Abstract
Fibroblast growth factors (FGF) and their receptors serve many functions in both the developing and adult organism. Humans contain 18 FGF ligands and four FGF receptors (FGFR). FGF ligands are polypeptide growth factors that regulate several developmental processes including cellular proliferation, differentiation, and migration, morphogenesis, and patterning. FGF-FGFR signaling is also critical to the developing axial and craniofacial skeleton. In particular, the signaling cascade has been implicated in intramembranous ossification of cranial bones as well as cranial suture homeostasis. In the adult, FGFs and FGFRs are crucial for tissue repair. FGF signaling generally follows one of three transduction pathways: RAS/MAP kinase, PI3/AKT, or PLCγ. Each pathway likely regulates specific cellular behaviors. Inappropriate expression of FGF and improper activation of FGFRs are associated with various pathologic conditions, unregulated cell growth, and tumorigenesis. Additionally, aberrant signaling has been implicated in many skeletal abnormalities including achondroplasia and craniosynostosis. The biology and mechanisms of the FGF family have been the subject of significant research over the past 30 years. Recently, work has focused on the therapeutic targeting and potential of FGF ligands and their associated receptors. The majority of FGF-related therapy is aimed at age-related disorders. Increased understanding of FGF signaling and biology may reveal additional therapeutic roles, both in utero and postnatally. This review discusses the role of FGF signaling in general physiologic and pathologic embryogenesis and further explores it within the context of skeletal development.
Keywords: Craniosynostosis, FGF signaling, Fibroblast growth factor, Fibroblast growth factor receptor, Genetics, Pathogenesis, Signal transduction, Skeletal development
Introduction
The fibroblast growth factor (FGF) family consists of structurally related polypeptides involved in several physiologic processes. Highly conserved, these growth factors are found in thousands of animal species, ranging from nematode and zebra fish to mouse and human.1 FGFs play a role in cellular proliferation, migration, and differentiation, mitogenesis, angiogenesis, embyrogenesis, and wound healing.2 It is by the activation of various signal transduction pathways that FGFs mediate multiple developmental processes.3
Mammals contain 18 FGF types (FGF1–FGF10 and FGF16–FGF23), which have been grouped into six distinct subfamilies based on phylogeny and sequence homology.4 FGFs share a similar internal core and have a characteristically high binding affinity for both heparin and fibroblast growth factor receptors (FGFRs). FGFRs are tyrosine kinase receptors that contain a heparin-binding sequence, three extracellular immunoglobulin-like domains (D1–D3), a hydrophobic transmembrane domain, and a split intracellular tyrosine kinase domain.5, 6, 7 The mammalian FGFR family consists of four members (FGFR1–FGFR4). The amino acid sequences of each receptor are highly conserved, with differentiation occurring only in their ligand affinity and tissue distribution.8 Characteristic of FGFRs is the acid box, which is a serine-rich, acidic sequence in the linker between D1 and D2.4 The acid box and D1 domain are thought to play a role in receptor autoinhibition.9 The D2–D3 fragment is required for ligand specificity and binding. In vertebrates, four genes encode the FGFRs (FGFR1-4), and undergo alternative splicing in their extracellular domain to produce many varieties of FGFR1-4 with varying affinities for their ligands.10
Many data suggest the role of FGF signaling in fundamental developmental pathways, including embryogenesis and the development of organ systems.11 Aberrations in this pathway have been associated with human disease. Cancers from various tissue types have been linked to dysregulated FGF signaling.12 Faulty signaling is also associated with many congenital syndromes. Many other conditions, including skeletal dysplasias,13 deafness,14 and lacrimo-auriculo-dento-digital syndrome,15 result from FGF signaling errors. Pathological conditions are mostly due to gain- or loss-of-function mutations in the ligands themselves or their receptors.4
The degree of involvement of FGF signaling in both normal and pathologic development has led to considerable research on the therapeutic applications and targeting of the FGF family. Recombinant FGFs and small-molecule FGF receptor kinase inhibitors have been used in the treatment of cancer and cardiovascular disease. Emerging research has also demonstrated their potential pharmacologic role in preventing chemotherapeutic side effects as well as treating metabolic syndrome.16 In this article, we review the current knowledge of FGF signaling in both physiologic and pathologic development and also address recent discoveries regarding its therapeutic potential.
The FGF signaling system
Positive regulation of signaling
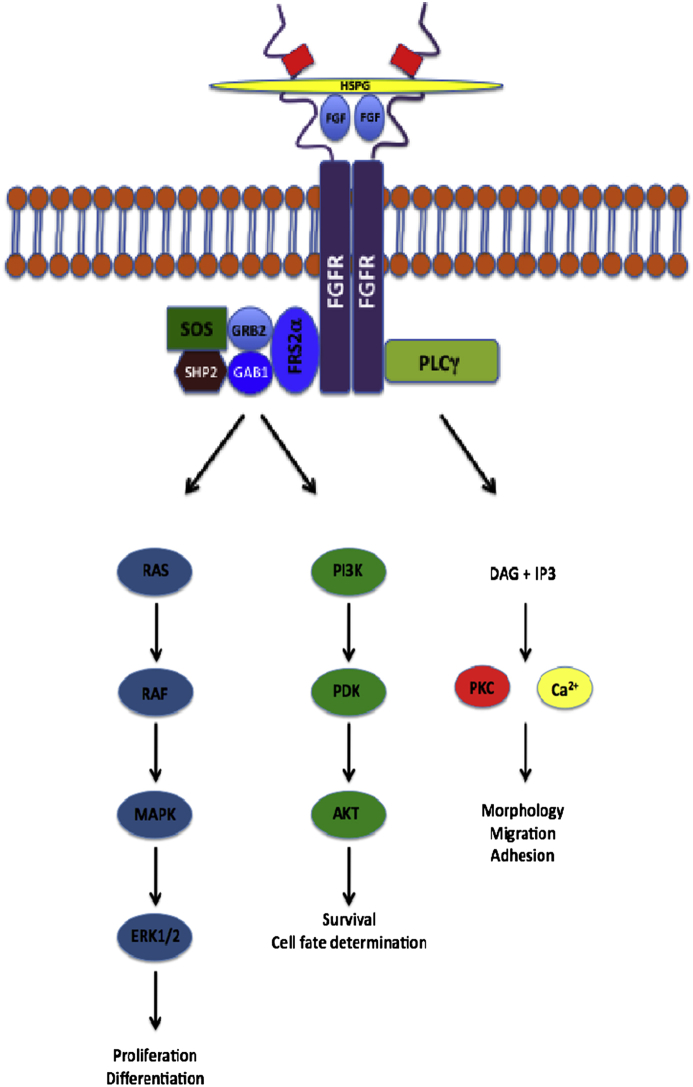
The FGF signaling cascade is initiated by the binding of FGF ligands to FGFRs. Following FGF binding, a ligand-dependent dimerization event takes place in which a complex is formed that consists of two FGFs, two heparin sulfate chains, and two FGFRs. Each ligand binds to both receptors, and the receptors make contact with one another via a patch on the D2 domain.4 This facilitates the transphosphorylation of each receptor monomer by an intrinsic tyrosine kinase domain. At least seven phosphorylation sites have been identified for FGFR1 (Tyr163, Tyr583, Tyr585, Tyr653, Tyr654, Tyr730, and Tyr766).17, 18, 19 Phosphotyrosine groups serve as docking sites for adaptor proteins that regulate downstream signaling.20 The FGF system is associated with several downstream signaling pathways; the best understood are the RAS/mitogen-activating protein (MAP) kinase pathway, the phosphoinositide 3 (PI3) kinase/AKT pathway, and the phospholipase C gamma (PLCγ) pathway (Fig. 1).21
Figure 1.
FGF-FGFR signaling pathway. The signaling cascade commences upon the formation of an FGF binding complex, consisting of two FGF ligands, two heparin sulfate chains, and two FGFRs. Signal transduction largely follows one of three pathways. The RAS/MAP kinase pathway, initiated upon the formation of an FRS2 complex, controls cell proliferation and differentiation. The PI3/AKT pathway is also initiated by the formation of an FRS2 complex, and regulates cell survival and fate determination. Finally, upon binding of PLCγ to the activated FGFR, DAG and IP3 are formed, activating PKC. The PLCγ pathway influences cell morphology, migration, and adhesion.
