Abstract
A novel exonuclease, designated as MszExo I, was cloned from Methylocaldum szegediense, a moderately thermophilic methanotroph. It specifically digests single-stranded DNA in the 3ʹ to 5ʹ direction. The protein is composed of 479 amino acids, and it shares 47% sequence identity with E. coli Exo I. The crystal structure of MszExo I was determined to a resolution of 2.2 Å and it aligns well with that of E. coli Exo I. Comparative studies revealed that MszExo I and E. coli Exo I have similar metal ion binding affinity and similar activity at mesophilic temperatures (25–47°C). However, the optimum working temperature of MszExo I is 10°C higher, and the melting temperature is more than 4°C higher as evaluated by both thermal inactivation assays and DSC measurements. More importantly, two thermal transitions during unfolding of MszExo I were monitored by DSC while only one transition was found in E. coli Exo I. Further analyses showed that magnesium ions not only confer structural stability, but also affect the unfolding of MszExo I. MszExo I is the first reported enzyme in the DNA repair systems of moderately thermophilic bacteria, which are predicted to have more efficient DNA repair systems than mesophilic ones.
Introduction
Exonuclease I (Exo I) from E. coli is an Mg2+ ion dependent exonuclease, which specifically degrades single-stranded DNA (ssDNA) from 3' to 5' in a highly processive manner [1,2]. It is involved in several important DNA repair systems in E. coli, such as RecBCD-dependent homologous recombination, methyl-dependent mismatch repair, and genomic tandem repeat maintenance [3,4].
Extensive research has focused on elucidating the mechanism for its high processivity [5,6]. Recently, the crystal structure of Exo I complexed with ssDNA revealed the topological linkage and two-site substrate binding underlying the high processivity [6].
Several lines of evidence indicate that Exo I digests ssDNA by a two-metal ion catalyzed mechanism [5–8], which was previously described for the 3'-5' exonuclease domain of DNA polymerase I [9,10]. Both Exo I and the exonuclease domain of DNA polymerase I belong to the DnaQ superfamily (or DEDD superfamily) based on sequence analysis [5,11,12]. According to the two-metal ion mechanism, two magnesium ions are essential for exonuclease activity, with MgB binding directly to the enzyme and MgA binding via the 3' end of the bound ssDNA substrate [6,9]. The contribution of two metal ions to thermal stability of Exo I has not been characterized.
Currently, E. coli Exo I is a popular commercial product for the removal of PCR primers prior to DNA sequencing. The potential use of Exo I in nanopore DNA sequencing has also been explored owning to its high processivity and unusual resistance to high salt [13]. However, E. coli Exo I is a mesophilic enzyme with limited tolerance to higher temperature. To expand the use of Exo I in various applications, more robust enzymes are required. Exo I is conserved in bacteria but not in archaea, which makes it difficult to find a thermophilic ortholog. PfuExo I was recently found in the hyperthermophilic archaeon, Pyrococcus furious. Although it works well at high temperature, PfuExo I hydrolyzes ssDNA at every two nucleotides [14], adopting a different mechanism to E. coli Exo I.
Few investigations have focused on the metabolism of nucleic acids in moderately thermophilic bacteria. It is of great interest to know whether their way of metabolizing nucleic acids is similar to that of mesophilic bacteria or hyperthermophilic archaea. Here, a moderately thermophilic Exo I, which shares sequence similarity to E. coli Exo I, was found in Methylocaldum szegediense [15], whose growth temperature is 57°C. It has similar metal ion binding affinity, and similar activity at mesophilic temperatures as E. coli Exo I. The x-ray crystal structure of MszExo I was determined at a resolution of 2.2 Å and it was similar to that of E. coli Exo I. However, MszExo I has a broader working temperature and higher thermal stability. More importantly a magnesium ion was found to modulate its thermal unfolding curve as measured by differential scanning calorimetry (DSC).
Materials and Methods
DNA substrates
The following oligonucleotides were synthesized by Sangon Biotech (Shanghai, China): FRET-dT67, 67-nt poly dT labeled by a BHQ1 quenchor [16] on dT1 which is the first nucleotide from 5’ end and a fluorescein (6-FAM) donor on dT15 which is the fifteenth nucleotide from 5’ end; dT67, 67-nt poly dT; d22, 5'-AATTCGTGCAGGCATGGTAGCT-3'; d27, 5-AGCTATGACCATGATTACGAATTGCTT-3'; 5FAM-d49, 5’-6-FAM-AGCTACCATGCCTGCACGAATTAAGCAATTCGTAATCATGGTCATAGCT-3'; 3FAM-d49, 5'-AGCTACCATGCCTGCACGAATTAAGCAATTCGTAATCATGGTCATAGCT-6-FAM-3’; 5FAM-dA28, 28-nt poly dA labeled by 6-FAM at 5’ end; 5FAM-dC28, 28-nt poly dC labeled by 6-FAM at 5’ end; 5FAM-dT28, 28-nt poly dT labeled by 6-FAM at 5’ end. The 5’-overhang DNA (d27 and 3FAM-d49), and 3’-overhang DNA (d22 and 5FAM-d49) were generated by annealing the oligonucleotides in the annealing buffer containing 10 mM Tris-HCl, pH 7.5, 50 mM NaCl.
Cloning MszExo I from Methylocaldum szegediense
The MszExo I gene (accession number: KM884769) was amplified from genomic DNA of Methylocaldum szegediense and 6*his-tag was attached to the C-terminus by PCR. The PCR amplifications were performed with Phusion DNA polymerase (New England Biolabs) for 28 cycles of 10 s at 98°C, 30 s at 60°C, and 30 s at 72°C. The final PCR product was digested with NdeI and SacI (New England Biolabs), ligated into placzz1 (a pUC19 derivative plasmid with a multiple-cloning site) vector, and transformed into E. coli strain BL21 (DE3). Positive clones were confirmed by DNA sequencing. The E. coli Exo I gene was amplified from the genomic DNA of E. coli strain BL21 (DE3) and ligated into pET21a overexpression vector (Invitrogen) with a 6*his-tag appended on the C-terminus of the protein.
