Significance
Mosquitoes transmit some of the most devastating human diseases. Acquisition of blood initiates a cascade of events in various tissues in the female mosquito. Here we describe our study using newly established genetic tools in mosquito biology and a comprehensive microRNA target identification analysis to characterize microRNA-8 (miR-8) functionally in Aedes aegypti female mosquitoes. miR-8 works in the fat body by regulating its target, secreted wingless-interacting molecule. miR-8 acts on the long-range Wingless signaling pathway and plays an essential role in the female mosquito fat body controlling the secretory activity of yolk protein precursors required for oocyte development. Thus, this study identifies the fat body specific action of miR-8 and its target in regulating mosquito reproduction.
Keywords: small RNA, microRNA, Wingless signaling, reproduction, mosquito
Abstract
Female mosquitoes require a blood meal for reproduction, and this blood meal provides the underlying mechanism for the spread of many important vector-borne diseases in humans. A deeper understanding of the molecular mechanisms linked to mosquito blood meal processes and reproductive events is of particular importance for devising innovative vector control strategies. We found that the conserved microRNA miR-8 is an essential regulator of mosquito reproductive events. Two strategies to inhibit miR-8 function in vivo were used for functional characterization: systemic antagomir depletion and spatiotemporal inhibition using the miRNA sponge transgenic method in combination with the yeast transcriptional activator gal4 protein/upstream activating sequence system. Depletion of miR-8 in the female mosquito results in defects related to egg development and deposition. We used a multialgorithm approach for miRNA target prediction in mosquito 3′ UTRs and experimentally verified secreted wingless-interacting molecule (swim) as an authentic target of miR-8. Our findings demonstrate that miR-8 controls the activity of the long-range Wingless (Wg) signaling by regulating Swim expression in the female fat body. We discovered that the miR-8/Wg axis is critical for the proper secretion of lipophorin and vitellogenin by the fat body and subsequent accumulation of these yolk protein precursors by developing oocytes.
Female hematophagous mosquitoes require a blood meal for reproduction, resulting in the transmission of many devastating vector-borne diseases in humans. Lack of vaccines, increasing drug resistance in pathogens, and insecticide resistance in vectors add to the urgency of exploring alternative strategies of vector control. Acquisition of blood initiates a cascade of events in several tissues in the female mosquito. The fat body, an adipose tissue analogous to the mammalian liver, plays a prominent role in energy metabolism, immunity, and reproduction. The adult female mosquito fat body undergoes dynamic changes before and after a blood meal to accommodate ovarian development and other physiological necessities (1–3). The fundamental step in these events is vitellogenesis, in which the intake of blood induces the synthesis and secretion of yolk protein precursors (YPPs) in the fat body and subsequent accumulation in the developing oocytes (1, 4). Despite the continuing efforts to identify factors controlling mosquito reproduction, studies regarding the role the fat body plays during ovarian development have not revealed the entire governing complexity of mosquito reproductive biology.
The discovery of small noncoding RNAs has revolutionized our understanding of complex gene networks. One class of endogenous small RNAs, known as microRNAs (miRNAs), plays key regulatory roles in gene expression at the posttranscriptional level (5). Animal miRNAs exert their action by base pairing of the miRNA “seed” to complementary sites within the 3′ UTR of mRNA, resulting in translational inhibition and mRNA decay (5, 6). Although miRNA function is a heavily studied topic in model organisms, such as the fruit fly Drosophila melanogaster, the exact functional role of miRNAs in mosquito biology remains largely unknown (5, 7). However, recent functional studies of specific miRNAs have linked miRNA action in mosquitoes to reproductive events (7, 8). Studying miRNA function in the female mosquito is crucial for developing a full understanding of the regulatory processes of mosquito reproduction and may pave the way toward the utilization of these small molecules as novel approaches to control.
The miR-8/miR-200 family of miRNAs is one of the most widely studied miRNA families, and its members have emerged as important regulators in animal development and disease (5, 9, 10). Previously, we reported that miR-8 exhibits high levels of expression in the female mosquito fat body post-blood meal (PBM) (11), suggesting that miR-8 might have a role in the regulation of female mosquito adult stages. Here, we describe our study to characterize miR-8 in the female mosquito fat body functionally and to establish an interaction between miR-8 and its target, secreted wingless-interacting molecule (swim). This study provides evidence that miR-8 acts on the Wingless (Wg) signaling pathway and functions in the fat body of the female mosquito, affecting reproductive processes. miR-8 functions as a regulator of the Aedes aegypti homolog of Drosophila Swim, thereby controlling the long-range Wg signaling activity. Thus, we identified the role of miR-8 and Wg/Swim in the secretory activity of the fat body in an adult female insect and elucidated their roles in reproduction.
Results
miR-8 Is Enriched in the Female Mosquito Fat Body After Blood Feeding.
To produce a thorough time-course expression analysis of mature miR-8 in the adult female mosquito fat body, we measured relative levels of mature miR-8 expression by quantitative real-time (qRT)-PCR analysis. Mature miR-8 levels were low in all tissues posteclosion (PE); however, expression levels increased substantially in the female mosquito fat body PBM (Fig. S1A). We monitored the abundance of mature miR-8 in the fat body using eight time points collected over the first reproductive cycle. We obtained total RNA samples from female mosquito fat bodies at 0–6, 24, 48, and 72 h PE and at 12, 24, 36, 48, and 60 h PBM. In this tissue, mature miR-8 was up-regulated significantly by 12 h PBM, reaching its peak by 48 h PBM and declining by 60 h PBM (Fig. S1B). These results suggest that the high expression of miR-8 in the female mosquito fat body may play an important role in tuning events associated with the adult life stage of the female mosquito.
Systemic miR-8 Depletion Results in Abnormal Ovarian Development.
