Abstract
Myxococcus xanthus displays a form of surface motility known as social (S) gliding. It is mediated by the type IV pilus (T4P) and requires the exopolysaccharide (EPS) to function. It is clear that T4P retraction powers S motility. EPS on a neighboring cell or deposited on a gliding surface is proposed to anchor the distal end of a pilus and trigger T4P retraction at its proximal end. Inversely, T4P has been shown to regulate EPS production upstream of the Dif signaling pathway. Here we describe the isolation of two Tn insertions at the stk locus which had been known to play roles in cellular cohesion and formation of cell groups. An insertion in stkA (MXAN_3474) was identified based on its ability to restore EPS to a pilA deletion mutant. The stkA encodes a DnaK or Hsp70 homolog and it is upstream of stkB (MXAN_3475) and stkC (MXAN_3476). A stkB insertion was identified in a separate genetic screen because it eliminated EPS production of an EPS+ parental strain. Our results with in-frame deletions of these three stk genes indicated that the stkA mutant produced increased level of EPS while stkB and stkC mutants produced less EPS relative to the wild type. S motility and developmental aggregation were affected by deletions of stkA and stkB but only minimally by the deletion of stkC. Genetic epistasis indicated that StkA functions downstream of T4P but upstream of the Dif proteins as a negative regulator of EPS production in M. xanthus.
Keywords: Myxococcus xanthus, Type IV pilus (T4P), Dif pathway, Exopolysaccharide (EPS), Social motility, StkA/DnaK/Hsp70
Introduction
Myxococcus xanthus, a gram negative bacterium, exhibits complex social interactions during its life cycles (Yang & Higgs, 2014). When nutrients are plentiful, M. xanthus cells grow, divide and move over solid surfaces as social swarms in a vegetative growth cycle. Upon nutrient limitation, M. xanthus initiates a developmental cycle wherein cells aggregate on solid surfaces by their gliding motility. When these aggregates mature into multicellular fruiting bodies, rod-shaped vegetative cells within morph into spherical myxospores. These metabolically dormant myxospores can endure adverse environmental elements such as heat, desiccation and UV radiation. When conditions become conducive for growth, myxospores germinate to reenter the vegetative cycle. Both developmental fruiting and vegetative swarming are multicellular behaviors which make M. xanthus a good model to study social or cell–cell interactions.
M. xanthus uses two genetically and morphologically distinct surface motility systems to facilitate its vegetative swarming and developmental aggregation (Mauriello et al., 2010). The adventurous (A) gliding system enables cells to move even when they are well isolated from one another. Social (S) gliding, on the other hand, only functions when cells are in close proximity or in groups. The bacterial type IV pilus (T4P) is known as the engine whose retraction powers M. xanthus S motility and bacterial twitching (Mauriello et al., 2010). Besides T4P, M. xanthus S motility requires the extracellular or exo-polysaccharide (EPS) to function (Yang et al., 2014). Available evidence supports a model wherein EPS on a neighboring cell or a surface triggers the T4P of M. xanthus to retract to actualize S motility (Li et al., 2003). This model explains why the function of S motility requires both T4P and EPS as well as cell proximity on most surfaces examined.
The T4P or Pil proteins as well as the Dif pathway play key roles in EPS regulation in M. xanthus (Black, Xu & Yang, 2006; Yang et al., 2014). pilA and other T4P−pil mutants have been found to be EPS−. A pilT mutant, which is T4P+ with non-retractable T4P, is EPS+. Therefore, there is a positive correlation between the presence of T4P and EPS. Genes at the dif locus encode products related to bacterial chemotaxis proteins (Yang et al., 1998). DifA is homologous to the chemoreceptor MCP, DifC to the coupling protein CheW, and DifE to the histidine kinase CheA. Two additional proteins DifD and DifG are similar to the response regulator CheY and the phosphatase CheC, respectively. Null mutations in difA, difC and difE led to EPS− and those in difD and difG to EPS overproduction (Black & Yang, 2004; Yang et al., 2000). Evidence indicated that DifE is a protein kinase and that it forms a ternary signaling complex with DifA and DifC (Black et al., 2010; Yang & Li, 2005). DifD and DifG influence the signaling strength of this DifACE complex by diverting phosphates from the kinase. Genetic studies showed that the T4P functions upstream of the Dif signaling proteins in EPS regulation (Black, Xu & Yang, 2006). The current model proposes that T4P function as physical sensors of other cells nearby. This sensory information is then relayed to the Dif pathway downstream to promote EPS production.
Of additional relevance to this work are the genes at the stk and the che7 loci. A frameshift mutation in cheW7 (cheW7-1) in the che7 gene cluster (Zusman et al., 2007) restored EPS production to a difA deletion (ΔdifA) mutant (Black et al., 2009). That is, a ΔdifA single mutant is EPS− but a ΔdifA cheW7-1 double mutant is EPS+. The Che7 chemosensory system likely plays an accessory role in EPS regulation in M. xanthus because a cheW7 null mutation by itself does not impact EPS production in an otherwise wild-type (WT) background (Black et al., 2009). The stk locus had been identified previously because its mutations enhanced cellular cohesion in liquid culture and increased group formation at colony edges (Dana & Shimkets, 1993). Here we describe the isolation of two transposon insertions at the stk locus and the genetic characterization of stkA (MXAN_3474), stkB (MXAN_3475) and stkC (MXAN_3476). A stkA insertion was found to suppress the EPS defect of a ΔpilA mutant whereas a stkB insertion was found to eliminate EPS production of a ΔdifA cheW7-1 strain. StkA is homologous to DnaK and HSP70 as described previously (Weimer et al., 1998). StkB shares similarity with the sterol carrier protein 2 (SCP2) or nonspecific lipid-transfer protein (NSLTP) (Lev, 2010; Schroeder et al., 2007). StkC is a small protein with limited homology to PhaE (Goldman et al., 2006), an enzyme involved in polyhydroxyalkanoate synthesis (Han et al., 2010). In-frame deletions were constructed for all three stk genes and their mutants were studied phenotypically. stkB and stkC deletions led to intermediate phenotypes in EPS production, motility and fruiting body development. Both stkA insertion and deletion restored EPS production to a ΔpilA mutant, but they failed to do so to a ΔdifA strain. These results support a model wherein StkA functions downstream of T4P but upstream of the Dif pathway in the regulation of M. xanthus EPS production as a negative regulator. StkB and StkC are required for EPS production at the wild-type level and the absence of either reduced but did not eliminate EPS production.
