Abstract
Ligation of tRNAs with their cognate amino acids, by aminoacyl-tRNA synthetases, establishes the genetic code. Throughout evolution, tRNAAla selection by alanyl-tRNA synthetase (AlaRS) has depended predominantly on a single wobble base pair in the acceptor stem, G3•U70, mainly on the kcat level. Here we report the crystal structures of an archaeal AlaRS in complex with tRNAAla with G3•U70 and its A3•U70 variant. AlaRS interacts with both the minor- and major-groove sides of G3•U70, widening the major groove. The geometry difference between G3•U70 and A3•U70 is transmitted along the acceptor stem to the 3′-CCA region. Thus, the 3′-CCA region of tRNAAla with G3•U70 is oriented to the reactive route that reaches the active site, whereas that of the A3•U70 variant is folded back into the “non-reactive route”. This novel mechanism enables the single wobble pair to dominantly determine the specificity of tRNA selection, by an approximate 100-fold difference in kcat.
The genetic code requires extremely high aminoacyl-tRNA synthesis specificity. Each aminoacyl-tRNA synthetase (aaRS) ligates its specific amino acid to the adenosine in the conserved 3′-terminal CCA sequence of tRNA. The identity, or the amino acid-accepting specificity, of each tRNA is determined by a particular set of structural features (identity set)1,2. The identity sets of most tRNAs consist of a limited number of nucleotides, such as anticodon nucleotides3–6.
The discovery that the identity set of alanine tRNA (tRNAAla) from Escherichia coli is just the single G•U wobble base pair, in the middle of the amino acid-acceptor stem (G3•U70)7,8, engendered the concept of the second genetic code9. A G3•U70-to-Watson-Crick mutation of E. coli tRNAAla and reverse mutations of non-alanine tRNAs drastically decreased and increased, respectively, the aminoacylation by alanyl-tRNA synthetase (AlaRS)7,8, reportedly on the kcat level10. Throughout evolution from bacteria to humans, AlaRS recognizes G3•U70 of tRNAAla as the dominant identity determinant11 (Fig. 1a, b).
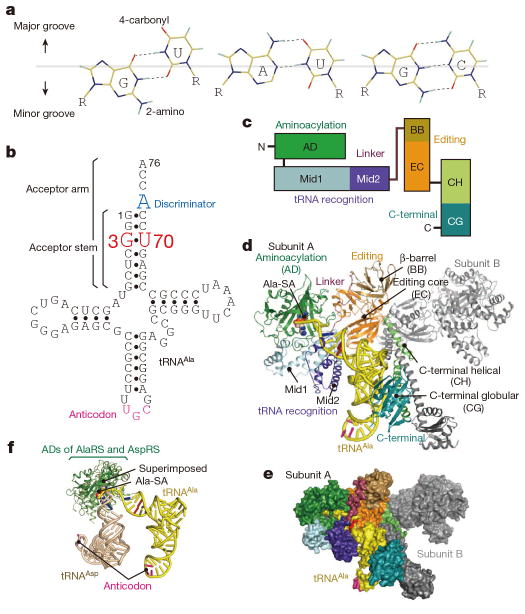
Figure 1. Structure of AlaRS in complex with tRNAAla.
a, G•U, A•U, and G•C pairs. b, Cloverleaf model of A. fulgidus tRNAAla. c, A diagram of the AlaRS domains/subdomains. d, e, The A. fulgidus AlaRS•tRNAAla•Ala-SA complex, presented by ribbon (AlaRS) and tube (tRNAAla) models (c) and by surface models (d) with the domains/subdomain of AlaRS subunit A colored as in c. f, Positions of tRNAAla and tRNAAsp relative to the class-II aminoacylation domains (superimposed) of AlaRS and AspRS, respectively.