To assess the function of miR-8 in the adult female mosquito, we achieved a knockdown using an miRNA-specific antisense oligonucleotide (antagomir). We designed an antagomir consisting of the reverse complement of miR-8 (miR-8Ant) and a randomly scrambled “missense” antagomir (MsAnt) for a control. Female mosquitoes were microinjected with miR-8Ant or MsAnt at a dose of 50 pmol per mosquito at 12 h PE. To evaluate the efficiency of the miR-8 depletion, we monitored endogenous levels of mature miR-8 by qRT-PCR. Female mosquitoes treated with the miR-8Ant displayed a depletion of mature miR-8 in the fat body by 24 h PBM compared to the MsAnt and noninjected control treatments (Fig. S1C). miR-8Ant had no effect on the relative expression levels of other miRNAs, such as mature miR-275 (Fig. S1D).
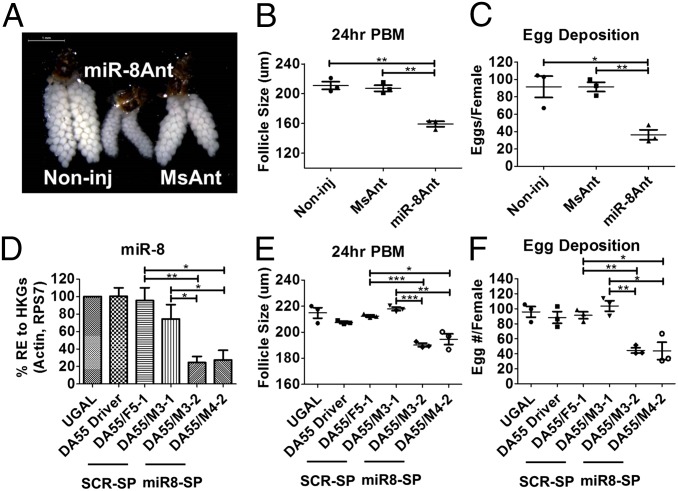
Female mosquitoes treated with miR-8Ant were screened for phenotypic manifestations. First, we evaluated the state of ovarian development in mosquitoes at 24 h PBM. Unlike the MsAnt and noninjected controls, blood-fed females treated with the miR-8Ant failed to develop fully vitellogenic ovaries during the first gonadotrophic cycle (Fig. 1A). The degree to which females treated with the miR-8Ant displayed this phenotype varied considerably across individuals. Ovarian follicle growth was inhibited drastically in miR-8–depleted females compared with control mosquitoes, with an average primary follicle length of 154.7 µm (Fig. 1B and Fig. S2C). Ovaries from MsAnt-treated females were similar to those in noninjected female mosquitoes at 24 h PBM, with primary follicles reaching an average length of 200–210 µm (Fig. 1B and Fig. S2 A and B). In addition, miR-8Ant–treated females displayed reduced fecundity, laying significantly fewer eggs (36.3 eggs per mosquito) than controls (Fig. 1C): MsAnt-treated and noninjected female mosquitoes laid a similar number of eggs, respectively 92.8 and 91.63 eggs per female on average (Fig. 1C). Previtellogenic ovary development (Fig. S2D), host-seeking behavior, and blood digestion were unaffected in miR-8Ant–treated females, and these females displayed no other adverse phenotypes when compared with MsAnt-treated and noninjected controls.
Fig. 1.
Systemic and tissue-specific depletion of miR-8 results in decreased follicle size and dramatically reduced egg number. (A) Female mosquito ovaries at 24 h PBM. Ovaries were visualized using the Leica M165FC stereo microscope. (Scale bar: 1 mm.) (B) Average follicle size of miR-8Ant– and MsAnt-treated and noninjected mosquitoes at 24 h PBM. (C) Egg numbers per female mosquito for miR-8Ant– and MsAnt-treated and noninjected mosquitoes. (D) Mature miR-8 levels are decreased in Vg-Gal4/UAS–miR-8-SP (DA55/M3-2; DA55/M4-2) female mosquitoes as compared with controls. (E) Average follicle size of Vg-Gal4/UAS–miR-8-SP (DA55/M3-2; DA55/M4-2) miR-8–depleted females compared with Vg-Gal4/UAS–Scr-SP (DA55/M3-1; DA55/F5-1), Vg-Gal4 (DA55) driver, and WT controls 24 h PBM. Data represent averages. (F) Egg number per female mosquito for Vg-Gal4/UAS–miR-8-SP (DA55/M3-2; DA55/M4-2) females compared with Vg-Gal4/UAS–Scr-SP (DA55/M3-1; DA55/F5-1), Vg-Gal4 (DA55) driver, and WT controls. Data represent three biological replicates with three technical replicates and are shown as average ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
Spatiotemporal Inhibition of miR-8 Reveals Function Specific to the Female Mosquito Fat Body.
Antagomir-mediated miRNA depletion is based on microinjections, creating systemic effects without a clear indication of tissue specificity. Therefore, we asked if the miR-8Ant phenotype is a result of miR-8 reduction in the fat body or in another tissue. To answer this question, we achieved a spatiotemporal inhibition of miR-8 using the miRNA sponge (miR-SP) transgenic method. Because of the high conservation of miR-8, a Drosophila upstream activating sequence (UAS)–miR-8-SP cassette was used (12) in combination with the previously established Aedes Vitellogenin (Vg)-linked yeast transcriptional activator Gal4 protein/UAS system (13). The fat body-specific system uses the Vg gene promoter to drive transgene expression (13), permitting precise in vivo spatiotemporal genetic testing of miR-8 functions in the female mosquito fat body PBM (Fig. S3A).