Materials & Methods
Bacterial strains and growth conditions
Escherichia coli DH5α was used for plasmid constructions while DH5αλpir was used to clone transposon insertions from M. xanthus mutants. They were grown and maintained on Luria Bertani (LB) agar plates or in LB liquid medium (Sambrook & Russell, 2001). M. xanthus strains used in this study are listed in Table 1 and were grown and maintained on Casitone yeast extract (CYE) agar plates or in its liquid form (Campos & Zusman, 1975). Clone-fruiting (CF) agar plates were used to examine fruiting body development (Hagen, Bretscher & Kaiser, 1978). Plates for general use contained 1.5% agar. Soft agar plates, which were used to examine S motility, contained 0.4% agar (Shi & Zusman, 1993). Whenever necessary, kanamycin and oxytetracycline were supplemented to media at 100 µg/ml and 15 µg/ml, respectively (Bellenger et al., 2002; Black & Yang, 2004).
Table 1. Strains and plasmids.
Plasmids and M. xanthus strains used in this study.
| Plasmid | Genotype or description | Source |
|---|---|---|
| pBJ113 | M. xanthus gene replacement vector | (Julien, Kaiser & Garza, 2000) |
| pBJ114 | M. xanthus gene replacement vector | (Julien, Kaiser & Garza, 2000) |
| pLZ407 | stkA in-frame deletion (ΔstkA) in pBJ113 | This study |
| pLZ429 | stkB in-frame deletion (ΔstkB) in pBJ114 | This study |
| pAM108 | stkC in-frame deletion (ΔstkC) in pBJ113 | This study |
| pMycoMar | magellan4 mutagenesis vector | (Rubin et al., 1999) |
| M. xanthus strain | ||
| BY129 | stkB1::Tn in YZ101 | This study |
| BY801 | ΔpilA stkA2::Tn | This study |
| BY1129 | stkB1::Tn | This study |
| BY1801 | stkA2::Tn | This study |
| DK1622 | Wild type (WT) | (Kaiser, 1979) |
| DK10407 | ΔpilA::Tet | (Wall & Kaiser, 1998) |
| YZ101 | ΔdifA cheW7-1 (ΔdifA suppressor strain) | (Black et al., 2009) |
| YZ601 | ΔdifA | (Xu et al., 2005) |
| YZ603 | ΔdifE | (Black & Yang, 2004) |
| YZ690 | ΔpilA | This study |
| YZ812 | ΔstkA | This study |
| YZ813 | ΔstkB | This study |
| YZ901 | ΔpilA ΔstkA | This study |
| YZ910 | ΔstkC | This study |
| YZ932 | ΔdifA ΔstkA | This study |
Transposon mutagenesis and identification of transposon insertions
Transposon mutagenesis was performed using the mariner-based magellan4 (Rubin et al., 1999) as previously described (Black et al., 2009). pMycoMar (containing magellan4) (Table 1) (Rubin et al., 1999) was electroporated into YZ101 (ΔdifA cheW7-1) or DK10407 (ΔpilA). Cells were allowed to recover for 4 h and plated on CYE plates with Congo Red at 30 µg/ml. About 20,000 colonies were visually screened for EPS phenotypes after 5–7 days of incubation at 32 °C (Black et al., 2009).
The site of a Tn insertion in a mutant of interest was identified by cloning and DNA sequencing as has been described (Black et al., 2009). Briefly, genomic DNA from a mutant was digested with SacII (New England Biolabs) and allowed to self ligate. The ligation was transformed into DH5αλpir. Two primers, MarR1 and/or MarL1 (Youderian et al., 2003) were used to sequence the plasmids that were recovered from the transformant.
Construction of plasmids and strains
Plasmids used in this study are listed in Table 1. In-frame deletion alleles of stk genes were constructed using a two-step overlap PCR procedure (Sambrook & Russell, 2001). PCR products with the in-frame deletion alleles of stkA and stkC were blunt-end ligated into SmaI-digested pBJ113 (Julien, Kaiser & Garza, 2000) create pLZ407 and pAM108, respectively. The PCR product with the stkB in-frame deletion was similarly ligated into SmaI-restricted pBJ114 (Julien, Kaiser & Garza, 2000) to create pLZ429. The mutant alleles in pLZ407, pLZ429 and pAM108 deleted codons 5 to 535 for StkA, 5 to 108 for StkB and 5 to 85 for StkC, respectively.