The main downstream pathway associated with FGF signaling is the RAS/MAP kinase pathway. This pathway is implicated during cellular proliferation and differentiation.22 MAP kinases are serine/threonine-specific protein kinases that act in response to extracellular stimuli and regulate various cellular processes. Examples of MAP kinase effectors include c-Jun N-terminal kinase (JNK), extracellular signal-regulated kinase (ERK), and p38 mitogen-activated kinase.23 After an FGF ligand binds to its receptor, an integral step in the signaling pathway is the phosphorylation of the tyrosine residues on the docking protein fibroblast growth factor receptor substrate 2 alpha (FRS2α). This permits binding of adaptor proteins that are associated with signal activation.24, 25 An FRS2 complex consisting of FRS2α, guanine nucleotide exchange factor 2 (GRB2), GRB2-associated binding protein 1 (GAB1), the son of sevenless (SOS), and tyrosine phosphatase (SHP2) is then formed that facilitates activation of the RAS/MAP kinase26 and also PI3 kinase/AKT pathways.27
The PI3 kinase/AKT pathway is associated with cellular survival and cell fate determination.26, 28 This pathway may also impact cell polarity.29 Like the RAS/MAP kinase pathway, the PI3 kinase/AKT pathway is initiated when an FRS2 signaling complex forms. GAB1 protein then links activated FGFRs with PI3 kinase. Downstream of PI3 kinase, phosphoinositide-dependent kinase and AKT (an anti-apoptotic protein kinase) are activated.21
Another target molecule of activated FGFR is PLCγ. This pathway is activated upon the binding of the PLCγ molecule to the phosphorylated Tyr766 of the receptor.21 Inositol triphosphate (IP3) and diacylglycerol (DAG) are then generated by the hydrolysis of activated PLCγ. DAG and cytoplasmic calcium released from the endoplasmic reticulum in response to IP3 together activate protein kinase C (PKC).21 Though it has not been completely elucidated, the PLCγ kinase pathway influences cell morphology, migration, and adhesion.26, 28
Negative regulation of signaling
Like signal activation, mechanisms that attenuate FGF-FGFR signaling are conserved among many animal species. These mechanisms, however, are less well understood than their positive regulation counterparts. In general, downstream signal attenuation occurs via the induction of MAPK phosphatases. First discovered in Drosophilia melanogaster,30 the sprouty (SPRY) family of proteins, which are MAPK phosphatases, inhibit receptor tyrosine kinase signaling by directly binding to RAF and blocking subsequent MAPK signaling31 or by competing for GRB2 binding, thereby preventing SOS-mediated RAS activation.12 Interestingly, FGF signaling activates SPRY proteins, which may be an example of autoinhibition. Additional negative modulators include the phosphatases MAPK phosphatase 3 (MKP3)32 and SEF.33 MKP3 attenuates the FGF cascade by dephosphorylating ERK1 and ERK2, molecules important for MAPK downstream signaling.34 SEF likely mediates its effects by inhibiting ERK phosphorylation and also by acting at various points along the signaling pathway to exert its function.34 Further control of signaling takes place at the level of the receptors. Following activation, FGFRs may be internalized and subsequently degraded or recycled.35
FGF signaling in physiologic development
FGF signaling in general embryonic development
The FGF signaling pathway plays many diverse and essential roles in orchestrating human embryonic development. Numerous studies, conducted in model organisms over the past 25 years, have demonstrated that FGF signaling is a widely utilized regulatory system in early vertebrate development and has been conserved throughout chordate evolution.36 FGFs control cell migration during gastrulation,37, 38, 39 epithelio-mesenchymal interactions during limb morphogenesis,11, 40, 41 and neural induction and patterning42, 43, 44, 45, 46, 47, 48 in later stages of development.49
An interesting role of FGFs to organize the migratory events of gastrulation by functioning as both chemoattractants and repellents has been documented.50, 51, 52, 53 FGF signaling is associated with the coordinated cellular movements of convergent extension and in the epithelial to mesenchymal transition that marks the onset of gastrulation.54, 55 Convergent extension involves the reorganization of cytoskeletal elements to ensure that cells become polarized along a similar axis. Polarized cells intercalate amongst one another, which causes the overall tissue layer to extend along the perpendicular axis.54 Both FGF and Wnt signaling have been shown to activate this planar cell polarity pathway and propagate the normal activities of convergent extension during gastrulation.56
In mammalian gastrulation, the process of epithelial-mesenchymal transition (EMT) takes place after formation of the primitive streak. During EMT, a fraction of tightly adherent cells of the epithelial layer lose contact with neighboring cells and migrate freely from this layer to enter the primitive streak in order to adopt a mesenchymal phenotype.36, 54 FGFR1 activation facilitates this transition by mediating down-regulation of E-cadherin-related cellular adhesions and stimulation of cell migration through the primitive streak.37, 38 This in part was elucidated by the fact that FGFR1−/− mice exhibit recessive embryonic lethality upon gastrulation57, 58 and display retarded migration of mesoderm precursor cells across the primitive streak.38, 59 Interestingly, FGF8-null mice also display embryonic lethality associated with a failure to develop through gastrulation.60
FGF signaling also contributes to tissue organization by directly promoting mesodermal formation while inhibiting endodermal development.61 During specification of germ layers, vegetal cells release signals through nodal and activin pathways to mediate mesoderm identity and patterning.62, 63 FGF2, FGF4, and FGF8 may regulate transcription factors downstream of the nodal and activin pathways that activate and maintain expression of mesoderm-specifying genes.64, 65, 66, 67
Neural induction is also mediated during gastrulation, marking a fundamental step of vertebrate central nervous system development. During neural induction, a subset of cells within the pluripotent dorsal ectoderm is selected to adopt a neural fate rather than an epidermal fate.49, 68 FGF signaling has been shown to facilitate neural induction in these cells by inhibiting the expression of bone morphogenetic proteins (BMP) that stimulate epidermal development.69 Specifically, FGF3 and FGF4 expression inhibits BMP4 and BMP7 in early neural tissues.52, 70, 71 FGF signaling also indirectly inhibits BMP signaling. Expression of Noggin (NOG) protein, which is a negative inhibitor of BMP signaling, is increased by FGF stimulation. Furthermore, FGF signaling results in the inhibitory phosphorylation of the SMAD1, SMAD5, and SMAD8 transcription factors, which blocks their ability to travel to the nucleus and activate the transcription of BMP target genes.36, 72, 73, 74, 75
FGF ligands also serve as posteriorizing factors during pattering of the neural plate and directly activate the transcription of a set of posterior neural genes.76 Studies manipulating ectopic FGF expression in developing central nervous systems demonstrate that FGFs convert anterior neural tissues to more posterior neural cell types.77, 78, 79, 80, 81, 82, 83, 84 Within the embryonic isthmic organizer, which is an important signaling center at the anatomical constriction of the vertebrate midbrain-hindbrain junction, expression of FGF8, FGF17, FGF18, and FGFR1 is observed.36, 85 There is strong evidence that FGF signaling modulates early patterning of the neural tissues via regulation of Hox genes, a family of homeobox transcription factors that dictate segmental identity.86 Ectopic application and overexpression of various FGFs during and after gastrulation increase the expression of key posterior Hox genes and also inhibit anterior development.75, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96 Additionally, deficient FGFR1 expression is correlated with repression of posterior Hox genes.87, 97
FGF signaling has also been implicated in limb bud development. In particular, FGF signaling mediates a positive feedback loop of paracrine signaling between mesenchymal and epithelial tissues that pattern the emerging limb bud and stimulate the outgrowth, morphogenesis, and maintenance of early limb structure.55 Before the induction of the apical ectodermal ridge (AER), FGFR1 is expressed in the underlying mesenchyme; FGFR2 is found in both the mesenchyme and ectoderm of the presumed future limb site.98, 99, 100 Expression of FGFR1 continues into the later stages of limb development, where it may play essential roles in mesodermal patterning of the distal limb fields and digit formation.101 After induction, the AER expresses FGF2, FGF4, FGF8, and FGF9, while limb bud mesoderm expresses FGF2 and FGF10.99 Upon the onset of limb development, FGF10 secreted by cells of the lateral plate mesoderm diffuses to the overlying surface ectoderm, and interacts with FGFR2b to induce formation of the AER superficially.99, 102 In response, the AER secretes FGF8, which acts upon the underlying mesenchymal cells through FGFR2c to maintain a proliferative state and continue FGF10 secretion.101, 103, 104, 105, 106 Secretion of additional FGF10 by the mesenchymal cells maintains AER expression of FGF8.107 This positive feedback loop drives further limb outgrowth by permitting the mesenchyme to maintain the organizational role of the AER at the propagating edge of the developing limb, while the AER reciprocally sustains the proximate mesenchyme of the progress zone in a mitotically active state.108
FGF signaling in cranial suture development
FGF signaling plays a critical role in the normal development and morphogenesis of the craniofacial skeleton during embryogenesis and postnatal growth.109 In fact, many events of normal craniofacial development have been elucidated during studies examining the etiologic relationship between FGF and FGFR mutations and various skeletal dysplasias.110, 111 FGF signaling is complex and likely interacts with additional signaling pathways during craniofacial development.109, 110, 112, 113, 114, 115, 116
In mammals, embryonic tissues of the facial and cranial bones are derived from neural crest cells.117, 118, 119, 120, 121, 122 FGF signaling induces cranial neural crest formation and is present in both the epithelia and mesenchyme of the facial primordia.112, 123, 124, 125, 126, 127 At six weeks of development, mesenchymal condensations begin to foreshadow bones of the basicranium, followed soon thereafter by those of the cranial vault bones.128 Delezoide et al observed FGFR1 gene expression throughout the entire mesenchyme in the head at this stage as well as strong FGFR2 expression in the epidermis, pre-bone mesenchymal condensations, and mesenchymal walls of the cranial vault.128 From the eighth through thirteenth week of gestation, skull bones develop via intramembranous ossification. Condensations of mesenchymal cells located between the dermal mesenchyme and the developing meninges simultaneously differentiate into osteoblasts to give rise to the major cranial bones. Newly differentiated osteoblasts then synthesize and deposit osteoid matrix radially outwards from these ossification centers. This osteoid matrix is primarily composed of type I collagen.129 During early intramembranous ossification, there is marked expression of FGFR1 and FGFR2 and, to a lesser extent, FGFR3 in the cells of the pre-bone mesenchyme and around the osteoid.128 Indeed, intramembranous ossification of the skull vault is characterized by co-expression of FGFR1-3 in osteoblast precursors and osteoblasts.
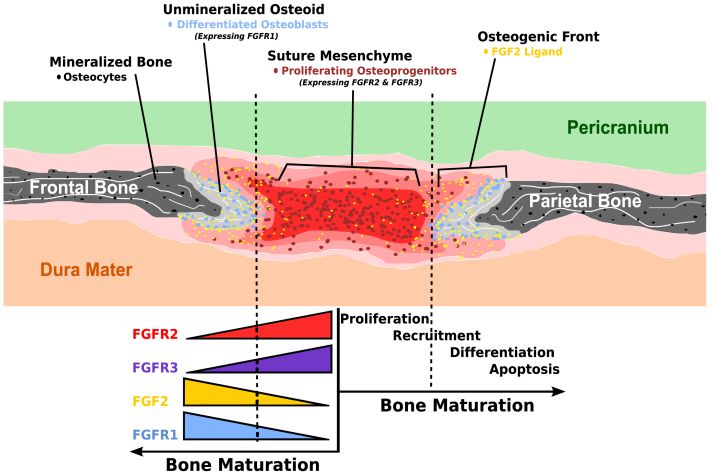
As mineralization of newly deposited osteoid matrix progresses outward from ossification centers, the periphery of the extending bone fields (osteogenic front) develops as a wedge-shaped proliferation of cells that invade and recruit intervening mesenchymal tissue into these advancing edges to increase the size of each cranial bone.112, 115, 129 By gestation week 18, bone fronts of adjacent cranial bones are in close proximity, and sutures develop along lines of approximation.112 The osteogenic fronts of neighboring cranial bones, undifferentiated mesenchyme between them, and the adjacent pericranium and dura mater function as a complex to maintain normal development along the suture. Morriss-Kay et al demonstrated that in normal coronal suture development, the maintenance of proliferating osteogenic cells at the margins of membrane bones forming the suture requires relatively low FGF levels.110 During normal development, cranial sutures remain in a patent, unossified state while new intramembranous bone is formed at the edges of osteogenic fronts. This requires mesenchymal cells within the suture to remain undifferentiated while cells of the osteogenic layer that line the bony front differentiate into osteoblasts and produce additional new bone.130, 131, 132 High FGF levels are associated with osteogenic differentiation. In other words, in the normal suture there is differential FGF expression with high levels found in the differentiated region and low levels found at the suture. At the time of suture fusion, increased FGF2 levels are observed.133 Cross-talk also exists between osteoblasts and osteoclasts in maintaining suture homeostasis on a molecular level as FGF2 directly upregulates RANKL mRNA and indirectly inhibits M-CSF production, thereby inducing osteoclast differentiation.134 In the setting of increased receptor activation, whether pathologic (see below) or experimental (by the addition of exogenous FGF), cellular proliferation ceases and suture fusion ensues.110 Additionally, increased signaling through FGFR1 is associated with premature fusion while FGFR2 and FGFR3 signaling maintain osteoprecursor cell proliferation (Fig. 2).130
Figure 2.
Schematic representation of developing coronal suture. In the presence of low concentrations of FGF2, undifferentiated osteoprogenitor cells expressing FGFR2 and FGFR3 proliferate within the suture mesenchyme between the two osteogenic fronts. At higher levels of FGF2, osteoprogenitor cells are recruited to differentiate into osteoblasts. This leads to increased of FGFR1 expression and deposition of osteoid matrix along the osteogenic fronts.
FGF signaling between dura mater and overlying cranial sutures during embryogenesis has implications on proper development of cranial sutures postnatally.115, 129, 135 Before the suture complex can sustain itself through intrinsic signals, an inductive stimulus of soluble factors emanating from the dura mater is required during early suture morphogenesis.129 Along these lines, Kim and colleagues found that signals from the dura mater regulate proper suture development prenatally, while signals within the osteogenic fronts dominate after birth.115 Many of these signals have been identified as FGF ligands and receptors.
