevents contact between dendrites and leads to excessive overlaps 100, 101. Several other molecules are involved in this pathway for 2D-restriction of dendrites, including the Tricornered kinase and Furry 102, and members of the TORC2 complex 100, 103. Thus, robust self-avoidance requires both interactions between dendrites, and interactions between dendrites and the ECM that enforce contact between sister dendrites.
Control of dendritic tiling
Tiling is observed in many neural systems, such as different cell types in the vertebrate and invertebrate retina 104 and neurons in the fly central nervous system 105, thus understanding the mechanisms that promote tiling are of great interest. Tiling is observed among class III and among class IV da sensory neurons 4 (Figures 4D, 4E, and 7). Cell-type specific boundaries could conceivably be explained by several different mechanisms, including competition for permissive cues in the environment or repulsive interactions between dendrites. Ablation of a class IV neuron results in the growth of neighboring class IV dendrites into the space previously occupied by the ablated cell 88, 89. By contrast if extra class IV cells are generated by mutations in the hamlet gene (introduced above as important for promoting es vs. md neuron identity) 57, then those cells integrate into the non-overlapping tiling pattern 88. These experiments indicate that tiling involves interactions between neighboring dendritic arbors. The precise mechanism of dendritic exclusion awaits identification of the molecular cues involved, which are currently not known.
Tiling and self-avoidance are analogous processes, but are likely to have at least partially distinct molecular underpinnings. For example, for self-avoidance, branches from the same cell must selectively recognize and repel each other, but for tiling, branches from neighboring, functionally similar neurons must interact and avoid crossing (Figures 6A and 7). Thus, self-avoidance requires recognition of “self”, whereas tiling requires recognition of “like”. Furthermore, mutations in Dscam1 do not disrupt tiling, while mutations in tricornered and furry do 95–97, 102. Thus, tiling likely depends on 2D restriction of arbors, but not on the surface identity provided by Dscam1 molecular diversity.
Sensory axon targeting to the central nervous system
Formation of functional sensory circuits requires precise positioning of axons in the CNS. Wiring of sensory axons requires that several sequential steps occur in precise spatial and temporal order. First, axons must successfully navigate a series of choice points presented to them en route to the CNS. Within the CNS, axons choose a broad zone within the neuropil in which to arborize, and then must form precise connections with specific second order neurons that reside within this zone. From the perspective of a sensory neuron residing along the body wall, finding the right target presents a formidable problem that requires sequential responses to many environmental guidance signals.
Axon navigation to the CNS
Axons navigate a complex, but compact, extracellular environment to reach the CNS, and appear to use a series of defined cellular landmarks to find their way. Depending on their cell body position along the dorsal-ventral axis, embryonic sensory axons project within either of two major peripheral nerves to the CNS. Sensory neurons located in ventral and ventral’ clusters travel to the CNS via the segmental nerve (SN), while neurons in the dorsal and lateral clusters merge with the inter-segmental nerve (ISN). Dorsal cluster neurons require signals of the Semaphorin family of axon guidance molecules to navigate an early choice point at the lateral body wall. The lateral neurons that comprise the lch5 organ also navigate two major choice points 106. A first choice point corresponds to the recognition of the substrate along which they grow (tracheal cells of the respiratory system), and another corresponds to the point at which axons meet and join the ISN. Once axons attach to their substrate, axon advance depends on the L1-type cell adhesion molecule Neuroglian (Nrg), which likely promotes adhesion between axonal growth cones and their substrate by acting in a heterophilic fashion 107. Disruption of adhesive interactions between axons and substrate generates stalling phenotypes, in which axons are unable to pass certain choice points. In the absence of the atypical protocadherin Flamingo lateral axons often stall before reaching the ISN. Notably, growth of the dbd pioneer axon does not require Flamingo or Neuorglian, suggesting differential control of growth in pioneer and follower axons 108. The LIM domain protein Prickle interacts with Flamingo to regulate axon advance at a specific choice point, the boundary between the PNS and CNS 109. Thus, as sensory axons navigate along peripheral tissue to the neuropil of the CNS they require multiple cues and adhesive interactions to ensure that they reach, and pass, successive landmarks.
Axon targeting within the CNS
The neuropil of insects refers to the dense collection of motor and sensory processes, interneurons and glia processes, and resides in the center of the CNS. Neuronal cell bodies reside in a region that surrounds the neuropil (Figure 8A). Axons of embryonic sensory neurons become precisely positioned within the neuropil by the action of a multitude of developmental signals. This targeting is likely critical for proper axon connectivity and larval behavior.
