Abstract
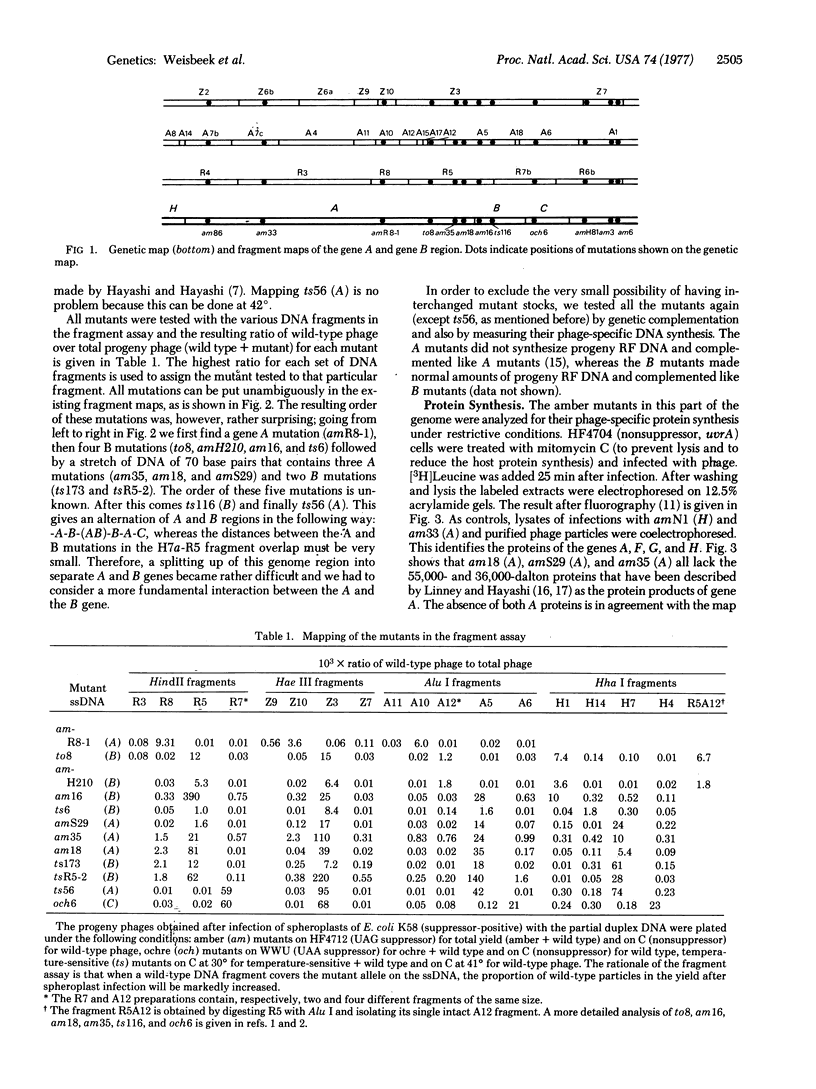
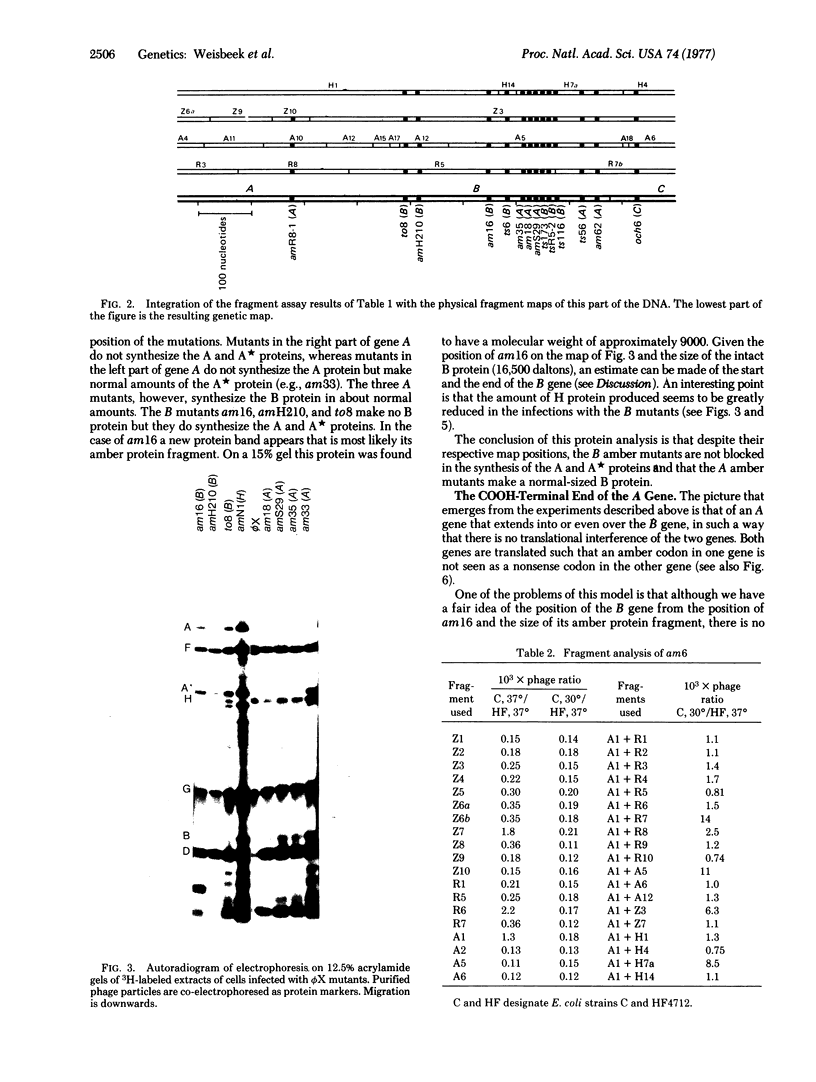
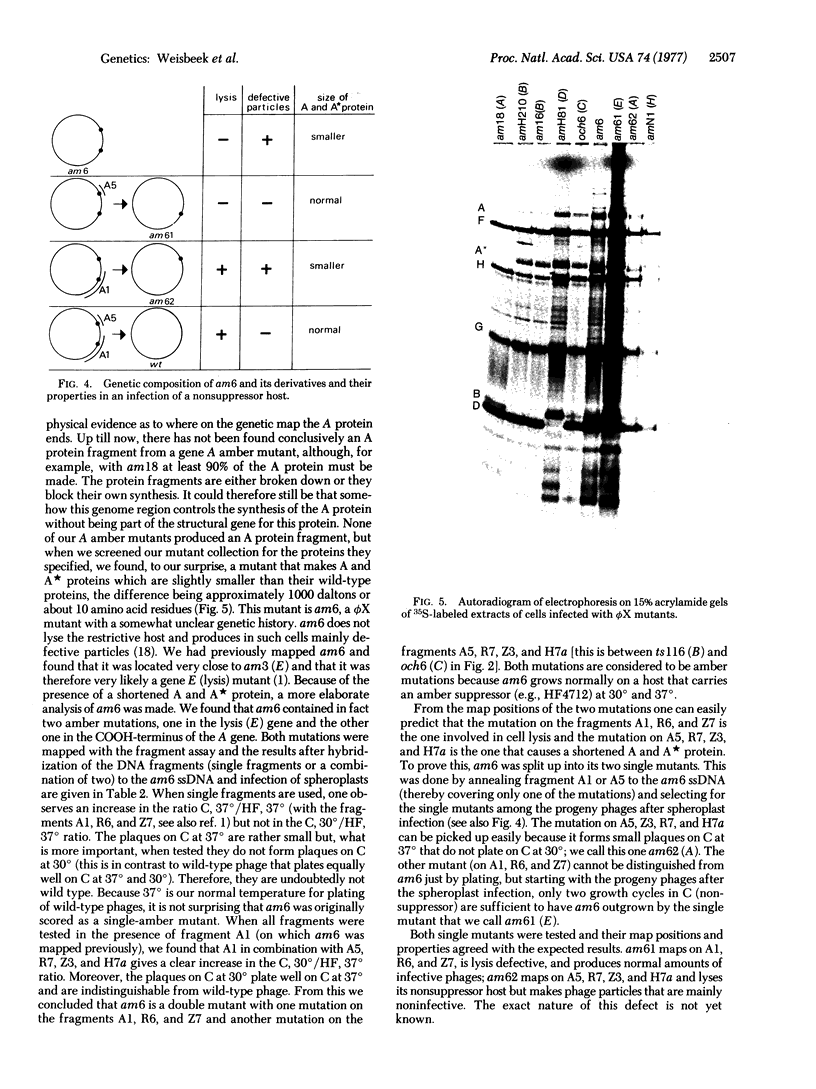
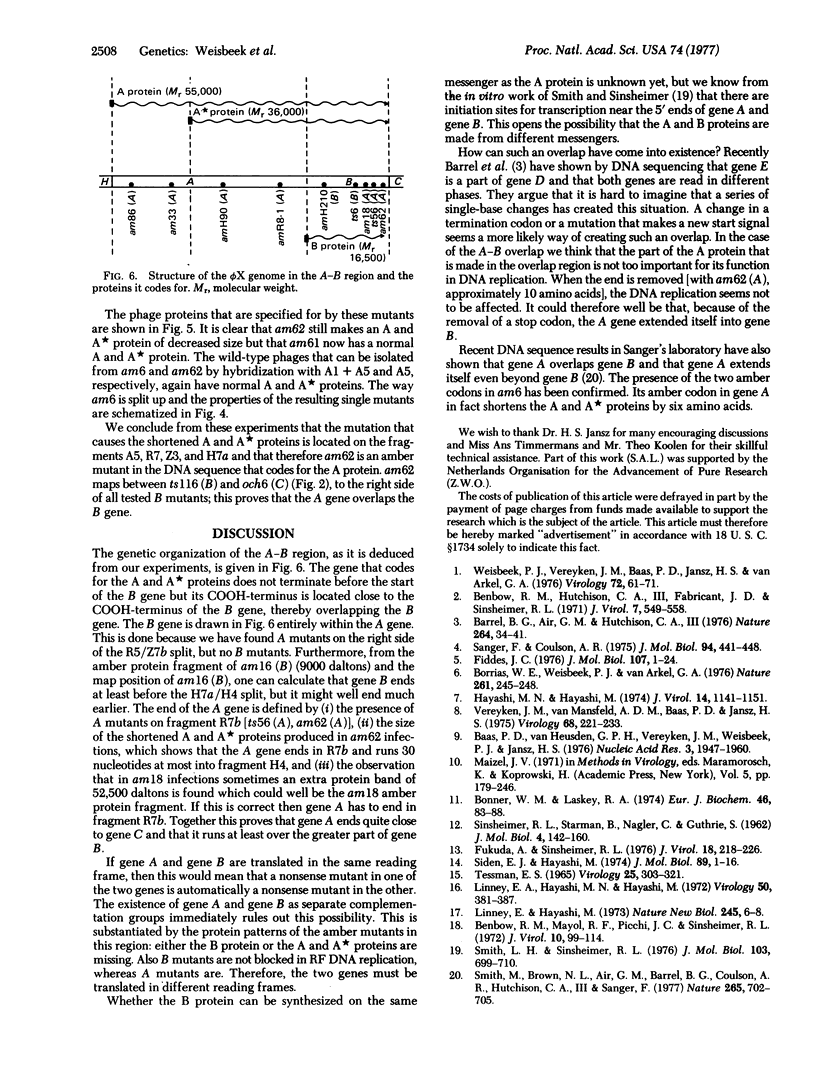
The map position of several phiX174 mutations in the genes A and B was determined by marker rescue with DNA fragments produced by the restriction enzymes Hha I, HindII, Hae III, and Alu I. All the gene B mutants were found to be located within gene A. Genetic complementation and analysis of phage-specific protein synthesis show that, under restrictive conditions, nonsense mutants in gene A do not block the synthesis and activity of the B protein and nonsense mutants in gene B do not affect the gene A function. The map position of the COOH-terminal end of gene A was determined using an amber mutant that synthesizes slightly shortened A and A proteins. It is concluded from these experiments that gene A overlaps gene B completely (or almost completely) and that the overlap region can be translated in two ways with different reading frames: one frame for the synthesis of the A and A proteins and another for the synthesis of the B protein.
