Abstract
Although great progress has been made in the characterization of off-target effects of engineered nucleases, sensitive and unbiased genome-wide methods for the detection of off-target cleavage events and potential collateral damage are still lacking. Here we describe a linear amplification–mediated modification of a previously published high-throughput, genome-wide translocation sequencing (HTGTS) method that robustly detects DNA double-stranded breaks (DSBs) generated by engineered nucleases across the human genome based on their translocation to other endogenous or ectopic DSBs. HTGTS with different Cas9:sgRNA or TALEN-nucleases revealed off-target hotspots for given nucleases that ranged from a few or none to dozens or more, and extended the number of known off-targets for certain previously characterized nucleases by more than 10-fold. We also identified translocations between bona fide nuclease targets on homologous chromosomes, an undesired collateral effect that has not been described. Finally, HTGTS confirmed that the Cas9D10A paired nickase approach suppresses off-target cleavage genome-wide.
Targeting endogenous loci in live cells with nucleases designed to generate DNA double-stranded breaks (DSBs) at specific endogenous sequences without the need for substrate integration has been very useful for introducing targeted mutations and holds great promise for targeted gene therapy in humans1–4. In this regard, the recently developed TALENs and Cas9:single guide RNA (sgRNA) endonucleases are particularly promising5–10. One continuing concern for employing TALENs and Cas9:sgRNAs for genome engineering, and for therapeutic human genome engineering in particular, is the potential for off-target DSB activity at non-consensus sites within the genome for any given enzyme2. Current assays for such off-target nuclease activity involve cytotoxicity11, prediction-based modeling12–14, select screening12,15,16, and viral vector DSB traps17,18. Such assays have been valuable for testing approaches designed to minimize undesired DNA cleavage activities of these enzymes1,2.
TALENs are dimeric site-specific nucleases with monomers consisting of an engineered DNA binding domain fused to a C-terminal FokI nuclease domain9,10. Specific TALEN activity requires the dimerization of the FokI domain from two TALEN subunits with each monomer providing half of the specific DNA recognition sequence2. The DNA-binding code for TALENs allows targeting of DSBs with 5' overhangs at nearly any position across different genomes2,19,20. For Cas9:sgRNA endonucleases, the Cas9 nuclease forms a complex with an engineered sgRNA comprised of a chimeric clustered, regularly interspaced, short palindromic repeat (CRISPR) RNA and trans-activating CRISPR RNA1. Cas9 sgRNA sequence specificity relies on hybridization of a 20nt targeting sequence on the 5’ end of the sgRNA to complementary DNA and recognition of an ‘NGG’ protospacer adjacent motif (PAM) on the non-complementary strand. Cas9:sgRNA complexes, which again can be designed to cleave a multitude of sites across the genome, generate blunt DSB ends 3bp into the 20nt target sequence proximal to the PAM1.
Chromosomal translocations can arise by fusion of ends of two DNA DSBs lying on heterologous chromosomes or on separated regions of a homologous chromosomes21,22. The high-throughput genome-wide translocation sequencing (HTGTS)23 and translocation-capture sequencing approaches24 were developed to identify translocations of yeast I-SceI meganuclease-generated ‘bait’ DSBs at target sites introduced into the genome of mouse cells to other ‘prey’ cellular DSBs genome-wide. Correspondingly, these methods also identified various classes of endogenous DSBs in primary and transformed B lymphocyte lineage cells23–27. HTGTS, which provides nucleotide-level resolution of junctions, further revealed I-SceI-generated DSBs at cryptic off-target sequences within the mouse genome23. Based on ability to detect off-target I-SceI meganuclease sites across the mouse genome, we proposed HTGTS might be developed into a robust general method for determining off-target activity of engineered nucleases23. We now describe the development of an enhanced HTGTS approach and its application in human cells for identifying nuclease-generated on-target and off-target DSBs and associated collateral chromosomal damage.
RESULTS
HTGTS Assay for Cas9-generated DSBs at the Human RAG1 Locus
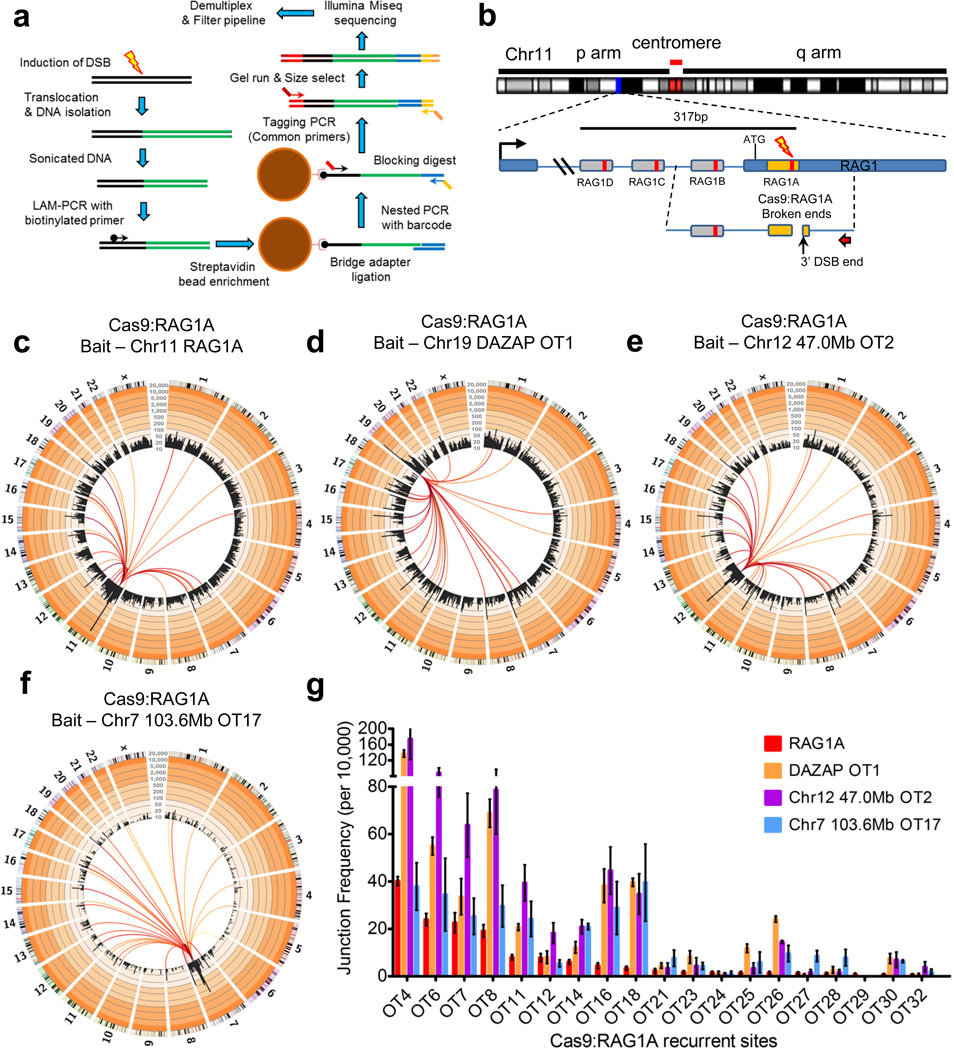
To evaluate use of HTGTS for identifying on- and off-target custom nuclease activity in human cells, we first performed HTGTS using Cas9:sgRNA-generated DSBs as ‘bait’ to capture ‘prey’ sequences genome-wide in 293T cells culture for 48 hours post-transfection with Cas9:sgRNA. For these studies, we have now developed a modified HTGTS approach based on linear-amplification-mediated PCR (LAM-PCR)28 that is more robust, cost-efficient and rapid than our prior emulsion-PCR23 HTGTS (Fig. 1a; Supplementary Fig. 1; Supplementary Table 1; Details in online methods). For initial studies, we selected the human RAG1 gene, a proposed target for gene correction therapy29,30. To induce RAG1 DSBs, we generated four sgRNAs that each targeted a distinct sequence within a 317bp region spanning the beginning of RAG1 exon 2; we refer to these four Cas9:sgRNA combinations as RAG1A, B, C, and D (Fig.1b; Supplementary Fig. 2a). We performed HTGTS from the 3' DSB end (with respect to RAG1 transcriptional orientation) of a given Cas9:RAG1-generated DSB, cloning from the A or B site via a specific primer positioned, respectively, 152bp and 194bp centromeric to them or from the C or D sites with a second specific primer positioned, respectively, 106bp and 227bp centromeric to them (Fig. 1b; Supplementary Fig. 2a). For each HTGTS library, recovered junctions fused uniquely mapped coordinates corresponding to bait sequence and genome-wide prey sequences (Supplementary Figs. 2b–e & 3a–c) and were mainly direct (‘blunt’) or had short micro-homologies (Supplementary Fig. 3d–g). On the bait-site side, junctions were enriched at or near the 3' DSB bait-site end with enrichment decreasing along the bait sequence length, consistent with variable end resection before joining (Supplementary Fig. 3a–c; see below).