The UAS–miR-8-SP and control “scramble” (UAS–Scr-SP) cassettes were subcloned into a piggyBac-based responder construct. Responder plasmids contained five repeat concatemers of the UAS, with a TATA box and hsp70 minimal promoter linked to the miR-SP cassette and an SV40 polyadenylation sequence (Fig. S3B). Responder transposons also included a dsRed-selectable marker gene under the 3xP3 eye-specific promoter (Fig. S3B). Responder constructs were sent to the Insect Transformation Facility at the University of Maryland for injection into Ae. aegypti Orlando strain. Three independent UAS–miR-8-SP responder lines and three independent UAS–Scr-SP lines were generated. Responder lines were selected based on a strong expression of eye-specific dsRed-selectable marker. Genomic PCR analysis was conducted to confirm the stable incorporation and the integrity of UAS–miR-8-SP and UAS–Scr-SP constructs into the Ae. aegypti genome (Fig. S3C). Vg-Gal4/UAS–miR-8-SP and Vg-Gal4/UAS–Scr-SP hybrid transgenic lines were produced by crossing homozygous responder lines with homozygous Vg-Gal4 driver. Genomic PCR analysis confirmed the presence of both transgenes in hybrid lines (Fig. S3D). Hybrid transgenic lines were selected by the simultaneous presence of both EGFP and dsRed eye-specific markers (Fig. S4).
For further analyses, we selected two Vg-Gal4/UAS–miR-8-SP and two Vg-Gal4/UAS–Scr-SP hybrid lines based on a strong expression of the eye-specific marker genes (Fig. S4). We analyzed mature miR-8 levels by means of qRT-PCR in the Vg-Gal4/UAS–miR-8-SP, Vg-Gal4/UAS–Scr-SP, Vg-Gal4 driver lines and in WT females. Mature miR-8 remained unaffected in the Vg-Gal4/UAS–Scr-SP hybrid lines, Vg-Gal4 driver, and WT females, whereas the Vg-Gal4/UAS–miR-8-SP hybrid lines displayed reduced expression of mature miR-8 24 h PBM (Fig. 1D). The Vg-Gal4/UAS–miR-8-SP, Vg-Gal4/UAS–Scr-SP, and Vg-Gal4 driver lines and WT females were subjected to phenotypic analysis. There were no detectable negative effects on mosquito development, fecundity, viability, or sex ratio in the Vg-Gal4/UAS–Scr-SP hybrid and Vg-Gal4 driver lines or WT females.
Similar to miR-8Ant–treated females, blood-fed Vg-Gal4/UAS–miR-8-SP females displayed an inhibition of ovarian development and reduced fecundity. Ovaries from the Vg-Gal4/UAS–Scr-SP hybrid lines and Vg-Gal4 driver lines were similar to those in WT female mosquitoes at 24 h PBM with primary follicles reaching 213.15 µm in length on average (Fig. 1E and Fig. S5 A–C). Ovarian follicle growth was reduced considerably in the Vg-Gal4/UAS–miR-8-SP hybrid lines at 24 h PBM compared with control mosquitoes, with an average primary follicle length of 192.4 µm (Fig. 1E and Fig. S5D). Vg-Gal4/UAS–miR-8-SP hybrid follicles were very heterogeneous in size compared with controls, ranging from 88–240 µm in length. Additionally, blood-fed Vg-Gal4/UAS–miR-8-SP hybrid females laid significantly fewer eggs (an average of 44.1 eggs per mosquito) than did controls (Fig. 1F). Additional striking phenotypes were detected in the Vg-Gal4/UAS–miR-8-SP hybrid lines. When female ovaries were observed at 5 d PBM, Vg-Gal4/UAS–miR-8-SP hybrid lines continued to retain eggs, which began to undergo melanization within the ovary, a process that normally is observed after oviposition (Fig. S5 E and F). Previtellogenic ovary development (Fig. S5G), host-seeking behavior, and blood digestion were unaffected in the Vg-Gal4/UAS–miR-8-SP hybrid females, and these females displayed no other adverse phenotypes when compared with controls. Together these results indicate that miR-8 action in the female mosquito fat body is responsible, in large measure, for the miR-8–depletion phenotype in the ovary.
Computational and in Vitro Analyses Indicate miR-8 Targets Swim.
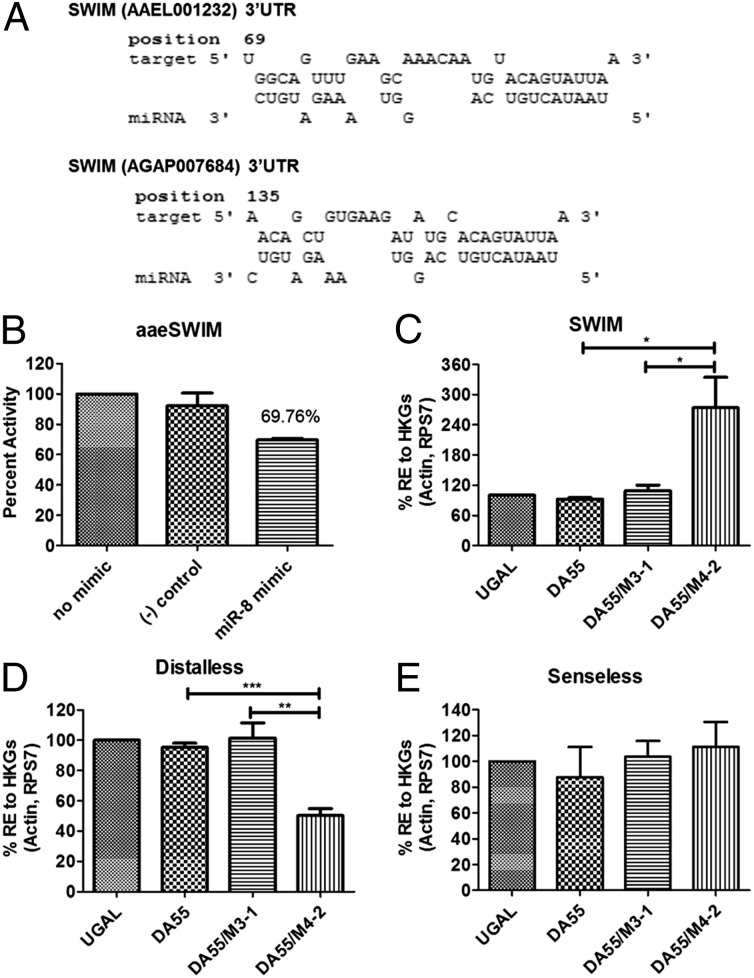
To identify the putative gene targets of miR-8, we took an in silico approach, as previously described (8), using five different target-prediction programs: TargetScan (14), PITA (15), miRanda (16), RNAhybrid (17), and a program developed in house (8). To assess conservation criteria, we predicted miR-8 targets using Ae. aegypti and Anopheles gambiae 3′ UTRs. We extracted the 3′ UTRs from the Ae. aegypti (AaegL1.3) and An. gambiae (AgamP3.7) genome genebuilds for use within each target-prediction program. Overlapping predicted targets between each program were assessed, and a list of top candidate targets was produced based on the predictions by multiple algorithms and conservation (Table S1). The Drosophila ortholog of AAEL001232, Swim, recently has been shown to function in the Wg signaling pathway to maintain the stability of the Wg ligand for long-range Wg signaling (18). Additionally, Drosophila miR-8 is known to target many players in the Wg signaling pathway (19). Because of this knowledge and conservation of the miR-8 binding site in An. gambiae (Fig. 2A), we chose AAEL001232 (aaeSwim) for further analysis.
