Abstract
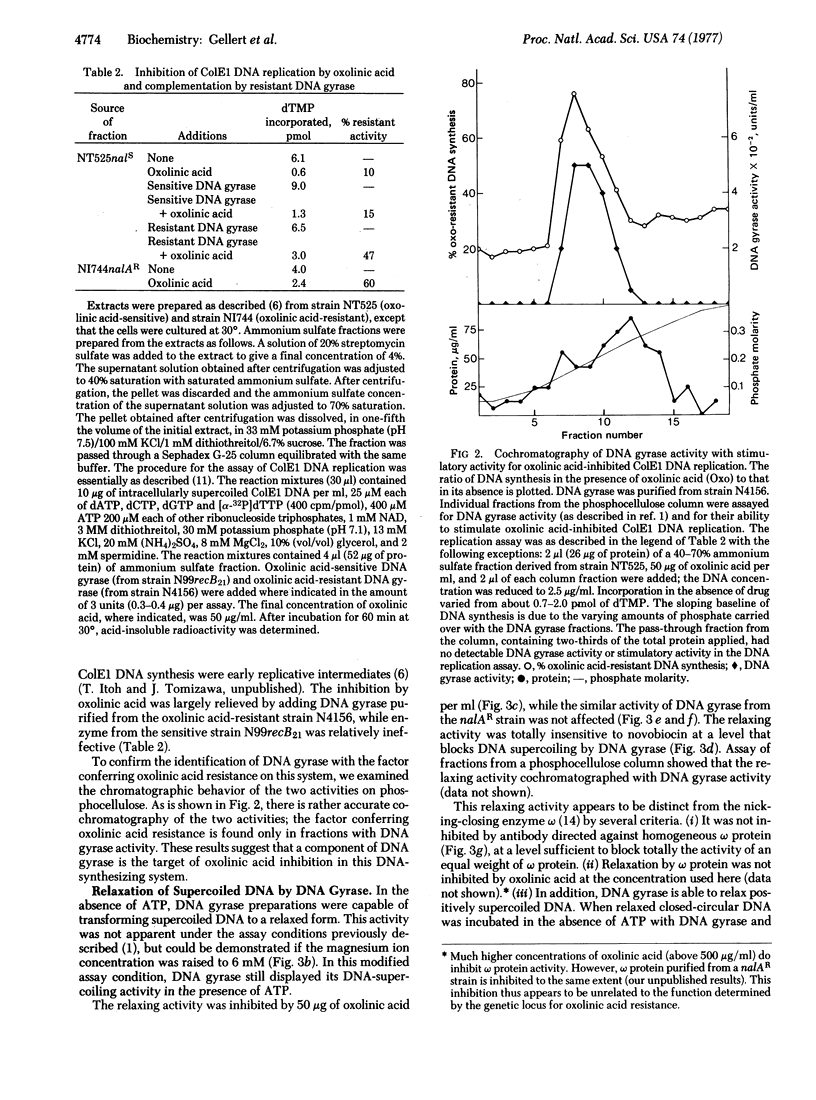
ATP-dependent DNA supercoiling catalyzed by Escherichia coli DNA gyrase was inhibited by oxolinic acid, a compound similar to but more potent than nalidixic acid and a known inhibitor of DNA replication in E. coli. The supercoiling activity of DNA gyrase purified from nalidixic acid-resistant mutant (nalAR) bacteria was resistant to oxolinic acid. Thus, the nalA locus is responsible for a second component needed for DNA gyrase activity in addition to the component determined by the previously described locus for resistance to novobiocin and coumermycin (cou). Supercoiling of λ DNA in E. coli cells was likewise inhibited by oxolinic acid, but was resistant in the nalAR mutant. The inhibition by oxolinic acid of colicin E1 plasmid DNA synthesis in a cell-free system was largely relieved by adding resistant DNA gyrase.
In the absence of ATP, DNA gyrase preparations relaxed supercoiled DNA; this activity was also inhibited by oxolinic acid, but not by novobiocin. It appears that the oxolinic acid-sensitive component of DNA gyrase is involved in the nicking-closing activity required in the supercoiling reaction. In the presence of oxolinic acid, DNA gyrase forms a complex with DNA, which can be activated by later treatment with sodium dodecyl sulfate and a protease to produce double-strand breaks in the DNA. This process has some similarities to the known properties of relaxation complexes.
Keywords: Escherichia coli, DNA supercoiling, oxolinic acid, colicin E1 DNA replication
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bourguignon G. J., Levitt M., Sternglanz R. Studies on the mechanism of action of nalidixic acid. Antimicrob Agents Chemother. 1973 Oct;4(4):479–486. doi: 10.1128/aac.4.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosloy S. D., Oishi M. Genetic transformation in Escherichia coli K12. Proc Natl Acad Sci U S A. 1973 Jan;70(1):84–87. doi: 10.1073/pnas.70.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray H. B., Jr, Upholt W. B., Vinograd J. A buoyant method for the determination of the superhelix density of closed circular DNA. J Mol Biol. 1971 Nov 28;62(1):1–19. doi: 10.1016/0022-2836(71)90127-6. [DOI] [PubMed] [Google Scholar]
- Marians K. J., Ikeda J. E., Schlagman S., Hurwitz J. Role of DNA gyrase in phiX replicative-form replication in vitro. Proc Natl Acad Sci U S A. 1977 May;74(5):1965–1968. doi: 10.1073/pnas.74.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisetsky D., Berkower I., Wickner R., Hurwitz J. Role of ATP in DNA synthesis in Escherichia coli. J Mol Biol. 1972 Nov 28;71(3):557–571. doi: 10.1016/s0022-2836(72)80023-8. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y., Tomizawa J. I. Replication of colicin E1 plasmid DNA in cell extracts. Proc Natl Acad Sci U S A. 1974 Mar;71(3):802–806. doi: 10.1073/pnas.71.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y., Tomizawa J. Replication of colicin E1 plasmid DNA in cell extracts. II. Selective synthesis of early replicative intermediates. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1403–1407. doi: 10.1073/pnas.71.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudenbauer W. L. Replication of Escherichia coli DNA in vitro: inhibition by oxolinic acid. Eur J Biochem. 1976 Mar 1;62(3):491–497. doi: 10.1111/j.1432-1033.1976.tb10183.x. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L. Replication of small plasmids in extracts of Escherichia coli. Mol Gen Genet. 1976 Jun 15;145(3):273–280. doi: 10.1007/BF00325823. [DOI] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumida-Yasumoto C., Yudelevich A., Hurwitz J. DNA synthesis in vitro dependent upon phiX174 replicative form I DNA. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1887–1891. doi: 10.1073/pnas.73.6.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. I., Sakakibara Y., Kakefuda T. Replication of colicin E1 plasmid DNA added to cell extracts. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1050–1054. doi: 10.1073/pnas.72.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]