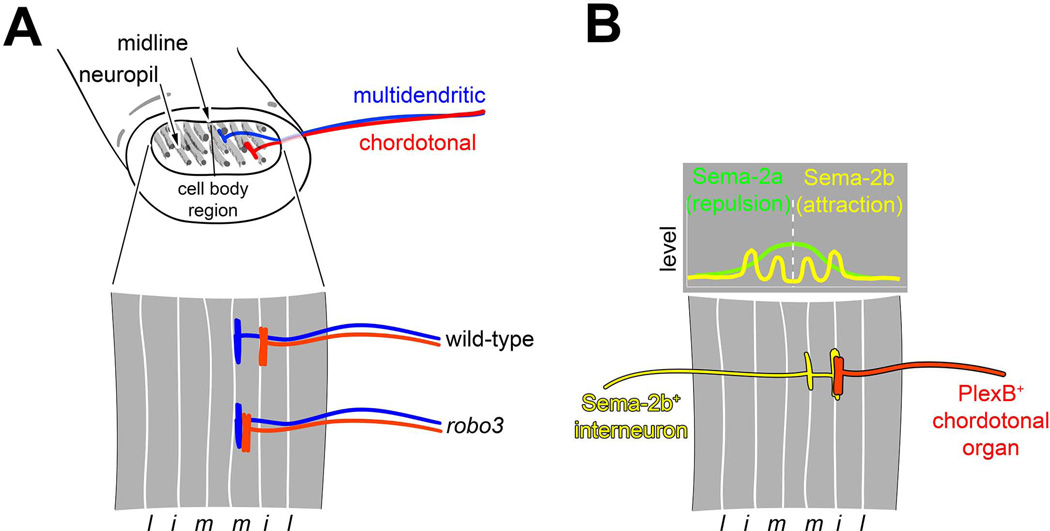
Figure 8. Sensory axon targeting in the CNS.
A. Organization and specification of axonal projections of md and ch organ sensory neurons. The ventral nerve cord is organized as an outer cell body region and an inner neuropil comprised of sensory axons, motor dendrites and interneuron processes. Axon tracts labeled by Fasciclin II (a cell adhesion molecule) are distributed along the dorsoventral and mediolateral axes and provide landmarks for description of axon targeting. The axons of some multidendritic neurons (blue) target a medial region of the neuropil. Chordotonal neurons (red) target intermediate neuropil. Mutations in robo3 shift chordotonal projections to a medial position that is characteristic of multidendritic neurons. l, lateral; i, intermediate; m, medial. Drawings based on data from Zlatic et al., 2003 113.
B. Semaphorin (Sema)/Plexin (Plex) signaling in the assembly of sensory circuitry. Top graph diagram shows relative levels of Sema 2a (green) and Sema 2b (yellow) across the mediolateral axis of the neuorpil. High levels of Sema-2a are seen at the midline, and lower levels laterally. Sema-2a mediates repulsion of PlexB-expressing axons from the midline region. Sema-2b is found at high levels in medial and intermediate axon tracts. Lower drawing shows projection pattern of Sema-2b-expressing interneuron to medial and intermediate tracts. This interneuron source of Sema-2b attracts PlexB-expressing chordotonal organs to the intermediate fascicle. Drawings adapted with permission from Wu et al., 2011 115.
Within the neuropil of the CNS, sensory axons show highly ordered arrangements. The positioning of sensory axon terminals correlates with soma position, modality, and proneural gene expression 110. For example, the axons of tactile and nociceptive neurons project ventrally, axons of chordotonal neurons project laterally, and axons of stretch responsive neurons, such as class I da neurons and bipolar dendrite neurons, project to more dorsal regions of the neuropil. The specification of these different types of axon projections correlates with expression of different proneural genes whose function is described above, with ASC-dependent sensory neurons comprising ventrally-projecting tactile and nociceptive subgroups, and Ato or Amos-dependent sensory neurons projecting axons laterally and dorsally 110. Axon positions of da neurons are related to their morphology and function 6. Class IV axons project to most medial regions of the neuropil with the dorsal and ventral cells also projecting axon branches across the midline, class III axons project slightly more laterally, and class II axons terminate adjacent to the class III axons. Such positioning could reflect modality specific connectivity, but this remains to be verified. Axon positioning relative to the midline is affected by misexpression of Roundabout 3 (Robo3) cell surface receptor, however it is unclear how precise relative positioning of class II-IV terminals is normally established. Midline crossing of class IV axons is promoted by the Tripartite motif protein Anomalies in sensory axon patterning (Asap), also known as Trim9. Asap is expressed highly in cells that cross the midline and functions to link the Netrin receptor, Frazzled, to downstream effectors 111. The transcription factor Mirror (Mirr) is also required for midline crossing of the ventral class IV neuron and ectopic Mirr expression in non-crossing neurons promotes contralateral projections 112. Correspondingly, both Mirror and Asap are important for behaviors that are mediated by class IV neurons 111, 112. Sensory axon terminals appear not to intermix with regions of motor neuropil, thus targets in the CNS are likely to be interneurons that await identification and characterization.