Protein expression and purification
The E. coli strains harboring the MszExo I (or E. coli Exo I) expression plasmid were grown at 37°C in LB medium containing 100 μg/ml ampicillin. When the OD600 value reached 0.6, the expression of recombinant protein was induced by 1 mM IPTG at 16°C for 16 hours. Cells were harvested and suspended in buffer A (50 mM Tris-HCl, pH7.5, 500 mM NaCl). Cells were disrupted by sonication on ice and the cell lysate was centrifuged (38,000 × g) for one hour at 4°C to remove insoluble materials. After centrifugation, the supernatant was collected, filtered by passing through a membrane filter (0.45 μm, Life Sciences) and loaded on a nickel column (Qiagen) equilibrated with buffer A. The protein was eluted with a linear gradient of 20–500 mM imidazole in buffer A. To get better purity, the resultant fractions containing target protein were collected and diluted 10-fold by buffer C (50 mM Tris-HCl, pH 7.5) before loading on a Q-sepharose anion exchange column (GE Healthcare Life Sciences) for E. coli Exo I or heparin agarose column (GE Healthcare Life Sciences) for MszExo I. The protein was eluted with a linear gradient of 50–1000 mM NaCl. Fractions containing the protein of interest were collected and dialyzed against buffer S (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5 mM EDTA, 50% Glycerol) for storage. Protein concentrations were determined by measuring its absorbance at 280 nm using a calculated extinction coefficient of 60850 M-1cm-1for MszExo I and 74830 M-1cm-1 for E. coli Exo I.
Exonuclease Assay
The exonuclease activity was measured by incubating 5 nM MszExo I with 50 nM various 6-FAM-labeled DNA substrates in the standard reaction buffer containing 50 mM Glycine-NaOH pH 9.5, 200 mM NaCl, 0.1 mg/ml BSA, 5 mM MgCl2 at 37°C. Reaction time is described in the figure legend. Reactions were quenched by 20 mM EDTA. The digestion products were visualized by Typhoon Trio variable mode imager (GE Healthcare Life Sciences) after electrophoretic separation on 20% (W/V) polyacrylamide-7 M urea gels.
Endonuclease Assay
MszExo I (100 nM) was incubated with 25 ng/μl M13mp18 circular ssDNA (New England Biolabs) or Φ174 plasmid (New England Biolabs) in the standard reaction buffer for 1 hour at 37°C. The reaction was quenched by 20 mM EDTA. The products were separated on a 1% agarose gel, and visualized by GoldView staining.
RNase Assay
The RNase activity was detected by using RNase contaminant kit ((New England Biolabs). MszExo I (100 nM) was incubated with 4 ng/μl 6-FAM-labeled RNA substrate from the kit in the standard reaction buffer for 1 hour at 37°C. The reaction was quenched by 20 mM EDTA. The products were visualized Typhoon Trio variable mode imager (GE Healthcare Life Sciences) after electrophoretic separation on 6% (W/V) polyacrylamide-7 M urea gels.
Crystallization and structure determination
For crystallization studies the MszExo I gene was synthesized with an N-terminal cleavable strep tag. The preparation of MszExo I protein for crystallization is described in supporting information (S1 File). Apo MszExo I crystals were grown by mixing the protein at a concentration of 8 mg/ml with 20% PEG6K, 0.1 M Tris-HCl pH8, 0.2 M calcium chloride at a protein to precipitant ratio of 1:1. Crystals appeared within 3 days, and grew to a suitable size for data collection after 10–14 days. MszExo I crystals were cryo-protected by dipping them into a solution consisting of the components used for crystallization supplemented with 20% glycerol for 2–5 sec followed by quick flash freezing in liquid nitrogen and storage for data collection.
Diffraction data were collected at beamline I02 of Diamond Light Source, United Kingdom. Data was processed using the XIA2 pipeline. The apo MszExo I structure was solved by molecular replacement (using PHASER) with E. coli Exo I structure as the starting model (PDB ID: 1FXX). COOT visualization software was used for model building and REFMAC5 for refinement. Selected data collection and refinement statistics are given in Table 1.
Table 1. Data collection and refinement statistics.
| Data collection | |
| Beamline | I02 |
| wavelength (Å) | 0.9795 |
| space group | P212121 |
| cell dimensions | |
| a, b, c (Å) | 60.95 97 111.39 |
| α, β, γ (o) | 90 90 90 |
| Resolution (Å) | 2.183 |
| Rmerge | 4.30% |
| Mean I/σI | 18.3 |
| completeness (%) | 99.22 |
| Redundancy | 4 |
| Refinement | |
| Resolution (Å) | 2.12 |
| No.reflections | 36277 |
| Rwork/Rfree | 22.2/25.3 |
| No.atoms | 4010 |
| B-factors (Å2): protein | 49.365 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (o) | 1.029 |
Fluorescence resonance energy transfer (FRET)-based exonuclease activity assay
The DNA substrate FRET-dT67 was dissolved in standard reaction buffer and diluted by dT67 at a ratio of 1:1. The FRET-based exonuclease activity was measured in a standard mixture containing 50 mM Glycine-NaOH pH 9.5, 200 mM NaCl, 0.1 mg/ml BSA, 5 mM MgCl2, 1 μM ssDNA (50% FRET-dT67 + 50% dT67), and 5–27 nM MszExo I or E. coli Exo I at 25 to 67°C. The reaction was triggered by the rapidly mixing of 90 μl of the enzyme in reaction buffer to 5 μl of substrate, which was preloaded in a 96-well plate. The degradation of ssDNA was instantly measured from continuous increase of fluorescence intensity at 522 nm excited by 492 nm for one minute on the Synergy Mx monochromator-based multi-mode microplate reader (Bioteck), controlled by Gene 5 software. The initial velocity of the reaction was obtained by calculating the slope of the initial linear portion of the curve.
Determination of metal ion binding affinity (Kobs,Mg) of E. coli Exo I and MszExo I by kinetic reaction
The initial velocities of exonuclease-catalyzed reactions were measured in the presence of 0 to 5 mM MgCl2. The temperature was controlled at 25°C by the microplate reader, and the protein was pre-incubated at 25°C for 30 sec on thermal cycler before assaying. The initial velocity data were fitted to the equation (2) to obtain parameters Kobs,Mg and Vmax. In this equation, the initial velocity is represented as “Vo”, the maximum of initial velocity represented as “Vmax”, and the observed dissociation constant for binding of two magnesium ions to the enzyme represented as Kobs,Mg. Here, Vmax is the initial velocity the enzyme would reach if both magnesium binding sites were fully saturated. The DNA substrate is already in a saturated condition.
Effect of temperature on exonuclease activity
Activities of E. coli Exo I and MszExo I were determined over a temperature range of 25–67°C. Samples were pre-incubated at a given temperature (from 25 to 67°C) on the thermal cycler for 30 sec before the activity assay. The reaction was triggered by rapidly mixing of enzymes to ssDNA substrate (50% FRET-dT67 + 50% dT67) preloaded in a 96-well plate. The increase of fluorescence intensity at 522 nm was continuously monitored by the microplate reader controlled at the given temperature.
Thermal inactivation assay
Enzymes were diluted in exonuclease reaction buffer to a final concentration of 15 nM. 100 μl of enzyme was incubated at fixed temperatures varying from 37 to 62°C for 10 min. Samples were immediately cooled on ice for 1 min, then incubated at 37°C for 30 sec followed by activity assay at this temperature. Data were normalized by taking the activity of Exo Is incubating at 37°C as 100%.
