Abstract
To investigate epigenetic contributions to Huntington's disease (HD) pathogenesis, we carried out genome-wide mapping of the transcriptional mark, trimethyl-histone H3-lysine 4 (H3K4me3) in neuronal nuclei extracted from prefrontal cortex of HD cases and controls using chromatin immunoprecipitation followed by deep-sequencing. Neuron-specific mapping of the genome-wide distribution of H3K4me3 revealed 136 differentially enriched loci associated with genes implicated in neuronal development and neurodegeneration, including GPR3, TMEM106B, PDIA6 and the Notch signaling genes hairy and enhancer of split 4 (HES4) and JAGGED2, supporting the view that the neuronal epigenome is affected in HD. Importantly, loss of H3K4me3 at CpG-rich sequences on the HES4 promoter was associated with excessive DNA methylation, reduced binding of nuclear proteins to the methylated region and altered expression of HES4 and HES4 targeted genes MASH1 and P21 involved in striatal development. Moreover, hypermethylation of HES4 promoter sequences was strikingly correlated with measures of striatal degeneration and age-of-onset in a cohort of 25 HD brains (r = 0.56, P = 0.006). Lastly, shRNA knockdown of HES4 in human neuroblastoma cells altered MASH1 and P21 mRNA expression and markedly increased mutated HTT-induced aggregates and cell death. These findings, taken together, suggest that epigenetic dysregulation of HES4 could play a critical role in modifying HD disease pathogenesis and severity.
Introduction
Huntington's disease (HD) is a devastating and progressive neurodegenerative disorder characterized by chorea, dystonia, cognitive impairment and behavioral changes (1,2). The CAG trinucleotide expansion in exon 1 of the huntingtin (HTT) gene (3) leads to widespread neuronal loss and gliosis and the appearance of intranuclear inclusions of the mutant huntingtin protein (HTT) in neurons, particularly in the striatum and cerebral cortex. While all HD patients have the same type of mutation (i.e. >35 CAG repeats) which accounts for two-thirds of the variance in age at disease onset, there is significant variation in motor and cognitive symptoms (4,5) and the remaining (one-third) variance of the age-of-onset is likely attributed to other genetic modifier factors (5,6). It is striking that the same genetic (same CAG repeats) architecture is associated with very different age-of-onset, and up to 30-year differences have been reported (5,6). Here, we hypothesize that differences in epigenetic regulation of specific promoter/regulatory sequences influences the degree of striatal degeneration and the disease age-of-onset. Epigenetic mechanisms, including the regulation of DNA methylation and various histone methylation and acetylation markings may contribute to dysregulated transcription, a central feature of HD pathogenesis (7,8). Epigenetic markers in HD may represent novel drug targets for the treatment of HD (9) and other neurodegenerative diseases (10).
Multiple lines of evidence support the possible contribution of epigenetic changes to HD pathogenesis. For example, global levels of histone acetylation are reduced in models of HD (11,12) and in human HD brains (13), and histone hypoacetylation in HD models can be reversed by treatment with histone deacetylase (HDAC) inhibitors which also arrest polyglutamine-dependent neurodegeneration (11) and counteract transcriptional and behavioral changes (14–16). Histone modifications were altered in HTT locus, with increased levels of H3K4 and H3K36 trimethylations and reduced level of H3K9 trimethylation in mouse R6/1 HD model (17). Furthermore, mutant HTT has been shown to interact with proteins for histone methylation, including HYPB, a novel human histone H3 lysine 36-specific methyltransferase associated with active genes (18) and the polycomb repressive complex 2 (PRC2), a multi-protein complex with histone methyltransferase activity specific to H3 lysine 27 (H3K27me3) (19). Mutant HTT can induce the repressive histone mark, histone H3 dimethyated at lysine 9 (H3K9me2), by a mechanism involving the transcription of alpha thalassemia/mental retardation X linked (ATRX) (20). In HD striatum, excessive levels of H3K9me2 (21) as well as the reduced level of H3 trimethylated at lysine K4 (H3K4me3) on the promoter for selective sets of the down-regulated genes such as dopamine receptor 2 (Drd2) and brain-derived neurotrophic factor (Bdnf) (9) have also been reported. However, to the best of our knowledge, no genome-wide study of histone methylation in human HD brains has been undertaken in a cell-type specific manner.
As the first step in probing genome-wide changes of histone methylation markers in HD brains, we mapped the genome-wide distribution of histone H3K4me3 with next generation sequencing technology. The H3K4me3 mark functions as the exclusive docking signal for RNA polymerase II and strongly correlates with activated RNA polymerase II occupancy and transcription rates (22). This epigenetic marker is localized to punctuated sites near the transcription start sites (TSS) of actively transcribed genes (23–26), correlates on a genome-wide scale broadly with gene expression activity, is sharply regulated at TSS and other regulatory sequences associated with the regulation of transcription (27) and may provide novel insights into transcriptional dysregulation in HD. In order to explore the epigenome in the cell type at risk in HD, the cortical and striatal neurons, (28), we employed fluorescent-activated nuclei sorting to isolate neuronal from non-neuronal nuclei residing in the prefrontal cortex (PFC) (29). This enabled a proper comparative analyses of histone marks in neuronal elements, which is an advantage of conventional studies on tissue homogenates because it bypasses the potential confound of degeneration and injury-induced increase in glia-to-neuron ratio (30). Our neuron-specific genome-wide mapping approach identified a large number of epigenetically altered loci in the neuronal HD genome, including loss of H3K4me3 and excessive DNA methylation on the HES4 promoter, as well as altered expression of HES4 and its target genes Mash1 and p21. We present multiple lines of evidence for a functional role of HES4 in HD pathogenesis, including a significant correlation between HES4 epigenetic dysregulation and clinical and histopathological parameters as well as mechanistic evidence such as mutant HTT-induced aggregates and cell death after shRNA knockdown of HES4 in human neuroblastoma cells.
Results
Histone H3K4me3 landscapes in prefrontal neurons of HD and control brains
We mapped the distribution of the H3K4me3 mark, which is sharply enriched around the 5′ end of genes and on a genome-wide scale broadly correlated with gene expression activity, in neuronal chromatin from dorsolateral PFC in six cases and five controls (Table 1). All but one of the HD postmortem brains were collected more than 15 years after onset of HD symptoms (mean = 17.2 years), at which time, striatal neurons would have largely degenerated, resulting in a dramatic decline of neuronal numbers in the caudate nucleus, accompanied by extensive gliosis (30). On the other hand, the PFC of HD brains displays pathological changes similar to the striatum (including mutant HTT aggregation), but with less severe degeneration that defines HD striatum (31,32). Thus, molecular changes detected in HD PFC may be more representative for HD pathology prior to widespread loss of neurons. Consistent with this, the overall FACS sorted NeuN+ cells (per gram tissue) were indistinguishable between HD (4980317 ± 661429) and control brains (5521686 ± 432578) (P > 0.05, n = 12–15, t-test), indicating that total number of NeuN+ neurons in cerebral cortex was not altered in HD brains.
Table 1.
Demographics of HD and control brains
| HD ID | Death | Onset | Duration | CAG repeat size | PMI (h) | Striatal score | Control ID | Death | PMI (h) |
|---|---|---|---|---|---|---|---|---|---|
| (A) Brain samples analyzed for FACS-ChIP-sequencing | |||||||||
| HD-1 | 55 | 44 | 11 | 45 | 37 | 2.66 | C-1 | 55 | 40 |
| HD-2 | 56 | 40 | 16 | 45 | 19 | 2.66 | C-2 | 56 | 17 |
| HD-3 | 71 | 52 | 19 | 43 | 21 | 2.43 | C-3 | 71 | 24 |
| HD-4 | 69 | 50 | 19 | 42 | 19 | 2.48 | C-4 | 69 | 18 |
| HD-5 | 43 | 28 | 15 | 49 | 21 | 2.70 | C-5 | 43 | 12 |
| HD-6 | 68 | 45 | 23 | 42 | 4 | 2.67 | |||
| (B) Brain samples analyzed for DNA methylation | |||||||||
| HD-2 | 56 | 40 | 16 | 45 | 19 | 2.66 | C-7 | 66 | 19 |
| HD-3 | 71 | 52 | 19 | 43 | 21 | 2.43 | C-8 | 69 | 15 |
| HD-4 | 69 | 50 | 19 | 42 | 19 | 2.48 | C-9 | 54 | 24 |
| HD-5 | 43 | 28 | 15 | 49 | 21 | 2.70 | C-10 | 61 | 10 |
| HD-6 | 68 | 45 | 23 | 42 | 4 | 2.67 | C-12 | 44 | 28 |
| HD-7 | 89 | 70 | 19 | 40 | 57 | 3.33 | C-13 | 53 | 24 |
| HD-8 | 69 | 63 | 6 | 41 | 6 | 2.64 | C-14 | — | — |
| HD-9 | 67 | 40 | 27 | 44 | 14 | 3.33 | C-15 | 57 | 20 |
| HD-10 | 61 | 35 | 26 | 46 | 25 | 3.58 | C-16 | 43 | 15 |
| HD-11 | 63 | 40 | 23 | 45 | 21 | 2.74 | C-17 | 52 | 23 |
| HD-12 | 62 | 40 | 22 | 45 | 28 | 3.58 | C-18 | 58 | 20 |
| HD-13 | 76 | 58 | 18 | 41 | 7 | — | C-19 | 70 | 21 |
| HD-14 | 48 | 25 | 23 | 48 | 19 | 3.82 | C-20 | 46 | 30 |
| HD-15 | 40 | 34 | 6 | 51 | — | 3.52 | C-21 | 66 | 17 |
| HD-16 | 55 | 31 | 24 | 47 | 24 | — | C-23 | 36 | 21 |
| HD-17 | 72 | 55 | 17 | 41 | 8 | 2.59 | C-24 | 60 | 24 |
| HD-18 | 67 | 48 | 19 | 43 | 22 | 2.74 | C-25 | 54 | 24 |
| HD-19 | 59 | 35 | 24 | 46 | 6 | 2.62 | C-27 | 61 | 17 |
| HD-20 | 72 | 55 | 17 | 42 | 12 | 2.74 | C-28 | 62 | 18 |
| HD-21 | 78 | 62 | 16 | 42 | 18 | — | C-29 | 55 | 26 |
| HD-22 | 68 | 52 | 16 | 42 | 13 | 2.66 | C-30 | 52 | 18 |
| HD-23 | 57 | 40 | 17 | 49 | 25 | 2.91 | C-31 | 69 | 26 |
| HD-24 | 53 | 40 | 13 | 48 | 23 | 3.60 | C-32 | 61 | 25 |
| HD-25 | 48 | 38 | 10 | 45 | 11 | 3.60 | C-33 | 64 | 19 |
| HD-26 | 36 | 24 | 12 | 54 | 21 | 2.91 | C-34 | 88 | 11 |
| C-35 | 71 | 40 | |||||||
| C-36 | 68 | 25 | |||||||
| (C) Brain samples analyzed for qPCR analysis of mRNA | |||||||||
| HD-2 | 56 | 40 | 16 | 45 | 19 | 2.66 | C-10 | 61 | 10 |
| HD-3 | 71 | 52 | 19 | 43 | 21 | 2.43 | C-11 | 68 | 19 |
| HD-4 | 69 | 50 | 19 | 42 | 19 | 2.48 | C-12 | 44 | 28 |
| HD-5 | 43 | 28 | 15 | 49 | 21 | 2.7 | C-13 | 53 | 24 |
| HD-6 | 68 | 45 | 23 | 42 | 4 | 2.67 | C-15 | 57 | 20 |
| HD-8 | 69 | 63 | 6 | 41 | 6 | 2.64 | C-18 | 58 | 20 |
| HD-16 | 55 | 31 | 24 | 47 | 24 | — | C-19 | 70 | 21 |
| HD-17 | 72 | 55 | 17 | 41 | 8 | 2.59 | C-22 | 73 | 19 |
| HD-19 | 59 | 35 | 24 | 46 | 6 | 2.62 | C-24 | 60 | 24 |
| HD-21 | 78 | 62 | 16 | 42 | 18 | — | C-26 | 76 | 26 |
| HD-22 | 68 | 52 | 16 | 42 | 13 | 2.66 | C-28 | 62 | 18 |
| HD-24 | 53 | 40 | 13 | 48 | 23 | 3.6 | C-31 | 69 | 26 |
| HD-25 | 48 | 38 | 10 | 45 | 11 | 3.6 | C-34 | 88 | 11 |
| HD-26 | 36 | 24 | 12 | 54 | 21 | 2.91 | C-37 | 93 | 13 |
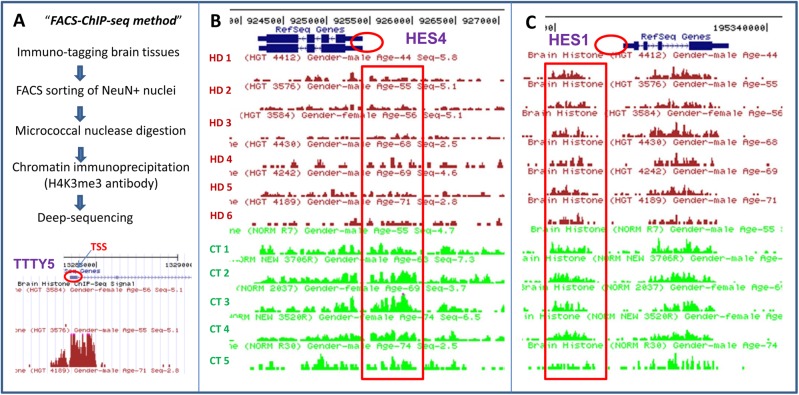
In our cohort, 85–90% of reads of HD and 82–90% of control cohorts were mapped to one unique location in the genome. Using Poisson statistics (see Materials and Methods) 136 H3K4me3 peaks were identified as differentially distributed between HD and control brain (Supplementary Material, Table S1), with 78 peaks maintaining significance (P < 0.05) after correction for False Discovery using the Benjamini and Hochberg method (Table 2). A total of 83 out of 136 peaks were overlapped within 2 kb of a TSS, consistent with previous studies (29,33). For example, there are clear dense H3K4me3 peaks around TSS of the TTTY15 gene in HD brain (Fig. 1A). Consistent with TTTY15′s location on the Y chromosome, there was complete lack of signal in the female sample (HD3584. Fig. 1A). The H3K4me3 distribution was highly consistent among the six HD samples, with >90% overlap with each sample, with few exceptions such as Y-chromosome specific sequences that were limited to male subjects only (Supplementary Material, Table S2).