A two-step procedure (Ueki, Inouye & Inouye, 1996) was performed to construct deletions of chromosomal stk genes. The three plasmids above were used to delete stkA, stkB andstkC from the WT strain (DK1622) to construct YZ812 (ΔstkA), YZ813 (ΔstkB) and YZ910 (ΔstkC) , respectively. In addition, pLZ407 was used to delete stkA from YZ690 (ΔpilA) and YZ601 (ΔdifA) to create strains YZ901 (ΔpilA ΔstkA) and YZ932 (ΔdifA ΔstkA).
Examination of EPS production
EPS production was examined by two different assays: one qualitative and one quantitative. The qualitative assay utilized plates with 50 µg/ml calcofluor white (CW), a fluorescent dye that binds to EPS (Black & Yang, 2004; Dana & Shimkets, 1993). Cells from overnight cultures were pelleted and resuspended in MOPS (morpholinepropanesulfonic acid) buffer (10 mM MOPS [pH 7.6], 2 mM MgSO4) at approximately 5 × 109 cells/ml. Then, 5 µl of the suspension were spotted onto CYE plates with CW and incubated at 32 °C for 6 days. Fluorescence under long-wavelength (∼365 nm) UV illumination was directly photographed with a digital camera (Black et al., 2009). The binding of trypan blue was used to quantify EPS in a liquid assay (Black & Yang, 2004). Cultures grown overnight in CYE to ∼ 3.5 × 108 cells/ml were harvested, washed and re-suspended to approximately 2.8 × 108 cells/ml in MOPS buffer with 5 µg/ml trypan blue. The control samples contained trypan blue in MOPS buffer without cells. The samples were vortexed and incubated with shaking at 300 rpm at 25 °C for 30 min. The absorbance of the supernatants after centrifugation was measured at 585 nm. EPS production of all strains was normalized to that of the WT strain which was arbitrarily set as 1 (Dana & Shimkets, 1993). Quantitative experiments with trypan blue were repeated at least three times with each sample analyzed in triplicates and a representative data set is shown in the paper.
Examination of motility and fruiting body development
Motility was examined by placing 5 µl of the cell suspension at 5 × 109 cells/ml onto the center of a standard (1.5% agar) or soft (0.4% agar) CYE plate. The standard agar plates were examined after 2 days and the soft agar plates after 5 days of incubation at 32 °C (Black & Yang, 2004; Shi & Zusman, 1993). For the examination of fruiting body formation, overnight cultures were harvested and resuspended in MOPS buffer at 5 × 109 cells/ml. Then, 5 µl of the suspension were spotted onto CF agar plates and development was observed after 5 days of incubation at 32 °C.
Results
Isolation of two M. xanthus transposon mutants with altered EPS production
To identify genes involved in the regulation and/or production of EPS, two genetic screens were carried out to search for mutants with altered EPS levels. In the first screen, a pilA deletion (ΔpilA) strain (DK10407), which is T4P− and EPS− (Black, Xu & Yang, 2006), was mutagenized by a transposon (Tn) and mutants were allowed to form colonies on agar plates with the dye Congo red (CR). M. xanthus EPS+ colonies appear red and EPS− ones are yellowish orange on these plates (See ‘Materials & Methods’). Among approximately 20,000 colonies screened, BY801 and BY802 were found to form red colonies, indicating that they contained suppressors of the pilA deletion. BY801 is discussed here and the work on BY802 has been published elsewhere (Wallace et al., 2014).
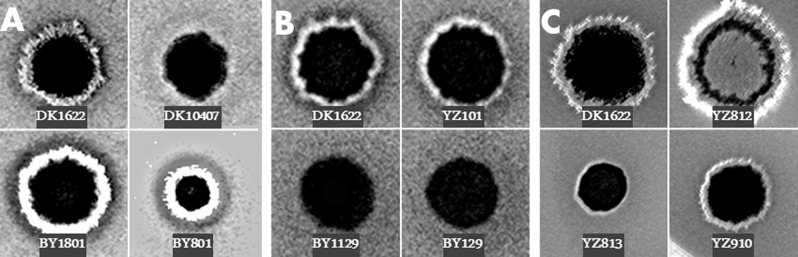
The suppressor phenotype of BY801 and its link to the Tn were confirmed by an alternative EPS assay and genetic linkage analysis, respectively. As shown in Fig. 1A, BY801 was verified to be EPS+ as indicated by the fluorescence on a plate containing the dye Calcofluor white (CW). The Tn insertion in BY801 was transferred to the parental ΔpilA mutant by genomic DNA transformation (Vlamakis, Kirby & Zusman, 2004). Sixteen of the resulting transformants were examined and all were found to be EPS+. This established that a single Tn insertion locus in BY801 was responsible for ΔpilA suppression instead of any additional mutations elsewhere. When the Tn insertion was transferred to the WT background, the resulting strain BY1801 showed enhanced EPS production as indicated by increased CW binding in comparison with the wild type (WT) (Fig. 1A). These observations demonstrate that the Tn insertion in BY801 altered the function of a gene or genes important for M. xanthus EPS production and/or regulation. It should be noted that despite its ability to produce EPS, the colonies of BY801 differ from those of the WT (Fig. 1A) because the latter is S+ while BY801 is S− without pilA.
Figure 1. EPS production of stk mutants.