Differential Scanning Calorimetry
DSC measurements were carried out on a MicroCal VP-capillary DSC differential scanning calorimeter (GE Healthcare Life Sciences) with an integrated autosampler. The DSC system was controlled by VPviewer program. Exo I samples (70 μM) were equilibrated in different buffers by dialysis before DSC measurement. Samples were analyzed by a programmed scan rate of 1.5°C/min over a temperature range of 10–110°C. Data were corrected by subtracting buffer baseline and normalized for protein concentration. Tm value was obtained by fitting E. coli Exo I data to a two-state model and MszExo I data to a non-two-state model or two-state model using the Microcal origin software. DSC data of MszExo I in buffers without magnesium ions were transformed into FU (fraction unfolded)-T plots by integration [17].
Results
MszExo I is an ssDNA-specific 3’ to 5’ exonuclease
Sequence identities between MszExo I and E. coli Exo I are 47%, which suggests that MszExo I is an ssDNA-specific exonuclease (S1 Fig.). To test this prediction, nuclease assays were performed using 3’and 5’-overhang DNAs as substrates. Highly purified MszExo I (S2 Fig.) showed poor activity and thermal stability in the commercial buffer for E. coli Exo I (67 mM Glycine-KOH pH 9.5, 6.7 mM MgCl2, 10 mM β-mercaptoethanol, 0.1 mg/ml BSA), which contains barely no salt (data not shown). The performance of MszExo I increased significantly in the standard reaction buffer (50 mM Glycine-NaOH pH 9.5, 5 mM MgCl2, 200 mM NaCl, 0.1 mg/ml BSA) and further increase of sodium chloride made little difference.
The degradation of substrates was only observed from the 3’-overhang DNA, and no cleavage was happened to the 5’-overhang DNA (Fig. 1A). This result supports the prediction that MszExo I is an ssDNA-specific exonuclease with 3’ to 5’ polarity. To further study substrate specificity of MszExo I, 5FAM-dA28, 5FAM-dT28 and 5FAM-dC28 were subjected to MszExo I treatment. The products were ladder-like bands at positions of lower than the 10-nt band for all three substrates (Fig. 1B). Probably, this was caused by the fact that the binding affinity of MszExo I to ssDNA decreased significantly when the length of ssDNA was shorter than 10 nucleotides (data not shown). These results indicate that the cleavage mode of MszExo I is processive. Interestingly, the ladder-like bands corresponded to every nucleotide judged from the position of the size marker bands, 10 nt and 5 nt. These results indicate the cleavage mode of MszExo I is similar to E. coli Exo I instead of PfuExo I.
Fig 1. Substrate specificity of MszExo I.
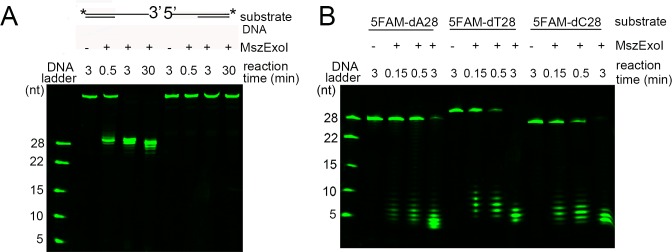
(A) MszExo I (5 nM) was incubated with 3’-overhang and 5’-overhang DNAs (50 nM) for indicated time at 37°C. The positions labeled by a 6-FAM are indicated by an asterisk for the DNA substrates. The 28-nt, 22-nt, 15-nt, 10-nt, and 5-nt poly dAs labeled by a 6-FAM at 5’-end were used as markers. Other details are described in Materials and Methods. (B) Time course experiments of the exonuclease activity of MszExo I were performed using 6-FAM labeled ssDNAs (5FAM-dA28, 5FAM-dT28 and 5FAM-dC28) as substrates. MszExo I (5 nM) was incubated with the DNA substrates (50 nM) at 37°C for indicated time. The 28-nt, 22-nt, 15-nt, 10-nt, and 5-nt poly dAs labeled by a 6-FAM at 5’-end were used as markers. Other details are described in Materials and Methods.
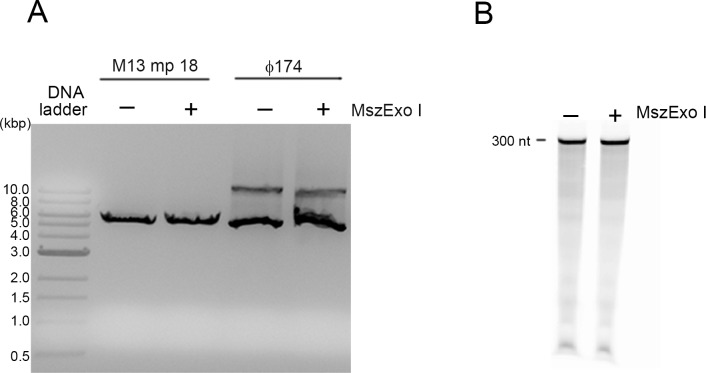
To determine whether MszExo I has endonuclease activity, endonuclease assays were performed using M13 mp18 which is a circular ssDNA and Φ174 which is a circular double-stranded DNA (dsDNA) as substrates. No cleavage was observed with either circular ssDNA or dsDNA after incubation at 37°C for 1 hour (Fig. 2A). To detect whether MszExo I has RNase activity, the RNase assay was performed using the 300-nt RNA internally labeled by 6-FAM as the substrate. There was no digestion even after incubation at 37°C for 1 hour (Fig. 2B). These results indicate MszExo I has no endonuclease activity, nor RNase activity.
Fig 2. Endonuclease and RNase assay of MszExo I.
(A) MszExo I (100 nM) was incubated with circular ssDNA (M13 mp18) or dsDNA (Φ174) in the standard reaction buffer at 37°C for 1 hour. The products were analyzed by electrophoresis on a 1% agarose gel which was then stained by GoldView. (B) In the RNase assay, the 300-nt RNA internally labeled by a 6-FAM was used as the substrate. The reaction was performed at 37°C for 1 hour in the standard reaction buffer. Samples were resolved on a 6% PAGE with 7 M urea. The products were visualized by Typhoon Trio variable mode imager.