Aberrant FGF signaling during development
Given the many integral actions controlled by FGF signaling, it is no surprise that disruption of the normal cascade has been implicated in many disease states. Most mutations that are familial in nature are inherited in autosomal dominant fashion. New mutations can occur sporadically, however. Therefore, each mutation is subject to significant variability with respect to genotypic mutation and phenotypic expression.8 Regarding FGFRs, many conditions are associated with mutations at specific gene locations. For example, craniosynostosis is often related to a mutation within the gene region responsible for the linker protein between the D2 and D3 extracellular domains of FGFRs. In addition, mutations around the N-terminal junction of the transmembrane domain are linked to skeletal disorders including thanatophoric dysplasia.8 Achondroplasia and other disorders of long bone growth are associated with mutations to the gene responsible for the FGFR tyrosine kinase domain expressed by chondrocytes at the physis.136 Several neoplastic conditions have also been linked to FGFR mutations.136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147
Numerous pathologic conditions are associated with mutated FGF ligands as well. FGF3, which plays a role in inner ear development, has been linked to inner ear agenesis and microtia.14 Myogenesis is regulated in part by FGF6; when mutated, defective muscle regeneration is present.148 An in depth discussion of the pathophysiology of mutations of each FGF ligand is beyond the scope of this review and has been reviewed elsewhere.4 In the following paragraphs, we detail aberrant FGF signaling in the context of skeletal disorders.
Achondroplasia
Achondroplasia is the commonest skeletal dysplasia and is characterized by short stature, rhizomelic limb shortening, limited elbow extension, frontal bossing, and midface deficiency. It is passed via autosomal dominant inheritance but can also occur sporadically. A gain-of-function mutation of the FGFR3 gene results in decreased inhibition of endochondral ossification.141 In the majority of patients, over activation of FGFR3 is due to substitution of arginine for glycine (G380R) within the transmembrane domain.136 The introduction of a hydrophilic residue into the hydrophobic receptor domain results in alteration of the signal transduction pathway due to disruption of the alpha helical structure of the transmembrane protein.140 Additionally, experimentally-induced under activation of FGFR3 results in elongation of the vertebral column and long bones.149 It is thought that mutations causing over activation of FGFR-3 impair chondrocytes within growth plates.136, 149 Studies are now underway regarding whether attenuation of FGFR3 signaling of chondrocytes located within physes in cases of increased FGFR3 activation can increase bone growth. Lorget and colleagues demonstrated that an analogue of C-natriuretic peptide (CNP), which antagonizes downstream effects of mutated FGFR3, has been used to enhance bone growth in mice.150, 151
Hypochondroplasia
Hypochondroplasia is essentially a milder form of achondroplasia that presents with similar radiographic findings in the limbs and spine.139, 141 It follows an autosomal dominant inheritance pattern. It is distinguished by normal facies and increased head circumference.141 Greater than half of hypochondroplasia cases are due to an FGFR3 gene missense mutation (N540K) in the first tyrosine kinase domain of the receptor.139, 142 This mutation causes a gain-of-function that results in premature fusion of the growth plates in the vertebral column and long bones.152 Severe phenotypes are associated with missense mutations at nucleotides encoding the extracellular region of FGFR3.139, 140 The mild phenotype and clinical heterogeneity of hypochondroplasia are likely due to the fact that there are several potential mutations that may cause it.140 Growth hormones have been used in an attempt to treat this condition but have been met with mixed results.152 Because they share a similar pathogenesis, therapeutic options for achondroplasia may be of benefit for patients with hypochondroplasia.150, 151.
Thanatophoric dysplasia
Thanatophoric dysplasia is a skeletal dysplasia often incompatible with life. It is characterized by shortened limbs with normal trunk length, excessive skin folding, a narrow thorax with shortened ribs, disproportionate macrocephaly, frontal bossing, and protruding eyes.141 Type I thanatophoric dysplasia is further characterized by micromelia with bowed femurs and a cloverleaf skull. Type II thanatophoric dysplasia is associated with micromelia with straight femurs, craniosynostosis, and a moderate-to-severe cloverleaf skull.141, 153 Patients with thanatophoric dysplasia often die of respiratory insufficiency soon after birth; however, survivors have been reported.141, 153 Though it follows an autosomal dominant inheritance pattern, virtually all cases are due to sporadic mutations.
Type I thanatophoric dysplasia can result from several different mutations affecting either extracellular or intracellular domains of FGFR-3. The most common mutation is the substitution of cysteine for arginine (R248C) in a polypeptide within the extracellular domain.143, 154, 155 Mutations allow for unimpaired cysteine residues to create disulfide bonds that enable the receptor to dimerize independent of the ligand.140 Type II thanatophoric dysplasia is caused by a point mutation (K650E) to the tyrosine kinase II domain of FGFR3 that is thought to also result in independent dimerization of the receptor.140, 143 The FGFR3 mutations associated with the aforementioned skeletal dysplasias support the notion that FGFR3 signaling is critical for bone growth regulation (Table 1).
Table 1.
Skeletal dysplasias associated with FGFR3 mutations.
| Disorder | Mutationa | Mechanism | Key features | Inheritanceb |
|---|---|---|---|---|
| Achondroplasia136, 140, 141 | G380R (transmembrane domain) | Gain-of-function mutation results in decreased inhibition of endochondral ossification | Short stature, rhizomelic limb shortening, short fingers and toes, large head with prominent forehead, small midface with flattened nasal bridge, spinal kyphosis/lordosis, varus/valgus deformities | Autosomal dominant; sporadic |
| Hypochondroplasia139, 140, 142, 152 | N540K (first tyrosine kinase domain); missense mutations (extra-cellular domain) | Gain-of-function mutations result in premature fusion of growth plates in vertebral column and long bones | Short stature, short limbs, increased head circumference, normal facies | Autosomal dominant; sporadic |
| Thanatophoric dysplasia140, 141, 143, 153, 154, 155 | R248C (extra-cellular domain; type I) K650E (second tyrosine kinase domain; type II) |
Gain-of-function mutations result in ligand-independent receptor activation | Early death, extremely short limbs, redundant skin folds, narrow chest with short ribs, underdeveloped lungs, large head, curved thigh bones (type I), cloverleaf skull (type II) | Autosomal dominant; sporadic |
FGFR3, fibroblast growth factor receptor 3.
Commonest mutation(s) is noted. Others may be documented.
Virtually all cases of thanatophoric dysplasia result from sporadic mutations because of its early lethality.
Craniosynostosis
Acrocephalosyndactyly, or craniosynostosis, was first described by Otto in 1830 as the premature fusion of the cranial sutures.156 Craniosynostosis can be classified as simple (one fused suture) or complex (two or more fused sutures).157 Simple or complex craniosynostosis can be further classified as primary (sutures prematurely fuse due to abnormal suture biology) or secondary (normal suture biology but sutures prematurely fuse due to abnormal external forces).157 Additional classification as syndromic or sporadic is based on FGFR gene mutations and associated phenotype. Syndromic craniosynostosis follows autosomal dominant inheritance but nearly 75% of craniosynostosis cases are of the sporadic variety.157 The overall incidence of craniosynostosis is one in 2500 live births.112, 156 In terms of nonsyndromic cases, sagittal suture synostosis occurs most frequently (scaphocephaly; 40–55% of cases) followed by coronal synostosis (anterior plagiocephaly; 20–25%), metopic synostosis (trigonocephaly; 5–15%), multiple synostoses (5–15%), and lambdoid synostosis (posterior plagiocephaly; 0–5%).156, 157, 158, 159
Most syndromic cases are the result of a gain-of-function mutation within the gene region responsible for the linker between the D2 and D3 domains of the FGFR.8 A mutation in this region may increase the receptor's affinity for its corresponding FGF ligand, resulting in increased signaling that stimulates cell differentiation and eventual suture fusion.160 However, several additional gene mutations have also been implicated in syndromic craniosynostosis.112, 156 Several craniosynostosis syndromes will be discussed (Table 2).
Table 2.
Syndromic craniosynostoses associated with FGFR mutations.
| Syndrome | Mutationa,b | Mechanism | Key features | Inheritance |
|---|---|---|---|---|
| Pfeiffer112, 161, 162, 163 | P252R (FGFR1); several sequence variants (FGFR2) | P252R gain-of-function mutation results in increased receptor affinity for ligand binding; FGFR2-related gain-of-function mutations result in ligand-independent receptor activation | Proptosis, hypertelorism, maxillary hypoplasia, beaked nose, developmental delay (types II and III), cloverleaf skull (type II), turribrachycephaly (type III) | Autosomal dominant; sporadic |
| Apert160, 165, 169 | S252W and P253R (FGFR2) | Gain-of-function mutations result in increased receptor affinity for ligand binding | Turribrachycephaly, midface hypoplasia, syndactyly of fingers and toes, varying degrees of developmental delay | Autosomal dominant; sporadic |
| Crouzon112, 161, 165 | Several missense mutations (FGFR2) | Gain-of-function mutations result in disulfide bond that stabilizes the D3 loop to allow for ligand-independent receptor activation | Proptosis, external strabismus, mandibular prognathism, normal extremities, normal intelligence | Autosomal dominant; sporadic |
| Beare-Stevenson cutis gyrata112, 161, 174, 175 | Y394C (FGFR2) | Gain-of-function mutation results in ligand-independent receptor activation | Midface hypoplasia, abnormal ears, natal teeth, widespread cutis gyrata, acanthosis nigricans, skin tags, developmental delay, pyloric stenosis, anterior anus | Autosomal dominant; sporadic |
| Jackson-Weiss161, 176 | C342S, C342R, Q289P, A344G (FGFR2) | Gain-of-function mutations result in ligand-dependent receptor overactivation | Mandibular prognathism, broad and medially deviated great toes, short first metatarsal, calcaneocuboid fusion, normal intellect | Autosomal dominant; sporadic |
| Muenke112, 170 | P250R (FGFR3) | Gain-of-function mutation results in increased receptor affinity for ligand binding | Uni- or bicoronal synostosis, megalencephaly, midface hypoplasia, hypertelorism, variable sensorineural hearing loss, osteochondroma | Autosomal dominant; sporadic |
FGFR, fibroblast growth factor receptor.
Commonest mutation(s) is noted. Others may be documented.
Type I Pfeiffer syndrome is associated with a P252R mutation in FGFR1 in 5% of cases. The majority of type I and all of types II and III Pfeiffer syndrome cases are associated with sequence variant mutations in FGFR2.
Pfeiffer syndrome
Pfeiffer syndrome is characterized by craniosynostosis, proptosis, hypertelorism, maxillary deficiency, and a beaked nose. Patients with Pfeiffer syndrome also have wide thumbs and great toes that bend away from other digits and may display brachydactyly or syndactyly. Three variants of Pfeiffer syndrome exist: type I is associated with FGFR1 and FGFR2 mutations; types II and III are associated with FGFR2 mutations.161 Significant genetic heterogeneity exists among patients with Pfeiffer syndrome. Approximately 5% of patients with type I Pfeiffer syndrome contain a gain-of-function P252R mutation of FGFR1.161, 162 This mutation occurs at the linker region between D2 and D3, resulting in a bulkier residue that increases the receptor's affinity for ligand binding and therefore excessive receptor activation.112 The other 95% of cases are thought to be due to sequence variants in the FGFR2 gene. Patients with type I Pfeiffer syndrome due to an FGFR1 mutation usually display a more favorable phenotype than patients with a mutation of FGFR2.