Figure 1. LAM-PCR HTGTS from Cas9:RAG1A on-target and off-target bait sites.
(a) LAM-PCR HTGTS method overview. Major modifications of prior HTGTS method include enriching for translocations directly from sonicated DNA via LAM-PCR and ssDNA ligation using a bridge adapter. See online methods for greater detail. (b) Positions of Cas9:RAG1A-D sgRNA target sites within the RAG1 locus on Chromosome (Chr) 11. Red arrow indicates the cloning primer used to capture the 3’ DSB ends from the RAG1A site (orange box); RAG1B uses the same primer but with longer bait. RAG1 C & D use a separate primer strategy (see Supplementary Fig. 2a). Cas9:RAG1 DSBs occur 3bp proximal to the PAM within the sgRNA target sequence (red bar inside sgRNA targets) (c) Circos plots31 of RAG1A HTGTS genome-wide prey junctions binned into 5Mb regions (black bars) are plotted on a log scale with indicated custom ticks; frequency ranges are colored from white (0–10) to increasingly darker orange colors (>10) by factors of 10. Colored lines connect the RAG1 bait site to the prey hotspot; line color range from dark red to yellow indicates high to low hotspot enrichment respectively (25,000 junctions; N=6). Hotspots are identified as significantly focal sites (see online methods for more details), contain junctions to both chromosomal orientations, and are found in multiple library replicates; all hotspots shown are related to nuclease off-target activity (See online methods for more details). (d–f) RAG1A off-target (OT) bait HTGTS circos plots using the (d) DAZAP OT1 on chr19, (e) 47.0Mb OT2 on chr12, and (f) 103.6Mb OT17 on chr7 (N=3 each; see Supplementary Table 1). (g) Bar graph of average junction frequency per 10,000 unique junctions for each off-target site not located on bait chromosomes 7, 11, 12, and 19. Error bars are displayed as S.E.M. The junction scale displayed for the figure circos plots is from 10–20,000.
For each set of HTGTS libraries from a particular break-site or under particular conditions, we used modified Circos plots of the human genome organized into individual chromosomes to visualize overall junction patterns and key features31. In these plots, translocation hotspots, bioinformatically identified in an unbiased fashion as focally enriched HTGTS junction clusters (Online Methods), are indicated by lines that connect the bait-site to a given hotspot and which range in color from dark red (highest junction enrichment) to yellow (lower junction enrichment) (Fig. 1c; Supplementary Fig. 2b–e). We also denote HTGTS junction frequency within 5-Mb bins across all chromosomes on the Circos plots by black bars plotted on a log scale with custom axes (see legend of Fig. 1). For each bait site analyzed in this study, at least 3 (and usually many more) separate HTGTS libraries were generated with individual libraries ranging in size from several thousand to 80,000 independent junctions (Supplementary table 1 and detailed for each experimental figure). Independent HTGTS libraries for a given site or condition gave reproducible overall results and conclusions (Supplementary Figure 2b–e; Supplementary Table 1; see below). Non-specific HTGTS background was estimated as described23 and found to be low (Supplementary Table 2).
Genome-Wide Off-Target Activities of Cas9:RAG1 sgRNAs
By convention, prey sequences are joined to bait DSBs as in ‘+’ (plus) orientation if they read from the junction in the p telomere to q telomere direction; correspondingly, junctions are in ‘−’ (minus) orientation if in the other direction (Supplementary Figure 4a). Other than break-site junctions (see below), genome-wide Cas9:RAG1A-D HTGTS junctions occurred across the genome at similar frequencies in both orientations (Supplementary Fig. 1). Frequent endogenous DSBs at two loci can dominate translocation landscapes27,32, due to cellular heterogeneity in three-dimensional (3-D) genome organization27,33,34. In this regard, we detected 33 highly significant, focally-enriched prey junction hotspots from RAG1A and two from RAG1B libraries (Fig. 1c; Supplementary Fig 2b,c; Supplementary table 3). In contrast, no hotspots were detected for RAG1C and RAG1D libraries, which based on random library size normalizations would have been readily detectable if they occurred at the level of RAG1A or RAG1B off-target sites (Supplementary Fig. 2d,e and legend). The RAG1A or RAG1B hotspot junctions showed expected characteristics for involvement of off-target DSBs, peaking precisely at predicted off-target break-sites based on their being highly related (with 2–7nt mismatches) to the respective bona fide on-target sequences of these two enzymes (Supplementary Table 3; Supplementary Fig. 4b). All junctions were fully consistent with DSB joining, displaying approximately equal numbers of (+) and (−) orientation joins that peaked at direct prey joins (no loss of nucleotides from predicted off-target break-site) and tailed off in both orientations into joins with up to 100bp of resection (Supplementary Figure 4c–e). Additional RAG1A studies at 24 hours and 96 hours after RAG1A transfection resulted in similar distributions of junctions genome-wide including RAG1A off-target sites (Supplementary Fig. 5). Also, RAG1B HTGTS libraries in a different human cell line (A549 lung carcinoma cells) revealed the same two off-target hotspots (Supplementary Fig. 6).
To unequivocally confirm that identified HTGTS off-target sites represented DSBs and further test the ability of HTGTS to identify off-target nuclease-generated DSBs genome-wide, we performed HTGTS using, respectively, RAG1A high level off-target sites on chromosomes 12 or 19 or a RAG1A lower level off-target site on chromosome 7 as bait (Fig. 1d–f). Strikingly, each bait produced HTGTS libraries with all characteristics expected for cloning from that specific off-target RAG1A bait DSB (Fig. 1d–f; see below). Moreover, all reproducibly captured the RAG1A on-target break-site as well as the vast majority of the off-target sites revealed by HTGTS from the on-target break-site (Fig. 1d–g). Indeed, the most highly enriched off-target translocation sites recovered when the bona fide RAG1A site was used as HTGTS bait were similarly highly-enriched when HTGTS was performed with each of the three off-target sites as bait (Fig. 1g; Supplementary Table 3). The ability of these different HTGTS bait DSBs on different chromosomes to robustly identify essentially the same set of recurrent DSBs genome-wide is consistent with our prior findings on the influences of cellular heterogeneity in 3D genome organization on the translocation landscape27. Likewise, the major difference in frequency of translocations captured by these 4 different RAG1A HTGTS baits was the enriched recovery of prey DSB junctions that fell onto the same chromosome as the particular bait-site (Supplementary Table 3), as also predicted by our prior studies27,33,34. Below, we present additional examples of such phenomena in the context of exploiting them to further facilitate HTGTS assays for nuclease off-target activity.