Table 2.
H3K4me3 is altered at 78 loci in HD cortical neurons, compared with control neuron
| TSS | bp to TSS | Mean CTR | Mean HGT | Log2(CTR/ HGT) | P-value (t-test) | FDR | Functions |
|---|---|---|---|---|---|---|---|
| ARC | 1715 | 0.0215 | 0.0097 | 2.543 | 0.0031 | 0.046 | Activity-regulated early response gene with key role in synaptic plasticity |
| BHLHE40 | 3320 | 0.0192 | 0.0077 | 1.774 | 0.0013 | 0.031 | A bHLH transcription factor and key component of the circadian clock |
| TMEM200B | 0 | 0.0050 | 0.0124 | 1.618 | 0.0011 | 0.029 | |
| MIR3675 | 11662 | 0.0140 | 0.0041 | 1.604 | 0.0035 | 0.048 | |
| MIR1257 | 11074 | 0.0290 | 0.0140 | 1.522 | 0.0004 | 0.021 | |
| FBXL16 | 2707 | 0.0106 | 0.0042 | 1.515 | 0.0001 | 0.014 | |
| NR4A1 | 10907 | 0.0101 | 0.0045 | 1.515 | 0.0000 | 0.011 | Nuclear receptor-related transcription factor implicated in neuroprotection; |
| SLC22A18 | 0 | 0.0105 | 0.0040 | 1.508 | 0.0003 | 0.018 | |
| DAB2IP | 0 | 0.0111 | 0.0047 | 1.448 | 0.0005 | 0.023 | A GTPase regulator involved in neuronal migration and growth |
| HBQ1 | 0 | 0.0101 | 0.0035 | 1.432 | 0.0001 | 0.012 | |
| PHLDA2 | 0 | 0.0176 | 0.0080 | 1.420 | 0.0010 | 0.028 | |
| RNF126 | 3401 | 0.0126 | 0.0051 | 1.376 | 0.0028 | 0.043 | |
| BAI1 | 46840 | 0.0122 | 0.0056 | 1.345 | 0.0014 | 0.032 | Angiogenesis inhibitor 1, interacts with LRRK2 kinase |
| CYP2S1 | 0 | 0.0184 | 0.0080 | 1.333 | 0.0001 | 0.014 | |
| LOXL4 | 0 | 0.0187 | 0.0087 | 1.324 | 0.0022 | 0.039 | |
| MIDN | 1136 | 0.0047 | 0.0126 | 1.324 | 0.0007 | 0.024 | Nucleolar protein with ubiquitin-like domain essential for mid-brain development |
| KIAA1274 | 0 | 0.0133 | 0.0292 | 1.321 | 0.0000 | 0.003 | |
| HES4 | 0 | 0.0122 | 0.0055 | 1.320 | 0.0000 | 0.011 | Regulator of neural stem-cell proliferation |
| FOS | 0 | 0.0325 | 0.0148 | 1.320 | 0.0010 | 0.029 | Early response gene involved in activity-regulated gene expression |
| RAMP3 | 0 | 0.0158 | 0.0391 | 1.313 | 0.0004 | 0.020 | |
| ETV4 | 0 | 0.0243 | 0.0098 | 1.307 | 0.0023 | 0.040 | |
| INF2 | 0 | 0.0235 | 0.0105 | 1.306 | 0.0001 | 0.013 | Inverted formin, a monogenic risk gene for Charcot–Marie–Tooth neuropathy |
| VRK1 | 235498 | 0.0108 | 0.0036 | 1.305 | 0.0003 | 0.018 | Monogenic causative gene for postnatal progressive microcephaly syndromes |
| DSG2 | 0 | 0.0159 | 0.0071 | 1.296 | 0.0001 | 0.011 | |
| BRSK2 | 19307 | 0.0190 | 0.0083 | 1.293 | 0.0014 | 0.033 | BRSK2/SAD defines neuronal polarization and axon growth in cerebral cortex |
| NPAS4 | 0 | 0.0144 | 0.0059 | 1.276 | 0.0017 | 0.035 | An activity-dependent TF critical for memory and inhibitory synape formation |
| ADRA1D | 0 | 0.0166 | 0.0074 | 1.268 | 0.0023 | 0.040 | Adrenergic receptor 1D, expressed in forebrain |
| SLC27A5 | 1597 | 0.0123 | 0.0056 | 1.265 | 0.0005 | 0.023 | |
| PPIC | 0 | 0.0112 | 0.0048 | 1.247 | 0.0001 | 0.013 | |
| KCNN1 | 0 | 0.0188 | 0.0401 | 1.237 | 0.0020 | 0.037 | Calcium-activated potassium channel SK-1, implicated in neuroprotection |
| GNG13 | 0 | 0.0170 | 0.0391 | 1.231 | 0.0017 | 0.035 | |
| AGRN | 12379 | 0.0118 | 0.0047 | 1.224 | 0.0001 | 0.014 | Synaptogenesis and plasticity in CNS, key neuromuscular junction protein |
| GOLT1A | 62360 | 0.0174 | 0.0378 | 1.216 | 0.0037 | 0.049 | |
| SLC26A1 | 3435 | 0.0142 | 0.0305 | 1.198 | 0.0010 | 0.028 | |
| C6orf27 | 0 | 0.0145 | 0.0067 | 1.196 | 0.0000 | 0.011 | |
| JAG2 | 395 | 0.0124 | 0.0056 | 1.192 | 0.0001 | 0.013 | Notch receptor ligand Jagged 2, implicated in generation of motor neurons. |
| HHATL | 0 | 0.0181 | 0.0066 | 1.190 | 0.0014 | 0.032 | |
| CLEC2L | 0 | 0.0353 | 0.0165 | 1.190 | 0.0000 | 0.008 | |
| C1orf187 | 0 | 0.0139 | 0.0057 | 1.179 | 0.0026 | 0.042 | Neural-specific antagonist to WNT signaling and axon guidance molecule |
| C19orf26 | 0 | 0.0215 | 0.0075 | 1.169 | 0.0010 | 0.029 | |
| CRHR2 | 0 | 0.0252 | 0.0116 | 1.167 | 0.0023 | 0.039 | Corticotropin releasing hormone receptor 2 |
| UNC5A | 6354 | 0.0122 | 0.0272 | 1.167 | 0.0016 | 0.034 | |
| FLJ37505 | 424827 | 0.0214 | 0.0092 | 1.166 | 0.0000 | 0.001 | |
| AGRN | 19672 | 0.0217 | 0.0101 | 1.164 | 0.0001 | 0.013 | Synaptogenesis and plasticity in CNS, key neuromuscular junction protein |
| NR4A1 | 0 | 0.0110 | 0.0047 | 1.163 | 0.0007 | 0.026 | Nuclear receptor-related transcription factor implicated in neuroprotection |
| IL2RB | 16371 | 0.0234 | 0.0094 | 1.161 | 0.0034 | 0.047 | |
| KIAA0182 | 146622 | 0.0219 | 0.0494 | 1.155 | 0.0005 | 0.022 | Interacting with the disrupted in schizophrenia (DISC1) protein |
| N4BP3 | 0 | 0.0173 | 0.0068 | 1.145 | 0.0000 | 0.009 | |
| TRAF7 | 2580 | 0.0126 | 0.0056 | 1.144 | 0.0019 | 0.037 | Encodes TNF receptor-associated protein that regulates apoptosis |
| MFSD10 | 0 | 0.0122 | 0.0040 | 1.143 | 0.0006 | 0.023 | |
| NCR2 | 91854 | 0.0147 | 0.0064 | 1.139 | 0.0008 | 0.026 | |
| LOC100128338 | 0 | 0.0372 | 0.0149 | 1.132 | 0.0000 | 0.009 | |
| SBK1 | 30099 | 0.0111 | 0.0046 | 1.132 | 0.0001 | 0.014 | |
| PDIA6 | 23739 | 0.0192 | 0.0079 | 1.129 | 0.0030 | 0.045 | An isomerase interacting with progranulin, involved in frontotemporal dementia |
| RCOR2 | 0 | 0.0102 | 0.0041 | 1.127 | 0.0001 | 0.012 | Rest Co-repressor 2, chromatin regulator in neuronal progenitor |
| WTIP | 0 | 0.0100 | 0.0043 | 1.119 | 0.0000 | 0.009 | |
| LGI2 | 0 | 0.0123 | 0.0053 | 1.118 | 0.0001 | 0.012 | |
| HAGHL | 0 | 0.0119 | 0.0048 | 1.115 | 0.0001 | 0.012 | |
| LINGO3 | 0 | 0.0108 | 0.0041 | 1.105 | 0.0031 | 0.046 | |
| GPR3 | 0 | 0.0148 | 0.0060 | 1.103 | 0.0001 | 0.012 | Orphan GPCR modulating beta-amyloid and neurodegeneration |
| LOC150381 | 0 | 0.0259 | 0.0115 | 1.102 | 0.0000 | 0.008 | |
| RHBDL1 | 0 | 0.0143 | 0.0064 | 1.101 | 0.0010 | 0.029 | |
| NFIX | 0 | 0.0069 | 0.0153 | 1.056 | 0.0012 | 0.030 | Nuclear protein regulating neural progenitor differentiation in hippocampus |
| GPM6B | 0 | 0.0121 | 0.0043 | −1.081 | 0.0009 | 0.028 | |
| SCN2A | 0 | 0.0121 | 0.0053 | −1.091 | 0.0009 | 0.027 | Sodium channel and monogenic neurodevelopmental risk gene |
| CHRNA1 | 76396 | 0.0209 | 0.0096 | −1.092 | 0.0013 | 0.031 | Nicotonic acetylcholine receptor important for axonal development |
| FLRT3 | 0 | 0.0160 | 0.0056 | −1.101 | 0.0029 | 0.044 | Repulsive axon guidance cue |
| PDZRN3 | 62936 | 0.0074 | 0.0209 | −1.119 | 0.0003 | 0.018 | |
| IL1RAPL1 | 0 | 0.0163 | 0.0074 | −1.135 | 0.0009 | 0.028 | IL 1 receptor accessory protein-like 1, a neurodevelopmental risk gene |
| SPRED2 | 0 | 0.0195 | 0.0033 | −1.151 | 0.0034 | 0.048 | |
| HMGN4 | 13970 | 0.0115 | 0.0048 | −1.156 | 0.0014 | 0.032 | |
| MTRF1 | 0 | 0.0176 | 0.0375 | −1.171 | 0.0009 | 0.028 | |
| COX7B | 0 | 0.0105 | 0.0039 | −1.198 | 0.0003 | 0.018 | |
| R3HDM1 | 54329 | 0.0118 | 0.0275 | −1.223 | 0.0020 | 0.037 | |
| KRT222 | 0 | 0.0124 | 0.0263 | −1.305 | 0.0020 | 0.038 | |
| SNRPN | 382 | 0.0176 | 0.0082 | −1.322 | 0.0033 | 0.047 | Small nuclear riboprotein-associated protein N highly expressed in neurons |
| ANXA1 | 71133 | 0.0150 | 0.0066 | −1.431 | 0.0011 | 0.030 | |
| ARHGAP21 | 100221 | 0.0159 | 0.0073 | −1.500 | 0.0018 | 0.036 |
Figure 1.