Patients with types II or III Pfeiffer syndrome generally have a more severe phenotype and worse prognosis. These conditions result from an FGFR2 mutation. The most common mutations occur in exons IIIa and IIIc, resulting in an unpaired cysteine residue that forms an intermolecular disulfide bond that causes ligand-independent receptor activation.161, 163
Apert syndrome
Apert syndrome is characterized by bilateral premature fusion of the coronal suture, developmental delay, midface hypoplasia, syndactyly of fingers and toes, and potentially other anomolies.160, 164, 165, 166, 167, 168 Activating mutations (S252W and P253R) in FGFR2 are thought to be responsible for the majority of cases and resultant brain dysmorphologies.160, 165 These mutations result in increased ligand affinity for the receptor and therefore trigger excessive activation. Evidence supports that the P253R mutation results in an indiscriminate increase in affinity of FGFR2 toward any FGF. The S252W mutation, however, selectively enhances FGFR2 affinity toward a subset of FGFs.169 These genotypic differences may also cause clinical variability in patient presentation.
Crouzon syndrome
Crouzon syndrome is due to a gain-of-function mutation in FGFR2 that results in ligand-independent, disulfide-mediated, covalent receptor dimerization and activation.165 The disulfide bond is formed between a cysteine-cysteine linkage that stabilizes the D3 loop, which allows for ligand-free activation of the receptor.112 Patients with Crouzon syndrome chiefly present with premature fusion of the coronal sutures, mandibular prognathism, strabismus, and proptosis.165 Unlike Apert syndrome, patients with Crouzon syndrome typically display normal cognitive development and normal-appearing extremities.112, 161, 170, 171, 172
A variant of Crouzon syndrome is Crouzonodermoskeletal syndrome. Patients present with many of the skeletal features of Crouzon syndrome but this condition is distinguished by the presence of acanthosis nigricans. Crouzonodermoskeletal syndrome is primarily associated with mutations to FGFR3. Specifically, an A391E mutation is thought to result in ligand-free activation of the receptor.112, 173
Beare-Stevenson cutis gyrata syndrome
Beare-Stevenson cutis gyrata syndrome is also a syndromic form of craniosynostosis caused by an FGFR2 mutation. It is characterized by cognitive impairment, moderate-to-severe midface hypoplasia, abnormal ears, cutis gyrata, acanthosis nigricans, prominent umbilical stump, accessory nipples, pyloric stenosis, and an anterior anus.112, 174 A Y394C mutation leads to a cysteine-cysteine disulfide bond that initiates ligand-free activation of the receptor.161, 175
Jackson-Weiss syndrome
Jackson-Weiss syndrome is characterized by mandibular prognathism, broad and medially deviated toes, short first metatarsals, calcaneocuboid fusions, and abnormal tarsals.176 It has been associated with several missense mutations of FGFR2, including C342S, C342R, Q289P, and A344G.161, 176 In each case, an amino acid substitution triggers excessive receptor activation.
Muenke syndrome
Muenke syndrome is an FGFR3-related craniosynostosis that generates isolated coronal synostosis and overlaps phenotypically with Pfeiffer and Jackson-Weiss syndromes.112, 161, 170 Patients have normal to mildly impaired intelligence, variable uni- or bicoronal synostosis, megalencephaly, midface hypoplasia, carpal-tarsal fusion, brachydactyly, sensorineural hearing loss, and osteochondroma.170, 177, 178 An FGFR3 P250R mutation occurs between the D2 and D3 domains, promoting excessive ligand binding through the substitution of a bulkier residue.112 This causes over activation of the receptor.
Nonsyndromic craniosynostosis
Nonsyndromic craniosynostosis is characterized by isolated premature suture fusion and no additional extra-cranial abnormalities. Classification is based on which sutures are fused. The genetic mechanisms of nonsyndromic variants are less well understood than those associated with syndromic craniosynostosis.179 It had been thought that mutations in FGFR3 may be responsible for particular sporadic cases. However, many of these cases were determined to be mild syndromic variants.157 Recently, evidence has shown a potential link between an A315Y mutation on FGFR2 and isolated sagittal synostosis.133, 157 Additionally, in utero factors may play a role in isolated synostosis. Intrauterine cranial constraints have been observed to induce over expression of FGFR2 at the osteogenic fronts of calvarial bones in the dura and osteoblasts.180
Concluding remarks and future directions
FGFs and their associated receptors have been studied extensively for the past 30 years. Recently, significant research has focused on the therapeutic potential of FGF-FGFR signaling. Beenken and Mohammadi provide an excellent review of traditional therapeutic applications of recombinant FGFs and small-molecule FGFR kinase inhibitors.4 Both in vitro and in vivo studies offer evidence as to the physiologic roles of each FGF ligand. Phenotypic studies of knockout mice in particular provide clues for the specific functions of each FGF ligand and whether a ligand is required for life. As discussed above, aberrant FGF-FGFR signaling is associated with many pathologic conditions including cancer and craniofacial and axial skeletal abnormalities. Aberrant signaling has also been implicated in cardiovascular disease, mood disorders, and potentially metabolic syndromes. To this end, the FGF-FGFR signaling system appears to an import therapeutic target.
To date, the therapeutic potential of FGFRs mostly relate to their role in tumorigenesis and cancer development. In particular, direct inhibition of FGFRs may improve outcomes of various cancers. Proof of principle with respect to effective treatment for malignancy comes from many lines of evidence. The small molecule inhibitors SU5402, PD173074, and nordihydroguaiaretic acid are efficacious in the treatment of multiple myeloma cell lines associated with deregulated FGFR3 expression.181, 182 PD173074 has also been shown to induce cell cycle arrest in FGFR2-mutated endometrial cancer cells.183 In addition, sunitinib, a receptor tyrosine inhibitor with activity against FGFRs, has received approval from the Food and Drug Administration (FDA) for use in patients with renal cell carcinoma and gastrointestinal stromal tumors.184 Interestingly, potential therapeutic targets have also been elucidated by experiments that involve the disruption of downstream signaling after FGFR activation. The blood cancer 8p11 myeloproliferative syndrome (EMS) is due to constitutive dimerization of the FGFR1 kinase domain in the setting of an inappropriate translocation.185 A mutation in the PLCγ1 binding site at Tyr766 attenuates EMS.186 A potential strategy in the treatment of EMS could therefore include the disruption of FGFR-PLCγ1 signaling. The interference of the interaction of FGFR and its downstream signaling pathways has become a conceivable therapeutic strategy.
Many potential roles for FGF ligands to be used as therapy also exist. FGF18 is key for physiologic bone growth and development. FGF18−/− mice demonstrate a wide variety of bone and cartilage malformations including a delay in osteogenic differentiation, closure of the calvarial sutures, and long-bone ossification; defective joint development; and enlargement of the proliferating and hypertrophic zones in long bone growth plates.187, 188 Recombinant FGF18 has been shown to stimulate growth of porcine and human articular chondrocytes and have an overall anabolic effect on cartilage.189 Clinically, it may stimulate the repair of cartilage damaged in the setting of progressive osteoarthritis.190
Many additional therapeutic uses of FGF ligands have been documented. FGF1 treatment can stimulate nerve repair and may enhance nerve graft take.191 FGF1, FGF2, and FGF4 have therapeutic promise in cardiovascular disease.192, 193, 194 Recombinant FGF2 may also provide benefit to patients with mood disorders.195 Recombinant FGF7 improves wound healing.196 Additionally, although further investigation is needed, FGF19 and -21 and FGF20 have potential roles in the treatment of diabetes and Parkinson's disease, respectively.197, 198, 199 Oppositely, inhibitors of FGF5 may aid in hair growth.200
Significant work has demonstrated the myriad developmental and homeostatic processes that involve FGF-FGFR signaling. Moving forward, ongoing research will almost certainly uncover additional functions of the FGF-FGFR signaling system. It is hopeful that the discovery of additional therapies that involve this system will parallel such findings. These may come in the form of recombinant proteins, small molecules, and gene therapy. At this time, the majority of FGF-based therapies are aimed at age-related disorders including osteoarthritis, diabetes, cardiovascular disorders, and Parkinson's disease. A substantial challenge is the prevention and treatment of FGF-related disorders that are present at birth. To this end, an even greater understanding of FGF biology will unlock additional strategies to treat disease, both in utero and postnatally. Finally, a new frontier in the investigation of FGF signaling relates to our recent understanding of the role of FGFs and FGFRs in inducing stem cell self-renewal and inhibiting stem cell senescence.201 Stem cells have tremendous potential in the treatment of human disease. The addition of targeted FGF therapy to this armamentarium could augment success.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
The authors apologize to the investigators whose original work was not cited due to space constraints. The reported work was supported in part by research grants from the NIH K08 Career Development Award (RRR, NIH 5K08DE20140-5) and the American Society of Plastic Surgeons/Plastic Surgery Foundation's (PSF) Pilot Research Grant Program (RRR). EMF and JR were recipients of the Pritzker Research Fellowship funded through a NIH T-35 training grant (NIDDK).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Ornitz D.M., Itoh N. Fibroblast growth factors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-reviews3005. REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seitz I.A., Teven C.M., Reid R.R., editors; Neligan P.C., Gurtner G.C., editors. Repair and Grafting of Bone. vol. 1. Elsevier Saunders; Philadelphia, PA: 2012. (Plastic Surgery). [Priciples] [Google Scholar]
- 3.Thisse B., Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. Nov 15 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Beenken A., Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. Mar 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh N., Ornitz D.M. Evolution of the Fgf and Fgfr gene families. Trends Genet. Nov 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Lee P.L., Johnson D.E., Cousens L.S., Fried V.A., Williams L.T. Purification and complementary DNA cloning of a receptor for basic fibroblast growth factor. Science. Jul 7 1989;245:57–60. doi: 10.1126/science.2544996. [DOI] [PubMed] [Google Scholar]