Five microliter aliquots of cell suspension at 5 × 109 cells/ml of an indicated strain were spotted onto CYE plates with the fluorescent dye calcofluor white (CW) and florescence was documented under UV illumination after 6 days of incubation at 32 °C (See ‘Materials & Methods’). (A) stkA insertion suppressed pilA deletion. (B) stkB insertion results in EPS defect. (C) Deletions of stk genes affect EPS levels to different extent. Strains: DK1622 (wild type), DK10407 (ΔpilA), BY801 (ΔpilA stkA2::Tn), BY1801 (stkA2::Tn), YZ101 (ΔdifA cheW7-1), BY129 (ΔdifA cheW7-1 stkB1::Tn), BY 1129 (stkB1::Tn), YZ812 (ΔstkA), YZ813 (ΔstkB), and YZ910 (ΔstkC).
In the second genetic screen, the EPS+ mutant YZ101 (cheW7-1 ΔdifA) (Black et al., 2009) was used for Tn mutagenesis to identify additional genes critical for M. xanthus EPS production. About 70 EPS− mutants were identified from approximately 20,000 colonies on CR plates. Some of these mutants were reported previously (Black et al., 2009; Lu et al., 2005). BY129, which showed no obvious CR binding in the initial screen, is described here. Examination on plates with CW confirmed that BY129 has negligible EPS levels in comparison with its parent and the WT (Fig. 1B). When the insertion was re-introduced into YZ101 by genomic DNA transformation, the resulting transformants displayed the same EPS− phenotype as BY129 (not shown). When the Tn insertion was introduced into the WT strain, the resulting mutant BY1129 did not bind CW in plate assays (Fig. 1B). The gene(s) mutated by the Tn insertion in BY129 must play a role in EPS production and/or reguation in M. xanthus.
Transposons in BY801 and BY129 inserted in two adjacent genes at the stk locus
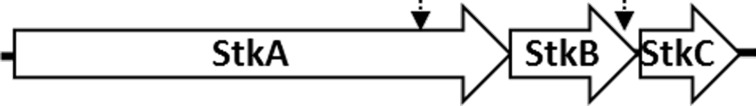
The Tn insertions in BY801 and BY129 were identified as previously described (Black et al., 2009). In BY801, the insertion occured in MXAN_3474, a gene known as stk because of the sticky phenotype of its mutant (Dana & Shimkets, 1993; Kim, Ramaswamy & Downard, 1999; Weimer et al., 1998) (Fig. 2). This gene will be designated as stkA and the insertion mutation here as stkA2::Tn hereafter (Table 1). stkA encodes a DnaK homologue of 540 amino acids (AAs) (Goldman et al., 2006) that was not found to be induced by heat shock (Otani et al., 2001). The stkA2::Tn insertion occurred in the 440th codon of stkA after a TA dinucleotide. In BY129, the Tn inserted into MXAN_3475, an open reading frame (ORF) of 141 codons 8 base pairs (bps) downstream of stkA. This ORF will be designated as StkB and the mutation as stkB1::Tn hereafter. StkB belongs to the superfamily of the sterol carrier protein 2 (SCP2) or nonspecific lipid-transfer protein (NSLTP) (Lev, 2010; Schroeder et al., 2007). Some members of this protein superfamily function in cholesterol trafficking and lipid metabolism as well as cell signaling in a variety of organisms (Lev, 2010; Schroeder et al., 2007). The stkB1::Tn insertion occurred after a TA dinucleotide in the 122nd codon. 12 bps downstream of stkB is MXAN_3476 (Goldman et al., 2006) or stkC. It encodes a protein of 89 AAs with limited homology to PhaE (Goldman et al., 2006), a polyhydroxyalkanoate synthetic enzyme (Han et al., 2010). The isolation of Tn insertions at the stk locus from independent genetic screens here and elsewhere (Dana & Shimkets, 1993) indicates that the stk genes are critical players in EPS production in M. xanthus.
Figure 2. M. xanthus stk locus X.
The stk region shown is 2.54 kb with the ORFs of StkA, StkB and StkC indicated by open arrows approximately to scale. The inverted arrows indicate positions of Tn insertions in stkA and stkB in BY801 and BY129, respectively.
ΔstkA produces more EPS while ΔstkB and ΔstkC produce less
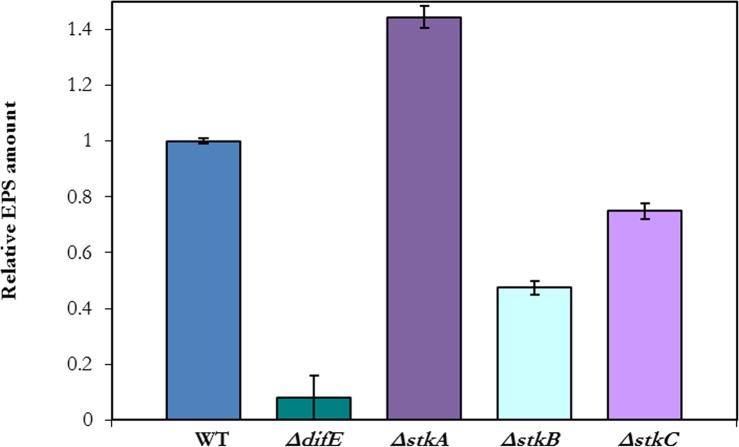
The transposons in BY801 and BY129 inserted at the 3′ ends of stkA and stkB, respectively (Fig. 2). As truncated StkA and StkB may retain part of their functions, these insertions could be leaky or even gain-of-function mutations. In addition, stkA, stkB and stkC may form an operon (Fig. 2) and both insertions could be polar on downstream genes. The previous stkA mutant harbors a Tn insertion as well (Dana & Shimkets, 1993). To clarify the roles of the stk genes in EPS production, in-frame deletions of these genes were constructed (See ‘Material & Methods’). YZ812, YZ813 and YZ910 deleted stkA, stkB and stkC, respectively (Table 1). EPS production by these strains was examined by CW binding (Fig. 1C); the ΔstkA strain exhibited more whereas ΔstkB and ΔstkC exhibited less fluorescence than the WT in this assay. In addition, EPS levels of the stk deletion mutants were quantified by a liquid dye binding assay (Black & Yang, 2004). As show in Fig. 3, the ΔstkA mutant increased EPS production over the wild type by about 50%. The ΔstkB and ΔstkC mutants produced about 50% and 25% less than the WT, respectively. The results in Figs. 1C and 3 with the in-frame deletion mutants clearly implicate stk genes in M. xanthus EPS production. StkA is likely a negative regulator as its absence results in EPS overproduction. The roles of StkB and StkC are less clear, as the deletion of their genes led to intermediate EPS phenotypes. Phenotypic comparisons also indicate that the stkB insertion in BY129 and BY1129 is likely polar because it led to a more severe EPS defect than the deletion of either stkB or stkC. There are two additional ORFs (MXAN_3471 and MXAN_3472) upstream of and in the same orientation as StkA (Goldman et al., 2006). Their in-frame deletions resulted in no alteration in M. xanthus EPS production or any other phenotype examined (results not shown) and these two genes are not discussed in this manuscript.