X-Ray crystal structure of MszExo I
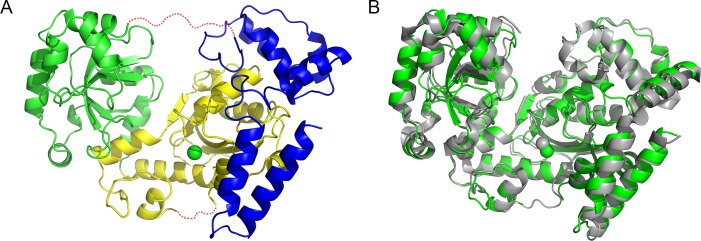
The results of activity assays and sequence alignment indicate MszExo I is an E. coli Exo I homolog in Methylocaldum szegediense. It would be interesting to know whether they have similar three-dimensional structures. The structure of MszExo I (PDB: 4RG8) was solved at 2.2 Å. The overall structure adopted a doughnut-like shape with a large groove through the center of the molecule, closely resembling E. coli Exo I (Fig. 3A). MszExo I is composed of three domains according to protein sequence alignment (Fig. 3A). The N-terminal exonuclease domain (residues 1–197) is located in the middle of the molecule. It is flanked by the SH3-like domain (residues 198–355) and C-terminal helical region (residues 356–479). A flexible region (residues 350–359) missing in the structure is predicted to behave as a linker to join the SH3-like domain and C-terminal domain (Fig. 3A). Residues at active site and anchor site, both of which are involved in substrate binding, are highly conserved among MszExo I and E. coli Exo I (S1 Fig). A single magnesium ion (MgB) was observed at the active site, coordinated by Asp11 and Asp104. Glu13 and Asp182 are predicted to bind the other magnesium ion (MgA), which was not present in the crystal structure probably due to the absence of substrate. Site-directed mutagenesis study showed that D104A and D182A mutants were devoid of exonuclease activity (data not shown), supporting the two-metal ion mechanism in 3' to 5' exonucleases.
Fig 3. Structural alignment between E. coli Exo I and MszExo I.
(A) Overall structure of MszExo I. Like E. coli Exo I, MszExo I can be divided into three domains, the N-terminal exonuclease domain (1–197) colored in yellow, the SH3-like domain (198–355) colored in green and the C-terminal helical region (356–479) colored in blue. The magnesium ion at the active site is represented as a green sphere. Two missing regions (173–177, 350–359) are represented as red dashed lines. (B) Structure alignment of E. coli Exo I and MszExo I. E. coli Exo I (PDB: 1FXX) is colored in grey, and MszExo I (PDB: 4RG8) colored in green. Images were generated using the Pymol program.
Structure alignment of two Exo Is showed they were very similar in overall structure. The superposition of MszExo I and E. coli Exo I yields RMSD value for α-carbon atoms of 0.98 Å over 358 residues (Fig. 3B).
Similar metal ion binding affinity is observed in MszExo I and E. coli Exo I
MgB was observed in the crystal structure of apo MszExo I. In E. coli Exo I, the corresponding magnesium ion is tightly bound, so that even incubation with 10 mM EDTA does not remove it entirely from the enzyme [6].
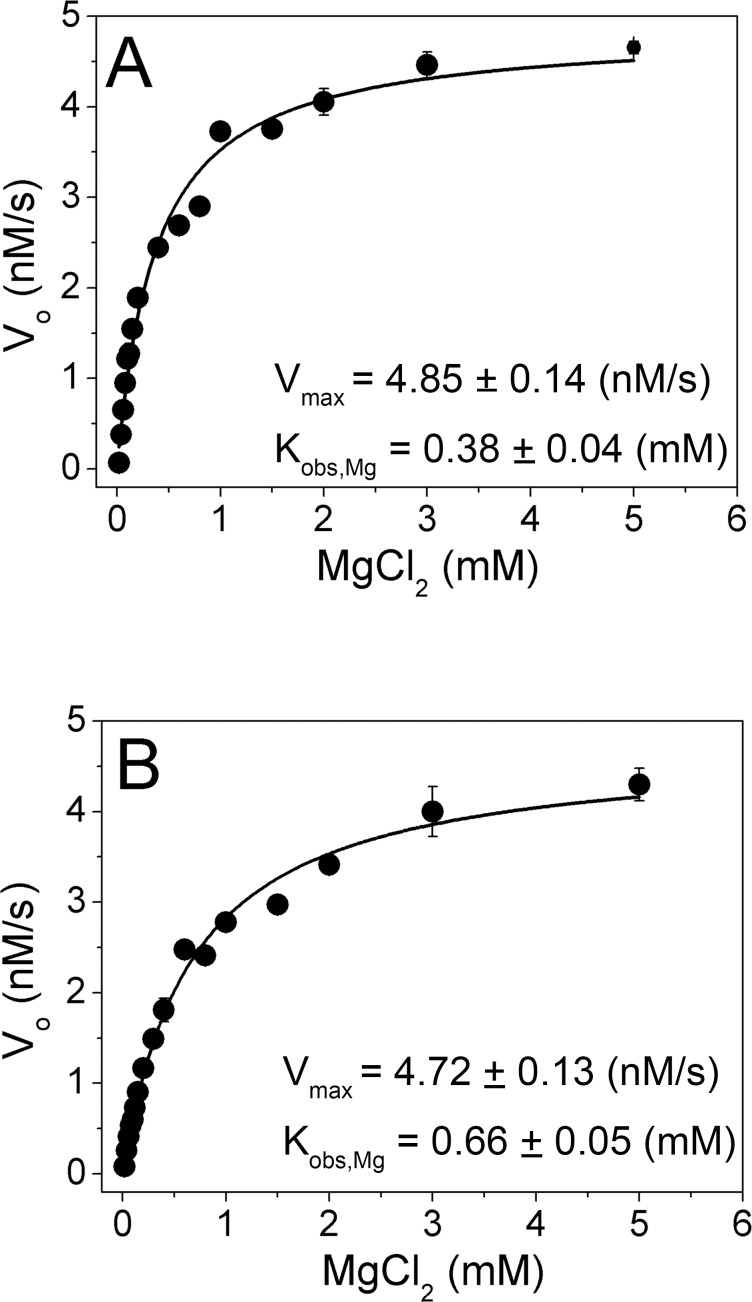
Sequence alignment suggests that Glu13 and Asp182 in MszExo I bind MgA, which is also essential for exonuclease activity. The value of Kobs,Mg (the observed dissociation constant for two magnesium ions from the Exo I complex), was used to compare metal ion binding affinities of MszExo I and E. coli Exo I. Activities of the two enzymes were measured in the presence of various concentrations of MgCl2 by FRET-based exonuclease activity assay [18]. Compared with the traditional gel electrophoresis-based assay using radiolabelled substrates, this FRET-based assay can provide kinetic parameters conveniently. The initial velocities were plotted as a function of the concentration of MgCl2. The data fitted well to the binding equation for a single binding site (Fig. 4). The value of Kobs,Mg for MszExo I is 0.66 mM, comparable to that for E. coli Exo I (Kobs,Mg = 0.38 mM). It indicates these two enzymes have similar metal ion binding affinities.
Fig 4. Determination of the metal ion binding affinity (Kobs,Mg) of E. coli Exo I and MszExo I by kinetic reaction.