A Common Class of Translocations for Engineered Nucleases
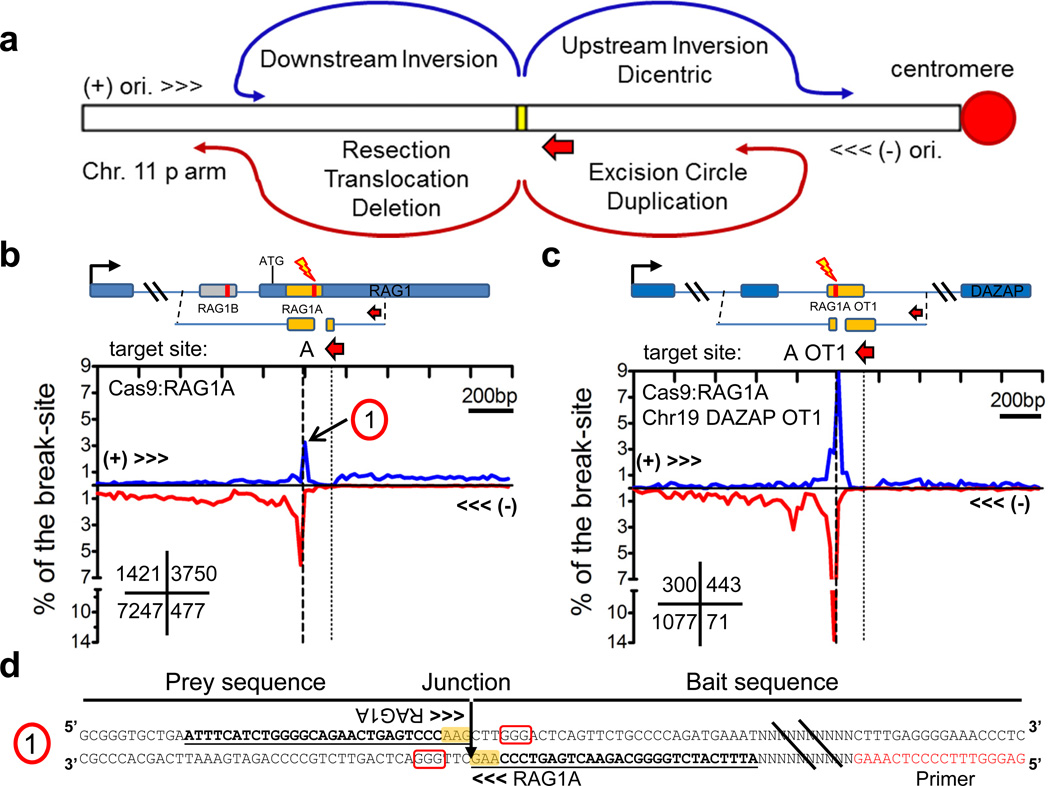
HTGTS junctions are highly-enriched in regions immediately around the break-site due to DSB rejoining following resection, as well as various types of break-site proximal translocations (deletions, inversions, and excision circles)23 that are enhanced due to spatial proximity27,33 (Fig. 2a; Supplementary Fig. 7a). Engineered endonucleases usually have target sites on both homologous chromosomes in diploid cells and primer sequences for detecting junctions from bait DSBs are usually present on both alleles (Supplementary Fig. 7b). Thus, most contributions from each of the homologous chromosome bait 3’ DSBs to break-site or genome-wide junctions cannot be distinguished. However, for the Cas9:RAG1A-D and RAG1A off-target bait break-sites (and others, see below), we find a high density of prey junctions at or very close to the break-site in the (+) orientation quadrant, of which many would correspond to inversional translocations that in cells would generate dicentric chromosomes (Fig. 2b,c; Supplementary Fig. 7c–e). In this regard, nucleotide sequences of such junctions confirms head-to-head inversional joins of the two break-sites, including perfect direct joins of the two Cas9 3' DSB ends (Fig. 2d), with additional junctions in this inversion/dicentric quadrant extending several 100bp or more upstream likely due to ‘prey’ sequence resection.
Figure 2. Distinct prey break-site junction joining to homologous chromosomes.
(a) Prey joining outcomes of 3’ DSB ends at the break-site. Yellow box indicates the break-site and the short red arrow indicates the (−) orientation and position of the cloning primer used. Most junctions represent rejoining of the processed 5’ DSB end (resection) followed by joins to other DSB ends which can result in deletions and excision circles and inversions to the (+) orientation (see Supplementary Fig. 7a). Additional types of joins can occur from the homologous chromosome (see Supplementary Fig. 7b) leading to tandem duplications, and translocations which can form dicentric chromosomes. (b,c) HTGTS libraries displaying prey junction enrichment within 1kb regions flanking the break-site for (b) RAG1A and (c) RAG1A OT1 on Chr 19. Diagrams above each graph indicate the target break-site position; the number within the lower left grid of each graph indicates total junctions for each quadrant. (d) The sequence of a RAG1A head-to-head inversion joined from the break-site on the homologous chromosome and corresponds to ‘1” indicated in panel b. PAM is boxed in red and yellow highlight indicate RAG1A target sequence. Identical sequences between the bait and prey are underlined.