Interactions between cell surface receptors and secreted signals are critical for guiding and positioning axons within specific regions of the neuropil. Spatial coordinates in the CNS can be assigned by the relative position of Fasciclin II-positive axon tracts (Figure 8). The axons of the ch neurons terminate at an intermediate fascicle while md neuron axons terminate at a more medial fascicle. This positioning is dependent on differential proneural gene expression in md and ch neurons, which determines expression status of Robo3 113. Robo3 restricts ch axons to the intermediate fascicle, and in the absence of Robo3 ch axons project to more medial regions of the neuropil (Figure 8A). Mediolateral projection patterns are controlled by a gradient of Slit, a ligand for Robo3 that is secreted by midline glia. Robo3 expression is normally restricted to ch organs due to regulation by the proneural gene ato, which as described above, is required for the specification of ch organs. Thus, a transcriptional program set up by proneural gene activity specifies different modality-specific axon projections through regulation of a specific cell surface receptor.
While Robo3 specifies the position of sensory axons along a medio-lateral axis, it does not affect positioning along the dorso-ventral axis. A high-to-low gradient of the secreted Semaphorin (Sema), Sema-2a, is found from central to dorsal and central to ventral 114. Sema1a and Sema2a function through the Sema receptors Plexin A and Plexin B (Plex A and Plex B) to repel axons from central (high Sema) regions of the neuropil, which restricts them to ventral regions. Thus, orthogonal Robo/Slit and Plexin/Sema systems restrict sensory axons to characteristic domains within the neuropil.
Sema and Robo guidance systems specify both sensory and interneuron targeting in the CNS, which helps to coordinate pre and post-synaptic positioning. Sema-2b/Plex B-mediated fasciculation establishes coherent axon fascicle organization in the CNS and also mediates an attractive response of sensory axons to the same zone (Figure 8B). On the other hand, Sema-2a is concentrated near the center of the CNS and Sema-2a/Plex B repulsive signaling prevents axons from innervating these regions of the CNS and helps in positioning of longitudinal tracts (Figure 8B). Thus, Plex B integrates repulsion and attraction mediated by different Sema ligands. Disruption of chordotonal connectivity in Sema-2b mutants leads to abnormal larval responses to vibration, indicating a key role for these guidance decisions in sensory circuit assembly and behavior 115.
Glia in PNS development
Glia are important for the development and function of the nervous system and are present throughout the PNS of Drosophila. Peripheral glia are a diverse cell population, deriving from both glial specific lineages and mixed lineages known as neuroglioblasts (which give rise to neurons and glia). Peripheral glia are of three major types: perineural glia, subperineural glia, and wrapping glia. Perineural glial cells form the outermost layer, subperineural glial cells form the blood brain barrier of the peripheral nerve and wrapping glial cells form the innermost layer that ensheaths axons. In total there are 12 embryonic peripheral glia (ePG) in each hemisegment, which are derived from the CNS and from SOPs 116. Glial cells derived from CNS precursors migrate to the periphery by associating with SN and ISN bundles. Following their migration, glia align with peripheral nerves at characteristic positions. Each of these cells has a characteristic identity that correlates with their origin, position, and gene expression pattern117.
We are starting to understand the role that glial cells play in the development of sensory neurons. Peripheral glial cells are important for neuronal survival and for guiding developing sensory axons to the CNS 118. Sensory axons exhibit stalling and targeting defects in animals where peripheral glial cells are ablated. In addition, peripheral nerves show defasiculation phenotypes in the absence of glia. Downregulation of EGFR and Ras signaling, which is activated by EGFR, in peripheral glia disrupts axon pathfinding, perhaps through regulation of Nrg expression119. Interestingly, glial cells also depend on neurons for their normal development. When sensory neurons are genetically ablated the number of peripheral glial cells is reduced. This loss of glial cells is not caused by migration defects as there was no increase in glial cell numbers in other parts of the nervous system 119.
There is also evidence that glial-neuron interactions can influence the development of sensory dendritic branches. In nrg mutants, neuron glia interaction is disrupted and as a result a subset of da neuron dendrites (Class I) fail to develop normal second order branches 120. Supplying nrg to neurons and glial cells in nrg mutants restores the number of second order branches suggesting that interactions between neurons and glia mediate dendritic development. Studies of developmental roles for peripheral glia are at an early stage and other roles are likely awaiting identification.