Fig. 2.
SWIM (AAEL001232) is a direct target of miR-8. (A) Putative miR-8 binding site within the AAEL001232 and AGAP007684 3′ UTRs. (B) Dual luciferase reporter assay for AAEL001232 (Swim). Data represent the percent activity (∆ fold activity × 100). Percentages are shown as the average ± SEM of triplicate samples. (C) AAEL001232 expression increases in Vg-Gal4/UAS–miR-8-SP (DA55/M4-2) female mosquitoes compared with controls (WT, DA55, and DA55/M3-1). (D) Long-range Wg signaling is reduced in Vg-Gal4/UAS–miR-8-SP (DA55/M4-2) female mosquitoes compared with controls (WT, DA55, and DA55/M3-1). (E) Short-range Wg signaling is unaffected in Vg-Gal4/UAS–miR-8-SP (DA55/M4-2) female mosquitoes compared with controls (WT, DA55, and DA55/M3-1). Data represent three biological replicates with three technical replicates and are shown as average ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
Following computational prediction, we assessed the 3′ UTR of aaeSwim for its response to miR-8 in vitro. The aaeSwim 3′ UTR was cloned downstream of the Renilla translational stop codon within the psiCheck-2 vector to generate 3′ UTR-fused luciferase reporters. When transfected into Drosophila Schneider 2 (S2) cells along with the miR-8 mimic, the luciferase reporter containing the full-length aaeSwim 3′ UTR yielded 69.75% luciferase activity compared with the negative control mimic and no-mimic control samples (Fig. 2B). Therefore, aaeSwim was validated as a potential target of miR-8 in vitro by means of a dual luciferase reporter assay in S2 cells. The luciferase reporter assay was optimized using a known Drosophila miR-8 target, dmUSH (Fig. S6A).
miR-8 Regulates Swim in Vivo.
Next, we monitored swim transcript levels in the fat body over the first reproductive cycle. We obtained total RNA samples from female mosquito fat bodies at 72 h PE and at 12, 24, 36, and 48 h PBM. Expression of swim declines early in PBM, increasing in expression by 24 h PBM and declining again by 36 h PBM—corresponding to the highest levels of miR-8 expression (Fig. S6B). To evaluate swim as an authentic miR-8 target in vivo, we measured swim transcript levels in the fat body of miR-8Ant, Vg-Gal4/UAS–miR-8-SP, and control females via qRT-PCR. We found that at 24 h PBM swim is enriched significantly in the fat body of miR-8–depleted female mosquitoes as compared with control females (Fig. 2C and Fig. S6C). In Drosophila, overexpression of swim interferes with long-range Wg signaling in fly imaginal discs (18). We assayed the level of the long-range Wg signaling-response gene distal-less (dll) and the short-range Wg signaling-response gene senseless (sens) in the fat body of miR-8Ant, Vg-Gal4/UAS–miR-8-SP, and control female mosquitoes. Fat body dll expression was significantly lower in miR-8–depleted females than in control females (Fig. 2D and Fig. S6D). Short-range Wg signaling remained largely unaffected in the fat body of the miR-8–depleted female mosquitoes (Fig. 2E and Fig. S6E). Expression of sens and dll was unaffected in the ovaries of miR-8–depleted female mosquitoes as compared with controls (Fig. S6 F and G). Together, these findings suggest that miR-8 depletion in female mosquitoes results in an impairment of long-range Wg signaling in the fat body.
Next, we conducted phenotypic rescue experiments through swim RNAi in miR-8–depleted female mosquitoes. It was expected that the RNAi-mediated knockdown of the physiologically relevant target of miR-8 would alleviate the adverse phenotypes caused by miR-8 depletion. dsRNA designed to target swim transcripts successfully depleted swim transcript levels in female mosquitoes (Fig. 3A). We coinjected 50 pmol of miR-8Ant or MSAnt and 0.5 μg of Swim or luciferase dsRNA into each female mosquito at 12 h PE. The inhibited ovary-development and egg-deposition phenotypes were not observed in the miR-8Ant/dsSwim-treated mosquitoes at 24 h PBM, whereas females treated with miR-8Ant and luciferase dsRNA control displayed the inhibited ovary-development and reduced egg-deposition characteristic of miR-8Ant–treated mosquitoes (Fig. S6 H and I). Additionally, the inhibited ovary-development and egg-deposition phenotypes were not observed in the MsAnt/dsSwim-treated and noninjected control mosquitoes at 24 h PBM (Fig. S6 H and I). Likewise, injection of Swim dsRNA into Vg-Gal4/UAS–miR-8-SP mosquitoes alleviated the inhibited egg-deposition phenotypes observed in Vg-Gal4/UAS–miR-8-SP females (Fig. 3B). Hence, Swim RNAi dramatically restored proper ovarian development in miR-8–depleted female mosquitoes, suggesting that swim is an authentic target of miR-8 in vivo.