Cas9 Paired Nickase Method Suppresses Off-Target Activity
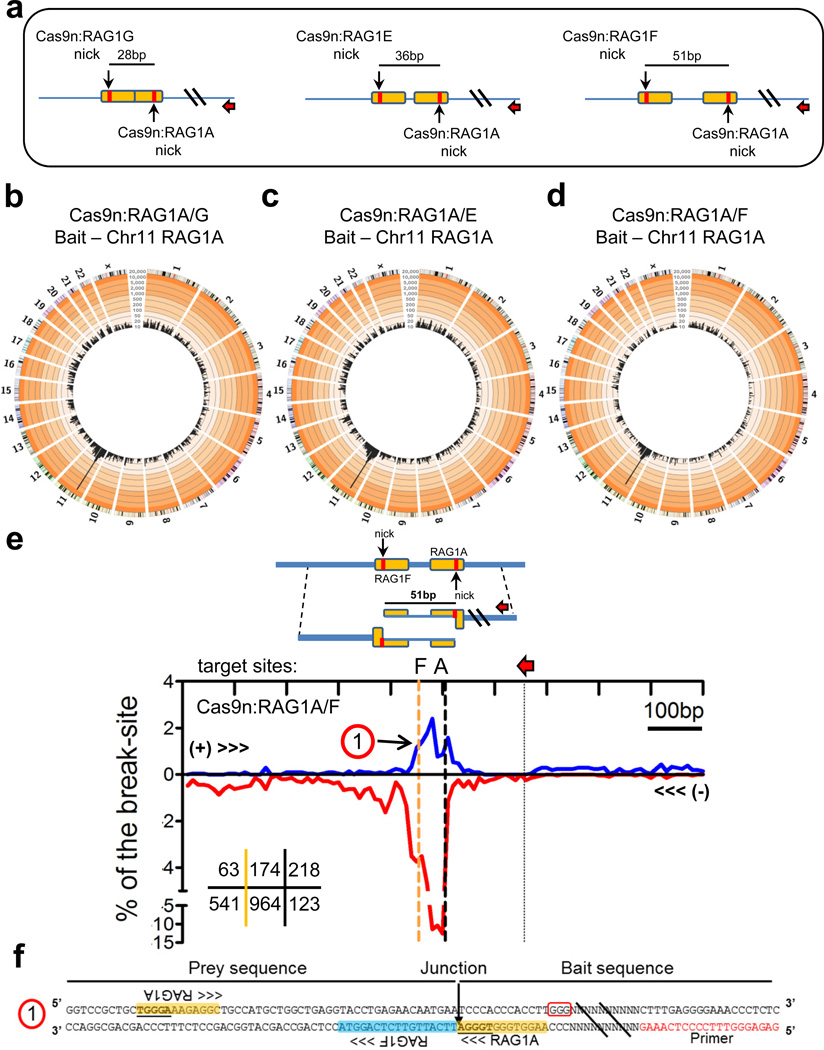
One approach to reduce Cas9:sgRNA off-target activity is to use Cas9 D10A mutation (Cas9n) which renders the endonuclease into a nickase that generates DSBs from off-set paired Cas9n:sgRNA combinations with variable length 5’ overhangs35,36. To test this approach via HTGTS, we paired the off-target-prone RAG1A sgRNA with nearby downstream sgRNA targets (RAG1G, E, and F), which would result in 5’ overhang DSBs of 28nt, 36nt, and 51nt, respectively, when used with Cas9n (Fig. 3a; Supplementary Fig. 8a). Cas9n:RAG1A/G, A/E, or A/F HTGTS libraries had similar genome-wide characteristics to standard Cas9:RAG1A HTGTS libraries except that they lacked hotspots (Fig. 3 b–d); although occasional junctions at RAG1A off-target sites were found upon inspection (Supplementary Table 3). Prey junctions around the break-site also revealed expected resection and translocation patterns (Fig. 3e; Supplementary Fig. 8b,c), including recurrent ‘dicentric’ (+) orientation junctions between break-sites on the two homologous chromosomes that encompassed the two off-set nick sites (Fig. 3e,f; Supplementary Fig. 8b,c).
Figure 3. Cas9n paired nick DSB HTGTS libraries.
(a) RAG1A is paired with nearby sgRNAs RAG1G, E, and F which target the opposing strand and generate DSBs with 5’ overhangs of 28, 36, and 51nt in length when used in combination with Cas9n. Red arrows indicate the orientation and position of the cloning primer used. Orange box indicates the sgRNA target sites with red bars indicating the position of Cas9 cleavage. (b–d) Circos plots (see Figure 1 legend) for HTGTS libraries of Cas9n (b) RAG1A/G (normalized to 20,000 junctions, N=6), (c) RAG1A/E (N=2), (d) and RAG1A/F (N=2) paired nicks using the RAG1A bait. See Supplementary Table 1 for junction frequencies. No hotspots relating to RAG1A off-target activity were identified. (e) Prey junction enrichment within 500bp of the RAG1A target site for Cas9n:RAG1A/F (N=2). Red arrows indicate position of the bait primer and the orange dashed line indicates the position of the opposing strand sgRNA target (RAG1F). Numbers in the lower left grid indicate total junctions for each region including the region between the paired off-set target sites (middle) (f) Sequence indicating a head-to-head inversion of the RAG1A bait to the RAG1F ‘prey” on the homologous chromosome as indicated as ‘1” on panel f of this figure. PAM is boxed in red and the yellow and blue highlights indicate RAG1A and RAG1F target sites respectively. Identical sequences between the two RAG1A target sites are underlined.
TALEN-generated bait DSB HTGTS libraries
To test ability of HTGTS to reveal TALEN off-target DSBs, we employed two previously described TALENs37 that, respectively, cleave the C-MYC gene on chromosome 8 or ATM gene on chromosome 11 (Supplementary Figure 9a,b). The ATM and C-MYC TALEN bait HTGTS libraries showed similar patterns of break-site proximal junctions as those generated with Cas9:sgRNAs, including readily detectable dicentric orientation joins between the TALEN break-sites on homologous chromosomes (Supplementary Fig. 9c–e). In addition, we detected a large number off-target sites for both the ATM (522 off-target sites) and C-MYC (384 off-target sites) TALENs, of which all were lower frequency than the most robust Cas9:RAG1A off-targets (Supplementary Figure 9a, b). Notably, many highly-enriched TALEN off-targets were pseudo-palindromic sequences that corresponded to variants of the recognition site of a single TALEN monomer (Supplementary Tables 4,5; Supplementary Fig. 9f, g; see Discussion). Both ATM and C-MYC TALEN bait libraries also reproducibly displayed a high enrichment of prey junctions along their respective break-site chromosomes (Supplementary Fig. 9a,b; see below).
Universal Donor Bait HTGTS Assay for Off-Target Detection
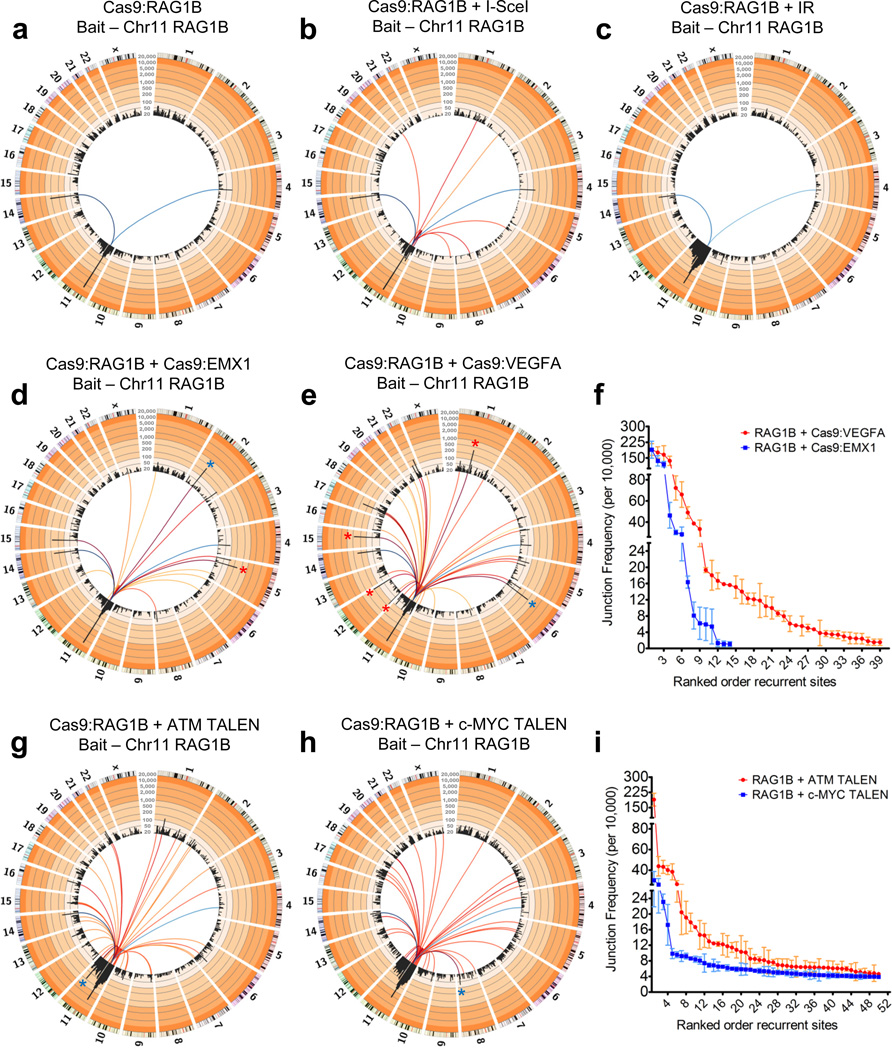
As illustrated by HTGTS from RAG1A off-target sites (Fig. 1), a fixed bait DSB from one nuclease should detect both on- and off-target DSBs of a second nuclease. To test the ability of this "universal donor bait" HTGTS approach to detect different types of potential nuclease generated DSBs, we co-expressed RAG1B with I-SceI in 293T cells and used the RAG1B bait-site to capture I-SceI off-target sites (Fig 4a–c). Indeed, beyond the two expected RAG1B off-target sites, we reproducibly identified 9 I-SceI off-target sites with 2–4nt mismatches from the consensus (Fig. 4a,b; Supplementary Figure 10a; Supplementary Table 6). I-SceI off-target sites displayed expected characteristics of such prey DSBs (Supplementary Fig. 10b,c) and were confirmed by in vitro I-SceI digestion (Supplementary Fig. 10d).