- 7.Johnson D.E., Lee P.L., Lu J., Williams L.T. Diverse forms of a receptor for acidic and basic fibroblast growth factors. Mol Cell Biol. Sep 1990;10:4728–4736. doi: 10.1128/mcb.10.9.4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Passos-Bueno M.R., Wilcox W.R., Jabs E.W., Sertie A.L., Alonso L.G., Kitoh H. Clinical spectrum of fibroblast growth factor receptor mutations. Hum Mutat. 1999;14:115–125. doi: 10.1002/(SICI)1098-1004(1999)14:2<115::AID-HUMU3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Wang F., Kan M., Yan G., Xu J., McKeehan W.L. Alternately spliced NH2-terminal immunoglobulin-like Loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J Biol Chem. Apr 28 1995;270:10231–10235. doi: 10.1074/jbc.270.17.10231. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X., Ibrahimi O.A., Olsen S.K., Umemori H., Mohammadi M., Ornitz D.M. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. Jun 9 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Moerlooze L., Spencer-Dene B., Revest J.M., Hajihosseini M., Rosewell I., Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. Feb 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 12.Turner N., Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. Feb 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 13.Kan S.H., Elanko N., Johnson D. Genomic screening of fibroblast growth-factor receptor 2 reveals a wide spectrum of mutations in patients with syndromic craniosynostosis. Am J Hum Genet. Feb 2002;70:472–486. doi: 10.1086/338758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tekin M., Hismi B.O., Fitoz S. Homozygous mutations in fibroblast growth factor 3 are associated with a new form of syndromic deafness characterized by inner ear agenesis, microtia, and microdontia. Am J Hum Genet. Feb 2007;80:338–344. doi: 10.1086/510920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohmann E., Brunner H.G., Kayserili H. Mutations in different components of FGF signaling in LADD syndrome. Nat Genet. Apr 2006;38:414–417. doi: 10.1038/ng1757. [DOI] [PubMed] [Google Scholar]
- 16.Spielberger R., Stiff P., Bensinger W. Palifermin for oral mucositis after intensive therapy for hematologic cancers. N Engl J Med. Dec 16 2004;351:2590–2598. doi: 10.1056/NEJMoa040125. [DOI] [PubMed] [Google Scholar]
- 17.Wesche J., Haglund K., Haugsten E.M. Fibroblast growth factors and their receptors in cancer. Biochem J. Jul 15 2011;437:199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 18.Mohammadi M., Dikic I., Sorokin A., Burgess W.H., Jaye M., Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol. Mar 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohammadi M., Honegger A.M., Rotin D. A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth factor receptor (Flg) is a binding site for the SH2 domain of phospholipase C-gamma 1. Mol Cell Biol. Oct 1991;11:5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eswarakumar V.P., Lax I., Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. Apr 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Yun Y.R., Won J.E., Jeon E. Fibroblast growth factors: biology, function, and application for tissue regeneration. J Tissue Eng. 2010;2010:218142. doi: 10.4061/2010/218142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorey K., Amaya E. FGF signalling: diverse roles during early vertebrate embryogenesis. Development. Nov 2010;137:3731–3742. doi: 10.1242/dev.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao D., Lazar M.A. Modulating nuclear receptor function: may the phos be with you. J Clin Invest. Jun 1999;103:1617–1618. doi: 10.1172/JCI7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong A., Lamothe B., Lee A., Schlessinger J., Lax I. FRS2 alpha attenuates FGF receptor signaling by Grb2-mediated recruitment of the ubiquitin ligase Cbl. Proc Natl Acad Sci U S A. May 14 2002;99:6684–6689. doi: 10.1073/pnas.052138899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lax I., Wong A., Lamothe B. The docking protein FRS2 alpha controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Mol Cell. Oct 2002;10:709–719. doi: 10.1016/s1097-2765(02)00689-5. [DOI] [PubMed] [Google Scholar]
- 26.Dailey L., Ambrosetti D., Mansukhani A., Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. Apr 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Lamothe B., Yamada M., Schaeper U., Birchmeier W., Lax I., Schlessinger J. The docking protein Gab1 is an essential component of an indirect mechanism for fibroblast growth factor stimulation of the phosphatidylinositol 3-kinase/Akt antiapoptotic pathway. Mol Cell Biol. Jul 2004;24:5657–5666. doi: 10.1128/MCB.24.13.5657-5666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. Oct 13 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 29.Katoh M. FGF signaling network in the gastrointestinal tract (review) Int J Oncol. Jul 2006;29:163–168. [PubMed] [Google Scholar]
- 30.Hacohen N., Kramer S., Sutherland D., Hiromi Y., Krasnow M.A. Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. Jan 23 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- 31.Casci T., Vinos J., Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. Mar 5 1999;96:655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y., Zhang Z.Y. The mechanism of dephosphorylation of extracellular signal-regulated kinase 2 by mitogen-activated protein kinase phosphatase 3. J Biol Chem. Aug 24 2001;276:32382–32391. doi: 10.1074/jbc.M103369200. [DOI] [PubMed] [Google Scholar]
- 33.Furthauer M., Lin W., Ang S.L., Thisse B., Thisse C. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol. Feb 2002;4:170–174. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- 34.Tsang M., Dawid I.B. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci STKE. Apr 13 2004;2004:pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- 35.Thien C.B., Langdon W.Y. Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol. Apr 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 36.Pownall M.E., Isaacs H.V. FGF Signalling in Vertebrate Development. Morgan & Claypool Life Sciences; San Rafael, CA: 2010. (Developmental Biology). [PubMed] [Google Scholar]
- 37.Ciruna B., Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. Jul 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 38.Ciruna B.G., Schwartz L., Harpal K., Yamaguchi T.P., Rossant J. Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: a role for FGFR1 in morphogenetic movement through the primitive streak. Development. Jul 1997;124:2829–2841. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- 39.Sun X., Meyers E.N., Lewandoski M., Martin G.R. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. Jul 15 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekine K., Ohuchi H., Fujiwara M. Fgf10 is essential for limb and lung formation. Nat Genet. Jan 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 41.Sun X., Mariani F.V., Martin G.R. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature. Aug 1 2002;418:501–508. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- 42.Gomez-Pinilla F., Cotman C.W. Distribution of fibroblast growth factor 5 mRNA in the rat brain: an in situ hybridization study. Brain Res. Mar 19 1993;606:79–86. doi: 10.1016/0006-8993(93)91572-a. [DOI] [PubMed] [Google Scholar]
- 43.Hattori Y., Miyake A., Mikami T., Ohta M., Itoh N. Transient expression of FGF-5 mRNA in the rat cerebellar cortex during post-natal development. Brain Res Mol Brain Res. Jul 1997;47:262–266. doi: 10.1016/s0169-328x(97)00065-x. [DOI] [PubMed] [Google Scholar]
- 44.Hattori Y., Yamasaki M., Konishi M., Itoh N. Spatially restricted expression of fibroblast growth factor-10 mRNA in the rat brain. Brain Res Mol Brain Res. Jul 1997;47:139–146. doi: 10.1016/s0169-328x(97)00044-2. [DOI] [PubMed] [Google Scholar]
- 45.Smallwood P.M., Munoz-Sanjuan I., Tong P. Fibroblast growth factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proc Natl Acad Sci U S A. Sep 3 1996;93:9850–9857. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stock A., Kuzis K., Woodward W.R., Nishi R., Eckenstein F.P. Localization of acidic fibroblast growth factor in specific subcortical neuronal populations. J Neurosci. Dec 1992;12:4688–4700. doi: 10.1523/JNEUROSCI.12-12-04688.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodward W.R., Nishi R., Meshul C.K., Williams T.E., Coulombe M., Eckenstein F.P. Nuclear and cytoplasmic localization of basic fibroblast growth factor in astrocytes and CA2 hippocampal neurons. J Neurosci. Jan 1992;12:142–152. doi: 10.1523/JNEUROSCI.12-01-00142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashita T., Yoshioka M., Itoh N. Identification of a novel fibroblast growth factor, FGF-23, preferentially expressed in the ventrolateral thalamic nucleus of the brain. Biochem Biophys Res Commun. Oct 22 2000;277:494–498. doi: 10.1006/bbrc.2000.3696. [DOI] [PubMed] [Google Scholar]
- 49.Dono R. Fibroblast growth factors as regulators of central nervous system development and function. Am J Physiol Regul Integr Comp Physiol. Apr 2003;284:R867–R881. doi: 10.1152/ajpregu.00533.2002. [DOI] [PubMed] [Google Scholar]
- 50.Yang X., Dormann D., Munsterberg A.E., Weijer C.J. Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev Cell. Sep 2002;3:425–437. doi: 10.1016/s1534-5807(02)00256-3. [DOI] [PubMed] [Google Scholar]
- 51.Kubota Y., Ito K. Chemotactic migration of mesencephalic neural crest cells in the mouse. Dev Dyn. Feb 2000;217:170–179. doi: 10.1002/(SICI)1097-0177(200002)217:2<170::AID-DVDY4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 52.Wilson R., Leptin M. Fibroblast growth factor receptor-dependent morphogenesis of the Drosophila mesoderm. Philos Trans R Soc Lond B Biol Sci. Jul 29 2000;355:891–895. doi: 10.1098/rstb.2000.0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montell D.J. The genetics of cell migration in Drosophila melanogaster and Caenorhabditis elegans development. Development. Jun 1999;126:3035–3046. doi: 10.1242/dev.126.14.3035. [DOI] [PubMed] [Google Scholar]
- 54.Bottcher R.T., Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. Feb 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- 55.Coumoul X., Deng C.X. Roles of FGF receptors in mammalian development and congenital diseases. Birth Defects Res C Embryo Today. Nov 2003;69:286–304. doi: 10.1002/bdrc.10025. [DOI] [PubMed] [Google Scholar]