Fig. 3.
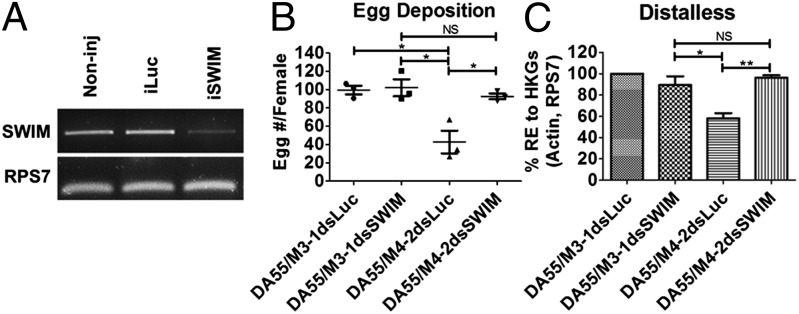
AAEL001232 RNAi rescues miR-8–depletion phenotypes. (A) Female mosquitoes were injected with 0.5 µg AAEL001232 (Swim) or luciferase dsRNA. Swim RNAi successfully knocks down swim transcripts in vivo. (B) Swim RNAi restores the egg-deposition phenotype of Vg-Gal4/UAS–miR-8-SP (DA55/M4-2) mosquitoes. (C) Swim RNAi restores long-range Wg signaling inhibition in Vg-Gal4/UAS–miR-8-SP (DA55/M4-2) mosquitoes. Data represent three biological replicates with three technical replicates and are shown as average ± SEM; *P < 0.05; **P < 0.01; NS, not significant.
For further in vivo analysis, we determined whether swim RNAi would restore dll expression to normal levels in the fat bodies of miR-8Ant and Vg-Gal4/UAS–miR-8-SP female mosquitoes. Although miR-8Ant/dsLuc–treated mosquitoes displayed the characteristic decrease in dll expression observed in miR-8–depleted female mosquitoes, miR-8Ant/dsSwim mosquitoes displayed restored levels of dll expression similar to those in the control treatments (Fig. S6J). Likewise, injection of Swim dsRNA into Vg-Gal4/UAS–miR-8-SP mosquitoes restored dll expression similar to levels in control mosquitoes (Fig. 3C). Together, these results suggest that elevated levels of the swim transcript in miR-8Ant–treated and Vg-Gal4/UAS–miR-8-SP female mosquitoes inhibit ovary development PBM and dramatically reduce fecundity, possibly because of a disruption of long-range Wg signaling.
To investigate further whether Wg signaling is functioning in the female mosquito fat body, we measured the relative expression levels of several putative Wg signaling pathway components from female mosquito fat bodies 72 h PE and 24 h PBM via qRT-PCR. Comparison of the 72-h PE and 24-h PBM samples revealed that transcript levels of the Wg signaling pathway receptors Frizzled-1 and Frizzled-2 and a downstream component of the Wg signaling pathway, Armadillo, displayed significant up-regulation in the female mosquito fat body at 24 h PBM (Fig. S7 A–D). Likewise, the expression of several Wg protein precursors tested also increased PBM (Fig. S7 E–J). These results suggest that Wg signaling components are indeed present in the female mosquito fat body PBM and further support our finding that Wg signaling plays a role in the female mosquito fat body.
miR-8 Depletion Results in Impaired YPP Secretion by the Fat Body and Lipid Accumulation in Developing Oocytes.
Overexpression of Wg in the Drosophila fat body results in lethality at the pupal stage and a reduction of fat body mass in third-instar larvae, demonstrating the role of Wg signaling in the fat body for fat regulation (20). Likewise, activation of the Wnt pathway in adipose tissue decreases fat mass in mammals (21, 22). Together with our findings, this effect may suggest that improper Wg signaling, such as the disruption of miR-8 expression, may interfere with normal fat body functions, including the synthesis and secretion of YPPs by the female mosquito fat body. To test this hypothesis, Vg and lipophorin (Lp) transcript levels were measured at 24 h PBM in the female fat body via qRT-PCR analysis. Treatment with miR-8Ant did not affect the expression of Vg (Fig. S8A) or Lp (Fig. S8B) as compared with MsAnt-treated and noninjected controls. Western blot analysis of Vg and Lp proteins revealed no significant change in protein levels in miR-8Ant female fat bodies as compared with controls (Fig. 4A), indicating that miR-8 is not needed for YPP gene expression in the fat body. However, treatment with miR-8Ant resulted in a drastic reduction of YPP protein levels in the ovary as compared with MsAnt-treated and noninjected controls (Fig. 4B), suggesting a disruption of the secretion into and/or uptake of YPP by miR-8–depleted female ovaries. To test this notion, we performed an in vitro fat body culture in which fat bodies were dissected from miR-8Ant–treated or control females 4 d after microinjection and were incubated in a complete culture medium supplemented with amino acids and 20-hydroxyecdysone. The culture medium was harvested to test proteins secreted by the fat body. Treatment with the miR-8Ant resulted in a drastic reduction of YPP protein levels in the culture medium as compared with MsAnt-treated and noninjected controls (Fig. 4C) with equal amounts of total protein (Fig. S8C), verifying a disruption of YPP secretion by the fat body. Nuclear (DAPI) staining of miR-8Ant–treated female ovaries revealed small primary follicles and large nurse cells (Fig. S8D), characteristic of impaired ovarian development, as compared with MsAnt-treated and noninjected controls (Fig. S8 E and F). Lipids that accumulate in developing oocytes originate in the fat body and are transported to the ovaries by Lp and Vg (23). Depletion of Lp in Drosophila imaginal discs causes a reduction in the transcriptional activity of long-range Wg target genes (24), suggesting a link between Lp and the Wg pathway. Nile Red staining of lipids in the miR-8Ant–treated mosquito ovaries depicted a drastic reduction in lipid content (Fig. S8G) compared with controls (Fig. S8 H and I). Together these results suggest that the miR-8/Wg axis is important for secretion of YPPs into and for lipid accumulation in the developing oocyte.