Detection and distribution of H3K4me3 peaks surrounding the HES4 and HES1 genes in HD and control subjects. (A) Flow chart of the FACS-ChIP-seq procedure as described in the Materials and Methods section for detecting genome-wide distribution of H3K4me3 marks from NeuN+ cortical nuclei of six HD and five control subjects. Low panel: detection of H3K4me3 peak signal for Y chromosome gene TTTY5 by FACS-ChIP-seq H3K4me3 peaks are distributed in punctuated pattern and highly enriched in TSS of TTTY5 gene (as indicated by red circle). H3K4me3 peaks surround TTS of TTTY5 were absent in a female subject (first line) but present in a male sample (second line), confirming specificity of the H3K4me3 peaks detected by FACS-ChIP-seq. (B) H3K4me3 peaks were detected by FACS-ChIP-seq in NeuN+ cortical nuclei from six HD and five control subjects as described in the Materials and Methods section. H3K4me3 peaks are clustered around TSS of the HES4 gene (as indicated red circle). Moreover, the H3K4me3 peak (tag) densities (ad indicated by red long square/box) in HD were lower, compared with controls. (C) In contrast, the H3K4me3 peak densities around HES1 gene were indistinguishable between HD and control subjects.
Among the 136 peaks, 85 peaks were decreased and 51 peaks increased in HD. At least 45 of the 136 peaks as defined by the nearest TSS were associated with neuronal genes important for connectivity and synaptic signaling (e.g. SHANK3, RIMS2, DLG2/PSD93) or neuronal transcription (ARC, RCOR2, MKL1) (Supplementary Material, Table S1), supporting the view that cortical circuitry is compromised in HD due to widespread alterations in the epigenetic architecture of cortical neurons. Gene ontology (GO) analysis of the 136 peaks, when corrected for multiple comparisons, showed enrichment for eight categories that were overwhelmingly related to neuronal compartments and synaptic signaling (Supplementary Material, Table S3). Notably, six out of eight over-represented GO categories are directly related to synaptic functions, a finding consistent with the fact that we used neuronal nuclei for our ChIP-seq analysis.
Furthermore, 14 of the 78 peaks (after correction of false discovery) altered in HD cortical neurons are ascribed with key roles in neurodegenerative conditions (Supplementary Material, Table S1). These include orphan G protein coupled receptors including GPR3 modulating gamma-secretase activity and beta-amyloid deposition (34) and GPR179 which, when mutated, lead to degeneration of bipolar neurons in the retina (35). The list also includes INF2, a monogenic cause for Charcot–Marie–Tooth neuropathy (36), VRK1, a monogenic cause for progressive postnatal microencephaly syndromes (37) and a transmembraneous protein, TMEM106B, implicated in frontotemporal dementia (38,39) and Alzheimer's diseases (40). In addition, multiple H3K4me3 peaks altered in HD neurons are located to the TSSs for genes having a key role in neuronal development and differentiation, such as TNFRSF18 and TRAF7, and for genes encoding two tumor necrosis factor (TNF) receptor-related molecules linked to the neurotrophin BDNF/TRKB signal cascade and developmental regulation of apoptosis (41,42). Notably, hairy and enhancer of split 4 (HES4) and JAGGED2, two components of the Notch signaling pathway implicated in the regulation of stem cells and neuronal progenitors (43,44) were identified. For the rest of the study, we focused on the HES4 epigenetic changes and function in HD for three experimental considerations: (i) our initial screening for 24 genes (randomly selected but with known biological functions related to HD pathogenesis) from 78 genes associated with altered H3K4me3 peaks showed that three genes (including HES4) exhibited altered DNA methylation in HD brains; (ii) HES4 mRNA was reduced reportedly by ∼50% in the diseased brains in an earlier transcriptome study on HD brains by Hodges et al. (45); and (iii) The HES gene family (and more broadly Notch signaling) is recognized for its role in forebrain neuronal development by controlling cell-fate determination in progenitor cells and induction of terminal differentiation (46–48).
HD cortical neurons show selective reduction of HES4 TSS-associated H3K4me3
Since HTT mutation exists from early development, neurodevelopmental defects have been suggested to contribute to HD pathogenesis (5). Given the recognized role of the HES gene family in development by controlling cell-fate determination in progenitor cells and induction of terminal differentiation (46–48), we performed additional targeted analysis of H3K4me3 signals and DNA methylation of the HES4 gene including its promoter. Figure 1B shows the altered H3K4me3 pattern of HES4 by FACS-ChIP-seq analysis. The H3K4me3 mark of HES4 in cortex was increased around the TSS site, while broader regions upstream of the promoter were also involved. As shown in Figure 1, H3K4me3 signals of HES4 were consistently reduced in all six HD brains compared with all five controls. Total tags of ChIP-seq signal were significantly reduced in HD compared with controls, and statistical analyses of tag densities in HD (0.0077) were statistically different from controls (FDR corrected P = 0.01).
Interestingly, the reduced H3K4me3 signal is specific to HES4 since careful analysis of this histone mark for other genes of the HES family (HES1-HES7) are not affected, as illustrative by the representative example of HES1 (Fig. 1C and Supplementary Material, Fig. S1). Recognizing that HES4 has no direct homologue in the mouse genome, we performed further detailed phylogenetic analysis of HES4 and HES family genes. The HES4 gene sequences are identified in humans and all analyzed primate species but HES4 is not specific for primates because close orthologs are found in other mammalian taxons. However, mammalian evolution is associated with occasional and independent losses of HES4. For example, rodent HES4 is lost in the ‘mouse-related’ clade (Mus musculus and Rattus norvegicus), but retained in the ‘squirrel-related’ clade (Ictidomys tridecemlineatus) (see Supplementary Material, Fig. S2A and B).
DNA methylation analysis uncovered an increase in intermediate methylation in the HES4 gene in HD brains
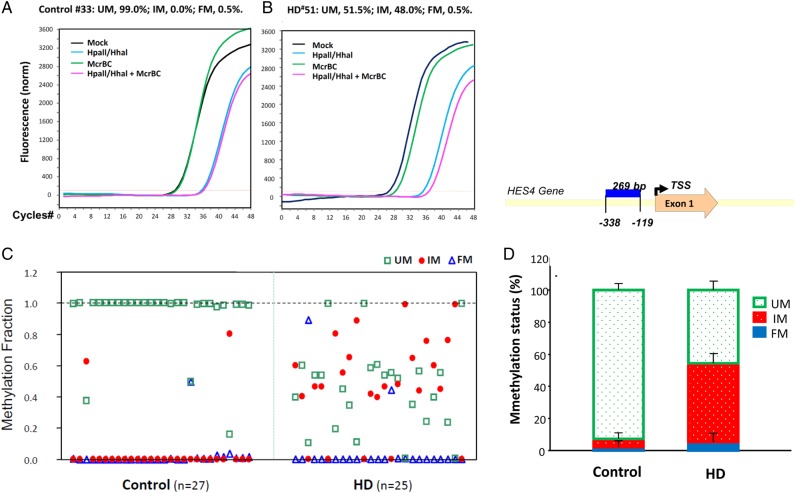
In consideration of the relationship between H3K4me3 and DNA methylation (49), we examined HES4 DNA methylation using the SABiosciences/Qiagen Methyl-Profiler method which assessed unmethylated (UM), fully methylated (FM) DNA and intermediately methylated (IM) DNA representing monoallelic DNA methylation as well as partial DNA methylation on one or both strands. We assessed DNA methylation states of selected CpG islands (CGIs) in the PFC of 27 control and 25 HD brains (Table 1B). Figure 2A shows examples of qPCR curves of all four reactions in one control (left) and in one HD (right) sample for HES4. The analysis showed that in the control brain, the HES4 promoter was largely unmethylated (∼95%, Fig. 2B, left panel), but in HD brain, the UM fraction in HES4 was significantly reduced (Fig. 2B, P < 0.01) and mostly converted to IM making the IM fraction significantly higher (P < 0.001) in HD. Specifically, IM is robustly increased from 5% of total input DNA in control to 49% in HD (Fig. 2B, right panel), indicating that most DNA methylation occurs heterogeneously on individual molecules. In contrast, FM of HES4 was not altered. After cloning the qPCR product from the DNA methylation assay, we obtained the sequence of this 269-bp fragment in the HES4 promoter, in which 33 CpG dinucleotides were identified on each strand proximate to the TSS (Fig. 2, upper, right panel).
Figure 2.
DNA methylation of the HES4 promoter of HD and control cortex. DNA methylation states of a 269-bp fragment of the HES4 promoter in the PFC of 27 controls and 25 HD were measured by Methyl-Profiler as described in the Materials and Methods section. (Left) Examples of qPCR curves of all four reactions in one control and in one HD for the HES4 gene. DNA methylation states for the HES4 promoter are expressed as fractions of unmethylated (UM), intermediate-methylated (IM) or fully methylated (FM) DNA. (Right) IM was robustly increased from 5% of total input DNA in control to 49% in HD while UM fraction in the HES4 promoter was reduced in HD. In contrast, FM of the HES4 gene did not exhibit significant change.