Conclusions
Studies of the insect PNS have opened up many fundamental questions in neural development to molecular genetic analysis, and have provided starting points for investigating conserved principles in neurogenesis and differentiation. Asymmetric localization of cell fate determinants has emerged as a major mechanism for cellular diversification during development. Many of the genes discussed above as important for specific cell fate decisions in the Drosophila PNS have been shown to have conserved roles in neural development in other organisms, including genes such as atonal and cut. In addition, genes and pathways that control the morphogenesis of peripheral sensory neurons, such as the secretory pathway and Dscams, have roles in analogous processes in vertebrate systems. Studies of the Drosophila PNS will therefore continue to serve as an important model for understanding conserved principles of nervous system development, connectivity, and function.
Acknowledgments
We thank Dr. Fabrice Roegiers for helpful comments and suggestions. The work in our lab is supported by NIH 5R01 NS61908.
References
- 1.McIver SB. Mechanoreception. In: Kerkut GA, Gilbert LI, editors. Comprehensive Insect Physiology Biochemistry and Pharmacology. Vol. 6. Oxford: Elsevier; 1985. pp. 71–132. [Google Scholar]
- 2.Hartenstein V. Development of Insect Sensilla. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 1. Elsevier; 2005. pp. 379–419. [Google Scholar]
- 3.Bodmer R, Jan YN. Morphological differentiation of the embryonic peripheral neurons in Drosophila. Roux's Arch Dev Biol. 1987;196:69–77. doi: 10.1007/BF00402027. [DOI] [PubMed] [Google Scholar]
- 4.Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- 5.Sweeney NT, Li W, Gao FB. Genetic manipulation of single neurons in vivo reveals specific roles of flamingo in neuronal morphogenesis. Dev Biol. 2002;247:76–88. doi: 10.1006/dbio.2002.0702. [DOI] [PubMed] [Google Scholar]
- 6.Grueber WB, Ye B, Yang CH, Younger S, Borden K, Jan LY, Jan YN. Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development. 2007;134:55–64. doi: 10.1242/dev.02666. [DOI] [PubMed] [Google Scholar]
- 7.Yan Z, Zhang W, He Y, Gorczyca D, Xiang Y, Cheng LE, Meltzer S, Jan LY, Jan YN. Drosophila NOMPC is a mechanotransduction channel subunit for gentle-touch sensation. Nature. 2012 doi: 10.1038/nature11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsubouchi A, Caldwell JC, Tracey WD. Dendritic Filopodia, Ripped Pocket, NOMPC, and NMDARs Contribute to the Sense of Touch in Drosophila Larvae. Curr Biol. 2012;22:2124–2134. doi: 10.1016/j.cub.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes CL, Thomas JB. A sensory feedback circuit coordinates muscle activity in Drosophila. Mol Cell Neurosci. 2007;35:383–396. doi: 10.1016/j.mcn.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song W, Onishi M, Jan LY, Jan YN. Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc Natl Acad Sci U S A. 2007;104:5199–5204. doi: 10.1073/pnas.0700895104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, Tracey WD. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, Jan YN. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature. 2010;468:921–926. doi: 10.1038/nature09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orgogozo V, Grueber WB. FlyPNS, a database of the Drosophila embryonic and larval peripheral nervous system. BMC Dev Biol. 2005;5:4. doi: 10.1186/1471-213X-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai EC, Orgogozo V. A hidden program in Drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev Biol. 2004;269:1–17. doi: 10.1016/j.ydbio.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 15.Orgogozo V, Schweisguth F. Evolution of the larval peripheral nervous system in Drosophila species has involved a change in sensory cell lineage. Dev Genes Evol. 2004;214:442–452. doi: 10.1007/s00427-004-0422-4. [DOI] [PubMed] [Google Scholar]
- 16.Orgogozo V, Schweisguth F, Bellaiche Y. Lineage, cell polarity and inscuteable function in the peripheral nervous system of the Drosophila embryo. Development. 2001;128:631–643. doi: 10.1242/dev.128.5.631. [DOI] [PubMed] [Google Scholar]
- 17.Jan YN, Jan LY. Genetic control of cell fate specification in Drosophila peripheral nervous system. Annu Rev Genet. 1994;28:373–393. doi: 10.1146/annurev.ge.28.120194.002105. [DOI] [PubMed] [Google Scholar]