- 56.Nutt S.L., Dingwell K.S., Holt C.E., Amaya E. Xenopus Sprouty2 inhibits FGF-mediated gastrulation movements but does not affect mesoderm induction and patterning. Genes Dev. May 1 2001;15:1152–1166. doi: 10.1101/gad.191301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng C.X., Wynshaw-Boris A., Shen M.M., Daugherty C., Ornitz D.M., Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. Dec 15 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 58.Yamaguchi T.P., Harpal K., Henkemeyer M., Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. Dec 15 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 59.Deng C., Bedford M., Li C. Fibroblast growth factor receptor-1 (FGFR-1) is essential for normal neural tube and limb development. Dev Biol. May 1 1997;185:42–54. doi: 10.1006/dbio.1997.8553. [DOI] [PubMed] [Google Scholar]
- 60.Meyers E.N., Martin G.R. Differences in left-right axis pathways in mouse and chick: functions of FGF8 and SHH. Science. Jul 16 1999;285:403–406. doi: 10.1126/science.285.5426.403. [DOI] [PubMed] [Google Scholar]
- 61.Mizoguchi T., Izawa T., Kuroiwa A., Kikuchi Y. Fgf signaling negatively regulates Nodal-dependent endoderm induction in zebrafish. Dev Biol. Dec 15 2006;300:612–622. doi: 10.1016/j.ydbio.2006.08.073. [DOI] [PubMed] [Google Scholar]
- 62.Nentwich O., Dingwell K.S., Nordheim A., Smith J.C. Downstream of FGF during mesoderm formation in Xenopus: the roles of Elk-1 and Egr-1. Dev Biol. Dec 15 2009;336:313–326. doi: 10.1016/j.ydbio.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 63.Kimelman D., Griffin K.J. Vertebrate mesendoderm induction and patterning. Curr Opin Genet Dev. Aug 2000;10:350–356. doi: 10.1016/s0959-437x(00)00095-2. [DOI] [PubMed] [Google Scholar]
- 64.Slack J.M., Darlington B.G., Heath J.K., Godsave S.F. Mesoderm induction in early Xenopus embryos by heparin-binding growth factors. Nature. Mar 12–18 1987;326:197–200. doi: 10.1038/326197a0. [DOI] [PubMed] [Google Scholar]
- 65.Cornell R.A., Kimelman D. Activin-mediated mesoderm induction requires FGF. Development. Feb 1994;120:453–462. doi: 10.1242/dev.120.2.453. [DOI] [PubMed] [Google Scholar]
- 66.Fletcher R.B., Harland R.M. The role of FGF signaling in the establishment and maintenance of mesodermal gene expression in Xenopus. Dev Dyn. May 2008;237:1243–1254. doi: 10.1002/dvdy.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amaya E., Musci T.J., Kirschner M.W. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. Jul 26 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- 68.Wilson P.A., Hemmati-Brivanlou A. Vertebrate neural induction: inducers, inhibitors, and a new synthesis. Neuron. May 1997;18:699–710. doi: 10.1016/s0896-6273(00)80311-6. [DOI] [PubMed] [Google Scholar]
- 69.Wilson S.I., Edlund T. Neural induction: toward a unifying mechanism. Nat Neurosci. Nov 2001;(4 suppl):1161–1168. doi: 10.1038/nn747. [DOI] [PubMed] [Google Scholar]
- 70.Marchal L., Luxardi G., Thome V., Kodjabachian L. BMP inhibition initiates neural induction via FGF signaling and Zic genes. Proc Natl Acad Sci U S A. Oct 13 2009;106:17437–17442. doi: 10.1073/pnas.0906352106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christofori G. Split personalities: the agonistic antagonist Sprouty. Nat Cell Biol. May 2003;5:377–379. doi: 10.1038/ncb0503-377. [DOI] [PubMed] [Google Scholar]
- 72.Pera E.M., Ikeda A., Eivers E., De Robertis E.M. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. Dec 15 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Branney P.A., Faas L., Steane S.E., Pownall M.E., Isaacs H.V. Characterisation of the fibroblast growth factor dependent transcriptome in early development. PLoS One. 2009;4:e4951. doi: 10.1371/journal.pone.0004951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Delaune E., Lemaire P., Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. Jan 2005;132:299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- 75.Pownall M.E., Isaacs H.V., Slack J.M. Two phases of Hox gene regulation during early Xenopus development. Curr Biol. May 21 1998;8:673–676. doi: 10.1016/s0960-9822(98)70257-x. [DOI] [PubMed] [Google Scholar]
- 76.Doniach T. Basic FGF as an inducer of anteroposterior neural pattern. Cell. Dec 29 1995;83:1067–1070. doi: 10.1016/0092-8674(95)90133-7. [DOI] [PubMed] [Google Scholar]
- 77.Umbhauer M., Penzo-Mendez A., Clavilier L., Boucaut J., Riou J. Signaling specificities of fibroblast growth factor receptors in early Xenopus embryo. J Cell Sci. Aug 2000;113(Pt 16):2865–2875. doi: 10.1242/jcs.113.16.2865. [DOI] [PubMed] [Google Scholar]
- 78.Lamb T.M., Harland R.M. Fibroblast growth factor is a direct neural inducer, which combined with noggin generates anterior-posterior neural pattern. Development. Nov 1995;121:3627–3636. doi: 10.1242/dev.121.11.3627. [DOI] [PubMed] [Google Scholar]
- 79.Ribisi S., Jr., Mariani F.V., Aamar E., Lamb T.M., Frank D., Harland R.M. Ras-mediated FGF signaling is required for the formation of posterior but not anterior neural tissue in Xenopus laevis. Dev Biol. Nov 1 2000;227:183–196. doi: 10.1006/dbio.2000.9889. [DOI] [PubMed] [Google Scholar]
- 80.Holowacz T., Sokol S. FGF is required for posterior neural patterning but not for neural induction. Dev Biol. Jan 15 1999;205:296–308. doi: 10.1006/dbio.1998.9108. [DOI] [PubMed] [Google Scholar]
- 81.Cox W.G., Hemmati-Brivanlou A. Caudalization of neural fate by tissue recombination and bFGF. Development. Dec 1995;121:4349–4358. doi: 10.1242/dev.121.12.4349. [DOI] [PubMed] [Google Scholar]
- 82.Sato T., Joyner A.L., Nakamura H. How does Fgf signaling from the isthmic organizer induce midbrain and cerebellum development? Dev Growth Differ. Dec 2004;46:487–494. doi: 10.1111/j.1440-169x.2004.00769.x. [DOI] [PubMed] [Google Scholar]
- 83.Sato T., Nakamura H. The Fgf8 signal causes cerebellar differentiation by activating the Ras-ERK signaling pathway. Development. Sep 2004;131:4275–4285. doi: 10.1242/dev.01281. [DOI] [PubMed] [Google Scholar]
- 84.Suzuki-Hirano A., Sato T., Nakamura H. Regulation of isthmic Fgf8 signal by sprouty2. Development. Jan 2005;132:257–265. doi: 10.1242/dev.01581. [DOI] [PubMed] [Google Scholar]
- 85.Crossley P.H., Martinez S., Martin G.R. Midbrain development induced by FGF8 in the chick embryo. Nature. Mar 7 1996;380:66–68. doi: 10.1038/380066a0. [DOI] [PubMed] [Google Scholar]
- 86.Pearson J.C., Lemons D., McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. Dec 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 87.Pownall M.E., Tucker A.S., Slack J.M., Isaacs H.V. eFGF, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development. Dec 1996;122:3881–3892. doi: 10.1242/dev.122.12.3881. [DOI] [PubMed] [Google Scholar]
- 88.Cho K.W., De Robertis E.M. Differential activation of Xenopus homeo box genes by mesoderm-inducing growth factors and retinoic acid. Genes Dev. Nov 1990;4:1910–1916. doi: 10.1101/gad.4.11.1910. [DOI] [PubMed] [Google Scholar]
- 89.Kolm P.J., Sive H.L. Regulation of the Xenopus labial homeodomain genes, HoxA1 and HoxD1: activation by retinoids and peptide growth factors. Dev Biol. Jan 1995;167:34–49. doi: 10.1006/dbio.1995.1005. [DOI] [PubMed] [Google Scholar]
- 90.Partanen J., Schwartz L., Rossant J. Opposite phenotypes of hypomorphic and Y766 phosphorylation site mutations reveal a function for Fgfr1 in anteroposterior patterning of mouse embryos. Genes Dev. Aug 1 1998;12:2332–2344. doi: 10.1101/gad.12.15.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bel-Vialar S., Itasaki N., Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. Nov 2002;129:5103–5115. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- 92.Isaacs H.V., Pownall M.E., Slack J.M. Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J. Jun 15 1998;17:3413–3427. doi: 10.1093/emboj/17.12.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Isaacs H.V., Pownall M.E., Slack J.M. eFGF regulates Xbra expression during Xenopus gastrulation. EMBO J. Oct 3 1994;13:4469–4481. doi: 10.1002/j.1460-2075.1994.tb06769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Keenan I.D., Sharrard R.M., Isaacs H.V. FGF signal transduction and the regulation of Cdx gene expression. Dev Biol. Nov 15 2006;299:478–488. doi: 10.1016/j.ydbio.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 95.Northrop J.L., Kimelman D. Dorsal-ventral differences in Xcad-3 expression in response to FGF-mediated induction in Xenopus. Dev Biol. Feb 1994;161:490–503. doi: 10.1006/dbio.1994.1047. [DOI] [PubMed] [Google Scholar]
- 96.Faas L., Isaacs H.V. Overlapping functions of Cdx1, Cdx2, and Cdx4 in the development of the amphibian Xenopus tropicalis. Dev Dyn. Apr 2009;238:835–852. doi: 10.1002/dvdy.21901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Godsave S.F., Durston A.J. Neural induction and patterning in embryos deficient in FGF signaling. Int J Dev Biol. Feb 1997;41:57–65. [PubMed] [Google Scholar]
- 98.Niswander L., Martin G.R. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development. Mar 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- 99.Martin G.R. The roles of FGFs in the early development of vertebrate limbs. Genes Dev. Jun 1 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- 100.Tickle C., Munsterberg A. Vertebrate limb development – the early stages in chick and mouse. Curr Opin Genet Dev. Aug 2001;11:476–481. doi: 10.1016/s0959-437x(00)00220-3. [DOI] [PubMed] [Google Scholar]
- 101.Xu X., Weinstein M., Li C. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. Feb 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- 102.Ohuchi H., Nakagawa T., Yamamoto A. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development. Jun 1997;124:2235–2244. doi: 10.1242/dev.124.11.2235. [DOI] [PubMed] [Google Scholar]
- 103.Mahmood R., Bresnick J., Hornbruch A. A role for FGF-8 in the initiation and maintenance of vertebrate limb bud outgrowth. Curr Biol. Jul 1 1995;5:797–806. doi: 10.1016/s0960-9822(95)00157-6. [DOI] [PubMed] [Google Scholar]
- 104.Vogel A., Rodriguez C., Izpisua-Belmonte J.C. Involvement of FGF-8 in initiation, outgrowth and patterning of the vertebrate limb. Development. Jun 1996;122:1737–1750. doi: 10.1242/dev.122.6.1737. [DOI] [PubMed] [Google Scholar]
- 105.Crossley P.H., Minowada G., MacArthur C.A., Martin G.R. Roles for FGF8 in the induction, initiation, and maintenance of chick limb development. Cell. Jan 12 1996;84:127–136. doi: 10.1016/s0092-8674(00)80999-x. [DOI] [PubMed] [Google Scholar]
- 106.Ros M.A., Lopez-Martinez A., Simandl B.K. The limb field mesoderm determines initial limb bud anteroposterior asymmetry and budding independent of sonic hedgehog or apical ectodermal gene expressions. Development. Aug 1996;122:2319–2330. doi: 10.1242/dev.122.8.2319. [DOI] [PubMed] [Google Scholar]
- 107.Goldfarb M. Functions of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev. Dec 1996;7:311–325. doi: 10.1016/s1359-6101(96)00039-1. [DOI] [PubMed] [Google Scholar]
- 108.Yu K., Ornitz D.M. FGF signaling regulates mesenchymal differentiation and skeletal patterning along the limb bud proximodistal axis. Development. Feb 2008;135:483–491. doi: 10.1242/dev.013268. [DOI] [PubMed] [Google Scholar]