Nuclear proteins binding to the HES4 promoter are reduced after DNA hypermethylation in vitro
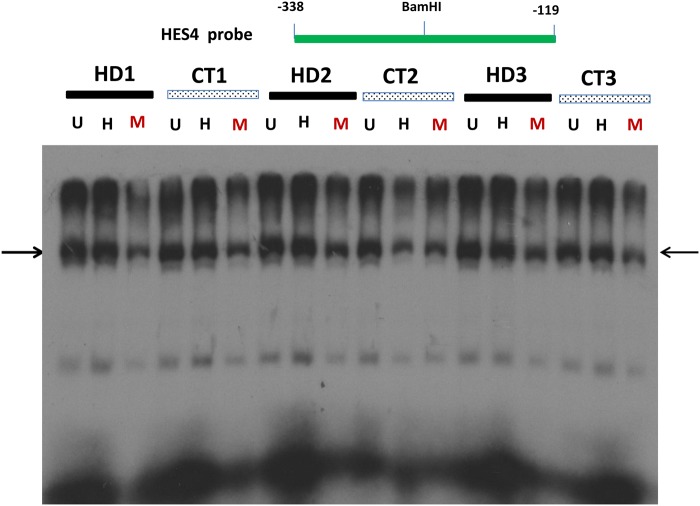
Dysfunctional transcription in HD may be associated either with altered DNA methylation in HTT-targeted gene promoters, or altered nuclear proteins that bind to these HTT-targeted gene promoters, or both. While Figure 2 indicates altered DNA methylation in the HES4 promoter, the abnormality of nuclear proteins in HD may play a role in altered transcription. To address this issue, we performed an electrophoretic mobility shift assay (EMSA) to analyze the interaction of nuclear proteins with this 269-bp fragment of the HES4 promoter (−338 to −119 bp upstream of TSS) after in vitro methylation. Since we cannot completely reproduce the DNA methylation status observed in human postmortem brains, we used double-stranded DNA fragment of the HES4 promoter as probes under conditions of non-methylation, hemi-methylation and full methylation to partially capture DNA methylation status. We tested unmethylated (U, both strands unmethylated), hemi-methylated (H, one strand methylated and other unmethylated) and fully methylated (M, both strands methylated) DNA in EMSA. Multiple bands were formed between nuclear proteins and the HES4 promoter fragment (Fig. 3). Interestingly, nuclear protein binding was significantly lower in all bands formed with the fully methylated HES4 promoter, compared with the unmethylated or hemi-methylated HES4 promoter, consistent with the notion that altered DNA methylation can affect nuclear proteins binding to the HES4 promoter. However, the binding to individual probes was indistinguishable between HD and control brains, indicating that there was no abnormality of nuclear proteins that bind to the HES4 promoter. Taken together with Figure 2 (increased DNA methylation) and 3 (reduced nuclear protein binding to fully methylated probe), it is likely that the DNA methylation status, but not the quality or quantity of nuclear proteins, may contribute to dysfunctional transcription in HD brains.
Figure 3.
The binding of nuclear proteins to the HES4 promoter is reduced after DNA hypermethylation in vitro. Binding of nuclear proteins from HD and control cortex to the 269-bp fragment of the HES4 promoter with in vitro DNA methylation by EMSA as described in the Materials and Methods section. This 269-bp fragment of the HES4 promoter was first digested BamHI into two identical DNA fragments and in vitro methylated and then re-annealed unmethylated (U), fully methylated (M) and hemi-methylated (H) double strand DNA probes for EMSA. Note that nuclear protein binding (indicated by arrows) was reduced and shifted to high molecular weight band at the fully methylated HES4 promoter compared with the un-methylated or hemi-methylated HES4 promoter.
The mRNA levels for HES4 and two down-stream target genes, Mash1 and p21, are reduced in HD versus control PFC
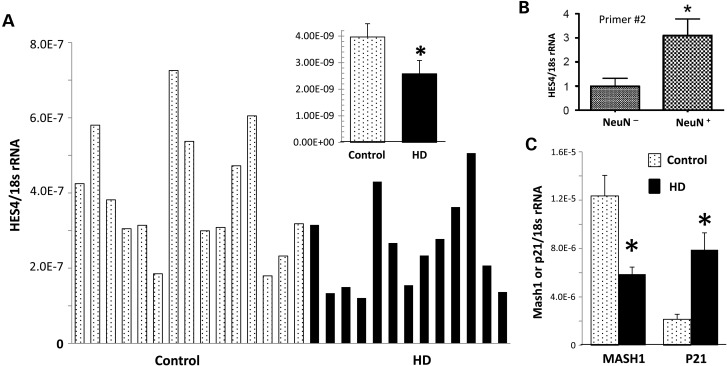
To examine the functional impact of the HES4 promoter IM increase, we determined the mRNA levels for HES4 in cortex by qPCR analysis in 14 HD and 14 control PFC samples (Table 1C) and found that HES4 mRNA is reduced ∼40% in HD cortex compared with control (Fig. 4A) (t-test, P < 0.05). This finding is consistent with an earlier transcriptome study in HD, with ∼50% reduction of HES4 mRNA in the diseased brains (45). Consistent with the strong H3K4me3 peaks associated with HES4 in NeuN+ nuclei, qPCR analysis of FACS sorted cells showed that HES4 mRNA was significantly enriched in neuronal (NeuN+) nuclei compared with non-neuronal (NeuN−) nuclei in human cortex, (Fig. 4B). Moreover, this decrease in HES4 mRNA is also consistent with the reduction in nuclear protein binding to fully methylated DNA, probably due to increased IM of symmetric and incomplete methylation of the HES4 promoter in HD brain. Considering that HES1 positively regulates expression of MASH1 (a proneuronal, forebrain-specific transcription factor) (50) and negatively regulates P21 (a cell cycle suppressor) (51–53), and that HES proteins share certain structural motifs (54), we hypothesized that HES4 modulates these HES targeted gene expression as MASH1 and P21. Indeed, our qRT–PCR results showed that the reduced HES4 mRNA was associated with down-regulation of MASH1 mRNA in HD cortex compared with the control. In contrast, P21 mRNA was increased in the cortex of HD compared with the control. Therefore, HES4 and its targeted genes MASH1 and P21 may play a role in the neurodegeneration of HD.
Figure 4.
The mRNA levels for HES4 as well as two down-stream target genes, MASH1 and P21, are reduced in the cortex of HD compared with controls. mRNA levels for HES4 (A) and its down-stream target MASH1 and P21 (C) in total cortex tissues were detected by qPCR analysis as described in the Materials and Methods section. We also measured relative heterogenous nuclear HES4 mRNA levels in NeuN− and NeuN+ nuclei FACS sorted from human brains using two different primer sets [primer #2, (B) and primer #3 (data not shown]. (A) HES4 mRNA is reduced ∼40% in HD cortex compared with control. (B) HES4 mRNA is enriched in human neuronal nuclei (N = 3, P = 0.05, Mann–Whitney, one-tailed). (C) MASH1 mRNA in HD cortex compared with the control while P21 mRNA was increased in the cortex of HD compared with control. *P < 0.05 (n = 14, t-test). All data are presented as relative ratio of targeted mRNA over 18s rRNA, and presented as mean ± SEM.
The extent of intermediate DNA methylation of the HES4 promoter is correlated with striatal degeneration and with age-of-onset in HD
The correlation of levels of un-methylated, intermediate methylation and hyper-methylation to the characteristics of the HD samples is presented in Table 3. The levels of FM and UM sites were not significantly correlated with any of the HD sample characteristics. The level of intermediate methylation was correlated with the level of striatal involvement (r = 0.56, P = 0.006) and was also correlated with the age at death (r = −0.47, P = 0.02), age at onset (r = −0.48, P = 0.02) and the size of the HD CAG repeat (r = 0.50, P = 0.01). The correlation between intermediate methylation and striatal involvement remained after removing the four samples with no intermediate methylation (r = 0.50, P = 0.02). No differences were seen between the HD cases and controls for age at death (t = −0.81, P = 0.42) or PMI (t = 1.21, P = 0.23).
Table 3.
Spearman correlation of methylation levels of the HES4 promoter with HD sample characteristics
| Hyper-methylation | Intermediate methylation | Un-methylated | |
|---|---|---|---|
| Striatal involvement | −0.22 | 0.56 | −0.36 |
| P-value | 0.32 | 0.006 | 0.10 |
| n | 22 | 22 | 22 |
| Cortical involvement | −0.33 | 0.09 | 0.06 |
| P-value | 0.13 | 0.69 | 0.79 |
| n | 22 | 22 | 22 |
| Onset age | −0.037 | −0.48 | 0.39 |
| P-value | 0.87 | 0.02 | 0.0661 |
| n | 23 | 23 | 23 |
| HD CAG repeat | −0.15 | 0.50 | −0.35 |
| P-value | 0.47 | 0.01 | 0.08 |
| n | 25 | 25 | 25 |
Because the intermediate methylation level was correlated with several different features of the HD samples, we sought to assess the main effect of the level of striatal involvement by multivariate analysis of the relationship of the level of intermediate methylation, controlling for the age-at-onset, the size of the HD repeat and the level of cortical involvement. The relationship of intermediate methylation to striatal involvement remained after adjustment for these other factors. Similar results were found consistently with other models including the level of cortical involvement or when removing onset age to avoid over parameterization.
shRNA knockdown of HES4 in human neuroblastoma cells alters MASH1 and P21 mRNA, and markedly increases mutant HTT-mediated aggregates and cell death
To probe a functional role of HES4 in development of mutant HTT-induced pathology, we further investigated the effect of shRNA-mediated knockdown of HES4 on mutant HTT-induced aggregates and cell deaths in human neuroblastoma HTB-11 cells. The effectiveness of shRNA-HES4 was confirmed by reduction of HES4 mRNA by ∼70% in cells transfected with shRNA-HES4-1 (selected from four distinct shRNAs against HES4) compared with cells transfected with shRNA-control (Fig. 5B). Furthermore, MASH1 mRNA was down-regulated and P21 mRNA was upregulated by HES4 knockdown with cognate shRNA while scramble (sc) shRNA did not alter these mRNAs (Fig. 5B). Thus, HES4 exerted similar control of the down-stream targeted gene, MASH1 and P21 in both mature neuron in the HD brain (Fig. 4) and cultured HTB-11 neuroblastoma cells.
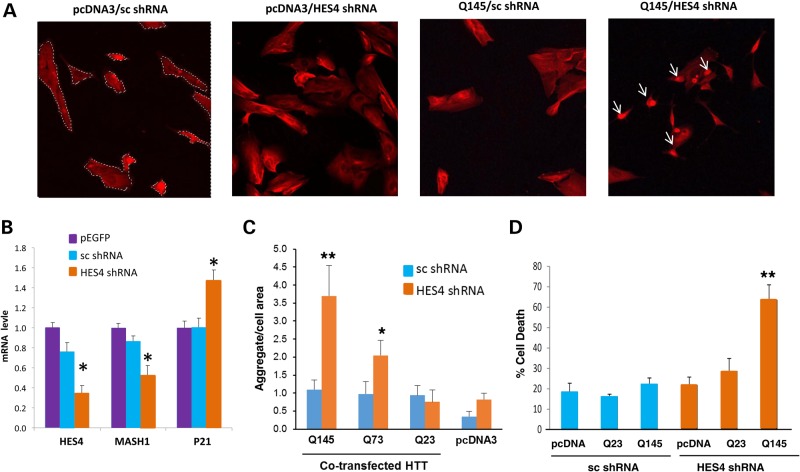
Figure 5.
shRNA knockdown of HES4 in human neuroblastoma cells alters mRNA levels of HES4, MASH1 and P21, and markedly increases mutant HTT-induced aggregates and cell death. HTB-11 cells were transfected with shRNA-HES4-1 or sc shRNA with or without different (23, 73 and 145) CAG repeats (HTT-Q23, HTT-Q73 and HTT-Q145), and qRT–PCR analysis, immunohistochemistry and cell death assays were performed as described in the Materials and Methods. (A) Ubiquitin immunohistochemistry showed that co-transfection of shRNA-HES4-1 and mutant HTT-Q145 for 24 h produced highly dense protein aggregates (aggregates = white arrow, cellular area = dash line) with no effect of shRNA-HES4-1 or HTT-Q145 alone compared with the cells transfected with shNRA-control/pcDNA3. (B) Quantitative analyses of HES4, MASH1 and P21 mRNAs in HBT-11 cells transfected with EGFP, sc shRNA or HES4-1 shRNA. Data are presented as mean values (plus s.e. from six transfection) of fold change relative to cells receiving EGFP after normalized to 18 s rRNA. HES4 shRNA decreased HES4 mRNA by ∼70%, MASH1 mRNA by ∼50% and increased P21 mRNA by ∼40%, compared with the sc shRNA. (C) Exacerbated HTT aggregates by shRNA knockdown of HES4 mRNA in HTB-11 cells. Cells were ubiquitin stained to detect the protein aggregation which was measured using automated image analysis as described in the Materials and Methods. Co-transfection of HES4 shRNA exacerbated HTT Q145-, Q73- (but not Q23-) induced cellular aggregates, as evident by increased relative size of cellular aggregates, in HTB-11 cells. (D) Trypan blue exclusion assay showed that shRNA-HES4-1 or HTT-Q145 alone did not have significant effect on cell death, but co-transfection of shRNA-HES4-1 and Q145 increased cell death rates compared with shRNA-control. **P < 0.01, Student's t-test or One-way ANOVA, post-hoc Dunn's test (B–D).