- 18.Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- 19.Huang ML, Hsu CH, Chien CT. The proneural gene amos promotes multiple dendritic neuron formation in the Drosophila peripheral nervous system. Neuron. 2000;25:57–67. doi: 10.1016/s0896-6273(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 20.Goulding SE, zur Lage P, Jarman AP. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron. 2000;25:69–78. doi: 10.1016/s0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 21.Marcellini S, Gibert JM, Simpson P. achaete, but not scute, is dispensable for the peripheral nervous system of Drosophila. Dev Biol. 2005;285:545–553. doi: 10.1016/j.ydbio.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 24.Cabrera CV, Alonso MC. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. Embo J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caudy M, Vassin H, Brand M, Tuma R, Jan LY, Jan YN. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell. 1988;55:1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- 26.Vaessin H, Brand M, Jan LY, Jan YN. daughterless is essential for neuronal precursor differentiation but not for initiation of neuronal precursor formation in Drosophila embryo. Development. 1994;120:935–945. doi: 10.1242/dev.120.4.935. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz-Gomez M, Ghysen A. The expression and role of a proneural gene, achaete, in the development of the larval nervous system of Drosophila. Embo J. 1993;12:1121–1130. doi: 10.1002/j.1460-2075.1993.tb05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Artavanis-Tsakonas S, Simpson P. Choosing a cell fate: a view from the Notch locus. Trends Genet. 1991;7:403–408. doi: 10.1016/0168-9525(91)90264-q. [DOI] [PubMed] [Google Scholar]
- 29.Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 30.Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 31.Jafar-Nejad H, Acar M, Nolo R, Lacin H, Pan H, Parkhurst SM, Bellen HJ. Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev. 2003;17:2966–2978. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- 33.Cachero S, Simpson TI, Zur Lage PI, Ma L, Newton FG, Holohan EE, Armstrong JD, Jarman AP. The gene regulatory cascade linking proneural specification with differentiation in Drosophila sensory neurons. PLoS Biol. 2011;9:e1000568. doi: 10.1371/journal.pbio.1000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma L, Jarman AP. Dilatory is a Drosophila protein related to AZI1 (CEP131) that is located at the ciliary base and required for cilium formation. J Cell Sci. 2011;124:2622–2630. doi: 10.1242/jcs.084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 36.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 37.Jan YN, Jan LY. Asymmetric cell division. Nature. 1998;392:775–778. doi: 10.1038/33854. [DOI] [PubMed] [Google Scholar]
- 38.Knoblich JA. The Drosophila nervous system as a model for asymmetric cell division. Symp Soc Exp Biol. 2001:75–89. [PubMed] [Google Scholar]
- 39.Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giebel B, Wodarz A. Notch signaling: numb makes the difference. Curr Biol. 2012;22:R133–R135. doi: 10.1016/j.cub.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 42.Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 43.Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, Di Fiore PP. Numb is an endocytic protein. J Cell Biol. 2000;151:1345–1352. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang H, Rompani SB, Atkins JB, Zhou Y, Osterwalder T, Zhong W. Numb proteins specify asymmetric cell fates via an endocytosis- and proteasome-independent pathway. Mol Cell Biol. 2005;25:2899–2909. doi: 10.1128/MCB.25.8.2899-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berdnik D, Torok T, Gonzalez-Gaitan M, Knoblich JA. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev Cell. 2002;3:221–231. doi: 10.1016/s1534-5807(02)00215-0. [DOI] [PubMed] [Google Scholar]
- 46.O'Connor-Giles KM, Skeath JB. Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev Cell. 2003;5:231–243. doi: 10.1016/s1534-5807(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 47.Couturier L, Mazouni K, Schweisguth F. Numb localizes at endosomes and controls the endosomal sorting of notch after asymmetric division in Drosophila. Curr Biol. 2013;23:588–593. doi: 10.1016/j.cub.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Cotton M, Benhra N, Le Borgne R. Numb inhibits the recycling of Sanpodo in Drosophila sensory organ precursor. Curr Biol. 2013;23:581–587. doi: 10.1016/j.cub.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 49.Upadhyay A, Kandachar V, Zitserman D, Tong X, Roegiers F. Sanpodo controls sensory organ precursor fate by directing Notch trafficking and binding gamma-secretase. J Cell Biol. 2013;201:439–448. doi: 10.1083/jcb.201209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- 51.Li HS, Wang D, Shen Q, Schonemann MD, Gorski JA, Jones KR, Temple S, Jan LY, Jan YN. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron. 2003;40:1105–1118. doi: 10.1016/s0896-6273(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 52.Kechad A, Jolicoeur C, Tufford A, Mattar P, Chow RW, Harris WA, Cayouette M. Numb is required for the production of terminal asymmetric cell divisions in the developing mouse retina. J Neurosci. 2012;32:17197–17210. doi: 10.1523/JNEUROSCI.4127-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blochlinger K, Bodmer R, Jan LY, Jan YN. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 1990;4:1322–1331. doi: 10.1101/gad.4.8.1322. [DOI] [PubMed] [Google Scholar]