Fig. 4.

miR-8–depleted females display a disruption in YPP secretion and lipid accumulation. (A) Western blot analyses using antibodies against Vg and Lp in fat bodies from miR-8Ant– and MsAnt-treated and noninjected female mosquitoes. β-Actin was used as a loading control. (B) Western blot analyses using antibodies against Vg and Lp in ovaries from miR-8Ant– and MsAnt-treated and noninjected female mosquitoes. β-Actin was used as a loading control. (C) Western blot analyses of proteins secreted during in vitro fat body culture using antibodies against Vg and Lp from miR-8Ant– and MsAnt-treated and noninjected female fat bodies.
Discussion
Our results show that depletion of the conserved miRNA, miR-8, in the female mosquito fat body PBM results in severe defects linked to ovary development and egg deposition. Using the Gal4–UAS system in combination with a miR-8 sponge in Ae. aegypti, this study represents a significant step toward defining the regulatory roles of miRNAs in a tissue-specific manner in mosquito disease vectors. Our work shows that miR-8 functions as a regulator of reproductive events in the female mosquito fat body through fine-tuning the expression of the Ae. aegypti homolog to Drosophila, Swim, thereby regulating the activity of long-range Wg signaling. Systemic antagomir and fat body-specific depletion of miR-8 resulted in dramatic impairment of ovarian development and egg deposition: Females displayed ovaries with drastically smaller primary follicles and failed to deposit eggs properly, suggesting impairment in signaling between the fat body and ovary.
A multialgorithm approach for miRNA target prediction was used to identify the physiologically relevant miR-8 target contributing to the miR-8–depletion phenotypes. We identified Swim as a direct target of miR-8 in vitro and in vivo. A dual luciferase reporter assay using a luciferase reporter vector containing Ae. aegypti swim 3′ UTR cotransfected along with the miR-8 mimic resulted in a decrease in Renilla luciferase activity in vitro, whereas swim RNAi rescue experiments recovered the miR-8–depletion phenotype in vivo. These results further confirmed Swim as an authentic miR-8 target in mosquitoes. Drosophila and Ae. aegypti Swim contain a Somatomedin B-like domain, implicating functions in protein–extracellular matrix interactions (2, 18). Likewise, the hydrophobic Wg morphogen has been shown to diffuse through the extracellular space and regulate long-range targets by associating with solubilizing molecules such as Swim (18). In Drosophila, overexpression of Swim in imaginal discs results in a disruption of Wg signaling, and in vitro analysis indicated that increased concentrations of Swim outcompete the Wg receptor for access to the Wg ligand (18). Indeed, depletion of miR-8 in the female mosquito fat body results in increased expression of swim transcript level and subsequent reduction in long-range Wg signaling activity in the fat body of the female mosquito. Rescue experiments also restored long-range Wg signaling in the female mosquito fat body. To our knowledge, the function of Wg signaling in the fat body of a reproducing female insect had not been uncovered previously. However, it has been shown that Wg signaling plays a role in regulating follicle stem cells in the Drosophila ovary (25, 26). More recently, it was shown that muscle-derived Wg controls fat mass within the fat body in Drosophila, suggesting that secreted Wg may transmit to and act on distant organs (20). However, systemic and fat body-specific depletion of miR-8 in the female mosquito did not appear to disrupt long-range Wg signaling in the ovary. Taken together, these results indicate that miR-8 regulates long-range Wg signaling through the Ae. aegypti Swim molecule in the fat body of the female mosquito and consequently affects ovarian development and egg deposition.
In Drosophila, overexpression of Wg in the fat body results in lethality at the pupal stage and decreased fat body mass in third-instar larvae, demonstrating the role of Wg signaling in the fat body for fat regulation (20). Likewise, activation of the Wnt pathway in adipose tissue decreases fat mass in mammals (21, 22, 27). Together with our findings, this effect may suggest that a distortion in proper Wg signaling may interfere with normal fat body functions, having consequences on mosquito reproductive processes. The drastically smaller primary ovarian follicles in the miR-8–depleted females suggested that YPP synthesis, secretion, and/or uptake were compromised. However, there was no significant difference in mRNA and protein expression of the two important YPP genes, Vg and Lp, in miR-8–depleted female mosquitoes. Instead, we identified a disruption of YPP secretion by the fat body. Swim and miR-8 are likely to fine tune Wg signaling in the fat body, allowing proper YPP secretion and leading to normal ovarian development.
In conclusion, our work has established a fundamental role for miR-8 in the female mosquito fat body. The investigation of miR-8 targets has revealed an intriguing role for the Wg signaling pathway in the female mosquito fat body and reproduction. Further investigation of the Wg signaling pathway in the adult mosquito fat body is warranted to determine the exact function of fat body Wg and its role in the regulation of the YPP secretory pathway.
Materials and Methods
Mosquito Rearing.
The Ae. aegypti WT UGAL/Rockefeller strain and transgenic lines were reared at 27 °C and 80% humidity, as described previously (28). Blood feeding of adult mosquitoes was performed using White Leghorn chickens.
DNA Transformation Constructs.
The Vg-Gal4 driver and pBac[3xP3-DsRed] transformation vectors were produced as previously described (13). The germline transformation vectors carrying the UAS–miR-8-SP and UAS–Scr-SP were constructed as described below. Drosophila miR-8-SP and nonspecific control (UAS–Scr-SP) cassettes in a pUAST-EGFP vector were provided by Tudor Fulga, University of Oxford, Oxford (12). The 4.0-kb UAS–miR-8-SP BamHI fragment was subcloned into the pBac[3xP3-DsRed] transformation vector using the BglII unique cloning site.
Germline Transformation.