Next, we evaluated the effect of shRNA-HES4-1 on mutant HTT-induced aggregates by immunohistochemistry. After co-transfection of shRNA-HES4-1 or control vector across three concentrations of HTT (0.5, 1.0 and 2.0 μg) with three different CAG repeats (23, 73 and 145 CAG repeats, Q23, Q73 and Q145, respectively), we found that co-transfection of shRNA-HES4-1 and HTT-Q145 (2.0 μg) produced highly dense nuclear aggregates with little effect of shRNA1-HES4-1 or HTT-Q145 alone compared with the cells transfected with sc shRNA /pcDNA3 (Fig. 5A). Quantitative evaluations of the relative size of cellular aggregates and the density of aggregation using an automated image analysis revealed that cells co-transfected with HTT-Q145 plus HES4 shRNA had significantly higher percent aggregation area than cell co-transfected with the HTT-Q145 plus sc shRNA (5.26% versus 0.75%; P = 0.002) while the density of aggregates were unaffected (P = 0.09) (Fig. 5C). The similar exacerbated effect was observed in HTT-Q73 transfected cells, which exhibited a significant increase in aggregation in cells co-transfected with HES4 shRNA compared with that co-transfected with sc shRNA (2.44% versus 0.82%, P = 0.02) but there was no significant change in aggregate density between the cells co-transfected with HES4 shRNA or sc shRNA (P = 0.99) (Fig. 5C). There was no significant effect of HES4 shRNA on HTT aggregates in cells transfected with HTT-Q23 or cells transfected with pcDNA3 vector.
Lastly, we evaluated the effect of shRNA-HES4-1 on HTT-induced cell death by trypan blue exclusion assay. Transfection of shRNA-HES4-1 or HTT-Q145 individually did not significantly affect cell death rate, but co-transfection of shRNA-HES4-1 and HTT-Q145 increased cell death rates by ∼3-folds compared with the pcDNA3-sc shRNA (Fig. 5D). This effect was selective for HTT-Q145 since co-transfection of shRNA-HES4-1 and HTT-Q23 did not affect the cell-death rate. Thus, down-regulation of HES4 by shRNA selectively increased HTT-Q145-induced ubiquitin-positive aggregates and cell death in HTB-11 cells. These findings support a pathological potential for HES4 in control of mutant HTT-induced HD pathogenesis.
Discussion
Transcriptional dysregulation is a central and early feature of HD pathogenesis, and includes a number of post-translational histone modifications (10,55,56). While histone acetylation is generally associated with open chromatin and active transcription, histone methylation is dependent on histone residue, degree of methylation (mono-, di- and tri-) and also specific gene locus and local chromatin environment, associated with repressive or facilitative effector proteins for the process of gene expression. For example, overall levels of a repressive mark, histone H3 trimethylated at lysine 9 (H3K9me3) and expression of H3K9 methyltransferase gene, ESET/SETDB1, are reportedly increased in HD patients and in transgenic R6/2 HD mice (21). Down-regulation of ESET expression and H3K9me3 by treatment with mithramycin and cystamine significantly ameliorated the behavioral and neuropathological phenotypes of R6/2 mice and extended survival by over 40% (57). A recent study in HD frontal cortex and striatal tissue extracts showed that H3K4me3 alterations at candidate gene promoters with down-regulated expression, including dopamine receptor 2 (DRD2), preproenkephalin (Penk1), brain-derived neurotrophic factor (BDNF) and synaptophysin (Syp) (9). To the best of our knowledge, our study is the first to embark on genome-scale unbiased mapping of H3K4me3 in a cell-type specific manner (FACS-sorted cortical neuronal nuclei) from postmortem HD and control brain. Consistent with H3K4me3 as a fingerprint for actively transcribed genes and a marker for transcription initiation sites (23–26), majority (83 out of 136) H3K4me3 peaks were mapped to genome positions within 2 kb of a TSS, with the highest peaks around 100 bp downstream of the TSS in both HD or control brains. Thus, mutant HTT protein is unlikely to be associated with a generalized distortion of histone methylation landscapes in diseased neurons. Instead, HD appears to be associated with highly specific defects at (according to our estimates) 78 loci (after correction for false discovery) in various portions of the genome. There was a striking enrichment for genes defining neuronal function and synaptic signaling (Table 2), confirming that the molecular pathology of HD is associated with severe defects in cortical neurons (58). At some of these loci, including the HES4 promoter, multiple types of epigenetic markings showed disease-associated changes, including DNA cytosine methylation which generally shows an opposing and largely non-overlapping distribution with H3K4me3. Importantly, altered H3K4me3 signaling in HD may relate to a strong inverse correlation between DNA methylation and the presence of H3K4me3 (49). Unmethylated CGIs have been shown to recruit the CxxC finger protein 1 (Cfp1) that associate with the H3K4 methyltransferase Setd1 (Set1/COMPASS or Set1B) (59,60) to create chromatin domains rich in H3K4me3 for enhanced gene expression (61). This is consistent with our finding that the reduced H3K4me3 signal for HES4 is associated with increased DNA methylation in the HES4 promoter. Furthermore, recent studies have demonstrated that normal HTT function facilitates epigenetic silencer polycomb repressive complex 2 (PRC2) which regulates methylation at histone H3-lysine 27 (19). Since H3K4me3 demethylase, namely Rbp2 (KDM5A or JARID1A), is recruited by PRC2 (62), mutant HTT may reduce H3K4me3 signaling by facilitating PCR2 function. In particular, a recent study by Vashishtha et al. (9) showed that knockdown of H3K4 demethylase SMCX/Jarid1c by shRNA in primary neurons reverses some of the down-regulation of key neuronal genes by HTT, such as BDNF. Furthermore, there is evidence for physical interactions, and functional crosstalk, between histone deacetylases and histone demethylases in intact cells (63,64). The Class I-specific HDAC inhibitors increase in histone acetylation as well as H3K4me2/H3K4me3 levels in cortical neurons and astrocytes (65). Since global levels of histone acetylation are reduced in models of HD (11,12) and HD brains (13), reduced histone acetylation by mutant HTT may also reduce H3K4me3. Thus, mutant HTT may specifically interact with molecules that regulate DNA methylation (CFP1) or histone methyltransferases associated with trithorax (TRXG) or polycomb 2 (PRC2) chromatin remodeling complexes, or HDAC to increase DNA methylation and reduce H3K4me2/3 in the HES4 promoter and a select subset of additional promoters and regulatory DNA sequences in HD cortical neurons. The alteration of H3K4me3 may directly (by interacting with histone modification enzymes such as PRC2 or Jarid1c, or indirectly by affecting chromatin structures and chromatin remodeling in general) produce HTT-induced transcriptional changes.
The involvements of the HES family in HD pathogenesis are largely unknown. Our finding of reduced H3K4me3 level, increased DNA methylation of the HES4 promoter and reduced HES4 mRNA levels in HD cortical neurons and increased mutant HTT-induced aggregates and cell death following shRNA knockdown of HES4 in human neuroblastoma cells provide evidence potentially linking the HES transcription factor family to HD pathogenesis. Interestingly, reduced H3K4me3 is specific for HES4 since analysis of this histone mark for other HES family members shows no significant changes. The HES4 gene, while present in many vertebrate genomes, is not found in Muridae (including mouse and rat) genomes. However, extensive cortical dysfunction in HD mouse models was noted, other notch signaling pathway (e.g. mouse Hes1, most closely related mouse homology to HES4 in human) may compensate for the dysfunction of HES4. HES4 mRNA is also significantly enriched in human neuronal nuclei as one of heterogeneous nuclear RNAs (Fig. 4B), raising a possibility of the involvement of hnHES4 RNA in transcriptional regulation and RNA processing. Importantly, we observed that a significant increase in intermediate methylated DNA of the HES4 promoter occurred in HD brain and this increase is associated with the reduced nuclear protein binding to the fully methylated HES4 promoter compared with the un-methylated or hemi-methylated HES4 promoter. It is likely that the increased intermediately methylated DNA (but not hemi-methylated DNA with only one strand methylated (66) can be attributed to increased asymmetric semi-methylation in HD in view of the similarity of its protein binding pattern to un-methylated and hemi-methylated DNA (Fig. 3). This type of asymmetric semi-DNA methylation is a mechanism that may be particular relevant in differentiated tissues in the context of disease (67,68), in contrast to hemimethylation which commonly is linked to the process of DNA replication. In HD brain, DNA methylation in the HES4 promoter, instead of alterations in nuclear protein function, is a key player in downregulating HES4 mRNA.
In addition to the altered epigenetic modifications of HES4 and reduced HES4 mRNA, our analysis uncovered that two HES4 targeted genes, MASH1 and P21, were dysregulated in HD cortex in opposite directions, down-regulation of MASH1 and up-regulation of P21. The opposite regulations of HES4 target genes were further confirmed by shRNA knockdown of HES4 in cultured neuroblastoma (Fig. 5A). Mash1 is a forebrain neuronal transcription factor and is critically involved in striatal development (46,50). HES4 can suppress MASH1 expression by disrupting the formation of E47 with the striatum-specific bHLH factor Mash1; HES4 can also interact with the Orange domain to remove the repression of transcription of P21. Blocking Hes1 (the closest rodent HES family to human HES4) expression stimulates the expression of cyclin-dependent kinase inhibitor P21 and modulates differentiation of GABAergic (striatal) neurons (51). Thus, the coordinate interplay of HES family proteins and its down-stream targets MASH1 and P21 play a critical role in guiding the phenotypic development of striatal GABAergic neurons. Thus, epigenetic changes of HES4 (i.e. reduced H3K4me3 signal and increased DNA methylation on the HES4 promoter) led to lower HES4 expression and dysregulation of putative HES4 target genes, including MASH1 and P21, to affect forebrain neuronal development and contribute to HD pathogenesis (69). Given the critical role of Notch signaling in forebrain neuronal development, the finding of dysregulated epigenetic modifications of HES4 and altered expression of Notch signaling molecules (MASH1 and P21) is consistent with the notion that HD may be a lifelong disease process (69) and suggests that abnormal neurodevelopment involving Notch signaling may contribute to HD pathogenesis.
The significance of epigenetic modifications of the HES4 function is substantiated by the finding that the degree of DNA methylation of the HES4 promoter is associated with striatal degeneration and age of onset of HD patients. Recently, we uncovered, among 523 HD patients, two classes of HD pathology with mainly striatal degeneration (class I) or cortical degeneration (class II) (31). We found that among 25 HD patients tested for DNA methylation in this study, the DNA intermediate methylation of the HES4 promoter is highly correlated with severity of striatal degeneration. Interestingly, this correlation is specific for striatal degeneration, but not cortical degeneration despite that the DNA methylation of HES4 was assessed in the cortex. The selective correlation between the degree of the intermediate methylation pattern for the HES4 promoter and striatal degeneration is in agreement with the primary striatal degeneration in HD, and with HES4 function to control the expression of forebrain neuron-specific transcriptional factor Mash1, and consequently striatal development (50,70). Functional significance of HES4 in development of HD was further supported by increased mutant HTT-induced nuclear aggregates and cell death following shRNA knockdown of HES4 in human neuroblastoma cells. While our HES4 shRNA data suggest pathological potential of HES4 by promoting mutant HTT aggregation and cell death in cells (and potentially in the human brain), future studies using HES4 shRNA in human stem cells derived from HD patients are needed to establish the causal role of HES4 in HD pathogenesis. Moreover, since mutant HTT produces broad changes in H3K4me3 patterns throughout the brain, we speculate that additional striatum-specific factors (such as striatum-specific HTT-interacting proteins) are recruited to act on the top of H3K4me3 changes to trigger striatal degeneration.