- 54.Bodmer R, Barbel S, Sheperd S, Jack JW, Jan LY, Jan YN. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell. 1987;51:293–307. doi: 10.1016/0092-8674(87)90156-5. [DOI] [PubMed] [Google Scholar]
- 55.Dambly-Chaudiere C, Jamet E, Burri M, Bopp D, Basler K, Hafen E, Dumont N, Spielmann P, Ghysen A, Noll M. The paired box gene pox neuro: a determinant of poly-innervated sense organs in Drosophila. Cell. 1992;69:159–172. doi: 10.1016/0092-8674(92)90127-x. [DOI] [PubMed] [Google Scholar]
- 56.Higashijima S, Michiue T, Emori Y, Saigo K. Subtype determination of Drosophila embryonic external sensory organs by redundant homeo box genes BarH1 and BarH2. Genes Dev. 1992;6:1005–1018. doi: 10.1101/gad.6.6.1005. [DOI] [PubMed] [Google Scholar]
- 57.Moore AW, Jan LY, Jan YN. hamlet, a binary genetic switch between single- and multiple- dendrite neuron morphology. Science. 2002;297:1355–1358. doi: 10.1126/science.1072387. [DOI] [PubMed] [Google Scholar]
- 58.Moore AW, Roegiers F, Jan LY, Jan YN. Conversion of neurons and glia to external-cell fates in the external sensory organs of Drosophila hamlet mutants by a cousin-cousin cell-type respecification. Genes Dev. 2004;18:623–628. doi: 10.1101/gad.1170904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corty MM, Matthews BJ, Grueber WB. Molecules and mechanisms of dendrite development in Drosophila. Development. 2009;136:1049–1061. doi: 10.1242/dev.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao FB, Kohwi M, Brenman JE, Jan LY, Jan YN. Control of dendritic field formation in Drosophila: the roles of flamingo and competition between homologous neurons. Neuron. 2000;28:91–101. doi: 10.1016/s0896-6273(00)00088-x. [DOI] [PubMed] [Google Scholar]
- 63.Brenman JE, Gao FB, Jan LY, Jan YN. Sequoia, a tramtrack-related zinc finger protein, functions as a pan-neural regulator for dendrite and axon morphogenesis in Drosophila. Dev Cell. 2001;1:667–677. doi: 10.1016/s1534-5807(01)00072-7. [DOI] [PubMed] [Google Scholar]
- 64.Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- 65.Nagel J, Delandre C, Zhang Y, Forstner F, Moore AW, Tavosanis G. Fascin controls neuronal class-specific dendrite arbor morphology. Development. 2012;139:2999–3009. doi: 10.1242/dev.077800. [DOI] [PubMed] [Google Scholar]
- 66.Sulkowski MJ, Iyer SC, Kurosawa MS, Iyer EP, Cox DN. Turtle functions downstream of Cut in differentially regulating class specific dendrite morphogenesis in Drosophila. PLoS One. 2011;6:e22611. doi: 10.1371/journal.pone.0022611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iyer SC, Wang D, Iyer EP, Trunnell SA, Meduri R, Shinwari R, Sulkowski MJ, Cox DN. The RhoGEF trio functions in sculpting class specific dendrite morphogenesis in Drosophila sensory neurons. PLoS One. 2012;7:e33634. doi: 10.1371/journal.pone.0033634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cubelos B, Sebastian-Serrano A, Beccari L, Calcagnotto ME, Cisneros E, Kim S, Dopazo A, Alvarez-Dolado M, Redondo JM, Bovolenta P, et al. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 2010;66:523–535. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li W, Wang F, Menut L, Gao FB. BTB/POZ-zinc finger protein abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron. 2004;43:823–834. doi: 10.1016/j.neuron.2004.08.040. [DOI] [PubMed] [Google Scholar]
- 70.Sugimura K, Satoh D, Estes P, Crews S, Uemura T. Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein abrupt. Neuron. 2004;43:809–822. doi: 10.1016/j.neuron.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 71.Jinushi-Nakao S, Arvind R, Amikura R, Kinameri E, Liu AW, Moore AW. Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 72.Hattori Y, Sugimura K, Uemura T. Selective expression of Knot/Collier, a transcriptional regulator of the EBF/Olf-1 family, endows the Drosophila sensory system with neuronal class-specific elaborated dendritic patterns. Genes Cells. 2007;12:1011–1022. doi: 10.1111/j.1365-2443.2007.01107.x. [DOI] [PubMed] [Google Scholar]