The germline transformation for the Vg-Gal4 driver line was performed as described previously (13, 29). PiggyBac-mediated germline transformation for the UAS–miR-8-SP and UAS–Scr-SP responder lines was carried out by the University of Maryland Institute for Bioscience and Biotechnology Research Insect Transformation Facility. The Vg-Gal4/UAS–miR-8-SP and Vg-Gal4/UAS–Scr-SP hybrid mosquito lines were established as previously described (13, 29).
Antagomir and dsRNA Treatments.
Antagomirs were obtained from Dharmacon using the RNA module for custom single-stranded RNA synthesis available at dharmacon.gelifesciences.com/rnai-and-custom-rna-synthesis/custom-rna-synthesis/single-strand-rna-synthesis/. Antagomir to miR-8 was 5′mG.*.mA.*.mC.mA.mU.mC.mU.mU.mU.mA.mC.mC.mU.mG.mA.mC.mA.mG.mU.mA.*.mU.*.mU.*.mA.*.mA. –Chl 3′, in which “*” is a phosphorothioate backbone instead of a PO backbone, and “m” is an OCH3 group on the 2′ end of the base instead of an OH group. A 3′ cholesterol (Chl) group was added to each RNA oligo for potency reasons. Antagomirs were constructed as previously described (8, 11). Mosquitoes were CO2 anesthetized 12 h after eclosion, and microinjections into the thorax were performed at a dose of 200 μM in a volume of 0.25 μL (50 pmol). Mosquitoes were allowed to recover for 3–4 d before blood feeding.
dsRNA was produced as described previously (28). In brief, dsRNA was synthesized using the MEGAscript kit (Ambion). At 12 h PE, 0.5 ug (0.25 μL of 2 μg/μL) dsRNA was injected into the thorax of female mosquitoes. For rescue experiments, mosquitoes were coinjected with 0.25 μL of an antagomir/dsRNA mixture with a final concentration of 200 μM antagomir and 2 μg/μL dsRNA.
Total RNA Extraction and Real-Time PCR.
Total RNA was extracted from tissue using the TRIzol method (Invitrogen) and treated with DNase I (Invitrogen) according to the manufacturer’s protocol. miRNA expression was measured as previously described (8, 11). In brief, cDNAs for miRNAs were produced using the miScript II RT Kit (Qiagen), and qRT-PCR was performed using the miScript SYBR Green PCR kit (Qiagen) according to the manufacturer’s protocol. For mRNA, cDNAs were synthesized from 2 µg total RNA SuperScript II Reverse Transcriptase (Invitrogen). qRT-PCR was performed using the QuantiFast SYBR Green PCR Kit (Qiagen). Quantitative measurements were performed in triplicate, and relative expression (RE) was measured as RE = 2-ΔΔCt. Normalization was performed against the housekeeping genes, S7 ribosomal protein (RPS7) and Actin, using the geometric average of the housekeeping genes’ Ct values. Percent relative expression (% RE) was calculated against WT (UGAL or Noninj) control.
Immunoblot.
Protein analysis of Vg and Lp was done as described previously (28). Tissues from blood-fed female mosquitoes were homogenized in lysis buffer [50 mM Tris HCl (pH 7.4), 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1× phosphatase inhibitor (Sigma catalog no. P2850), and 1× protease inhibitor (Sigma catalog no. P8340)]. Then 10 μg protein was boiled in LDS (4×) NuPage sample buffer (Invitrogen) with 10× sample reducing agent (Invitrogen) and was run on 4–12% Tris-Glycine gels (Invitrogen) before being transferred to PVDF membranes. For detection of Vg, Vg monoclonal antibodies (30) were used at a 1:5,000 dilution followed by the secondary anti–mouse-HRP (Sigma) at a 1:2,000 dilution. For detection of Lp, apolipoprotein-I polyclonal antibody (31) was used at a 1:10000 dilution followed by the secondary anti–rabbit-HRP (Roche) at a 1:10000 dilution. For detection of Actin, β-actin monoclonal antibody (Sigma) was used at a 1:5,000 dilution followed by the secondary anti–mouse-HRP (Sigma) at a 1:2,000 dilution.
In Vitro Fat Body Culture.
Fat bodies were dissected from mosquitoes 4 d after microinjection in Aedes physiological saline buffer and were incubated in a complete culture medium supplemented with amino acids and 20-hydroxyecdysone (1 μM concentration) (Sigma) for 6 h, as described previously (28). An aliquot of complete culture medium was removed and was trichloroacetic acid-precipitated using the ProteoPrep Protein Precipitation Kit (Sigma). Precipitants were dissolved in 1× Laemmli Sample Buffer (Sigma). Total protein was quantified using the Bio-Rad Protein Assay. Two micrograms of protein were used for immunoblotting as described above.
Nile Red and DAPI Staining.
Ovaries were harvested from mosquitoes at 24 h PBM and were fixed in 4% (vol/vol) paraformaldehyde at room temperature for 1 h with shaking, as previously described (8). Fixed tissues were incubated in a solution [20% glycerol in PBS, with a 1:10,000 dilution of 10% Nile Red (Sigma), in DMSO] for 2 h at room temperature. Tissue was mounted using ProLong Diamond Antifade Mountant with DAPI (Molecular Probes) and was visualized on a Leica SP5 confocal microscope using the 20× objective lens.
Computational Target Prediction.
Computational target prediction was performed as previously described (8). The readily available miRNA target-prediction programs used were TargetScan, PITA, miRANDA, and RNAhybrid. In addition, an in-house miRNA target-prediction program was used also (8). The 3′ UTRs from the Ae. aegypti AaegL1.3 and An. gambiae Agam3.7 genebuilds were used.
Cell Culture and Luciferase Assay.