Age-at-onset in HD is variable by as much as 25 years even with the same number of CAG repeats in the HTT gene, suggesting that non-genetic factors, such as the epigenetic factors identified here, may play a role here. Supporting this possibility, we found that there was a strong correlation between DNA intermediate methylation on the HES4 promoter and age-of-onset of HD. Importantly, this correlation is independent of CAG repeat, indicating that HES4 may represent an epigenetic modifier of HD. Therefore, it is reasonable to speculate that certain environmental exposures alter DNA methylation of the HES4 promoter, leading to abnormal HES targeted gene expression in some individuals. Such epigenetic modifications may in turn interact with other genetic susceptibility and facilitate HD pathogenesis. In view of the fact that most promoters are unmethylated in mature tissues including the brain (71,72), methylation of the HES4 promoter can be considered abnormal and specific to HD pathology although underlying mechanism remains unknown. Our finding identifies the epigenetic modulation of the HES4 promoter as a potential therapeutic target to postpone HD the age-of-onset. If similar patterns of HES4 DNA methylation changes in peripheral blood cells are confirmed as in human brains, the HES4 epigenetic change may also represent a novel biomarker to predict the age-of-onset of the disease.
In summary, our neuron-specific, genome-wide mapping approach identified a large number of epigenetically altered loci in the neuronal HD genome, including loss of H3K4me3 and excessive DNA methylation on the HES4 promoter, as well as altered expression of HES4 and its target genes MASH1 and P21. The functional role of HES4 in HD pathogenesis is supported by a significant correlation between HES4 epigenetic dysregulation and age-of-onset and striatal degeneration of HD, and by the exacerbation of mutant HTT-induced aggregates and cell death after shRNA knockdown of HES4 in human neuroblastoma cells. Our findings implicate imbalances in histone and DNA methylation on HES4 and other gene promoters as important modifiers of neuronal degeneration and age-of-onset in HD. Thus, epigenetic changes at HES4 gene may represent a biomarker for predicting clinical and histopathological outcomes and a novel avenue for therapeutic intervention of HD.
Materials and Methods
HD and control brain samples
Fifty-seven postmortem brains, (25 HD and 32 control), were obtained from the Harvard Brain Tissue Resource Center, McLean Hospital (Table 1). All ChIP-sequencing, qPCR and DNA methylation studies were conducted on frozen (never fixed) tissue collected from the rostral dorsolateral portion of the frontal lobe (Brodmann 9). HD brains were selected from a restricted CAG repeat size between 40 and 54 repeats, representative of common repeat sizes in adult onset HD. To increase sample homogeneity (73), each specimen was micro-dissected, avoiding the surface and layer 1 and taking as uniform a sample from the cortical grey matter (II–VI) as possible.
ChIP-seq
Table 1A summarizes the demographics of the six HD and five control brains used for FACS-ChIP sequencing. Postmortem intervals were within the time window in which H3 trimethylation is stable (74–76).
DNA methylation
For DNA methylation analysis, genomic DNA was extracted from 25 HD (including four from ChIP-sequencing) and 27 control brains (Table 1B).
Quantitative reverse-transcriptase polymerase chain reaction
For qRT–PCR, RNA was extracted from a subset of the cohort used for DNA methylation assays (14 HD and 14 control brains, Table 1C).
FACS-ChIP-seq protocol
Neuronal and non-neuronal nuclei were separated (77,78) by fluorescence-based nuclei sorting (FACS), followed by chromatin immunoprecipitation and genome-wide histone methylation mapping via next generation sequencing (ChIP-Seq, see Fig. 1A) (29,76). (i) Nuclei extraction and FACS: ∼750 mg of tissue was homogenized in 5 ml of lysis buffer. Lysates were loaded on a sucrose solution and centrifuged at 24 400 rpm for 2.5 h at 4°C. Nuclei pellets were resuspended in 500 μl PBS and incubated in staining mix containing 1:1200 anti-NeuN (Millipore), 1:1400 Alexa488 goat anti-mouse secondary antibody (Invitrogen)] for 45 min. FACS was done at the Boston University Medical School Flow Cytometry Core Lab on a FACSVantage SE flow cytometer. (ii) The sorted nuclei (3–5 million) were digested with micrococcal nuclease (4 U/ml) at 37°C for 5 min. The reaction was stopped and nuclei were lysed and pre-cleared by Protein G Agarose. Chromatin immunoprecipitation was carried out by incubating digested nuclei with anti-H3K4me3 (1:315; Upstate; 07–473) at 4°C overnight. Immunoprecipitated chromatin was incubated with Protein G Agarose for 1 h, and beads were washed by a series of low and high salt buffer, lithium chloride buffer, TE buffer and then eluted in 0.1 M NaHCO3 and 1% SDS. The eluted DNA was digested with proteinase K and then purified. (iii) ChIP-Seq Library Construction was carried out according to the Illumina protocol using Genomic Adaptor Oligo Mix (Illumina) by Fast-link DNA Ligation Kit (Epicentre) the Genomic PCR Primers (Illumina) according to the Illumina protocol. PCR product was cleaned and correct size of PCR product was confirmed by gel electrophoresis. (iv) The smaller smear was gel purified and libraries were sent for deep-sequencing on the Illumina Genome Analyzer of Deep Sequencing core facilities at University of Massachusetts School of Medicine.
Computational analysis of H3 trimethylation landscapes
All sequencing libraries were single-end 36 bp reads which were mapped to the gender appropriate human genome (HG19) by Bowtie (version 0.11.3), allowing up to one mismatch. Reads that mapped to multiple locations were discarded. The MACS software (version 1.3.5) was used to identify statistically enriched H3K4me3 regions (termed ‘peaks’ hereafter). We contrasted each sample against the input sample using bw = 230 and tsize = 36, and default values for the remaining parameters in MACS. To identify differentially expressed H3K4me3 peaks between controls and HD cases, all peaks were combined and overlapping peaks merged, resulting in 33 148 peaks. H3K4me3 peaks that were significantly decreased in the HD samples were defined as follows: (i) minimum peak size of 1 Kb with pseudo-count 0.001 for average densities; (ii) average read density in control samples ≥0.01, (iii) the ratio of average read densities Control:HD ≥2 and (iv) the t-test P-value ≤0.05. A Benjamini Hochberg false discovery rate was calculated. Reciprocal criteria were used to define H3K4me3 peaks significantly increased in HD.
GO term enrichment analysis
We used the getEnrichedGO function in the ChIPpeakAnno R package (79) to test whether certain loci of H3K4me3 (associated with specific genes) were overrepresented than would be expected by chance (adjusted P < 0.05, according to Benjamini & Hochberg (1995) step-up FDR controlling procedure).
Phylogenetic analysis of HES family genes
HES gene family and protein sequences were obtained from NCBI and Ensembl databases. Multiple sequence alignment of protein sequences was performed using ClustalW algorithm and edited in GeneDoc program using Blosum62 as similarity scoring matrix.
DNA methylation detection protocol
Genomic DNA (gDNA) was extracted from frozen brain using the Blood & Cell Culture DNA kit (Qiagen) and quantified by NanoDrop 2000 and 0.7% agarose gel electrophoresis for DNA integrity. Samples showing a A260/A280 ratio >1.7 and a major band around 30 kb were included in methylation analysis. DNA methylation was measured by the Methyl-Profiler PCR Array according to manufacturer's instructions (SABiosciences/Qiagen). This assay is based on MethylScreen (80,81) with combined digestion of methylation-sensitive type II enzyme (HpaII/HhaI) and methylation-dependent type IV enzyme (McrBC) (EpiTect Methyl DNA Restriction kit, Qiagen) followed by real-time PCR analysis of remaining gDNA. Primers were designed, evaluated and provided by SABiosciences/Qiagen for human HES4 (catalog# MePH00010-2A). Briefly, 1 µg of gDNA from each case or control was divided among four digestion-conditions: mock, HpaII/HhaI, McrBC and HpaII/HhaI+ McrBC. Over-night digestion at 37°C with qPCR was conducted with gene-specific primers for equal quantities (1/25th) of differentially treated genomic DNAs on an ABI Prism 7000 system. Cycle threshold (Ct) values for each condition were used to calculate un-methylated (UM), fully methylated (FM) and intermediately methylated (IM) DNA such that UM, FM and IM sum to 1.0 for a given sample. All experiments and data analyses were done in double blind.
RNA isolation and gene expression analyses (qRT–PCR)
Total RNA was extracted from frozen human HD and control brain with Trizol reagent and cleaned with an RNeasy micro kit (Qiagen). Total RNA was reverse transcribed to cDNA using SuperScript II Reverse Transcriptase Kit (Invitrogen). The qRT–PCR was performed using Taqman Gene Expression Assays on 7500 Real-Time PCR System. Probes and primers specific for human HES4 and 18S RNA (Hs00368353_g1 and Hs99999901_s1, respectively) were used according to the manufacturer's protocol. Averaged threshold-cycle (Ct) values of the 18S RNA were used to normalize the target gene (HES4), which then were used to determine the relative expression of the gene for HD versus control samples by the ΔΔCt method.
To analyze HES4 mRNA in Neu+ (neuronal) and Neu− (non-neuronal) nuclei, total RNA was extracted from 1–3 million of FACS sorted human brain nuclei using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. cDNA was synthesized using Script™ cDNA Synthesis Kit (Bio-Rad #170-8891), following the manufacturer's instructions. Quantitative real-time PCR was performed in triplicate by using Power SYBR® Green PCR Master Mix (AB applied biosystem, #4367659) on LightCycler 96 Real-Time PCR System from Roche. The mRNA level was normalized by gene 18 s rRNA. Primers used for HES4 primer set #2: forward TCAGCTCAAAACCCTCATCC, reverse TGTCTCACGGTCATCTCCAG; HES4 primer set #3: forward ATCCTGGAGATGACCGTGAG, reverse CGGTACTTGCCCAGAACG; 18s rRNA forward GTTGGTGGAGCGATTTGTCT, reverse GAACGCCACTTGTCCCTCTA.
Electrophoretic mobility shift assay
The promoter region of the HES4 gene was obtained by cloning the qPCR product of the HES4 DNA methylation assay into pGEM3zf at the HincII site followed by DNA sequencing of the insert for confirmation. This qPCR amplicon expands a 269 bp region −387/−118 upstream of the human HES4 gene TSS. To test its binding capability under different methylation states to nuclear proteins in brain, we excised this fragment from vector using EcoRI and HindIII, and digested it further with BamHI sites to yield two fragments of identical size (134–135 bp) and then treated the fragments with or without SssI DNA methylase. Complementary genome DNA strands were annealed at room temperature for 30 minutes after heated to 80°C by the following different combinations (i) ‘unmethylated’ probe from two strands without treatment of Sssi, (ii) ‘methylated’ probe from two strands with treatment of Sssi, (iii) ‘hemi-methylated probe’ from one strand with treatment of Sssi and one strand without treatment of Sssi. The BamHI-digested, un-/hemi-/fully-methylated double-stranded DNA then were filled in with 32P-labeled dCTP as described previously (82). EMSA was performed as described previously with modifications (82). Briefly, lysate in 20 μg proteins was pre-incubated on ice for 10 min in binding buffer (15 µl volume) before 32P-labeled double-strand probe in 1 ng was added. After 10 min incubation at 23°C reaction mixtures were fractionated on 4% non-denaturing polyacrylamide gel in 0.25 × TBE and autoradiography was completed.