Figure 3. The stkA mutant produced more EPS whereas the stkB and stkC mutants produced less.
EPS levels were quantified by a trypan blue binding assay (See ‘Materials & Methods’). The amount of EPS for each strain was compared to that of the WT (DK1622) which was normalized to a value of one. Other strains are YZ812 (ΔstkA), YZ813 (ΔstkB), and YZ910 (ΔstkC) with the EPS− strain YZ603 (ΔdifE) as a control.
StkA functions downstream of PilA but upstream of Dif Proteins
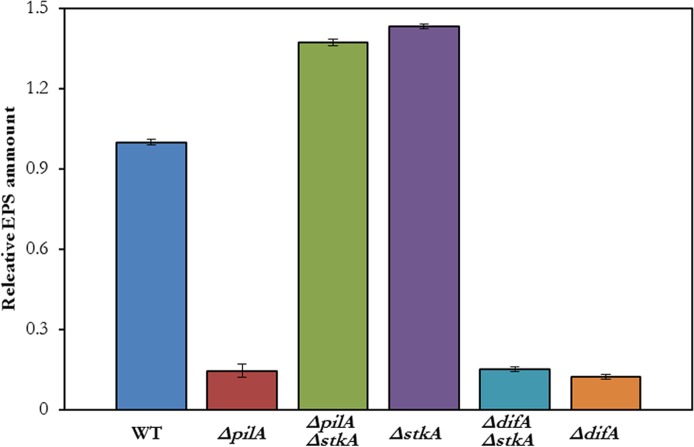
EPS production is regulated in part by the Dif pathway (Black, Xu & Yang, 2006; Yang et al., 2014). T4P are proposed to perceive and relay signals downstream to Dif proteins to promote EPS production (Black, Xu & Yang, 2006). The relationship of StkA with Dif was examined by the construction of a ΔdifA ΔstkA mutant. In addition, a ΔpilA ΔstkA mutant was constructed to confirm the suppression of ΔpilA by the stkA deletion. As shown in Fig. 4, the ΔpilA ΔstkA double mutant (YZ901) produced more EPS similarly as the Δ stkA single mutant (YZ812), indicating the suppression of ΔpilA by stkA null mutations. On the other hand, the ΔdifA ΔstkA double mutant (YZ932) appeared similar to the ΔdifA single mutant (YZ601) with both producing very little EPS (Fig. 4). The finding that the ΔstkA mutation is epistatic to a ΔpilA but not a ΔdifA mutation led to the conclusion that StkA functions between T4P and the Dif pathway in the regulation of EPS production in M. xanthus.
Figure 4. ΔstkA suppresses ΔpilA but not ΔdifA.
EPS levels were quantified by a trypan blue binding assay as in Fig. 4. The strains are DK1622 (WT), YZ690 (ΔpilA), YZ901 (ΔpilA ΔstkA), YZ812 (ΔstkA), YZ932 (ΔdifA ΔstkA) and YZ601 (ΔdifA).
stk mutants show defects in motility
The surface motility of the stk deletions were examined first on regular agar plates (1.5% agar) which allow both A and S motility to contribute (Shi & Zusman, 1993) (Fig. 5A). Compared to the WT, the colony of the ΔstkC mutant (YZ910) appears only slightly smaller, consistent with the slight effect of the stkC deletion on EPS levels (Figs. 1C and 3). The colony morphology of the ΔstkC mutant was also highly similar to that of WT in its yellow pigmentation, high opacity as well as its rough surface and jagged edges. The ΔstkA and especially ΔstkB mutants showed a more diminished ability to spread on hard agar surfaces as their swarming colonies appeared smaller than that of the WT. The surface of the ΔstkA mutant colony is rougher and more elevated than that of the WT. The colony of ΔstkB is smoother, glossier and flatter than the WT. These observations are consistent with the ΔstkA mutant overproducing and the ΔstkB mutants significantly underproducing EPS in comparison with the WT.