The reaction conditions were 27 nM enzyme, 1 μM ssDNA substrate (half FRET-dT67 with half dT67), 50 mM Glycine-NaOH buffer, pH 9.5, 200 mM NaCl, 0.1 mg/ml BSA and 0–5 mM MgCl2 at 25°C. Initial velocity data were fitted to binding equation (2) as described in Materials and Methods, and then parameters Vmax and Kobs,Mg were determined. Their values were indicated in the figure. (A) E. coli Exo I, (B) MszExo I. Errors were calculated from three parallel experiments.
Broader working temperature and higher thermal stability of MszExo I
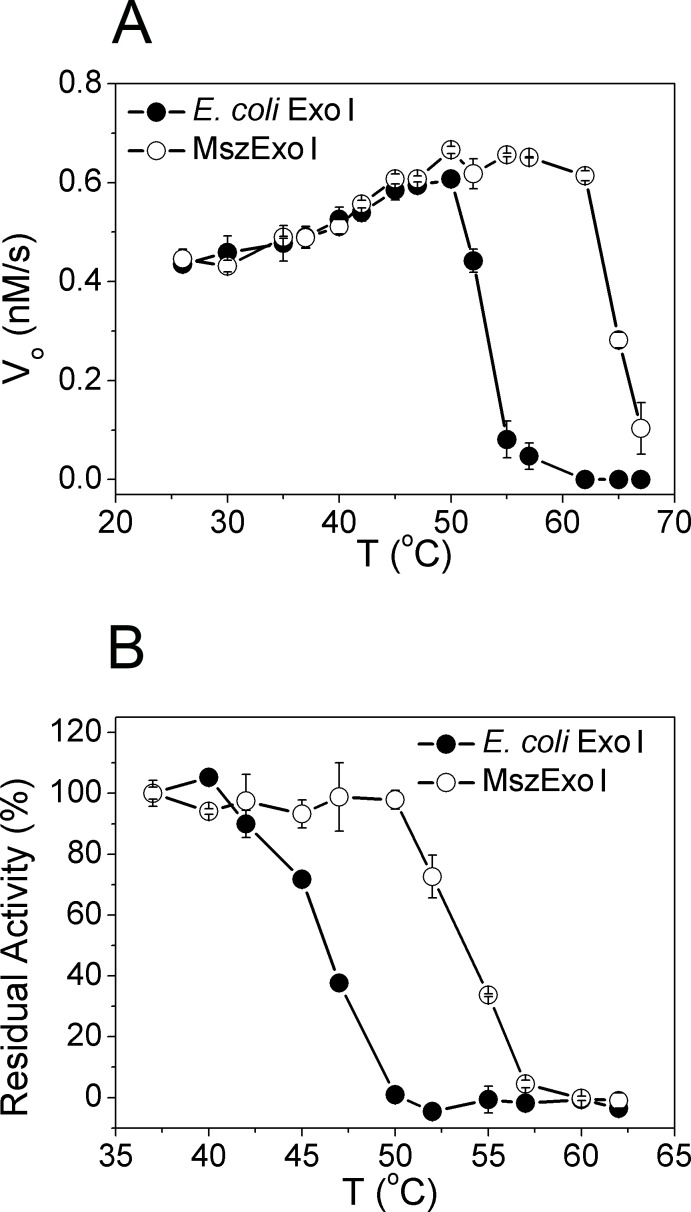
MszExo I is predicted to have better thermal stability than its E. coli counterpart since it was cloned from a moderately thermophilic strain. To test this idea, exonuclease activities were measured over a temperature range of 25–67°C (Fig. 5A) to derive the optimum working temperature for the respective exonucleases. The protein concentration was 5 nM. Up to 47°C, MszExo I showed similar activity to E. coli Exo I. The activity of E. coli Exo I dropped rapidly at temperatures over 50°C, and was completely inactivated at around 60°C. However, during this range (50–60°C), MszExo I maintained high exonuclease activity, with activity decreasing only at temperatures above 60°C.
Fig 5. Thermal profiling of E. coli Exo I and MszExo I.
(A) Determining optimum working temperature for two Exo Is. The reaction conditions were 5 nM enzyme, 1 μM ssDNA substrate (half FRET-dT67 with half dT67), 50 mM Glycine-NaOH buffer, pH 9.5, 200 mM NaCl, 0.1 mg/ml BSA and 5mM MgCl2 at given temperature varying from 25 to 67°C. Other details are described in Materials and Methods. MszExo I (open circle) showed broader working temperature than E. coli Exo I (filled circle). Errors were calculated from three parallel experiments. (B) Thermal stability of two Exo Is evaluated by thermal inactivation assay. In this assay, 15 nM enzyme in standard reaction buffer was incubated at given temperature varying from 25 to 62°C for 10 min, then cooled on ice, and finally activity was determined at 37°C. The data were normalized by taking the activity of enzymes incubated at 37°C as 100%. Errors came from three parallel experiments. Thermal stability of MszExo I was by 5°C higher than that of E. coli Exo I.
To further characterize MszExo I, thermal inactivation assays were performed. Though the half-life of E. coli Exo I at 37°C was longer than six hours (data not shown), its activity began to decrease after incubating at 42°C for 10 minutes (Fig. 5B). It was completely inactivated after being incubated at 50°C for 10 min. In the case of MszExo I, higher thermal stability was observed. MszExo I retained most of its activity even when incubated at 50°C for 10 min, and was completely inactivated at nearly 60°C.
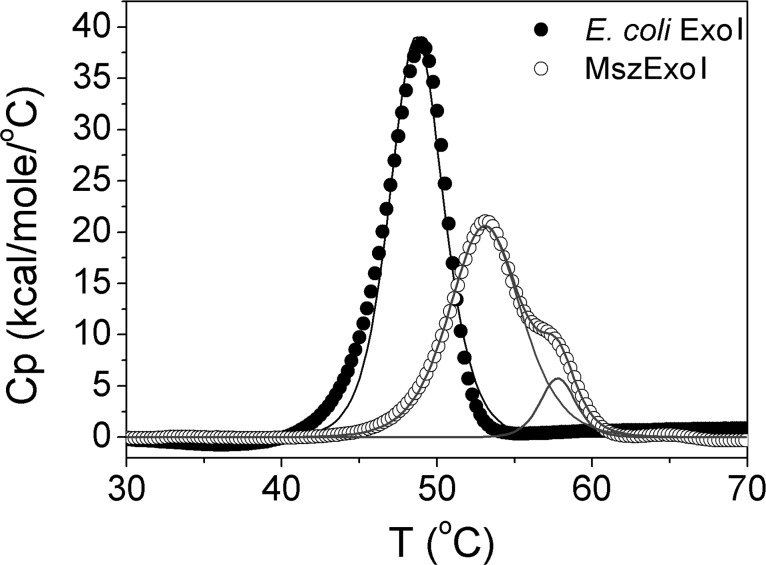
Thermal stabilities of the two enzymes were further compared by DSC measurements. E. coli Exo I exhibited a cooperative two-state transition at 49.0°C (Tm) while MszExo I followed a non-two-state transition with calculated Tm values at 53.1°C and 57.4°C (Fig. 6). Higher Tm value of MszExo I indicates a higher thermal stability, consistent with the result from thermal inactivation assays. MszExo I has two unfolding entities melting at different temperatures, as indicated by the fitted data while E. coli Exo I only has one (Fig. 6). It is interesting to observe that the two enzymes displayed different unfolding profile, especially considering the sequence similarity and 3D structure similarities.