- 109.Nie X., Luukko K., Kettunen P. FGF signalling in craniofacial development and developmental disorders. Oral Dis. Mar 2006;12:102–111. doi: 10.1111/j.1601-0825.2005.01176.x. [DOI] [PubMed] [Google Scholar]
- 110.Morriss-Kay G.M., Iseki S., Johnson D. Genetic control of the cell proliferation-differentiation balance in the developing skull vault: roles of fibroblast growth factor receptor signalling pathways. Novartis Found Symp. 2001;232:102–116. doi: 10.1002/0470846658.ch8. discussion 116–121. [DOI] [PubMed] [Google Scholar]
- 111.Hatch N.E. FGF signaling in craniofacial biological control and pathological craniofacial development. Crit Rev Eukaryot Gene Expr. 2010;20:295–311. doi: 10.1615/critreveukargeneexpr.v20.i4.20. [DOI] [PubMed] [Google Scholar]
- 112.Wilkie A.O. Craniosynostosis: genes and mechanisms. Hum Mol Genet. 1997;6:1647–1656. doi: 10.1093/hmg/6.10.1647. [DOI] [PubMed] [Google Scholar]
- 113.Nuckolls G.H., Shum L., Slavkin H.C. Progress toward understanding craniofacial malformations. Cleft Palate Craniofac J. Jan 1999;36:12–26. doi: 10.1597/1545-1569_1999_036_0012_ptucm_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- 114.Marie P.J., Debiais F., Hay E. Regulation of human cranial osteoblast phenotype by FGF-2, FGFR-2 and BMP-2 signaling. Histol Histopathol. 2002;17:877–885. doi: 10.14670/HH-17.877. [DOI] [PubMed] [Google Scholar]
- 115.Kim H.J., Rice D.P., Kettunen P.J., Thesleff I.F.G.F.- BMP- and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development. Apr 1998;125:1241–1251. doi: 10.1242/dev.125.7.1241. [DOI] [PubMed] [Google Scholar]
- 116.Rice D.P., Aberg T., Chan Y. Integration of FGF and TWIST in calvarial bone and suture development. Development. May 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- 117.Noden D.M. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev Biol. Mar 1983;96:144–165. doi: 10.1016/0012-1606(83)90318-4. [DOI] [PubMed] [Google Scholar]
- 118.Noden D.M. Origins and patterning of craniofacial mesenchymal tissues. J Craniofac Genet Dev Biol Suppl. 1986;2:15–31. [PubMed] [Google Scholar]
- 119.Schaaf P. Histochemical behavior of Paneth's cells in Central American bat species with varying nutritional habits. Results of the 1st Cuban-German “Alexander-von-Humbolt” expedition 1967–68. 4. Anat Anz. 1970;126:275–277. [PubMed] [Google Scholar]
- 120.Couly G.F., Le Douarin N.M. Mapping of the early neural primordium in quail-chick chimeras. II. The prosencephalic neural plate and neural folds: implications for the genesis of cephalic human congenital abnormalities. Dev Bio. Mar 1987;120:198–214. doi: 10.1016/0012-1606(87)90118-7. [DOI] [PubMed] [Google Scholar]
- 121.Couly G.F., Coltey P.M., Le Douarin N.M. The developmental fate of the cephalic mesoderm in quail-chick chimeras. Development. Jan 1992;114:1–15. doi: 10.1242/dev.114.1.1. [DOI] [PubMed] [Google Scholar]
- 122.Couly G.F., Coltey P.M., Le Douarin N.M. The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development. Feb 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- 123.Baker C.V., Bronner-Fraser M. The origins of the neural crest. Part I: embryonic induction. Mech Dev. Dec 1997;69:3–11. doi: 10.1016/s0925-4773(97)00132-9. [DOI] [PubMed] [Google Scholar]
- 124.Villanueva S., Glavic A., Ruiz P., Mayor R. Posteriorization by FGF, Wnt, and retinoic acid is required for neural crest induction. Dev Biol. Jan 15 2002;241:289–301. doi: 10.1006/dbio.2001.0485. [DOI] [PubMed] [Google Scholar]
- 125.Monsoro-Burq A.H., Fletcher R.B., Harland R.M. Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development. Jul 2003;130:3111–3124. doi: 10.1242/dev.00531. [DOI] [PubMed] [Google Scholar]
- 126.Monsoro-Burq A.H., Wang E., Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell. Feb 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 127.Bachler M., Neubuser A. Expression of members of the Fgf family and their receptors during midfacial development. Mech Dev. Feb 2001;100:313–316. doi: 10.1016/s0925-4773(00)00518-9. [DOI] [PubMed] [Google Scholar]
- 128.Delezoide A.L., Benoist-Lasselin C., Legeai-Mallet L. Spatio-temporal expression of FGFR 1, 2 and 3 genes during human embryo-fetal ossification. Mech Dev. Sep 1998;77:19–30. doi: 10.1016/s0925-4773(98)00133-6. [DOI] [PubMed] [Google Scholar]
- 129.Opperman L.A. Cranial sutures as intramembranous bone growth sites. Dev Dyn. Dec 2000;219:472–485. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1073>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 130.Iseki S., Wilkie A.O., Morriss-Kay G.M. Fgfr1 and Fgfr2 have distinct differentiation- and proliferation-related roles in the developing mouse skull vault. Development. Dec 1999;126:5611–5620. doi: 10.1242/dev.126.24.5611. [DOI] [PubMed] [Google Scholar]
- 131.Pritchard J.J., Scott J.H., Girgis F.G. The structure and development of cranial and facial sutures. J Anat. Jan 1956;90:73–86. [PMC free article] [PubMed] [Google Scholar]
- 132.Opperman L.A., Chhabra A., Nolen A.A., Bao Y., Ogle R.C. Dura mater maintains rat cranial sutures in vitro by regulating suture cell proliferation and collagen production. J Craniofac Genet Dev Biol. Jul–Sep 1998;18:150–158. [PubMed] [Google Scholar]
- 133.Mehrara B.J., Mackool R.J., McCarthy J.G., Gittes G.K., Longaker M.T. Immunolocalization of basic fibroblast growth factor and fibroblast growth factor receptor-1 and receptor-2 in rat cranial sutures. Plast Reconstr Surg. Nov 1998;102:1805–1817. doi: 10.1097/00006534-199811000-00001. discussion 1818–1820. [DOI] [PubMed] [Google Scholar]
- 134.Chikazu D., Katagiri M., Ogasawara T. Regulation of osteoclast differentiation by fibroblast growth factor 2: stimulation of receptor activator of nuclear factor kappaB ligand/osteoclast differentiation factor expression in osteoblasts and inhibition of macrophage colony-stimulating factor function in osteoclast precursors. J Bone Miner Res. Nov 2001;16:2074–2081. doi: 10.1359/jbmr.2001.16.11.2074. [DOI] [PubMed] [Google Scholar]
- 135.Opperman L.A., Passarelli R.W., Morgan E.P., Reintjes M., Ogle R.C. Cranial sutures require tissue interactions with dura mater to resist osseous obliteration in vitro. J Bone Miner Res. Dec 1995;10:1978–1987. doi: 10.1002/jbmr.5650101218. [DOI] [PubMed] [Google Scholar]
- 136.Aviezer D., Golembo M., Yayon A. Fibroblast growth factor receptor-3 as a therapeutic target for Achondroplasia – genetic short limbed dwarfism. Curr Drug Targets. Jul 2003;4:353–365. doi: 10.2174/1389450033490993. [DOI] [PubMed] [Google Scholar]
- 137.Shiang R., Thompson L.M., Zhu Y.Z. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. Jul 29 1994;78:335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- 138.Bellus G.A., Hefferon T.W., Ortiz de Luna R.I. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet. Feb 1995;56:368–373. [PMC free article] [PubMed] [Google Scholar]
- 139.Heuertz S., Le Merrer M., Zabel B. Novel FGFR3 mutations creating cysteine residues in the extracellular domain of the receptor cause achondroplasia or severe forms of hypochondroplasia. Eur J Hum Genet. Dec 2006;14:1240–1247. doi: 10.1038/sj.ejhg.5201700. [DOI] [PubMed] [Google Scholar]
- 140.Bonaventure J., Rousseau F., Legeai-Mallet L., Le Merrer M., Munnich A., Maroteaux P. Common mutations in the fibroblast growth factor receptor 3 (FGFR 3) gene account for achondroplasia, hypochondroplasia, and thanatophoric dwarfism. Am J Med Genet. May 3 1996;63:148–154. doi: 10.1002/(SICI)1096-8628(19960503)63:1<148::AID-AJMG26>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 141.Cohen M.M., Jr. Achondroplasia, hypochondroplasia and thanatophoric dysplasia: clinically related skeletal dysplasias that are also related at the molecular level. Int J Oral Maxillofac Surg. Dec 1998;27:451–455. doi: 10.1016/s0901-5027(98)80036-2. [DOI] [PubMed] [Google Scholar]
- 142.Bellus G.A., McIntosh I., Smith E.A. A recurrent mutation in the tyrosine kinase domain of fibroblast growth factor receptor 3 causes hypochondroplasia. Nat Genet. Jul 1995;10:357–359. doi: 10.1038/ng0795-357. [DOI] [PubMed] [Google Scholar]
- 143.Noe E.J., Yoo H.W., Kim K.N., Lee S.Y. A case of thanatophoric dysplasia type I with an R248C mutation in the FGFR3 gene. Korean J Pediatr. Dec 2010;53:1022–1025. doi: 10.3345/kjp.2010.53.12.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Parker B.C., Engels M., Annala M., Zhang W. Emergence of FGFR family gene fusions as therapeutic targets in a wide spectrum of solid tumours. J Pathol. Jan 2014;232:4–15. doi: 10.1002/path.4297. [DOI] [PubMed] [Google Scholar]
- 145.Dodurga Y., Tataroglu C., Kesen Z., Satiroglu-Tufan N.L. Incidence of fibroblast growth factor receptor 3 gene (FGFR3) A248C, S249C, G372C, and T375C mutations in bladder cancer. Genet Mol Res. 2011;10:86–95. doi: 10.4238/vol10-1gmr923. [DOI] [PubMed] [Google Scholar]
- 146.Pollock P.M., Gartside M.G., Dejeza L.C. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. Nov 1 2007;26:7158–7162. doi: 10.1038/sj.onc.1210529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Naski M.C., Ornitz D.M. FGF signaling in skeletal development. Front Biosci. 1998;3:d781–794. doi: 10.2741/a321. [DOI] [PubMed] [Google Scholar]
- 148.Floss T., Arnold H.H., Braun T. A role for FGF-6 in skeletal muscle regeneration. Genes Dev. Aug 15 1997;11:2040–2051. doi: 10.1101/gad.11.16.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Matsui Y., Yasui N., Kimura T., Tsumaki N., Kawabata H., Ochi T. Genotype phenotype correlation in achondroplasia and hypochondroplasia. J Bone Joint Surg Br. Nov 1998;80:1052–1056. doi: 10.1302/0301-620x.80b6.9277. [DOI] [PubMed] [Google Scholar]
- 150.Ireland P.J., Pacey V., Zankl A., Edwards P., Johnston L.M., Savarirayan R. Optimal management of complications associated with achondroplasia. Appl Clin Genet. 2014;7:117–125. doi: 10.2147/TACG.S51485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lorget F., Kaci N., Peng J. Evaluation of the therapeutic potential of a CNP analog in a Fgfr3 mouse model recapitulating achondroplasia. Am J Hum Genet. Dec 7 2012;91:1108–1114. doi: 10.1016/j.ajhg.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bober M.B., Bellus G.A., Nikkel S.M., Tiller GE. 1993. Hypochondroplasia. [PubMed] [Google Scholar]
- 153.Karczeski B., Cutting G.R. 1993. Thanatophoric Dysplasia. [Google Scholar]
- 154.Rousseau F., el Ghouzzi V., Delezoide A.L. Missense FGFR3 mutations create cysteine residues in thanatophoric dwarfism type I (TD1) Hum Mol Genet. Apr 1996;5:509–512. doi: 10.1093/hmg/5.4.509. [DOI] [PubMed] [Google Scholar]
- 155.Tavormina P.L., Rimoin D.L., Cohn D.H., Zhu Y.Z., Shiang R., Wasmuth J.J. Another mutation that results in the substitution of an unpaired cysteine residue in the extracellular domain of FGFR3 in thanatophoric dysplasia type I. Hum Mol Genet. Nov 1995;4:2175–2177. doi: 10.1093/hmg/4.11.2175. [DOI] [PubMed] [Google Scholar]