With a specific dependence on G3•U70, E. coli AlaRS alanylates small RNAs recapitulating the tRNAAla acceptor stem (7–12 base pair stem-and-loop or duplex RNAs)1,7,12–17. These model RNAs were used for analyses with nucleotide analogues, such as inosine and deoxyribonucleotides, to elucidate which functional groups in the tRNAAla acceptor stem are important for alanylation. First, unlike the Watson-Crick pairs, G and U in the G•U pair (Fig. 1a) are shifted toward the minor and major groove sides, respectively, and form two hydrogen bonds without using the 2-amino group of G and the 4-carbonyl group of U18. Actually, the non-hydrogen-bonded nature of the 2-amino group in the G3•U70 wobble pair is important for aminoacylation19. Furthermore, the 2′-hydroxyl groups of U70 and C71 and the base of A73 contribute to alanylation. It is likely, therefore, that a number of nucleotides in the acceptor stem are involved, directly or indirectly, in interactions with AlaRS. However, it is still mysterious how just a single pair at positions 3•70 plays the dominant role among the interacting nucleotides, to determine the strict specificity for tRNAAla (refs 7,8,20).
Here, we report two crystal structures of AlaRS from the archaeon Archaeoglobus fulgidus, in complexes with tRNAAla bearing G3•U70 (tRNAAla/GU) and with a variant bearing A3•U70 (tRNAAla/AU), together with an alanyl-AMP analogue. The tRNA recognition domain of AlaRS widens the major groove, and thereby contacts both the minor- and major-grooves of the acceptor stem. Between the AlaRS•tRNAAla/GU and AlaRS•tRNAAla/AU structures, the local geometrical difference between G3•U70 and A3•U70 (Fig. 1a) propagates into the drastically distinct orientation of the CCA region, so that the 3′-end A76 of tRNAAla/AU is kept far from the aminoacylation active site. This unprecedented mechanism explains why just a small difference in the sequence (the single base pair at positions 3 and 70) results in the specific selection of tRNAAla, predominantly on the kcat level.
The AlaRS•tRNAAla complex structures
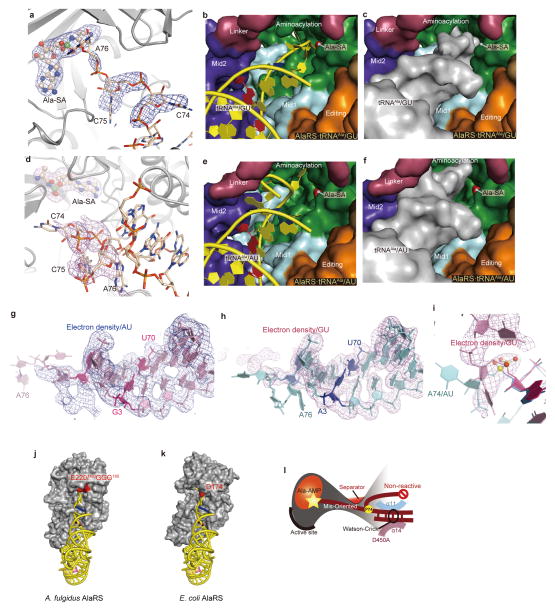
The crystal structure of the complex of A. fulgidus AlaRS, tRNAAla, and an alanyl-adenylate analog, 5′-O-[N-(L-alanyl)sulfamoyl]adenosine (Ala-SA)21, was solved at 3.3 Å resolution (Fig. 1c–e and Extended Data Table 1). Furthermore, we determined the 3.5 Å-resolution structure of a non-reactive complex of AlaRS with a tRNAAla variant possessing A3•U70 instead of G3•U70 (tRNAAla/AU). In both structures, the crystallographic asymmetric unit contains one homodimer of A. fulgidus AlaRS, which is consistent with the results from gel-filtration chromatography of A. fulgidus AlaRS22 and sedimentation equilibrium analyses of A. fulgidus and E. coli AlaRSs (ref. 23, Methods).
In both structures, AlaRS is composed of the aminoacylation, tRNA-recognition, editing, and C-terminal domains, among which the latter three consist further of the Mid1/Mid2, β-barrel/editing-core, and helical/globular subdomains, respectively24–31 (Fig. 1c, d). The C-terminal and editing domains constitute the dimer interface (Fig. 1d, e). Both aminoacylation domains of the dimer bind Ala-SA, while only subunit A binds tRNAAla using its tRNA-recognition and C-terminal domains. Isothermal titration calorimetry revealed that one AlaRS dimer binds one tRNAAla with a Kd value of 0.14± 0.3 μM at 20 °C. Between the two subunits, the corresponding subdomains exhibit the same folds. However, the tRNAAla-bound (A) and tRNAAla-free (B) subunits are in the closed and open forms, respectively, because the Mid2 subdomain and the C-terminal domain are differently rearranged relative to the other (sub)domains (Fig. 1d, e and Extended Data Fig. 1a). The canonical L-shaped structure of the tRNA is snugly embedded in a large cavity, formed by the closed-form subunit (Fig. 1d, e). The aminoacylation domain of AlaRS adopts the typical class-II aminoacylation-domain fold32,33, but the position of tRNAAla relative to the fold is completely different from that conserved in all of the other class-II aaRSs (Fig. 1f and Extended Data Fig. 2). The acceptor–T arm moiety of tRNAAla interacts with the aminoacylation, tRNA-recognition, and C-terminal domains of AlaRS, whereas the anticodon is free of interactions (Fig. 1d, e). A summary of the AlaRS•tRNAAla interactions is provided in Extended Data Fig. 3a.