Fig 6. Thermal stability of E. coli Exo I and MszExo I evaluated by DSC.
Comparison of DSC curve of E. coli Exo I (filled circle) and MszExo I (open circle). The protein concentration was 70 μM. The E. coli Exo I data (filled circle) were fitted to two-state transition model (black solid line). The Tm value for E. coli Exo I was 49°C. The MszExo I data (open circle) were fitted to non-two-state transition model (grey solid line). Two peaks indicated the two individual transitions. Tm values of these two transitions were 53.1°C and 57.4°C, respectively.
Magnesium ion modulates thermal unfolding curve of MszExo I
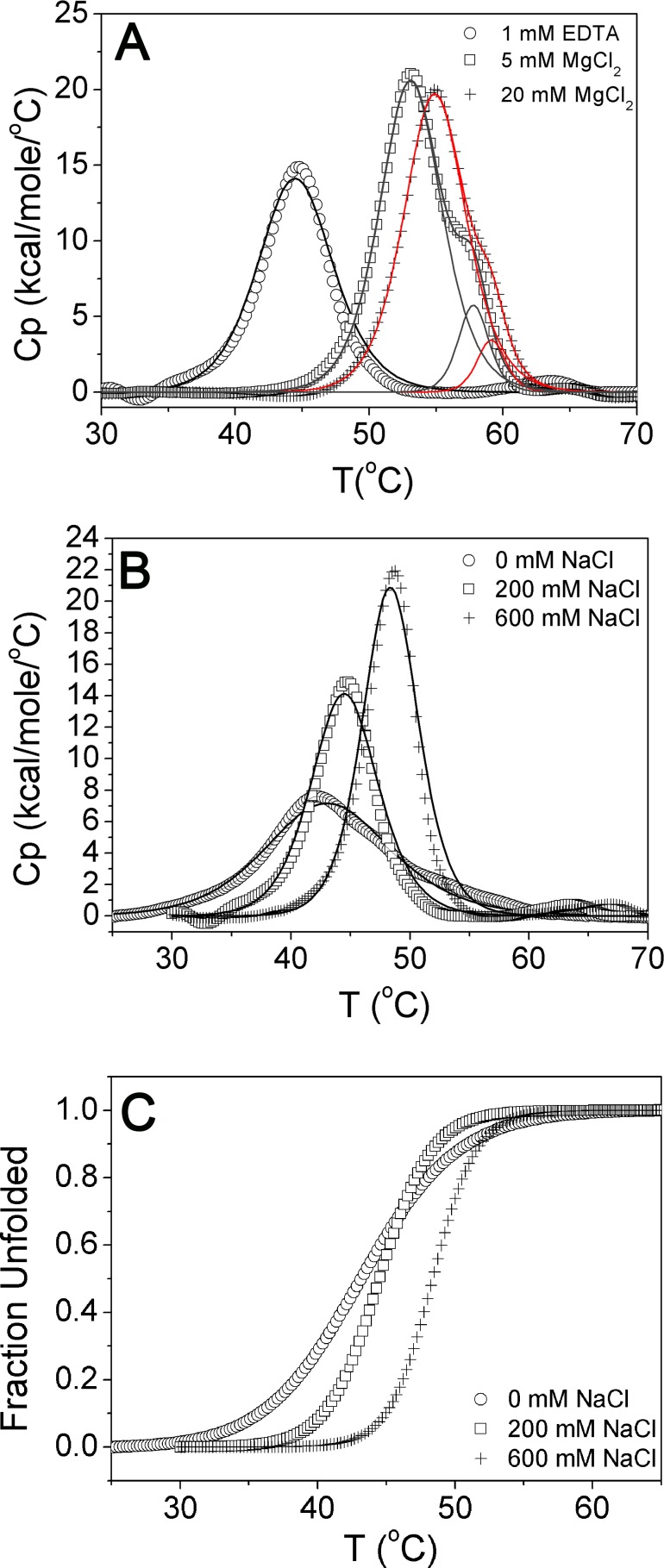
To further investigate the unfolding transitions of MszExo I, DSC measurements were performed as a function of concentration of magnesium ions. In the presence of 1 mM EDTA, single thermal transition was observed (Fig. 7A). In the presence of 5 mM MgCl2, the Tm value increased by more than 8°C, and what is more, two peaks representing two unfolding transitions were clearly observed. If the concentration of MgCl2 was further increased to 20 mM, the second peak became smaller and appeared as a shoulder. According to the fitted data, two unfolding entities were also clearly observed under this condition (Fig. 7A). Meanwhile, the Tm value of the first transition increased by 1.7 degrees (Table 2). These results indicate the magnesium ion plays multiple roles during thermal unfolding of MszExo I. It could not only stabilize the whole structure of MszExo I, but also affect unfolding entities or unfolding intermediates as well.
Fig 7. DSC measurements of MszExo I under different conditions.
(A) DSC measurements of MszExo I as a function of magnesium ions concentration. DSC measurements of MszExo I in three different buffers were performed. The base buffer was 50 mM Glycine-NaOH, 200 mM NaCl, pH 9.5. Open circle represents the base buffer in the presence of 1mM EDTA, and black line represents the best fit to two-state transition model. Open square represents the base buffer in addition of 5 mM MgCl2, and grey line represents the best fit to non-two-state transition model. Plus sign represents the base buffer in addition of 20 mM MgCl2, and red line represents the best fit to non-two-state transition model. All Tm values are shown in Table 2. (B) DSC measurements of MszExo I as a function of salt concentration. DSC measurements of MszExo I in three different buffers were performed. The base buffer was 50 mM Glycine-NaOH, 1 mM EDTA, pH 9.5. Open circle represents the base buffer, open square represents the base buffer in addition of 200 mM NaCl, and plus sign represents the base buffer in addition of 600 mM NaCl. DSC data were fitted to two-state transition model. Tm values are shown in Table 2. (C) The unfolded fraction of MszExo I monitored by the transition enthalpy. DSC Data in Fig. 7B were transformed into FU-T plots by integration of the Cp versus T curve.