Figure 4. Identification of off-target sites from other nucleases using the universal Cas9:RAG1B bait.
(a,b) Circos plots of (a) RAG1B only (N=6) and (b) RAG1B + I-SceI (N=3) HTGTS libraires. (c) Circos plot of IR-treated Cas9:RAG1B (N=3) libraries. Note the enrichment on the break-site chromosome. (d,e) Circos plots of RAG1B + (d) Cas9:EMX1 (N=3) or (e) Cas9:VEGFA (N=3) HTGTS libraries. Red asterisks indicate previously identified sites15 and blue asterisks indicate the on-target prey site. (f) Plot of on-target and off-target frequencies per 10,000 unique junctions from Cas9:EMX1 and Cas9:VEGFA (N=3 each) using Cas9:RAG1B as the bait. (g,h) Circos plots of RAG1B + (g) ATM or (h) c-MYC TALEN HTGTS libraries. Note that only the top 10% of identified recurrent sites—including the on-target site (blue asterisk)—are shown for TALEN off-target sites. (i) Average junction frequencies for the top 50 recurrent ATM and c-MYC TALEN sites are listed. Blue colored lines for all circos plots in this figure indicate RAG1B off-target sites (dark blue to light blue = high to low enrichment), whereas red to yellow colored lines (dark red to yellow = high to low enrichment) indicate the on and off-target sites of the co-expressed nuclease. (a–e,g,h) All circos plots shown are normalized to 25,000 junctions and presented as similarly described in Fig. 1 (see legend). Junction scale for all Figure 4 circos plots: 20–20,000. (f,i) Error bars are displayed as S.E.M. See Figure 1 legend for further description of circos plots.
We used the RAG1B universal donor bait HTGTS assay to evaluate previously described Cas9 EMX1 and VEGFA sgRNAs15. For both EMX1 and VEGFA sgRNA targets, RAG1B bait HTGTS identified, respectively, the single and the four off-target sites previously documented by the established T7 endonuclease I (T7EI) cleavage assay15, and also identified, respectively, an additional 12 and 34 novel off-target sites (Fig. 4d–f). Notably, all HTGTS-detected Cas9 EMX1 or VEGFA off-target sequences that we identified were related to the corresponding on-target sites and the majority were previously predicted but not confirmed (Supplementary Table 7; Supplementary Fig. 11a)12,15,16,38. When tested by the T7EI assay, we also detected the two on-target break-sites and three previously described off-target sites; but only one of four tested off-target sites revealed by HTGTS (Supplementary Fig. 11b). Consistent with these findings, a prior T7EI assay study failed to detect 23 previously predicted Cas9 EMX1 or VEGFA sgRNA off-target sites15 that were clearly identified by our unbiased HTGTS assay. The RAG1B co-expression HTGTS assays also identified a large number of ATM and C-MYC TALEN off-target sites including all of the approximately 100 most dominant off-target sites detected by HTGTS using the individual TALEN break-sites as bait (Fig. 4g–i; Supplementary Fig. 9a,b; Supplementary Tables 4 & 5).
Low-level DSBs that occur widely across the genome can greatly influence translocation profiles of a given bait DSB. Specifically, treating cells with ionizing radiation (IR) to generate random ectopic DSBs ‘normalizes’ DSB frequency genome-wide27, leading to diminution of dominant endogenous DSB hotspots and causing the length of a given break-site chromosome in cis to be a translocation hotspot region due to a larger contribution of proximity effects27,33. To test ability of the RAG1B HTGTS assay to detect increased levels of wide-spread DSBs that do not qualify as hotspots individually ("wide-spread low-level DSBs"), we introduced RAG1B into 293T cells for 24 hours, then treated them with 7Gy of IR (to introduce approximately 140 random DSBs per cell39), further cultured for 24 hours and performed HTGTS from the RAG1B bait-site. As predicted from prior mouse cell studies27, IR-treatment enhanced generation of HTGTS junctions that were greatly enriched across the entire RAG1B bait-site chromosome 11, with little or no increase on other chromosomes and a diminished recovery of break-site and recurrent off-target junctions (Fig.4c; compare with Fig. 4a). Furthermore, similar IR-treatment results were obtained with RAG1A break-site bait HTGTS libraries for the on-target site on chromosome 11 and also with two tested RAG1A off-target sites on chromosomes 12 and 19 (Supplementary Fig. 12). Thus, subsequent to induction of wide-spread IR-generated DSBs genome-wide, each chromosome containing a particular on-target or off-target DSB hotspot becomes a hotspot region for the targeted DSBs they harbor. Cas9 EMX1 and VEGFA sgRNAs did not contribute markedly to increased junctions on the bait containing chromosome when used at the assayed concentration (Fig. 4d,e; see discussion). However, the ATM and C-MYC TALENs each generated substantially increased HTGTS junctions along the RAG1B break-site chromosome, reminiscent of that observed in the IR-treated RAG1B bait libraries (Fig. 4g,h; Supplementary Fig. 13; see Discussion).