Statistical analysis of the relationship of HES4 DNA methylation with level of striatal involvement and age-of-onset in HD
Twenty-two of the 25 HD samples studied had been evaluated previously for levels of striatal and cortical involvement (31). Briefly, each brain sample was reviewed by gross and microscopic examination for the level of involvement for 50 brain regions. Cluster analysis reduced the data to two main measures of involvement: (i) striatal and (ii) cortical. The striatal cluster represented a synthesis of 28 brain measures and the cortical cluster constituted 13 brain measures. Comparisons between HD cases and controls to assess possible differences in age at death and postmortem interval were analyzed by Student's t-test. The relationship of the level of UM, IM or FM to the level of striatal involvement was studied by Spearman correlation, and by a general linear model controlling for the effect of the size of the expanded CAG repeat size and the level of cortical involvement. The t-tests, Spearman correlation and general linear models were performed by SAS version 9.3.
HTB-11 cell culture, transfection of shRNA-HES4 and qRT–PCR of HES4 mRNA
HTB-11 cells (SK-N-SH, human neuroblastoma) were obtained from ATCC and cultured in DMEM medium plus 10% fetal bovine serum, glutamine, high glucose and sodium pyruvate. Constructs expressing human HTT fragment (30 amino acid residues at the N-terminus) with varied length (23, 73 or 145) of N-terminal Q repeats in pcDNA3 plasmid were obtained from Coriell Institute for Medical Research via GenScript (Piscataway, NJ). Constructs expressing one control shRNA and four different shRNAs to the human HES4 gene were purchased from Qiagen. Cell transfection was conducted by Lipofectamine 2000 (Life Technologies). Successful transfection was indicated by green fluorescence of GFP expressed by co-transfected shRNA vector.
Twenty-six hours after transfecting with three different amounts of individual shRNA constructs, we extracted total RNAs and converted them into cDNA using SuperScript III kit (Life Technologies). Human HES4 mRNA level was determined by qRT–PCR analysis using the TaqMan kit (Life Technologies) as described previously (83).
Detection of mutant HTT-induced aggregates and cell death in HBT-11 cells
To test the effect of shRNA-HES4 on mutant HTT-induced aggregates and cell death, we determined ubiquitin-positive aggregates by immunocytochemistry and cell death by trypan blue exclusion assay for HTB-11 cells cultured on glass surface coated with d-poly-lysine using the protocol described previously (84). HTB-11 cells were transfected with one of four distinct shRNA-HES4 (shRNA-HES4–1, shRNA-HES4–2, shRNA4–3 and shRNA4–4) or scramble shRNA (control shRNA for non-specific and off-target effects) with or without HTT constructs at different ratio for 24 h before. HTB-11 cells were incubated with polyclonal antibody against ubiquitin (Dako, diluted 1:100) at 4°C overnight and visualized by a second antibody conjugated with Cy3. GFP expressed from shRNA constructs was used as indicator of transfection efficiency. Nuclear DNA was highlighted by DAPI staining (1 ng/ml) for 5 min before mounting. Signals of ubiquitin, GFP and DAPI were collected from cells as described previously using fluorescent microscope (84). Cell death was tested by trypan blue exclusion 40 hrs after transfection, according to the manufacturer protocol (Sigma-Aldrich, St Louis, MO). Briefly, cells were exposed to a mixture of 0.4% trypan blue reagent:HBSS = 1:1 for 5 min at 37°C before cell images were taken for the same view on bright field for blue staining of dying cells, and under fluorescence for GFP (Nikon Eclipse TE200 system equipped with QICAM CCD camera). Cell images at ×10 object lens were taken randomly from triple transfected wells. Blue cells were labeled by Photo Shop software and counted by ImageJ software after alignment of blue cells with GFP positive cells. Each view contained 60∼120 GFP-positive cells. Percentage of cell death was calculated by blue cell number against GFP cells. Mean values were further calculated from five views.
Quantitative analysis of HTT aggregates
To assess the effect of HES4 shRNA knockdown, we used an automated using ImageJ version 1.49 g analysis with a custom script for analysis of HTT aggregation in cells stained with ubiquitin after transfection with HTT and shRNA. Non-cellular background was first removed from the ubiquitin-RFP image using Gaussian blur with a sigma of 20, thresholding by mean signal. Next, the cellular area was identified based on the mask. Each selected area was separately analyzed and using ‘max entropy’ thresholding, particles 102 pixels or greater were counted as an aggregate. We quantified the relative size of cellular aggregates by calculating from the proportion of the aggregate to the cellular area being assessed. We quantified the density of aggregation by the measurement of pixel intensity, which was calculated by multiplying the total aggregate area by the mean signal intensity. Unequal variance Welch's t-tests were used to assess statistical differences in protein aggregation between different HTT cotransfected with sc shRNA or HES4 shRNA.
Authors' Contributions
(i) Conducted experiments (I.C., M.J., J.E.C., P.L., Y.W., Y.J.: nuclei sorting and ChIP-seq assays; G.B., K.P., M.Z.: DNA methylation assays, HTB-11 HES4 knock-down, Ubiquitin/HTT aggregates, cell death, EMSA; P.L., Y.W., Y.J.: EMSA, RNA quantification and qPCR assays); (ii) bioinformatical analyses (H.S., X.D., A.G., E.R., Z.W.); (iii) provided resources and materials (R.H.M., S.A.); (iv) designed the study and wrote the paper (J.F.-C., R.H.M., Z.W., S.A., G.B.).
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
This work was supported by the National Institutes of Health (1R01NS073947 Epigenetic Markers in Huntington's disease Brain, NS041083-07, R21NS076958) and The Jerry McDonald Huntington's Disease Research Fund. We thank Adam Labadorf for his analysis of the correlation between HES4 mRNA and striatal degeneration score in HD brains.
Supplementary Material
References
- 1.Martin J.B., Gusella J.F. Huntington's disease. Pathogenesis and management. N. Engl. J. Med. 1986;315:1267–1276. doi: 10.1056/NEJM198611133152006. [DOI] [PubMed] [Google Scholar]
- 2.Vonsattel J.P., DiFiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Group, T.H.s.D.C.R. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 4.MacDonald M.E., Gines S., Gusella J.F., Wheeler V.C. Huntington's disease. Neuromol. Med. 2003;4:7–20. doi: 10.1385/NMM:4:1-2:7. [DOI] [PubMed] [Google Scholar]
- 5.Gusella J.F., MacDonald M.E. Huntington's disease: the case for genetic modifiers. Genome Med. 2009;1:80. doi: 10.1186/gm80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djousse L., Knowlton B., Hayden M., Almqvist E.W., Brinkman R., Ross C., Margolis R., Rosenblatt A., Durr A., Dode C., et al. Interaction of normal and expanded CAG repeat sizes influences age at onset of Huntington disease. Am. J. Med. Genet. A. 2003;119A:279–282. doi: 10.1002/ajmg.a.20190. [DOI] [PubMed] [Google Scholar]
- 7.Cha J.H. Transcriptional dysregulation in Huntington's disease. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- 8.Dunah A.W., Jeong H., Griffin A., Kim Y.M., Standaert D.G., Hersch S.M., Mouradian M.M., Young A.B., Tanese N., Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 9.Vashishtha M., Ng C.W., Yildirim F., Gipson T.A., Kratter I.H., Bodai L., Song W., Lau A., Labadorf A., Vogel-Ciernia A., et al. Targeting H3K4 trimethylation in Huntington disease. Proc. Natl Acad. Sci. USA. 2013;110:E3027–E3036. doi: 10.1073/pnas.1311323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakovcevski M., Akbarian S. Epigenetic mechanisms in neurological disease. Nat. Med. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steffan J.S., Bodai L., Pallos J., Poelman M., McCampbell A., Apostol B.L., Kazantsev A., Schmidt E., Zhu Y.Z., Greenwald M., et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 12.Steffan J.S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y.Z., Gohler H., Wanker E.E., Bates G.P., Housman D.E., Thompson L.M. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl Acad. Sci. USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dompierre J.P., Godin J.D., Charrin B.C., Cordelieres F.P., King S.J., Humbert S., Saudou F. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington's disease by increasing tubulin acetylation. J. Neurosci.: the Official Journal of the Society for Neuroscience. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrante R.J., Kubilus J.K., Lee J., Ryu H., Beesen A., Zucker B., Smith K., Kowall N.W., Ratan R.R., Luthi-Carter R., et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J. Neurosci.: the Official Journal of the Society for Neuroscience. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryu H., Lee J., Olofsson B.A., Mwidau A., Deodeoglu A., Escudero M., Flemington E., Azizkhan-Clifford J., Ferrante R.J., Ratan R.R. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc. Natl Acad. Sci. USA. 2003;100:4281–4286. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas E.A., Coppola G., Desplats P.A., Tang B., Soragni E., Burnett R., Gao F., Fitzgerald K.M., Borok J.F., Herman D., et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington's disease transgenic mice. Proc. Natl Acad. Sci. USA. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goula A.V., Stys A., Chan J.P., Trottier Y., Festenstein R., Merienne K. Transcription elongation and tissue-specific somatic CAG instability. PLoS Genet. 2012;8:e1003051. doi: 10.1371/journal.pgen.1003051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun X.J., Wei J., Wu X.Y., Hu M., Wang L., Wang H.H., Zhang Q.H., Chen S.J., Huang Q.H., Chen Z. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J. Biol. Chem. 2005;280:35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 19.Seong I.S., Woda J.M., Song J.J., Lloret A., Abeyrathne P.D., Woo C.J., Gregory G., Lee J.M., Wheeler V.C., Walz T., et al. Huntingtin facilitates polycomb repressive complex 2. Hum. Mol. Genet. 2010;19:573–583. doi: 10.1093/hmg/ddp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J., Hong Y.K., Jeon G.S., Hwang Y.J., Kim K.Y., Seong K.H., Jung M.K., Picketts D.J., Kowall N.W., Cho K.S., et al. ATRX induction by mutant huntingtin via Cdx2 modulates heterochromatin condensation and pathology in Huntington's disease. Cell Death Diff. 2012;19:1109–1116. doi: 10.1038/cdd.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu H., Lee J., Hagerty S.W., Soh B.Y., McAlpin S.E., Cormier K.A., Smith K.M., Ferrante R.J. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc. Natl Acad. Sci. USA. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruthenburg A.J., Allis C.D., Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G.A., Stewart R., Thomson J.A. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell. Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 27.Zhou V.W., Goren A., Bernstein B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genetics. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 28.Han I., You Y., Kordower J.H., Brady S.T., Morfini G.A. Differential vulnerability of neurons in Huntington's disease: the role of cell type-specific features. J. Neurochem. 2010;113:1073–1091. doi: 10.1111/j.1471-4159.2010.06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung I., Shulha H.P., Jiang Y., Matevossian A., Wang J., Weng Z., Akbarian S. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc. Natl Acad. Sci. USA. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers R.H., Vonsattel J.P., Paskevich P.A., Kiely D.K., Stevens T.J., Cupples L.A., Richardson E.P., Jr, Bird E.D. Decreased neuronal and increased oligodendroglial densities in Huntington's disease caudate nucleus. J. Neuropathol. Exp. Neurol. 1991;50:729–742. doi: 10.1097/00005072-199111000-00005. [DOI] [PubMed] [Google Scholar]