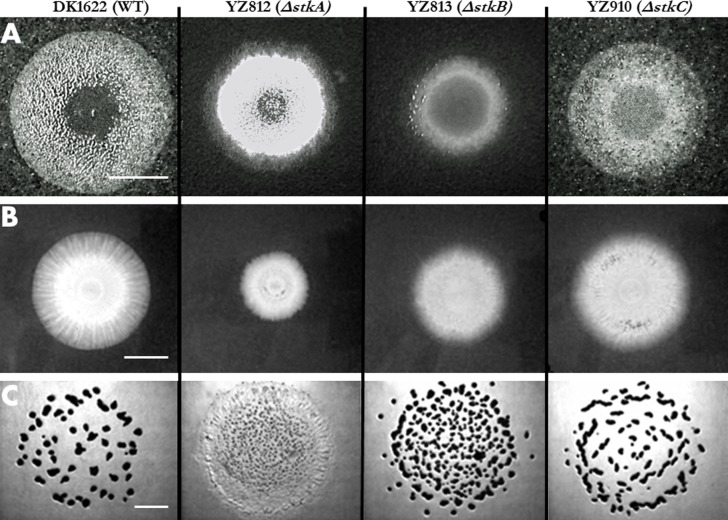
Figure 5. Motility and developmental aggregation of stk mutants.
A 5 µl aliquot of the cell suspension at 5 × 109 cells/ml for each strain were plated in the center of a CYE plate with 1.5% (A) or 0.4% agar (B) to examine motility. The same amount of cells of a strain was spotted onto a CF plate to examine development (C). Results were documented after incubation at 32 °C (See Materials and Methods). The scale bars in all three panels represent 1 cm. Indicated on the top of the figure are the strains used for all three panels: DK1622 (WT), YZ812 (ΔstkA), YZ813 (ΔstkB), and YZ910 (ΔstkC).
Because EPS is essential for the T4P-mediated S motility, plates with 0.4% agar (soft agar plates) were used to examine S motility more specifically (Shi & Zusman, 1993) (Fig. 5B). The size of the swarming colony of the ΔstkC mutant is similar to that of the WT, indicating no obvious S-motility defect. The spreading of ΔstkB and ΔstkA mutants, especially the latter, was defective in comparison with the WT. Interestingly, except the size, the colony morphology of the ΔstkA mutant more closely resembled that of the WT. There were obvious swarming zones or rings for strains of the WT and the ΔstkA, but not for those of the ΔstkB and the ΔstkC. Overall, these results indicate that StkA and StkB are more important players in S-motility whereas StkC may influence the organization of swarms but not the overall rate of swarming by S motility.
The stk mutants show defects in developmental aggregation consistent with their EPS phenotypes
The stk mutants were examined for development under nutrient limitation (Fig. 5C). With respect to both fruiting body morphology and the completeness of aggregation, the ΔstkA mutant was the most defective followed by ΔstkB and ΔstkC. While some of the aggregates of ΔstkC are elongated, their distribution and number are the most similar to those of the WT. The aggregates formed by the ΔstkB mutant darkened as those of the WT, but they appeared less well organized and more variable in size and number. While the ΔstkA mutant showed signs of aggregation in the center of the bacterial lawn, these aggregates are smaller and more numerous. On the edge of the lawn, ΔstkA cells appeared to move outward with no signs of aggregation. These results are consistent with the varying degrees of EPS defects of stk mutants as ΔstkA had the most severe EPS phenotype, followed by ΔstkB and ΔstkC.
Discussion & Conclusions
To summarize, two stk insertion mutants were isolated in two separate genetic screens based on their altered EPS phenotypes. Further analysis indicated that both a stkA in-frame deletion and an insertion resulted in EPS overproduction in the WT background. While stkA mutations suppressed ΔpilA in EPS regulation, they failed to restore EPS production to a ΔdifA mutant. Both stkB and stkC deletions resulted in varying reductions in EPS production and surface motility, consistent with the correlation between EPS and motility observed previously (Xu et al., 2005). These results established that the three genes at the stk locus are important for EPS production in M. xanthus, albeit to different degrees. The observation that the stkA mutant displayed reduced swarming by S motility (Fig. 5B) indicates that optimal S motility requires a fine balance or a tight regulation of EPS production; too little or too much apparently results in reduced efficiency of spreading through S motility (Xu et al., 2005).
StkA is a member of the Hsp70 protein family (Weimer et al., 1998). The prototype Hsp70 is the E. coli chaperone DnaK (Genevaux, Georgopoulos & Kelley, 2007). It functions as part of a molecular machine with DnaJ and GrpE, its partners or co-chaperones, to facilitate the folding of nascent polypeptides and the refolding of denatured or misfolded proteins. These proteins are induced by heat shock and confer thermotolerance to E. coli once induced. Multiple lines of evidence indicate that StkA is a negative regulator of EPS production in M. xanthus. Previously, StkA was not found to be induced by heat shock (Otani et al., 2001) and thus not a typical bacterial Hsp70. Instead, our genetic epistasis results here support a model wherein StkA functions as a negative regulator downstream of T4P but upstream of the Dif chemotaxis protein in the EPS regulatory pathway. Previous results left little doubt that StkA is critical for the production of fibrils (Dana & Shimkets, 1993), of which EPS is a major constituent (Behmlander & Dworkin, 1994). The results here demonstrate that StkA itself is a negative regulator of EPS and lies downstream of T4P and upstream of Dif in the EPS regulatory pathway in M. xanthus.