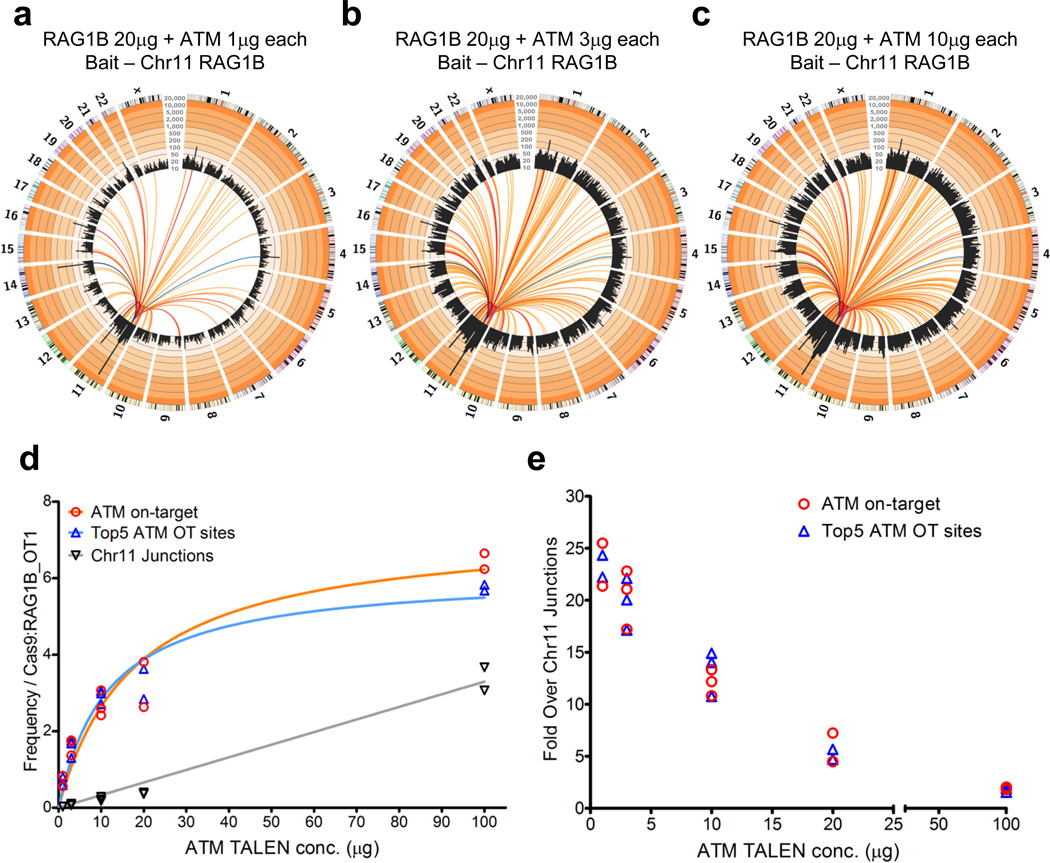
Titration of Engineered Nuclease Genome-wide Activities
Increasing levels of ATM TALEN over a 10-fold range revealed additional lower level off-target sites and suggested an apparent increase in wide-spread, low-level DSBs (Supplementary Fig. 14). Assaying a single nuclease over increasing levels is not optimal for titrating on-target versus the various types of potential off-target activities, since both bait and prey breaks are influenced. Therefore, we employed the RAG1B bait assay to determine whether HTGTS could help assess optimal ATM TALEN nuclease levels for on-target versus off-target DSB activities. Frequencies of recovered ATM TALEN on-target versus the top 5 ATM off-target hotspot sites from RAG1B bait remained constant over a 100-fold tested range of ATM TALEN (Fig. 5a–d; Supplementary Fig. 15). As both on-target and off-target sites occurred at similar relative levels even at the lowest ATM TALEN concentrations, where on-target activity is low, relative off-target activities roughly corresponded to on-target activity (Fig. 5d, e). In contrast, the relative level of on-target and off-target activity to wide-spread, low-level DSB activity decreased as ATM TALEN concentration increased over the 100-fold range tested, indicating that HTGTS can be used to optimize relative levels of on-target versus the wide-spread, low-level activity of the ATM TALEN (Fig. 5e). Increasing the amount of Cas9:RAG1B over an approximately 10-fold range did not reveal additional off-target sites or increase the potentially very low-level, wide-spread DSB activity (Supplementary Fig. 16).
Figure 5. Nuclease titration to measure on-target/background efficiency.
(a–c) Circos plots (see Figure 1 legend) of fixed concentration RAG1B bait + titrated ATM TALEN prey HTGTS libraries at (a) 1µg, (b) 3µg, (c) 10µg of each ATM TALEN plasmid; total junctions for each library are shown (N=1 each) See Supplementary Fig. 15 for additional replicates and concentrations. Dark blue to light blue colored lines indicate RAG1B off-target sites, dark red to yellow colored lines designate ATM TALEN recurrent sites. (d) Graph of multiple RAG1B bait ATM TALEN titration HTGTS libraries plotting the ATM on-target frequency, the sum of the top 5 ATM TALEN off-target sites and the average of junction frequencies along the break-site chromosome (100 regions minus bait break-site region and known ATM off-target sites: 174 sites on chromosome 11, 880bp mean size/site) ranging from 1µg to 100µg each (1µg N=2; 3–10µg N=3; 20–100µg N=2). Libraries were normalized to RAG1B OT1 junction enrichment. (e) The fold enrichment of the ATM TALEN on-target and the sum of the top 5 ATM off-target sites over the ATM TALEN wide-spread, low level DSB activity junctions is indicated for each transfected ATM TALEN plasmid concentration assayed.
DISCUSSION
Robust and accessible methods to test for off-target DSB inducing activities of engineered nucleases are important as this class of enzymes continues to be groomed for human therapeutic purposes2. We demonstrate that LAM-PCR-based HTGTS employing Cas9:sgRNAs, Cas9 paired nickases, or TALEN bait DSBs provides a robust assay for identifying endogenous cellular DSB targets of these enzymes genome-wide in human cells. Thus, LAM-PCR-based HTGTS readily revealed off-target sites for a variety of different engineered nucleases. With respect to sensitivity, the off-target sites that we reproducibly detected in these studies included numerous sites predicted for previously tested nucleases but that had failed to be documented by existing methods, as well as a large number of off-target sites that were not predicted, but which were highly specific for each individually tested nuclease. Beyond off-target activities, we also found that HTGTS revealed apparently wide-spread, but very low-level DSB-inducing activities of some nucleases. Further studies will be needed to determine whether this type of activity is generated by a large number of very infrequently cleaved off-target sites (i.e. requiring some degree of sequence recognition), an even more random DSB-generating activity, or a combination of the two. Whatever the case, this latter application of the HTGTS assay also should be useful for testing non-specific DSB activity of chemotherapeutic and other agents.
LAM-PCR-based HTGTS is a versatile assay that goes beyond simply detecting nuclease off-target sites by also revealing collateral damage in the form of recurrent translocations between on-target DSBs and off-target DSBs, as well as translocations between different off-target DSBs. Although not an ‘off-target’ event, HTGTS also revealed that a major translocation hotspot for on-target Cas9:sgRNA, Cas9 paired nickases, and TALEN-induced DSBs is the corresponding on-target DSB on the homologous chromosome that would lead to dicentric chromosome formation. While likely being controlled substantially by cellular checkpoint responses in normal cells40,41, dicentrics have the potential to generate additional genomic instability via breakage-fusion-bridge cycles25. Many such dicentric junctions likely originate from translocations between break-sites on the two homologues23 that could be eliminated via engineered nucleases that recognize sequence nucleotide polymorphisms; but some conceivably could also result from stalled replication-fork-mediated template switching42. Finally, our HTGTS findings indicate that wide-spread, low-level nuclease-generated DSBs can make each chromosome in a cell a marked hotspot region for translocations of on-target and/or off-target sites within it. Overall, HTGTS not only reveals all of these complex patterns of collateral damaged generated in the context of certain nucleases, but also provides an approach to estimate their relative frequency.