Table 2. Tm value of MszExo I in different buffers.
| Buffer | Tm1 (°C) | Tm2 (°C) |
|---|---|---|
| 1 mM EDTA + B1* | 44.6 | No |
| 5 mM MgCl2 + B1 | 53.1 | 57.4 |
| 20 mM MgCl2 + B1 | 54.8 | 58.4 |
| 0 mM NaCl + B2** | 43.1 | No |
| 200 mM NaCl + B2 | 44.6 | No |
| 600 mM NaCl + B2 | 48.5 | No |
*, B1 is the buffer which is composed of 50 mM Glycine-NaOH, 200 mM NaCl, pH 9.5.
**, B2 is the buffer which is composed of 50 mM Glycine-NaOH, 1 mM EDTA, pH 9.5.
To rule out the possibility that the appearance of two unfolding transitions of MszExo I was caused by increased ionic strength of buffers with 5 mM MgCl2 or 20 mM MgCl2, DSC measurements were performed as a function of salt concentration (Fig. 7B). As the figure shows, MszExo I always exhibited single peak during thermal unfolding in buffers with different concentration of salts, though higher Tm value with increased salt concentration was also observed (Fig. 7B & Table 2). Besides, the unfolding transition of MszExo I in the presence of 200 mM NaCl and 600 mM NaCl was highly cooperative. However, in the absence of sodium chloride, the unfolding transition became less cooperative (Fig. 7C), indicating salts are essential for the stability of MszExo I. MszExo I began to unfold after 35°C without sodium chloride, which is consistent with the fact that MszExo I showed poor activity and thermal stability in the Exo I commercial buffer which contains barely no salt.
Taken together, the role of magnesium ions in thermal unfolding of MszExo I is unique and could not be replaced by salts.
Discussion
Exo I was first characterized in E. coli by Lehman and Nussbaum [1]. It is conserved in bacteria, but not in archaea and eukarya. Recently, a hyperthermophilic exonuclease I, PfuExo I, was identified, though with limited sequence similarity to E. coli Exo I [14]. It digests ssDNA at every two nucleotides, different from the catalytic mechanism of E. coli Exo I. Considering this fact, moderately thermophilic bacteria seemed a productive source for finding robust alternatives to E. coli Exo I.
In bacteria, the nucleic acids metabolic pathways of E. coli and Bacillus subtilis have been well studied [19,20], and those of archaea are emerging [21–23]. However, few reports on the DNA metabolic pathways of moderately thermophilic bacteria have been published, probably due to the small population of moderately thermophilic bacteria being found. However, more and more moderately thermophilic bacteria have been described recently, in many cases as potential sources of useful enzymatic activities [24–28]. For example, the keratinolytic protease from Meiothermus ruber H328 shows extraordinary tolerance to denaturants [28]. Several genomes of moderately thermophilic bacteria have been completely sequenced [29,30]. Therefore, more studies of moderately thermophilic bacteria are expected.
Our report is the first characterizing an enzyme involved in a DNA repair system from a moderately thermophilic bacteria. At higher temperatures the amount of DNA damage will increase, hence repair systems are required to be more efficient to ensure the faithful transmission of genomic DNA to the next generation. More research is needed to elucidate the differences in DNA repair systems from moderately thermophilic bacteria and mesophilic ones, as well as the similarities. In our study, we found that MszExo I exhibits similar metal ion binding affinity and specific activity as E. coli Exo I. However, during thermal denaturation different unfolding pathways were observed even though their crystal structures are highly conserved. This may be related to the high performance of MszExo I at moderately thermophilic temperatures. Obviously, in vivo studies on the role of MszExo I in DNA repair systems of Methylocaldum szegediense would be informative to demonstrate whether MszExo I functions similarly to its E. coli counterpart or has extra functions.
Supporting Information
(DOCX)
Identical residues are labeled by ‘*’, and similar residues are labeled by ‘:’ or ‘.’. Residues at the active site are highlighted in yellow, and those at the anchor site are highlighted in red. Sequence alignment was performed by CLUSTALW program.
(TIF)
The purified MszExo I protein (4 μg) was subjected to 10% SDS-PAGE followed by Commassie Brilliant Blue staining. The yield of MszExo I was around 1–2 mg per liter culture. Marker, color protein standard (broad range) (New England Biolabs).
(TIF)
Acknowledgments
We would like to thank Professor Victor L. Davidson from University of Central Florda, Dr. William Jack from New England Biolabs, and Professor Yongting Wang from Shanghai Jiao Tong University for critical reviews. We also acknowledge scientists from University of Washington that sent us the genomic DNA of Methylocaldum szegediense and scientists from JGI who sent us the raw reads of the genomic DNA sequence.
Data Availability
MszExo I gene sequence is available from the genebank database (accession number: KM884769). Structure of MszExo I protein is available from Protein Data Bank database (PDB code: 4RG8).