In vitro target validation was performed as previously described (8). Drosophila S2 cells (Invitrogen) were kept at 28 °C in Schneider’s Drosophila medium (Gibco, Life Technologies) supplemented with 10% heat-inactivated FBS (Gibco, Life Technologies) and 1× Antibiotic-Antimycotic (Gibco, Life Technologies). Luciferase constructs were made by inserting the Ae. aegypti miR-8 putative target 3′ UTRs into the multiple cloning region located downstream of the Renilla translational stop codon within the psiCheck-2 vector (Promega). Then 100 ng of psiCheck-2 reporters and synthetic aae-miR-8 miScript miRNA Mimic (Qiagen) or AllStars Negative Control siRNA (Qiagen) at a final concentration of 100 nm were cotransfected into Drosophila S2 cells using Attractene Transfection Reagent (Qiagen). A no-mimic treatment was performed also. The dual luciferase reporter assay was completed 48 h posttransfection using the dual luciferase reporter assay System (Promega). Firefly luciferase was used for normalization of Renilla luciferase expression. Treatments were made in triplicate, and transfections were repeated three times.
Supplementary Material
Acknowledgments
We thank Dr. Tudor Fulga for providing miR-8-SP and scramble constructs, and the University of Maryland Insect Transformation Facility for genetic transformation. This work was supported by National Institutes of Health Grant R01 AI113729 (to A.S.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424408112/-/DCSupplemental.
References
- 1.Raikhel AS, et al. Molecular biology of mosquito vitellogenesis: From basic studies to genetic engineering of antipathogen immunity. Insect Biochem Mol Biol. 2002;32(10):1275–1286. doi: 10.1016/s0965-1748(02)00090-5. [DOI] [PubMed] [Google Scholar]
- 2.Price DP, et al. The fat body transcriptomes of the yellow fever mosquito Aedes aegypti, pre- and post- blood meal. PLoS ONE. 2011;6(7):e22573. doi: 10.1371/journal.pone.0022573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou Z, et al. Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proc Natl Acad Sci USA. 2013;110(24):E2173–E2181. doi: 10.1073/pnas.1305293110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raikhel AS, Dhadialla TS. Accumulation of yolk proteins in insect oocytes. Annu Rev Entomol. 1992;37:217–251. doi: 10.1146/annurev.en.37.010192.001245. [DOI] [PubMed] [Google Scholar]
- 5.Lucas K, Raikhel AS. Insect microRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem Mol Biol. 2013;43(1):24–38. doi: 10.1016/j.ibmb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucas KJ, Myles KM, Raikhel AS. Small RNAs: A new frontier in mosquito biology. Trends Parasitol. 2013;29(6):295–303. doi: 10.1016/j.pt.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Lucas KJ, Roy S, Ha J, Raikhel AS. Mosquito-specific microRNA-1174 targets serine hydroxymethyltransferase to control key functions in the gut. Proc Natl Acad Sci USA. 2014;111(40):14460–14465. doi: 10.1073/pnas.1416278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trümbach D, Prakash N. The conserved miR-8/miR-200 microRNA family and their role in invertebrate and vertebrate neurogenesis. Cell Tissue Res. 2015;359(1):161–177. doi: 10.1007/s00441-014-1911-z. [DOI] [PubMed] [Google Scholar]
- 10.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5(3):115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant B, Macdonald W, Raikhel AS. microRNA miR-275 is indispensable for blood digestion and egg development in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2010;107(52):22391–22398. doi: 10.1073/pnas.1016230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6(12):897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kokoza VA, Raikhel AS. Targeted gene expression in the transgenic Aedes aegypti using the binary Gal4-UAS system. Insect Biochem Mol Biol. 2011;41(8):637–644. doi: 10.1016/j.ibmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 16.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(web server issue):W451-4. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulligan KA, et al. Secreted Wingless-interacting molecule (Swim) promotes long-range signaling by maintaining Wingless solubility. Proc Natl Acad Sci USA. 2012;109(2):370–377. doi: 10.1073/pnas.1119197109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc Natl Acad Sci USA. 2008;105(40):15417–15422. doi: 10.1073/pnas.0807763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, Bassel-Duby R, Olson EN. Heart- and muscle-derived signaling system dependent on MED13 and Wingless controls obesity in Drosophila. Proc Natl Acad Sci USA. 2014;111(26):9491–9496. doi: 10.1073/pnas.1409427111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longo KA, et al. Wnt10b inhibits development of white and brown adipose tissues. J Biol Chem. 2004;279(34):35503–35509. doi: 10.1074/jbc.M402937200. [DOI] [PubMed] [Google Scholar]
- 22.Wright WS, et al. Wnt10b inhibits obesity in ob/ob and agouti mice. Diabetes. 2007;56(2):295–303. doi: 10.2337/db06-1339. [DOI] [PubMed] [Google Scholar]
- 23.Arrese EL, Soulages JL. Insect fat body: Energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panáková D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435(7038):58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- 25.Sahai-Hernandez P, Nystul TG. A dynamic population of stromal cells contributes to the follicle stem cell niche in the Drosophila ovary. Development. 2013;140(22):4490–4498. doi: 10.1242/dev.098558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song X, Xie T. Wingless signaling regulates the maintenance of ovarian somatic stem cells in Drosophila. Development. 2003;130(14):3259–3268. doi: 10.1242/dev.00524. [DOI] [PubMed] [Google Scholar]
- 27.Ross SE, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289(5481):950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 28.Roy SG, Hansen IA, Raikhel AS. Effect of insulin and 20-hydroxyecdysone in the fat body of the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2007;37(12):1317–1326. doi: 10.1016/j.ibmb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kokoza V, et al. Engineering blood meal-activated systemic immunity in the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci USA. 2000;97(16):9144–9149. doi: 10.1073/pnas.160258197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raikhel AS, Pratt L, Lea AO. Monoclonal antibodies as probes for processing of yolk protein in the mosquito; production and characterization. J Insect Physiol. 1986;32:879–890. [Google Scholar]
- 31.Sun J, Hiraoka T, Dittmer NT, Cho KH, Raikhel AS. Lipophorin as a yolk protein precursor in the mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2000;30(12):1161–1171. doi: 10.1016/s0965-1748(00)00093-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.