Consistent with cellular heterogeneity in 3D genome organization allowing dominant DSB sites across the genome to drive recurrent translocations to each other27,33, we identified the same large set of Cas9:RAG1A off-target DSBs in HTGTS assays that employed as bait, respectively, either the RAG1A on-target DSB site or three different RAG1A off-target DSB sites (each on a different chromosome). Based on this finding, we further improved the HTGTS assay by using the RAG1B DSB as a universal donor bait to identify on-target, off-target and low-level wide-spread DSB activities of co-expressed nucleases. Indeed, this approach identified the known EMX1 and VEGFA sgRNA on-target and off-target sites, as well as many additional off-target sites. Thus, this modification of the HTGTS assay should facilitate rapid evaluation of on-target, off-target and low-level wide-spread DSB generating activities of candidate nucleases from fixed bait DSB sites without the need for generating and optimizing bait-site primers. Indeed, this approach can be used to identify engineered nuclease endogenous target sequences genome-wide even in cells that lack a known ‘on-target’ site for the nuclease tested as we showed for I-SceI.
The frequency of off-target sites for the four RAG1 Cas9:sgRNAs tested varied considerably, with two showing no detectable off-target activity. If desired, HTGTS could be scaled-up for even greater sensitivity and sensitivity also could be enhanced by performing HTGTS from target sites on each individual chromosome to increase identification of off-target sites on given chromosomes due to 3D proximity effects27,33. HTGTS confirmed that off-target activity of the RAG1A sgRNA was dramatically suppressed genome-wide via the Cas9 D10 nickase approach35,36; but also revealed that this approach does not suppress translocations involving DSBs on both bait-site chromosomes. While two tested TALENs had numerous off-target sites, a large fraction appeared to be generated by TALEN homodimers; thus, emerging approaches to enforce TALEN heterodimerization43 should greatly reduce TALEN off-target activity. Finally, we find that HTGTS also may be used to optimize specific versus wide-spread, low-level DSB-inducing activities via ‘titration’ of engineered nuclease levels. Given the wide-ranging variations in engineered nuclease on-target versus off-target activities, such titration could greatly facilitate specific custom nuclease design.
ONLINE METHODS
Plasmid DNA construction
Cas9 gRNAs targeting the RAG1 locus were cloned into pX330 or pX335 (Addgene plasmids 42230 and 42335 respectively; Feng Zhang) as described5. The following gRNA targeting sequences (PAM) were used for Cas9 targeting: RAG1A: GCCTCTTTCCCACCCACCTT (GGG), RAG1B: GACTTGTTTTCATTGTTCTC (AGG), RAG1C: GCACCTAACATGATATATTA (AGG), RAG1D: GACCTTAAGGTTTTTGTGGA (AGG), RAG1E: GCCATGCTGGCTGAGGTACCT (GAG), RAG1F: GTACCTGAGAACAATGAAAAC (AAG), RAG1G: GAAAGAGGCTGCCATGCTGGCTG (AGG). Guide RNAs for EMX1: GAGTCCGAGCAGAAGAAGAA (GGG) and VEGFA: GGGTGGGGGGAGTTTGCTCC (TGG) corresponded to T4 and T1 respectively in the prior study15. The EcoRI/XhoI-cleaved I-SceI cDNA from the pMX-I-SceI vector was cloned into the EcoRI/SalI-cleaved pHR’-IRES-eGFP vector to generate pHR’-I-SceI-IRES-eGFP.
Cell lines and transfection
293T and A549 cells were maintained at 37°C, 5% CO2 and cultured in DMEM with glutamine supplemented with 10% FCS and 0.5% penicillin/streptomycin (Invitrogen). 293T translocation libraries were prepared by CaPO4 co-transfection in 10cm dishes of either 20µg pX330 or 20µg each pX335 gRNA combinations with 5µg pCMX-eGFP followed by FACS analysis of GFP and DNA isolation 48 hours post-transfection. A549 cells (Courtesy of David Weinstock, Dana-Farber Cancer Institute) were nucleofected with 2µg or 10µg per 2 ×106 cells using the SF cell line kit (Lonza) and the CM130 program. A549 cells were cultured for 48 hours prior to DNA isolation. For Cas9/I-SceI co-expression studies, pX330 gRNA vectors were co-transfected with pHR-I-SceI followed by FACS analysis 48 hours post-transfection. TALEN pairs (previously generated by FLASH asssembly37) corresponding to ATM and MYC targeting (Addgene plasmids 36805/36806 and 36713/36714 respectively; Keith Joung)37 were co-transfected into 293T using 20µg each—or otherwise indicated—with 5µg pCMX-eGFP and cultured for 48 hours prior to FACS and DNA isolation. In some experiments 293T cells were gamma irradiated with 7Gy 24hrs post-transfection.
High-Throughput Genome-Wide Translocation Sequencing
General overview of the original emulsion-PCR method is described previously23. Junction cloning involved sonication (700bp-1.5kb fragment target size), end-repair (T4 DNA polymerase, Polynucleotide kinase, and Klenow Fragment DNA polymerase; New England Biolabs - NEB), A-tailing, adapter ligation, blocking digest to remove germline sequence, and locus-specific nested priming coupled with step-out adapter priming to both enrich for captured junctions while suppressing adapter-ended fragments (Supplementary Table 8). Libraries included illumina paired-end sequence for MiSeq along with the inclusion of extra nucleotides of variable length to enhance diversity of reads from the same locus. Illumina paired-end sequence-specific primer tails (I5, I7 sequence added to nested and adapter primers respectively) were used for emulsion PCRII followed by PCRIII primers, P5 and P7, recognizing I5 and I7 sequences respectively to reconstruct the requisite P5I5 and P7I7 sequences necessary for Miseq sequencing. Primers for HTGTS are listed (Supplementary Table 8). Taq polymerase (Qiagen) was used for all PCR translocation cloning steps. RAG1A-D translocation cloning used EcoNI (NEB) to minimize amplification of germline fragments. RAG1 A/B HTGTS cloning conditions included the following: Biotin-PCRI: 94°C 120s; 94°C 20s; 66°C 60s; 72°C 60s; 20 cycles; 72°C 600s. Emulsion PCRII: 94°C 120s; 94°C 20s; 66°C 30s; 72°C 60s; 30 cycles; 72°C 600s. PCRIII: 94°C 120s; 94°C 20s; 64°C 60s; 72°C 60s; 12 cycles; 72°C 600s. RAG1 C/D HTGTS PCR conditions were identical to RAG1 A/B cloning but instead used 60°C annealing for all steps.
HTGTS libraries were also generated using a modified protocol which involved linear amplification-mediated (LAM)-PCR28 and bridge adapter ligation44 to bypass end-repair and A-tailing steps followed by nested illumina sequence-tailed PCR, blocking digest, and a step-out tagging PCR to suppress germline sequence and to incorporate illumina sequence tags for sequencing. This method is typically performed over 2 days with very little hands-on time the first day. Together with other HTGTS components (transfection, genomic DNA isolation, sequencing, filtering, and analysis) the entire process is complete in less than 1 week. Briefly, sonicated DNA was subjected to LAM-PCR using Taq polymerase and the single biotinylated primer for 50 cycles. More Taq polymerase was added to the reaction mixture and proceeded for an additional 50 cycles. The 50 cycle PCR for all sites tested consisted of the following conditions: 94°C 180s; 94°C 30s; 58°C 30s; 72°C 90s; 50 cycles; 72°C 600s. Biotinylated DNA fragments were bound to MyOne C1 streptavidin beads (Invitrogen) prior to overnight on-bead ligation with bridge adapters (Supplementary Table 8) in the presence of 15% PEG-8000 (Sigma) and 1mM hexammine cobalt chloride (Sigma). Ligation conditions were as follows: 25°C, 60min; 22°C, 120min; 16°C 8–12 hours. Adapter-ligated products were subjected to nested-locus PCR for 15 cycles with primer tails corresponding to illumina I5 and I7 sequences, digested with EcoNI to block germline sequence accumulation, and a final PCR for another 10–15 cycles with P5 and P7 primers to fully reconstruct Miseq sequence tags (Supplementary Table 6).