Full text
PDF
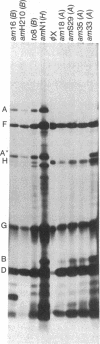
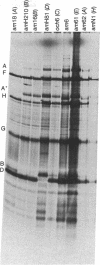
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. D., van Heusden G. P., Vereijken J. M., Weisbeek P. J., Jansz H. S. Cleavage map of bacteriophage phiX174 RF DNA by restriction enzymes. Nucleic Acids Res. 1976 Aug;3(8):1947–1960. doi: 10.1093/nar/3.8.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell B. G., Air G. M., Hutchison C. A., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976 Nov 4;264(5581):34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- Benbow R. M., Hutchison C. A., Fabricant J. D., Sinsheimer R. L. Genetic Map of Bacteriophage phiX174. J Virol. 1971 May;7(5):549–558. doi: 10.1128/jvi.7.5.549-558.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbow R. M., Mayol R. F., Picchi J. C., Sinsheimer R. L. Direction of Translation and Size of Bacteriophage phiX174 Cistrons. J Virol. 1972 Jul;10(1):99–114. doi: 10.1128/jvi.10.1.99-114.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Borrias W. E., Weisbeek P. J., Van Arkel G. A. An intracistronic region of gene A of bacteriophage piX174 not involved in progeny RF DNA synthesis. Nature. 1976 May 20;261(5557):245–248. doi: 10.1038/261245a0. [DOI] [PubMed] [Google Scholar]
- Fiddes J. C. Nucleotide sequence of the intercistronic region between genes G and F in bacteriophage phiX174 DNA. J Mol Biol. 1976 Oct 15;107(1):1–24. doi: 10.1016/s0022-2836(76)80014-9. [DOI] [PubMed] [Google Scholar]
- Fukuda A., Sinsheimer R. L. Process of infection with bacteriophage phi chi 174. XL. Viral DNA replication of phi chi 174 mutants blocked in progeny single-stranded DNA synthesis. J Virol. 1976 Apr;18(1):218–226. doi: 10.1128/jvi.18.1.218-226.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M. Fragment maps of phiX-174 replicative DNA produced by restriction enzymes from haemophilus aphirophilus and haemophilus influenzae H-I. J Virol. 1974 Nov;14(5):1142–1151. doi: 10.1128/jvi.14.5.1142-1151.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linney E. A., Hayashi M. N., Hayashi M. Gene A of X174. 1. Isolation and identification of its products. Virology. 1972 Nov;50(2):381–387. doi: 10.1016/0042-6822(72)90389-3. [DOI] [PubMed] [Google Scholar]
- Linney E., Hayashi M. Two proteins of gene A of psiX174. Nat New Biol. 1973 Sep 5;245(140):6–8. doi: 10.1038/newbio245006a0. [DOI] [PubMed] [Google Scholar]
- SINSHEIMER R. L., STARMAN B., NAGLER C., GUTHRIE S. The process of infection with bacteriophage phi-XI74. I. Evidence for a "replicative form". J Mol Biol. 1962 Mar;4:142–160. doi: 10.1016/s0022-2836(62)80047-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Siden E. J., Hayashi M. Role of the gene beta-product in bacteriophage phi-X174 development. J Mol Biol. 1974 Oct 15;89(1):1–16. doi: 10.1016/0022-2836(74)90159-4. [DOI] [PubMed] [Google Scholar]
- Smith L. H., Sinsheimer R. L. The in vitro transcription units of bacteriophage phiX174. II. In vitro initiation sites of phiX174 transcription. J Mol Biol. 1976 Jun 5;103(4):699–710. doi: 10.1016/0022-2836(76)90204-7. [DOI] [PubMed] [Google Scholar]
- Smith M., Brown N. L., Air G. M., Barrell B. G., Coulson A. R., Hutchison C. A., 3rd, Sanger F. DNA sequence at the C termini of the overlapping genes A and B in bacteriophage phi X174. Nature. 1977 Feb 24;265(5596):702–705. doi: 10.1038/265702a0. [DOI] [PubMed] [Google Scholar]
- TESSMAN E. S. COMPLEMENTATION GROUPS IN PHAGE S13. Virology. 1965 Feb;25:303–321. doi: 10.1016/0042-6822(65)90208-4. [DOI] [PubMed] [Google Scholar]
- Vereijken J. M., Van Mansfeld A. D., Baas P. D., Jansz H. S. Arthrobacter luteus restriction endonuclease cleavage map of phi chi 174 RF DNA. Virology. 1975 Nov;68(1):221–233. doi: 10.1016/0042-6822(75)90163-4. [DOI] [PubMed] [Google Scholar]
- Weisbeek P. J., Vereijken J. M., Baas P. D., Jansz H. S., Van Arkel G. A. The genetic map of bacteriophage phiX174 constructed with restriction enzyme fragments. Virology. 1976 Jul 1;72(1):61–71. doi: 10.1016/0042-6822(76)90311-1. [DOI] [PubMed] [Google Scholar]