- 156.Senarath-Yapa K., Chung M.T., McArdle A. Craniosynostosis: molecular pathways and future pharmacologic therapy. Organogenesis. Oct–Dec 2012;8:103–113. doi: 10.4161/org.23307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ursitti F., Fadda T., Papetti L. Evaluation and management of nonsyndromic craniosynostosis. Acta Paediatr. Sep 2011;100:1185–1194. doi: 10.1111/j.1651-2227.2011.02299.x. [DOI] [PubMed] [Google Scholar]
- 158.Rice D.P. Craniofacial sutures. Development, disease and treatment. Preface. Front Oral Bio. 2008;12:xi. [PubMed] [Google Scholar]
- 159.Cohen M.M., Jr. Craniosynostosis update 1987. Am J Med Genet Suppl. 1988;4:99–148. doi: 10.1002/ajmg.1320310514. [DOI] [PubMed] [Google Scholar]
- 160.Anderson J., Burns H.D., Enriquez-Harris P., Wilkie A.O., Heath J.K. Apert syndrome mutations in fibroblast growth factor receptor 2 exhibit increased affinity for FGF ligand. Hum Mol Genet. Sep 1998;7:1475–1483. doi: 10.1093/hmg/7.9.1475. [DOI] [PubMed] [Google Scholar]
- 161.Robin N.H., Falk M.J., Haldeman-Englert C.R. 1993. FGFR-related Craniosynostosis Syndromes. [Google Scholar]
- 162.Rossi M., Jones R.L., Norbury G., Bloch-Zupan A., Winter R.M. The appearance of the feet in Pfeiffer syndrome caused by FGFR1 P252R mutation. Clin Dysmorphol. Oct 2003;12:269–274. doi: 10.1097/00019605-200310000-00012. [DOI] [PubMed] [Google Scholar]
- 163.Cornejo-Roldan L.R., Roessler E., Muenke M. Analysis of the mutational spectrum of the FGFR2 gene in Pfeiffer syndrome. Hum Genet. May 1999;104:425–431. doi: 10.1007/s004390050979. [DOI] [PubMed] [Google Scholar]
- 164.Holmes G., Basilico C. Mesodermal expression of Fgfr2S252W is necessary and sufficient to induce craniosynostosis in a mouse model of Apert syndrome. Dev Biol. Aug 15 2012;368:283–293. doi: 10.1016/j.ydbio.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yeh E., Fanganiello R.D., Sunaga D.Y. Novel molecular pathways elicited by mutant FGFR2 may account for brain abnormalities in Apert syndrome. PLoS One. 2013;8:e60439. doi: 10.1371/journal.pone.0060439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rouzier C., Soler C., Hofman P., Brennetot C., Bieth E., Pedeutour F. Ovarian dysgerminoma and Apert syndrome. Pediatr Blood Cancer. Mar 2008;50:696–698. doi: 10.1002/pbc.21156. [DOI] [PubMed] [Google Scholar]
- 167.Renier D., Arnaud E., Cinalli G., Sebag G., Zerah M., Marchac D. Prognosis for mental function in Apert's syndrome. J Neurosurg. Jul 1996;85:66–72. doi: 10.3171/jns.1996.85.1.0066. [DOI] [PubMed] [Google Scholar]
- 168.Zarate Y.A., Putnam P.E., Saal H.M. Intestinal malrotation in a patient with Pfeiffer syndrome type 2. Cleft Palate Craniofac J. Nov 2010;47:638–641. doi: 10.1597/09-115. [DOI] [PubMed] [Google Scholar]
- 169.Ibrahimi O.A., Eliseenkova A.V., Plotnikov A.N., Yu K., Ornitz D.M., Mohammadi M. Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome. Proc Natl Acad Sci U S A. Jun 19 2001;98:7182–7187. doi: 10.1073/pnas.121183798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rutland P., Pulleyn L.J., Reardon W. Identical mutations in the FGFR2 gene cause both Pfeiffer and Crouzon syndrome phenotypes. Nat Genet. Feb 1995;9:173–176. doi: 10.1038/ng0295-173. [DOI] [PubMed] [Google Scholar]
- 171.Lapunzina P., Fernandez A., Sanchez Romero J.M. A novel insertion in the FGFR2 gene in a patient with Crouzon phenotype and sacrococcygeal tail. Birth Defects Res A Clin Mol Teratol. Jan 2005;73:61–64. doi: 10.1002/bdra.20093. [DOI] [PubMed] [Google Scholar]
- 172.Murdoch-Kinch C.A., Ward R.E. Metacarpophalangeal analysis in Crouzon syndrome: additional evidence for phenotypic convergence with the acrocephalosyndactyly syndromes. Am J Med Genet. Nov 28 1997;73:61–66. [PubMed] [Google Scholar]
- 173.Chen L., Deng C.X. Roles of FGF signaling in skeletal development and human genetic diseases. Front Biosci. 2005;10:1961–1976. doi: 10.2741/1671. [DOI] [PubMed] [Google Scholar]
- 174.Roscioli T., Flanagan S., Mortimore R.J. Premature calvarial synostosis and epidermal hyperplasia (Beare-Stevenson syndrome-like anomalies) resulting from a P250R missense mutation in the gene encoding fibroblast growth factor receptor 3. Am J Med Genet. Jul 1 2001;101:187–194. doi: 10.1002/ajmg.1369. [DOI] [PubMed] [Google Scholar]
- 175.Percival C.J., Wang Y., Zhou X., Jabs E.W., Richtsmeier J.T. The effect of a Beare-Stevenson syndrome Fgfr2 Y394C mutation on early craniofacial bone volume and relative bone mineral density in mice. J Anat. Nov 2012;221:434–442. doi: 10.1111/j.1469-7580.2012.01555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tartaglia M., Di Rocco C., Lajeunie E., Valeri S., Velardi F., Battaglia P.A. Jackson-Weiss syndrome: identification of two novel FGFR2 missense mutations shared with Crouzon and Pfeiffer craniosynostotic disorders. Hum Genet. Nov 1997;101:47–50. doi: 10.1007/s004390050584. [DOI] [PubMed] [Google Scholar]
- 177.Honnebier M.B., Cabiling D.S., Hetlinger M., McDonald-McGinn D.M., Zackai E.H., Bartlett S.P. The natural history of patients treated for FGFR3-associated (Muenke-type) craniosynostosis. Plast Reconstr Surg. Mar 2008;121:919–931. doi: 10.1097/01.prs.0000299936.95276.24. [DOI] [PubMed] [Google Scholar]
- 178.Barbosa M., Almeida Mdo R., Reis-Lima M., Pinto-Basto J., dos Santos H.G. Muenke syndrome with osteochondroma. Am J Med Genet A. Feb 2009;149A:260–261. doi: 10.1002/ajmg.a.32616. [DOI] [PubMed] [Google Scholar]
- 179.David L., Glazier S., Pyle J., Thompson J., Argenta L. Classification system for sagittal craniosynostosis. J Craniofac Surg. Mar 2009;20:279–282. [PubMed] [Google Scholar]
- 180.Hunenko O., Karmacharya J., Ong G., Kirschner R.E. Toward an understanding of nonsyndromic craniosynostosis: altered patterns of TGF-beta receptor and FGF receptor expression induced by intrauterine head constraint. Ann Plast Surg. May 2001;46:546–553. doi: 10.1097/00000637-200105000-00015. discussion 553–544. [DOI] [PubMed] [Google Scholar]
- 181.Meyer A.N., McAndrew C.W., Donoghue D.J. Nordihydroguaiaretic acid inhibits an activated fibroblast growth factor receptor 3 mutant and blocks downstream signaling in multiple myeloma cells. Cancer Res. Sep 15 2008;68:7362–7370. doi: 10.1158/0008-5472.CAN-08-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Grand E.K., Chase A.J., Heath C., Rahemtulla A., Cross N.C. Targeting FGFR3 in multiple myeloma: inhibition of t(4;14)-positive cells by SU5402 and PD173074. Leukemia. May 2004;18:962–966. doi: 10.1038/sj.leu.2403347. [DOI] [PubMed] [Google Scholar]
- 183.Byron S.A., Gartside M.G., Wellens C.L. Inhibition of activated fibroblast growth factor receptor 2 in endometrial cancer cells induces cell death despite PTEN abrogation. Cancer Res. Sep 1 2008;68:6902–6907. doi: 10.1158/0008-5472.CAN-08-0770. [DOI] [PubMed] [Google Scholar]
- 184.Chow L.Q., Eckhardt S.G. Sunitinib: from rational design to clinical efficacy. J Clin Oncol. Mar 1 2007;25:884–896. doi: 10.1200/JCO.2006.06.3602. [DOI] [PubMed] [Google Scholar]
- 185.Cross N.C., Reiter A. Fibroblast growth factor receptor and platelet-derived growth factor receptor abnormalities in eosinophilic myeloproliferative disorders. Acta Haematol. 2008;119:199–206. doi: 10.1159/000140631. [DOI] [PubMed] [Google Scholar]
- 186.Roumiantsev S., Krause D.S., Neumann C.A. Distinct stem cell myeloproliferative/T lymphoma syndromes induced by ZNF198-FGFR1 and BCR-FGFR1 fusion genes from 8p11 translocations. Cancer Cell. Mar 2004;5:287–298. doi: 10.1016/s1535-6108(04)00053-4. [DOI] [PubMed] [Google Scholar]
- 187.Ohbayashi N., Shibayama M., Kurotaki Y. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. Apr 1 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu Z., Xu J., Colvin J.S., Ornitz D.M. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. Apr 1 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ellsworth J.L., Berry J., Bukowski T. Fibroblast growth factor-18 is a trophic factor for mature chondrocytes and their progenitors. Osteoarthr Cartilage. Apr 2002;10:308–320. doi: 10.1053/joca.2002.0514. [DOI] [PubMed] [Google Scholar]
- 190.Moore E.E., Bendele A.M., Thompson D.L. Fibroblast growth factor-18 stimulates chondrogenesis and cartilage repair in a rat model of injury-induced osteoarthritis. Osteoarthr Cartilage. Jul 2005;13:623–631. doi: 10.1016/j.joca.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 191.Cheng H., Liao K.K., Liao S.F., Chuang T.Y., Shih Y.H. Spinal cord repair with acidic fibroblast growth factor as a treatment for a patient with chronic paraplegia. Spine (Phila Pa 1976) Jul 15 2004;29:E284–E288. doi: 10.1097/01.brs.0000131217.61390.2c. [DOI] [PubMed] [Google Scholar]
- 192.Schumacher B., Pecher P., von Specht B.U., Stegmann T. Induction of neoangiogenesis in ischemic myocardium by human growth factors: first clinical results of a new treatment of coronary heart disease. Circulation. Feb 24 1998;97:645–650. doi: 10.1161/01.cir.97.7.645. [DOI] [PubMed] [Google Scholar]
- 193.Ruel M., Laham R.J., Parker J.A. Long-term effects of surgical angiogenic therapy with fibroblast growth factor 2 protein. J Thorac Cardiovasc Surg. Jul 2002;124:28–34. doi: 10.1067/mtc.2002.121974. [DOI] [PubMed] [Google Scholar]
- 194.Henry T.D., Grines C.L., Watkins M.W. Effects of Ad5FGF-4 in patients with angina: an analysis of pooled data from the AGENT-3 and AGENT-4 trials. J Am Coll Cardiol. Sep 11 2007;50:1038–1046. doi: 10.1016/j.jacc.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 195.Turner C.A., Gula E.L., Taylor L.P., Watson S.J., Akil H. Antidepressant-like effects of intracerebroventricular FGF2 in rats. Brain Res. Aug 11 2008;1224:63–68. doi: 10.1016/j.brainres.2008.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Werner S. Keratinocyte growth factor: a unique player in epithelial repair processes. Cytokine Growth Factor Rev. Jun 1998;9:153–165. doi: 10.1016/s1359-6101(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 197.Fu L., John L.M., Adams S.H. Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology. Jun 2004;145:2594–2603. doi: 10.1210/en.2003-1671. [DOI] [PubMed] [Google Scholar]
- 198.Kharitonenkov A., Shiyanova T.L., Koester A. FGF-21 as a novel metabolic regulator. J Clin Invest. Jun 2005;115:1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Takagi Y., Takahashi J., Saiki H. Dopaminergic neurons generated from monkey embryonic stem cells function in a Parkinson primate model. J Clin Invest. Jan 2005;115:102–109. doi: 10.1172/JCI21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hebert J.M., Rosenquist T., Gotz J., Martin G.R. FGF5 as a regulator of the hair growth cycle: evidence from targeted and spontaneous mutations. Cell. Sep 23 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 201.Coutu D.L., Galipeau J. Roles of FGF signaling in stem cell self-renewal, senescence and aging. Aging (Albany NY) Oct 2011;3:920–933. doi: 10.18632/aging.100369. [DOI] [PMC free article] [PubMed] [Google Scholar]