- 73.Crozatier M, Vincent A. Control of multidendritic neuron differentiation in Drosophila: the role of Collier. Dev Biol. 2008;315:232–242. doi: 10.1016/j.ydbio.2007.12.030. [DOI] [PubMed] [Google Scholar]
- 74.Stewart A, Tsubouchi A, Rolls MM, Tracey WD, Sherwood NT. Katanin p60-like1 promotes microtubule growth and terminal dendrite stability in the larval class IV sensory neurons of Drosophila. J Neurosci. 2012;32:11631–11642. doi: 10.1523/JNEUROSCI.0729-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hattori Y, Usui T, Satoh D, Moriyama S, Shimono K, Itoh T, Shirahige K, Uemura T. Sensory-neuron subtype-specific transcriptional programs controlling dendrite morphogenesis: genome-wide analysis of Abrupt and Knot/Collier. Dev Cell. 2013;27:530–544. doi: 10.1016/j.devcel.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 76.Kim MD, Jan LY, Jan YN. The bHLH-PAS protein Spineless is necessary for the diversification of dendrite morphology of Drosophila dendritic arborization neurons. Genes Dev. 2006;20:2806–2819. doi: 10.1101/gad.1459706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye B, Kim JH, Yang L, McLachlan I, Younger S, Jan LY, Jan YN. Differential regulation of dendritic and axonal development by the novel Kruppel-like factor Dar1. J Neurosci. 2011;31:3309–3319. doi: 10.1523/JNEUROSCI.6307-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iyer EP, Cox DN. Laser capture microdissection of Drosophila peripheral neurons. J Vis Exp. 2010 doi: 10.3791/2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stone MC, Roegiers F, Rolls MM. Microtubules have opposite orientation in axons and dendrites of Drosophila neurons. Mol Biol Cell. 2008;19:4122–4129. doi: 10.1091/mbc.E07-10-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rolls MM. Neuronal polarity in Drosophila: sorting out axons and dendrites. Dev Neurobiol. 2011;71:419–429. doi: 10.1002/dneu.20836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Satoh D, Sato D, Tsuyama T, Saito M, Ohkura H, Rolls MM, Ishikawa F, Uemura T. Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat Cell Biol. 2008;10:1164–1171. doi: 10.1038/ncb1776. [DOI] [PubMed] [Google Scholar]
- 83.Zheng Y, Wildonger J, Ye B, Zhang Y, Kita A, Younger SH, Zimmerman S, Jan LY, Jan YN. Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat Cell Biol. 2008;10:1172–1180. doi: 10.1038/ncb1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 85.Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ori-McKenney KM, Jan LY, Jan YN. Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron. 2012;76:921–930. doi: 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kramer AP, Stent GS. Developmental arborization of sensory neurons in the leech Haementeria ghilianii. II. Experimentally induced variations in the branching pattern. J Neurosci. 1985;5:768–775. doi: 10.1523/JNEUROSCI.05-03-00768.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 89.Sugimura K, Yamamoto M, Niwa R, Satoh D, Goto S, Taniguchi M, Hayashi S, Uemura T. Distinct developmental modes and lesion-induced reactions of dendrites of two classes of Drosophila sensory neurons. J Neurosci. 2003;23:3752–3760. doi: 10.1523/JNEUROSCI.23-09-03752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 91.Neves G, Chess A. Dscam-mediated self- versus non-self-recognition by individual neurons. Cold Spring Harb Symp Quant Biol. 2004;69:485–488. doi: 10.1101/sqb.2004.69.485. [DOI] [PubMed] [Google Scholar]
- 92.Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 93.Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, Schmucker D. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 97.Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, Jan YN. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miura SK, Martins A, Zhang KX, Graveley BR, Zipursky SL. Probabilistic splicing of Dscam1 establishes identity at the level of single neurons. Cell. 2013;155:1166–1177. doi: 10.1016/j.cell.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488:517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Han C, Wang D, Soba P, Zhu S, Lin X, Jan LY, Jan YN. Integrins regulate repulsion-mediated dendritic patterning of drosophila sensory neurons by restricting dendrites in a 2D space. Neuron. 2012;73:64–78. doi: 10.1016/j.neuron.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim ME, Shrestha BR, Blazeski R, Mason CA, Grueber WB. Integrins establish dendrite-substrate relationships that promote dendritic self-avoidance and patterning in drosophila sensory neurons. Neuron. 2012;73:79–91. doi: 10.1016/j.neuron.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 103.Koike-Kumagai M, Yasunaga K, Morikawa R, Kanamori T, Emoto K. The target of rapamycin complex 2 controls dendritic tiling of Drosophila sensory neurons through the Tricornered kinase signalling pathway. EMBO J. 2009;28:3879–3892. doi: 10.1038/emboj.2009.