Funding Statement
The authors received no specific funding for this work. Li Fei, SiSi Tian, and Dapeng Sun are employees of New England Biolabs, Shanghai R&D Center. Tom Evans is an employee of New England Biolabs Inc. Ruth Moysey and Mihaela Misca are employees of Oxford Nanopore Technologies Ltd. John J. Barker, Myron A Smith, Paul A McEwan, Ewa S Pilka, and Lauren Crawley are employees of Evotec (UK) Ltd. New England Biolabs Shanghai R&D Center, Oxford Nanopore Technologies Ltd., Evotec (UK) Ltd and New England Biolabs provided support in the form of salaries, supplies, equipment, and infrastructure support for authors LF, ST, DS, TE,RM, MM, JJB, MAS, PAM, ESP and LC, provided editorial suggestions on content and data presentation for the manuscript and approved submission of the manuscript for publication, but did not have any additional role in control of the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1. Lehman IR, Nussbaum AL (1964) The Deoxyribonucleases of Escherichia Coli. V. On the Specificity of Exonuclease I (Phosphodiesterase). J Biol Chem 239: 2628–2636. [PubMed] [Google Scholar]
- 2. Brody RS, Doherty KG, Zimmerman PD (1986) Processivity and kinetics of the reaction of exonuclease I from Escherichia coli with polydeoxyribonucleotides. J Biol Chem 261: 7136–7143. [PubMed] [Google Scholar]
- 3. Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P (2001) In vivo requirement for RecJ, ExoVII, Exo I, and ExoX in methyl-directed mismatch repair. Proc Natl Acad Sci U S A 98: 6765–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feschenko VV, Rajman LA, Lovett ST (2003) Stabilization of perfect and imperfect tandem repeats by single-strand DNA exonucleases. Proc Natl Acad Sci U S A 100: 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breyer WA, Matthews BW (2000) Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nat Struct Biol 7: 1125–1128. [DOI] [PubMed] [Google Scholar]
- 6. Korada SK, Johns TD, Smith CE, Jones ND, McCabe KA, et al. (2013) Crystal structures of Escherichia coli exonuclease I in complex with single-stranded DNA provide insights into the mechanism of processive digestion. Nucleic Acids Res 41: 5887–5897. 10.1093/nar/gkt278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Busam RD (2008) Structure of Escherichia coli exonuclease I in complex with thymidine 5'-monophosphate. Acta Crystallogr D Biol Crystallogr 64: 206–210. 10.1107/S090744490706012X [DOI] [PubMed] [Google Scholar]
- 8. Steitz TA, Steitz JA (1993) A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A 90: 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beese LS, Steitz TA (1991) Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J 10: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang W, Lee JY, Nowotny M (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell 22: 5–13. [DOI] [PubMed] [Google Scholar]
- 11. Liu WF, Zhang A, Cheng Y, Zhou HM, Yan YB (2007) Effect of magnesium ions on the thermal stability of human poly(A)-specific ribonuclease. FEBS Lett 581: 1047–1052. [DOI] [PubMed] [Google Scholar]
- 12. Bernad A, Blanco L, Lazaro JM, Martin G, Salas M (1989) A conserved 3'——5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59: 219–228. [DOI] [PubMed] [Google Scholar]
- 13. Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, et al. (2009) Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol 4: 265–270. 10.1038/nnano.2009.12 [DOI] [PubMed] [Google Scholar]
- 14. Tori K, Ishino S, Kiyonari S, Tahara S, Ishino Y (2013) A novel single-strand specific 3'-5' exonuclease found in the hyperthermophilic archaeon, Pyrococcus furiosus. PLoS One 8: e58497 10.1371/journal.pone.0058497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bodrossy L, Holmes EM, Holmes AJ, Kovacs KL, Murrell JC (1997) Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch Microbiol 168: 493–503. [DOI] [PubMed] [Google Scholar]
- 16. Didenko VV (2001) DNA probes using fluorescence resonance energy transfer (FRET): designs and applications. Biotechniques 31: 1106–1116, 1118, 1120–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takacs E, Grolmusz VK, Vertessy BG (2004) A tradeoff between protein stability and conformational mobility in homotrimeric dUTPases. FEBS Lett 566: 48–54. [DOI] [PubMed] [Google Scholar]
- 18. Ghosh SS, Eis PS, Blumeyer K, Fearon K, Millar DP (1994) Real time kinetics of restriction endonuclease cleavage monitored by fluorescence resonance energy transfer. Nucleic Acids Res 22: 3155–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wigley DB (2013) Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat Rev Microbiol 11: 9–13. 10.1038/nrmicro2917 [DOI] [PubMed] [Google Scholar]
- 20. Lenhart JS, Schroeder JW, Walsh BW, Simmons LA (2012) DNA repair and genome maintenance in Bacillus subtilis. Microbiol Mol Biol Rev 76: 530–564. 10.1128/MMBR.05020-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Wolferen M, Ajon M, Driessen AJ, Albers SV (2013) How hyperthermophiles adapt to change their lives: DNA exchange in extreme conditions. Extremophiles 17: 545–563. 10.1007/s00792-013-0552-6 [DOI] [PubMed] [Google Scholar]
- 22. Li Z, Kelman LM, Kelman Z (2013) Thermococcus kodakarensis DNA replication. Biochem Soc Trans 41: 332–338. 10.1042/BST20120303 [DOI] [PubMed] [Google Scholar]
- 23. Frols S, White MF, Schleper C (2009) Reactions to UV damage in the model archaeon Sulfolobus solfataricus. Biochem Soc Trans 37: 36–41. 10.1042/BST0370036 [DOI] [PubMed] [Google Scholar]
- 24. Slobodkina GB, Kolganova TV, Tourova TP, Kostrikina NA, Jeanthon C, et al. (2008) Clostridium tepidiprofundi sp. nov., a moderately thermophilic bacterium from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol 58: 852–855. 10.1099/ijs.0.65485-0 [DOI] [PubMed] [Google Scholar]
- 25. Iino T, Nakagawa T, Mori K, Harayama S, Suzuki K (2008) Calditerrivibrio nitroreducens gen. nov., sp. nov., a thermophilic, nitrate-reducing bacterium isolated from a terrestrial hot spring in Japan. Int J Syst Evol Microbiol 58: 1675–1679. 10.1099/ijs.0.65714-0 [DOI] [PubMed] [Google Scholar]
- 26. Ueda J, Yamamoto S, Kurosawa N (2013) Paenibacillus thermoaerophilus sp. nov., a moderately thermophilic bacterium isolated from compost. Int J Syst Evol Microbiol 63: 3330–3335. 10.1099/ijs.0.048090-0 [DOI] [PubMed] [Google Scholar]
- 27. You ZQ, Li J, Qin S, Tian XP, Wang FZ, et al. (2013) Georgenia sediminis sp. nov., a moderately thermophilic actinobacterium isolated from sediment. Int J Syst Evol Microbiol 63: 4243–4247. 10.1099/ijs.0.051714-0 [DOI] [PubMed] [Google Scholar]
- 28.Kataoka M, Yamaoka A, Kawasaki K, Shigeri Y, Watanabe K (2013) Extraordinary denaturant tolerance of keratinolytic protease complex assemblies produced by Meiothermus ruber H328. Appl Microbiol Biotechnol. [DOI] [PubMed]
- 29. Anderson I, Chertkov O, Chen A, Saunders E, Lapidus A, et al. (2012) Complete genome sequence of the moderately thermophilic mineral-sulfide-oxidizing firmicute Sulfobacillus acidophilus type strain (NAL(T)). Stand Genomic Sci 6: 1–13. 10.4056/sigs.2736042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goker M, Saunders E, Lapidus A, Nolan M, Lucas S, et al. (2012) Genome sequence of the moderately thermophilic, amino-acid-degrading and sulfur-reducing bacterium Thermovirga lienii type strain (Cas60314(T)). Stand Genomic Sci 6: 230–239. 10.4056/sigs.2726028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Identical residues are labeled by ‘*’, and similar residues are labeled by ‘:’ or ‘.’. Residues at the active site are highlighted in yellow, and those at the anchor site are highlighted in red. Sequence alignment was performed by CLUSTALW program.
(TIF)
The purified MszExo I protein (4 μg) was subjected to 10% SDS-PAGE followed by Commassie Brilliant Blue staining. The yield of MszExo I was around 1–2 mg per liter culture. Marker, color protein standard (broad range) (New England Biolabs).
(TIF)
Data Availability Statement
MszExo I gene sequence is available from the genebank database (accession number: KM884769). Structure of MszExo I protein is available from Protein Data Bank database (PDB code: 4RG8).