Sequence Analysis and Hotspot Identification
Miseq reads were de-multiplexed and adapter sequence trimmed using the fastq-multx tool from ea-utils (http://code.google.com/p/ea-utils/) and the SeqPrep utility (https://github.com/jstjohn/SeqPrep) respectively. Reads were mapped to the hg19 reference genome using Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml) with the top fifty alignments reported that had an alignment score above 50, representing a perfect 25nt local alignment. On average, 94% of de-multiplexed reads per library harbored a bait sequence alignment with <10% of these reads containing an alignable prey junction. We used a best-path searching algorithm to select the optimal sequence of alignments that describe the read’s composition, typically finding the bait and prey alignments. This approach was inspired by the YAHA read aligner and breakpoint detector45. Aligned reads were filtered on the following conditions: (1) reads must include both a bait alignment and a prey alignment and (2) the bait alignment cannot extend more than 10 nucleotides beyond the targeted site. For vector controls and off-set nicking with multiple sites, the distal targeted site was used. We compared discarded alignments to the selected prey alignment; if any of the discarded alignments surpassed both a coverage and score threshold with respect to the prey alignment, the read was filtered due to low mapping quality. To remove possible mispriming events and other artifacts, the bait alignment must extend 10 nucleotides past the primer. We removed potential duplicates by comparing the coordinates of the end of the bait alignment and the start of the prey alignment across all reads. A read will be marked as a duplicate if it has a bait alignment off-set within 2nt and a prey alignment offset within 2nt of another read’s bait and prey alignments. Post-filter stringency was applied to remove junctions with gaps larger than 30nt and bait sequences shorter than 50nt. Reads with prey alignments to telomere repeat sequences were also removed. Genome mixing experiments were similarly filtered as described above but with using a combined hg19/mm9 reference.
Identification of enriched regions was performed using the MACS2 software46, designed for ChIP-seq peak calling, which gave similar results to the previously described method23. Junctions associated with MACS-defined peaks (FDR-adjusted p-value enrichment threshold of 10−9) were extracted for further analysis. Hotspots were defined as having significant focal enrichment and present in more than one biological replicate library. Hotspots proximal to the breaksite (~100kb) were excluded from analysis. Off-target sites were defined as hotspots that contained genomic sequence differing from the on-target sequence by less than or equal to ½ the targeted sequence length.
Code Availability
Programs/scripts used for this manuscript are listed above and described elsewhere. Details of additional parameters not described above are available upon request.
I-SceI Off-Target in vitro Digestion and T7EI Cleavage Assay
I-SceI off-target sites were amplified from 293T DNA using phusion polymerase (NEB) and standard PCR conditions. Purified amplicons were digested with I-SceI for 1 hour at 37°C. T7EI assays were performed as described previously37 using phusion polymerase and standard PCR conditions followed by ethanol precipitation prior to denaturation, reannealing, T7EI digestion of amplicons and agarose gel electrophoresis. Primers for each site are listed (Supplementary Table 8). EMX1 and VEGFA on- and off-target site primers are described elsewhere15. To derive the fraction cleaved we measured germline (amplicon) band intensities (quantified by ImageJ) and used the following formula:
where IC+ is the intensity of T7EI digested nuclease-expressed sample, IN+ is the intensity of mock digested (no T7EI) nuclease-expressed sample, IC− is the intensity of T7EI digested nuclease-deficient sample, and IN− is the intensity of the mock digested nuclease-deficient sample.
Western analysis
293T cell protein lysates were separated via SDS-PAGE. Antibodies to detect FLAG-Cas9 (FLAG; Sigma F7425) and Tubulin (loading control; Sigma T5168) were used at 1:2000 and 1:5000 respectively.
Nucleic Acid Multiple Sequence Alignment
Sequence logos for Cas9:gRNA and I-SceI off-target sites were generated by using the weblogo interface at weblogo.berkeley.edu.
Statistical Analysis
Where appropriate, data were expressed as mean ± S.E.M.. Two-way ANOVA and Tukey post-tests were performed to compare the same individual OT site frequencies from two different libraries. P-values less than 0.05 were considered significant.
Supplementary Material
Acknowledgements
This work is supported by the National Institutes of Health Grants P01CA109901 and P01AI076210. R.L.F. was supported by the National Institutes of Health NRSA T32CA070083 and is supported by the National Health Institutes of Health NRSA T32AI007512. J.H. is supported by Robertson Foundation/Cancer Research Institute Irvington Fellowship. F.W.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions: R.L.F, J.H. and F.W.A. designed the research; R.L.F., J.H., and E.K. performed the research; R.L.F., J.H., R.M.M., Y.J.H., and E.K. analyzed the data; R.L.F. and F.W.A. wrote the paper.
Competing Financial Interest: None
References
- 1.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 3.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 11.Mussolino C, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle EL, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao A, et al. CasOT: a genome-wide Cas9/sgRNA off-target searching tool. Bioinformatics. 2014;30:1180–1182. doi: 10.1093/bioinformatics/btt764. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 18.Petek LM, Russell DW, Miller DG. Frequent endonuclease cleavage at off-target locations in vivo. Mol. Ther. 2010;18:983–986. doi: 10.1038/mt.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung YH, et al. Knockout mice created by TALEN-mediated gene targeting. Nat. Biotechnol. 2013;31:23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 21.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 22.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein IA, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Tepsuporn S, Meyers RM, Gostissa M, Alt FW. Developmental propagation of V(D)J recombination-associated DNA breaks and translocations in mature B cells via dicentric chromosomes. Proc. Natl Acad. Sci. USA. 2014;111:10269–10274. doi: 10.1073/pnas.1410112111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barlow JH, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt M, et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 29.Lee YN, et al. A systematic analysis of recombination activity and genotype-phenotype correlation in human recombination-activating gene 1 deficiency. J. Allergy Clin. Immunol. 2014;133:1099–1108. e1012. doi: 10.1016/j.jaci.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munoz IG, et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 2011;39:729–743. doi: 10.1093/nar/gkq801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gostissa M, et al. IgH class switching exploits a general property of two DNA breaks to be joined in cis over long chromosomal distances. Proc. Natl Acad. Sci. USA. 2014;111:2644–2649. doi: 10.1073/pnas.1324176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asaithamby A, Chen DJ. Cellular responses to DNA double-strand breaks after low-dose gamma-irradiation. Nucleic Acids Res. 2009;37:3912–3923. doi: 10.1093/nar/gkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franco S, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Molecular cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Ramiro AR, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nature reviews. Genetics. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guilinger JP, et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat. Methods. 2014;11:429–435. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou ZX, et al. Mapping genomic hotspots of DNA damage by a single-strand-DNA-compatible and strand-specific ChIP-seq method. Genome Res. 2013;23:705–715. doi: 10.1101/gr.146357.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faust GG, Hall IM. YAHA: fast and flexible long-read alignment with optimal breakpoint detection. Bioinformatics. 2012;28:2417–2424. doi: 10.1093/bioinformatics/bts456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.