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grueber WB, Sagasti A. Self-avoidance and tiling: Mechanisms of dendrite and axon spacing. Cold Spring Harb Perspect Biol. 2010;2:a001750. doi: 10.1101/cshperspect.a001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vonhoff F, Duch C. Tiling among stereotyped dendritic branches in an identified Drosophila motoneuron. J Comp Neurol. 2010;518:2169–2185. doi: 10.1002/cne.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Harris KL, Whitington PM. Pathfinding by sensory axons in Drosophila: substrates and choice points in early lch5 axon outgrowth. J Neurobiol. 2001;48:243–255. doi: 10.1002/neu.1054. [DOI] [PubMed] [Google Scholar]
- 107.Martin V, Mrkusich E, Steinel MC, Rice J, Merritt DJ, Whitington PM. The L1-type cell adhesion molecule Neuroglian is necessary for maintenance of sensory axon advance in the Drosophila embryo. Neural Dev. 2008;3:10. doi: 10.1186/1749-8104-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Steinel MC, Whitington PM. The atypical cadherin Flamingo is required for sensory axon advance beyond intermediate target cells. Dev Biol. 2009;327:447–457. doi: 10.1016/j.ydbio.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 109.Mrkusich EM, Flanagan DJ, Whitington PM. The core planar cell polarity gene prickle interacts with flamingo to promote sensory axon advance in the Drosophila embryo. Dev Biol. 2011;358:224–230. doi: 10.1016/j.ydbio.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 110.Merritt DJ, Whitington PM. Central projections of sensory neurons in the Drosophila embryo correlate with sensory modality, soma position, and proneural gene function. J Neurosci. 1995;15:1755–1767. doi: 10.1523/JNEUROSCI.15-03-01755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morikawa RK, Kanamori T, Yasunaga K, Emoto K. Different levels of the Tripartite motif protein, Anomalies in sensory axon patterning (Asap), regulate distinct axonal projections of Drosophila sensory neurons. Proc Natl Acad Sci U S A. 2011;108:19389–19394. doi: 10.1073/pnas.1109843108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Karim MR, Moore AW. Convergent local identity and topographic projection of sensory neurons. J Neurosci. 2011;31:17017–17027. doi: 10.1523/JNEUROSCI.2815-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zlatic M, Landgraf M, Bate M. Genetic specification of axonal arbors: atonal regulates robo3 to position terminal branches in the Drosophila nervous system. Neuron. 2003;37:41–51. doi: 10.1016/s0896-6273(02)01131-5. [DOI] [PubMed] [Google Scholar]
- 114.Zlatic M, Li F, Strigini M, Grueber W, Bate M. Positional cues in the Drosophila nerve cord: semaphorins pattern the dorso-ventral axis. PLoS Biol. 2009;7:e1000135. doi: 10.1371/journal.pbio.1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu Z, Sweeney LB, Ayoob JC, Chak K, Andreone BJ, Ohyama T, Kerr R, Luo L, Zlatic M, Kolodkin AL. A combinatorial semaphorin code instructs the initial steps of sensory circuit assembly in the Drosophila CNS. Neuron. 2011;70:281–298. doi: 10.1016/j.neuron.2011.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stork T, Bernardos R, Freeman MR. Analysis of glial cell development and function in Drosophila. Cold Spring Harb Protoc. 2012;2012:1–17. doi: 10.1101/pdb.top067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.von Hilchen CM, Beckervordersandforth RM, Rickert C, Technau GM, Altenhein B. Identity, origin, and migration of peripheral glial cells in the Drosophila embryo. Mech Dev. 2008;125:337–352. doi: 10.1016/j.mod.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 118.Sepp KJ, Schulte J, Auld VJ. Developmental dynamics of peripheral glia in Drosophila melanogaster. Glia. 2000;30:122–133. doi: 10.1002/(sici)1098-1136(200004)30:2<122::aid-glia2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 119.Sepp KJ, Auld VJ. Reciprocal interactions between neurons and glia are required for Drosophila peripheral nervous system development. J Neurosci. 2003;23:8221–8230. doi: 10.1523/JNEUROSCI.23-23-08221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yamamoto M, Ueda R, Takahashi K, Saigo K, Uemura T. Control of axonal sprouting and dendrite branching by the Nrg-Ank complex at the neuron-glia interface. Curr Biol. 2006;16:1678–1683. doi: 10.1016/j.cub.2006.06.061. [DOI] [PubMed] [Google Scholar]
- 121.Castro B, Barolo S, Bailey AM, Posakony JW. Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development. 2005;132:3333–3344. doi: 10.1242/dev.01920. [DOI] [PubMed] [Google Scholar]