As a component of the EPS signaling pathway, StkA may modulate the function of other EPS regulators in a chaperone-like capacity or it may act directly as a signaling protein in an unknown manner. In this context, it is noted that SglK, another M. xanthus Hsp70 homologue, has the opposite function in EPS regulation when compared with StkA (Weimer et al., 1998; Yang, Geng & Shi, 1998). That is, a sglK mutant is EPS− and has no S motility. It is surprising that there are 15 Hsp70-like proteins encoded by the M. xanthus genome (Goldman et al., 2006). This is in contrast to E. coli which codes three Hsp70 members on its genome (Genevaux, Georgopoulos & Kelley, 2007). Besides the canonical heat shock protein DnaK, HscA and HscC are the other two Hsp70-like proteins in this enteric bacterium. HscA is a specialized chaperone that facilitates the assembly and maturation of iron-sufur [Fe-S] proteins. HscC appears to be involved in response to more general stress including UV exposure through mechanisms that is not entirely clear. If StkA functions as a chaperone like HscA, it may facilitate a negative regulator of EPS to attain or maturate to its native and active state. If StkA is a signaling protein, it may function in a similar fashion as Ssz1, a regulatory Hsp70 in yeast (Prunuske et al., 2012). The identification of the direct target of StkA will provide insights into the mechanisms of EPS regulation by this member of Hsp70 proteins in M. xanthus.
It is unclear whether StkB and StkC function in a regulatory or a biosynthetic capacity. In comparison with the stkB and the stkC deletions, the stkB1::Tn mutant has a more severe EPS phenotype (Fig. 1). This suggests that the stkB1::Tn mutation is polar on stkC (Fig. 2) and that StkB and StkC have overlapping or redundant functions. The homology of StkC to PhaE (Han et al., 2010) may be taken as circumstantial evident that StkC as well as StkB are EPS biosynthetic enzymes. However, the homology of StkB to NSLTPs (Lev, 2010; Schroeder et al., 2007) leads to ambiguities on whether there is indeed an overlapping function for these two proteins. NSLTPs are involved not only in lipid metabolism but also in cell signaling (Lev, 2010; Schroeder et al., 2007), which could mean both function in cell signaling instead of the biosynthetic process. Further investigations are necessary to better understand the roles of StkB and StkC in M. xanthus EPS production.
Acknowledgments
RAW was partially supported by the Post Baccalaureate Research and Education Program (PREP) and the Multicultural Academic Opportunities Program (MAOP) at Virginia Tech.
Funding Statement
This work was partially supported by the National Science Foundation Grant MCB-1417726, the National Institute of Health Grant GM071601, and the Fralin Life Science Institute to Zhaomin Yang. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Pamela L. Moak conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Wesley P. Black conceived and designed the experiments, performed the experiments, reviewed drafts of the paper.
Regina A. Wallace performed the experiments, prepared figures and/or tables, reviewed drafts of the paper.
Zhuo Li performed the experiments.
Zhaomin Yang conceived and designed the experiments, analyzed the data, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
References
- Behmlander & Dworkin (1994).Behmlander RM, Dworkin M. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. Journal of Bacteriology. 1994;176:6295–6303. doi: 10.1128/jb.176.20.6295-6303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellenger et al. (2002).Bellenger K, Ma X, Shi W, Yang Z. A CheW homologue is required for Myxococcus xanthus fruiting body development, social gliding motility, and fibril biogenesis. Journal of Bacteriology. 2002;184:5654–5660. doi: 10.1128/JB.184.20.5654-5660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black et al. (2010).Black WP, Schubot FD, Li Z, Yang Z. Phosphorylation and dephosphorylation among Dif chemosensory proteins essential for exopolysaccharide regulation in Myxococcus xanthus. Journal of Bacteriology. 2010;192:4267–4274. doi: 10.1128/JB.00403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black et al. (2009).Black WP, Xu Q, Cadieux CL, Suh S-J, Shi W, Yang Z. Isolation and characterization of a suppressor mutation that restores Myxococcus xanthus exopolysaccharide production. Microbiology. 2009;155:3599–3610. doi: 10.1099/mic.0.031070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, Xu & Yang (2006).Black WP, Xu Q, Yang Z. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Molecular Microbiology. 2006;61:447–456. doi: 10.1111/j.1365-2958.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- Black & Yang (2004).Black WP, Yang Z. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. Journal of Bacteriology. 2004;186:1001–1008. doi: 10.1128/JB.186.4.1001-1008.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos & Zusman (1975).Campos JM, Zusman DR. Regulation of development in Myxococcus xanthus: effect of 3’:5’-cyclic AMP, ADP, and nutrition. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:518–522. doi: 10.1073/pnas.72.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana & Shimkets (1993).Dana JR, Shimkets LJ. Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. Journal of Bacteriology. 1993;175:3636–3647. doi: 10.1128/jb.175.11.3636-3647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevaux, Georgopoulos & Kelley (2007).Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Molecular Microbiology. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- Goldman et al. (2006).Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, Halling C, Hinkle G, Iartchuk O, Kim HS, Mackenzie C, Madupu R, Miller N, Shvartsbeyn A, Sullivan SA, Vaudin M, Wiegand R, Kaplan HB. Evolution of sensory complexity recorded in a myxobacterial genome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, Bretscher & Kaiser (1978).Hagen DC, Bretscher AP, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Developmental Biology. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Han et al. (2010).Han J, Hou J, Liu H, Cai S, Feng B, Zhou J, Xiang H. Wide distribution among halophilic archaea of a novel polyhydroxyalkanoate synthase subtype with homology to bacterial type III synthases. Applied and Environmental Microbiology. 2010;76:7811–7819. doi: 10.1128/AEM.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien, Kaiser & Garza (2000).Julien B, Kaiser AD, Garza A. Spatial control of cell differentiation in Myxococcus xanthus. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser (1979).Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Ramaswamy & Downard (1999).Kim SH, Ramaswamy S, Downard J. Regulated exopolysaccharide production in Myxococcus xanthus. Journal of Bacteriology. 1999;181:1496–1507. doi: 10.1128/jb.181.5.1496-1507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev (2010).Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nature Reviews Molecular Cell Biology. 2010;11:739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- Li et al. (2003).Li Y, Sun H, Ma X, Lu A, Lux R, Zusman D, Shi W. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5443–5448. doi: 10.1073/pnas.0836639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2005).Lu A, Cho K, Black WP, Duan XY, Lux R, Yang Z, Kaplan HB, Zusman DR, Shi W. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Molecular Microbiology. 2005;55:206–220. doi: 10.1111/j.1365-2958.2004.04369.x. [DOI] [PubMed] [Google Scholar]
- Mauriello et al. (2010).Mauriello EM, Mignot T, Yang Z, Zusman DR. Gliding motility revisited: how do the myxobacteria move without flagella? Microbiology and Molecular Biology Reviews. 2010;74:229–249. doi: 10.1128/MMBR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani et al. (2001).Otani M, Tabata J, Ueki T, Sano K, Inouye S. Heat-shock-induced proteins from Myxococcus xanthus. Journal of Bacteriology. 2001;183:6282–6287. doi: 10.1128/JB.183.21.6282-6287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunuske et al. (2012).Prunuske AJ, Waltner JK, Kuhn P, Gu B, Craig EA. Role for the molecular chaperones Zuo1 and Ssz1 in quorum sensing via activation of the transcription factor Pdr1. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:472–477. doi: 10.1073/pnas.1119184109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin et al. (1999).Rubin EJ, Akerley BJ, Novik VN, Lampe DJ, Husson RN, Mekalanos JJ. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook & Russell (2001).Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd edition. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schroeder et al. (2007).Schroeder F, Atshaves BP, McIntosh AL, Gallegos AM, Storey SM, Parr RD, Jefferson JR, Ball JM, Kier AB. Sterol carrier protein-2: new roles in regulating lipid rafts and signaling. BBA Molecular and Cell Biology of Lipids. 2007;1771:700–718. doi: 10.1016/j.bbalip.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi & Zusman (1993).Shi W, Zusman DR. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki, Inouye & Inouye (1996).Ueki T, Inouye S, Inouye M. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene. 1996;183:153–157. doi: 10.1016/S0378-1119(96)00546-X. [DOI] [PubMed] [Google Scholar]
- Vlamakis, Kirby & Zusman (2004).Vlamakis HC, Kirby JR, Zusman DR. The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Molecular Microbiology. 2004;52:1799–1811. doi: 10.1111/j.1365-2958.2004.04098.x. [DOI] [PubMed] [Google Scholar]
- Wall & Kaiser (1998).Wall D, Kaiser D. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3054–3058. doi: 10.1073/pnas.95.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace et al. (2014).Wallace RA, Black WP, Yang X, Yang Z. A CRISPR with roles in Myxococcus xanthus development and Exopolysaccharide production. Journal of Bacteriology. 2014;196:4036–4043. doi: 10.1128/JB.02035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer et al. (1998).Weimer RM, Creighton C, Stassinopoulos A, Youderian P, Hartzell PL. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. Journal of Bacteriology. 1998;180:5357–5368. doi: 10.1128/jb.180.20.5357-5368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu et al. (2005).Xu Q, Black WP, Ward SM, Yang Z. Nitrate-dependent activation of the Dif signaling pathway of Myxococcus xanthus mediated by a NarX-DifA interspecies chimera. Journal of Bacteriology. 2005;187:6410–6418. doi: 10.1128/JB.187.18.6410-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Geng & Shi (1998).Yang Z, Geng Y, Shi W. A DnaK homolog in Myxococcus xanthus is involved in social motility and fruiting body formation. Journal of Bacteriology. 1998;180:218–224. doi: 10.1128/jb.180.2.218-224.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (1998).Yang Z, Geng Y, Xu D, Kaplan HB, Shi W. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Molecular Microbiology. 1998;30:1123–1130. doi: 10.1046/j.1365-2958.1998.01160.x. [DOI] [PubMed] [Google Scholar]
- Yang & Higgs (2014).Yang Z, Higgs PI. Myxobacteria. Norfolk: Caister Academic Press; 2014. p. 235. [Google Scholar]
- Yang & Li (2005).Yang Z, Li Z. Demonstration of interactions among Myxococcus xanthus Dif chemotaxis-like proteins by the yeast two-hybrid system. Archives of Microbiology. 2005;183:243–252. doi: 10.1007/s00203-005-0767-8. [DOI] [PubMed] [Google Scholar]
- Yang et al. (2014).Yang Z, Li CY, Friedrich C, Sogaard-Andersen L. Type IV pili and exopolysaccharide-dependent motility in Myxococcus xanthus. In: Yang Z, Higgs PI, editors. Myxobacteria: genomics, cellular and molecular biology. Norfolk: Caister Academic Press; 2014. pp. 183–198. Chapter 10. [Google Scholar]
- Yang et al. (2000).Yang Z, Ma X, Tong L, Kaplan HB, Shimkets LJ, Shi W. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. Journal of Bacteriology. 2000;182:5793–5798. doi: 10.1128/JB.182.20.5793-5798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian et al. (2003).Youderian P, Burke N, White DJ, Hartzell PL. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Molecular Microbiology. 2003;49:555–570. doi: 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- Zusman et al. (2007).Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nature Reviews Microbiology. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]