- 31.Hadzi T.C., Hendricks A.E., Latourelle J.C., Lunetta K.L., Cupples L.A., Gillis T., Mysore J.S., Gusella J.F., MacDonald M.E., Myers R.H., et al. Assessment of cortical and striatal involvement in 523 Huntington disease brains. Neurology. 2012;79:1708–1715. doi: 10.1212/WNL.0b013e31826e9a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Roon-Mom W.M., Hogg V.M., Tippett L.J., Faull R.L. Aggregate distribution in frontal and motor cortex in Huntington's disease brain. Neuroreport. 2006;17:667–670. doi: 10.1097/00001756-200604240-00022. [DOI] [PubMed] [Google Scholar]
- 33.Shulha H.P., Cheung I., Whittle C., Wang J., Virgil D., Lin C.L., Guo Y., Lessard A., Akbarian S., Weng Z. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch. Gen. Psychiatr. 2012;69:314–324. doi: 10.1001/archgenpsychiatry.2011.151. [DOI] [PubMed] [Google Scholar]
- 34.Thathiah A., Horre K., Snellinx A., Vandewyer E., Huang Y., Ciesielska M., De Kloe G., Munck S., De Strooper B. beta-arrestin 2 regulates Abeta generation and gamma-secretase activity in Alzheimer's disease. Nat. Med. 2013;19:43–49. doi: 10.1038/nm.3023. [DOI] [PubMed] [Google Scholar]
- 35.Peachey N.S., Ray T.A., Florijn R., Rowe L.B., Sjoerdsma T., Contreras-Alcantara S., Baba K., Tosini G., Pozdeyev N., Iuvone P.M., et al. GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2012;90:331–339. doi: 10.1016/j.ajhg.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez P.Q., Lohkamp B., Celsi G., Mache C.J., Auer-Grumbach M., Wernerson A., Hamajima N., Tryggvason K., Patrakka J. Novel INF2 mutation p. L77P in a family with glomerulopathy and Charcot-Marie-Tooth neuropathy. Pediatr. Nephrol. 2013;28:339–343. doi: 10.1007/s00467-012-2299-1. [DOI] [PubMed] [Google Scholar]
- 37.Paciorkowski A.R., Darras B.T. Making sense of genetic heterogeneity: emergence of pathways in developmental brain disorders. Neurology. 2013;80:426–427. doi: 10.1212/WNL.0b013e31827f101c. [DOI] [PubMed] [Google Scholar]
- 38.Wood H.B. TMEM106B is a susceptibility locus for Ftld. Nat. Rev. Neurology. 2010;6:184. doi: 10.1038/nrneurol.2010.22. [DOI] [PubMed] [Google Scholar]
- 39.Finch N., Carrasquillo M.M., Baker M., Rutherford N.J., Coppola G., Dejesus-Hernandez M., Crook R., Hunter T., Ghidoni R., Benussi L., et al. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76:467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutherford N.J., Carrasquillo M.M., Li M., Bisceglio G., Menke J., Josephs K.A., Parisi J.E., Petersen R.C., Graff-Radford N.R., Younkin S.G., et al. TMEM106B risk variant is implicated in the pathologic presentation of Alzheimer disease. Neurology. 2012;79:717–718. doi: 10.1212/WNL.0b013e318264e3ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Keeffe G.W., Gutierrez H., Pandolfi P.P., Riccardi C., Davies A.M. NGF-promoted axon growth and target innervation requires GITRL-GITR signaling. Nat. Neurosci. 2008;11:135–142. doi: 10.1038/nn2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu L.G., Li L.Y., Shu H.B. TRAF7 potentiates MEKK3-induced AP1 and CHOP activation and induces apoptosis. J. Biol. Chem. 2004;279:17278–17282. doi: 10.1074/jbc.C400063200. [DOI] [PubMed] [Google Scholar]
- 43.El Yakoubi W., Borday C., Hamdache J., Parain K., Tran H.T., Vleminckx K., Perron M., Locker M. Hes4 controls proliferative properties of neural stem cells during retinal ontogenesis. Stem Cells. 2012;30:2784–2795. doi: 10.1002/stem.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabadan M.A., Cayuso J., Le Dreau G., Cruz C., Barzi M., Pons S., Briscoe J., Marti E. Jagged2 controls the generation of motor neuron and oligodendrocyte progenitors in the ventral spinal cord. Cell Death Diff. 2012;19:209–219. doi: 10.1038/cdd.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hodges A., Strand A.D., Aragaki A.K., Kuhn A., Sengstag T., Hughes G., Elliston L.A., Hartog C., Goldstein D.R., Thu D., et al. Regional and cellular gene expression changes in human Huntington's disease brain. Human Molecular Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 46.Kageyama R., Ohtsuka T., Kobayashi T. Roles of Hes genes in neural development. Dev. Growth Diff. 2008;50(Suppl. 1):S97–103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- 47.Bertrand N., Castro D.S., Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neuroscience. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 48.Jhas S., Ciura S., Belanger-Jasmin S., Dong Z., Llamosas E., Theriault F.M., Joachim K., Tang Y., Liu L., Liu J., et al. Hes6 inhibits astrocyte differentiation and promotes neurogenesis through different mechanisms. J. Neurosci. 2006;26:11061–11071. doi: 10.1523/JNEUROSCI.1358-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D., Johnson B.E., Hong C., Nielsen C., Zhao Y., et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casarosa S., Fode C., Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 51.Diguet E., Fernagut P.O., Scherfler C., Wenning G., Tison F. Effects of riluzole on combined MPTP + 3-nitropropionic acid-induced mild to moderate striatonigral degeneration in mice. J. Neural. Transm. 2005;112:613–631. doi: 10.1007/s00702-004-0206-z. [DOI] [PubMed] [Google Scholar]
- 52.Ryman-Rasmussen J.P., Griffith A., Oloff S., Vaidehi N., Brown J.T., Goddard W.A., III, Mailman R.B. Functional selectivity of dopamine D1 receptor agonists in regulating the fate of internalized receptors. Neuropharmacology. 2007;52:562–575. doi: 10.1016/j.neuropharm.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katritch V., Cherezov V., Stevens R.C. Structure-function of the G protein-coupled receptor superfamily. Ann. Rev. Pharmacol. Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajagopal S., Rajagopal K., Lefkowitz R.J. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat. Rev. Drug Discovery. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ooi L., Wood I.C. Chromatin switching and transcriptional regulation in disease. Biochem. Soc. Trans. 2008;36:599–602. doi: 10.1042/BST0360599. [DOI] [PubMed] [Google Scholar]
- 56.Urdinguio R.G., Sanchez-Mut J.V., Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet. Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 57.Stack E.C., Del Signore S.J., Luthi-Carter R., Soh B.Y., Goldstein D.R., Matson S., Goodrich S., Markey A.L., Cormier K., Hagerty S.W., et al. Modulation of nucleosome dynamics in Huntington's disease. Hum. Mol. Genet. 2007;16:1164–1175. doi: 10.1093/hmg/ddm064. [DOI] [PubMed] [Google Scholar]
- 58.Eidelberg D., Surmeier D.J. Brain networks in Huntington disease. J. Clin. Invest. 2011;121:484–492. doi: 10.1172/JCI45646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tai Y.F., Scherfler C., Brooks D.J., Sawamoto N., Castiello U. The human premotor cortex is ‘mirror’ only for biological actions. Curr. Biol. 2004;14:117–120. doi: 10.1016/j.cub.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 60.Brooks D.J. Detection of preclinical Parkinson's disease with PET. Geriatrics. 1991;46(Suppl. 1):25–30. [PubMed] [Google Scholar]
- 61.Scherfler C., Khan N.L., Pavese N., Eunson L., Graham E., Lees A.J., Quinn N.P., Wood N.W., Brooks D.J., Piccini P.P. Striatal and cortical pre- and postsynaptic dopaminergic dysfunction in sporadic parkin-linked parkinsonism. Brain: J. Neurol. 2004;127:1332–1342. doi: 10.1093/brain/awh150. [DOI] [PubMed] [Google Scholar]
- 62.Pasini D., Hansen K.H., Christensen J., Agger K., Cloos P.A., Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Gene Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toffolo E., Rusconi F., Paganini L., Tortorici M., Pilotto S., Heise C., Verpelli C., Tedeschi G., Maffioli E., Sala C., et al. Phosphorylation of neuronal Lysine-Specific Demethylase 1LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. J. Neurochem. 2014;128:603–616. doi: 10.1111/jnc.12457. [DOI] [PubMed] [Google Scholar]
- 64.Lilja T., Heldring N., Hermanson O. Like a rolling histone: epigenetic regulation of neural stem cells and brain development by factors controlling histone acetylation and methylation. Biochimica et Biophysica Acta. 2013;1830:2354–2360. doi: 10.1016/j.bbagen.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 65.Audet M., Bouvier M. Restructuring G-protein- coupled receptor activation. Cell. 2012;151:14–23. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 66.Adams R.L., Lindsay H. What is hemimethylated DNA? FEBS Lett. 1993;320:243–245. doi: 10.1016/0014-5793(93)80595-l. [DOI] [PubMed] [Google Scholar]
- 67.Gao Z.G., Verzijl D., Zweemer A., Ye K., Goblyos A., Ijzerman A.P., Jacobson K.A. Functionally biased modulation of A(3) adenosine receptor agonist efficacy and potency by imidazoquinolinamine allosteric enhancers. Biochem. Pharmacol. 2011;82:658–668. doi: 10.1016/j.bcp.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verzijl D., Ijzerman A.P. Functional selectivity of adenosine receptor ligands. Purinergic Signal. 2011;7:171–192. doi: 10.1007/s11302-011-9232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gusella J.F., MacDonald M.E. Huntington's disease: seeing the pathogenic process through a genetic lens. Trend Biochem. Sci. 2006;31:533–540. doi: 10.1016/j.tibs.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 70.Cussac D., Newman-Tancredi A., Duqueyroix D., Pasteau V., Millan M.J. Differential activation of Gq/11 and Gi(3) proteins at 5-hydroxytryptamine(2C) receptors revealed by antibody capture assays: influence of receptor reserve and relationship to agonist-directed trafficking. Mol. Pharmacol. 2002;62:578–589. doi: 10.1124/mol.62.3.578. [DOI] [PubMed] [Google Scholar]
- 71.Illingworth R., Kerr A., Desousa D., Jorgensen H., Ellis P., Stalker J., Jackson D., Clee C., Plumb R., Rogers J., et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bird A. DNA methylation patterns and epigenetic memory. Gene Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 73.Petretto E., Mangion J., Dickens N.J., Cook S.A., Kumaran M.K., Lu H., Fischer J., Maatz H., Kren V., Pravenec M., et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akbarian S., Huang H.S. Epigenetic regulation in human brain-focus on histone lysine methylation. Biol. Psychiatr. 2009;65:198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang H.S., Matevossian A., Jiang Y., Akbarian S. Chromatin immunoprecipitation in postmortem brain. J. Neurosci. Method. 2006;156:284–292. doi: 10.1016/j.jneumeth.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 76.Huang H.S., Matevossian A., Whittle C., Kim S.Y., Schumacher A., Baker S.P., Akbarian S. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J. Neurosci. 2007;27:11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang Y., Matevossian A., Huang H.S., Straubhaar J., Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neuroscience. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matevossian A., Akbarian S. Neuronal nuclei isolation from human postmortem brain tissue. J. Visual Exp. 2008;20:e914. doi: 10.3791/914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S., Green M.R. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holemon H., Korshunova Y., Ordway J.M., Bedell J.A., Citek R.W., Lakey N., Leon J., Finney M., McPherson J.D., Jeddeloh J.A. MethylScreen: DNA methylation density monitoring using quantitative PCR. BioTechniques. 2007;43:683–693. doi: 10.2144/000112597. [DOI] [PubMed] [Google Scholar]
- 81.Brooks D.J. The clinical role of PET in cerebrovascular disease. Neurosurg. Rev. 1991;14:91–96. doi: 10.1007/BF00313030. [DOI] [PubMed] [Google Scholar]
- 82.Bai G., Kusiak J.W. Functional analysis of the proximal 5′-flanking region of the N-methyl-D-aspartate receptor subunit gene, NMDAR1. J. Biol. Chem. 1995;270:7737–7744. doi: 10.1074/jbc.270.13.7737. [DOI] [PubMed] [Google Scholar]
- 83.Bai G., Ambalavanar R., Wei D., Dessem D. Downregulation of selective microRNAs in trigeminal ganglion neurons following inflammatory muscle pain. Mol. Pain. 2007;3:15. doi: 10.1186/1744-8069-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang W., Zhang Y., Xiao L., Van Cleemput J., Ji S., Bai G., Zhang X. Cannabinoids promote embryonic and adult hippocampus neurogenesis and produce anxiolytic- and antidepressant-like effects. J. Clin. Invest. 2005;115:3104–3116. doi: 10.1172/JCI25509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.