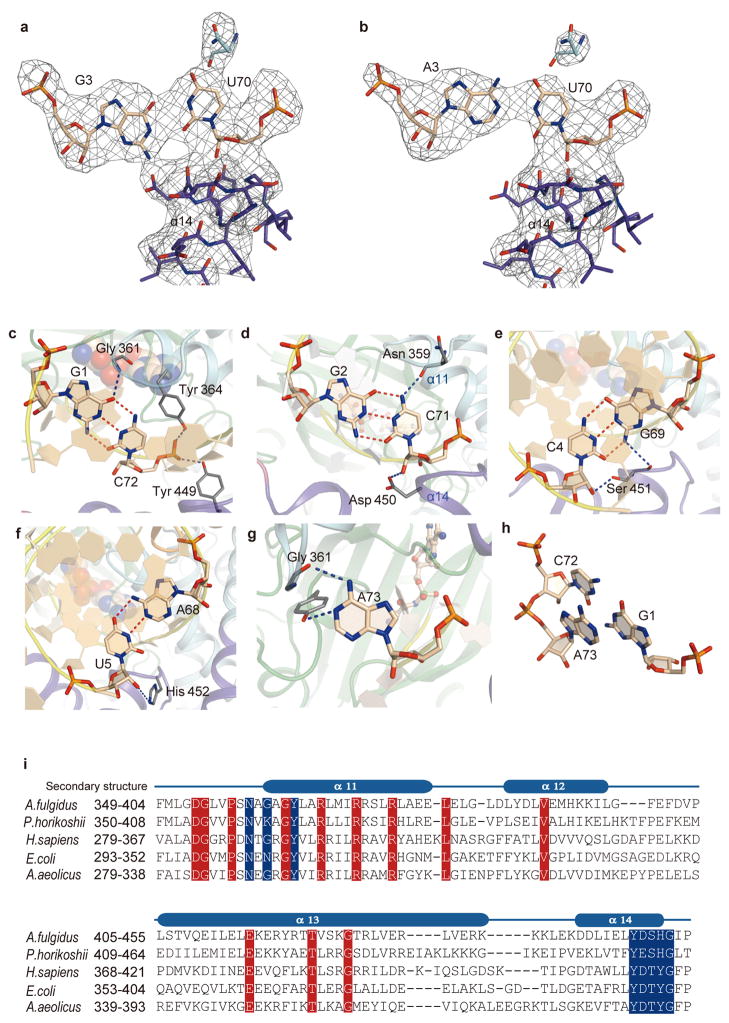
The G3•U70 pair exhibits the wobble geometry: G3 is shifted to the minor-groove side and the two bases are slightly rotated, as compared with the Watson-Crick geometry of A3•U70 (Figs. 1a and 2a, b and Extended Data Fig. 4a, b). These geometrical differences are also observed for the model RNA hairpins that recapitulate the tRNAAla acceptor stem34,35. Surprisingly, the two complexes further exhibit a drastic difference in the orientation of the 3′-end CCA region (positions 74–76). The 3′-CCA region of the wild-type tRNAAla/GU extends toward the aminoacylation catalytic site, so that A76 can reach the bound Ala-SA molecule (Fig. 2c and Extended Data Fig. 5a–c, g, h). However, the entire 3′-CCA region of tRNAAla/AU folds back into a completely different site (Fig. 2d and Extended Data Fig. 5d–f, g, h), thus placing A76 20 Å away from the active site. This is an unprecedented mechanism for keeping the bound non-cognate tRNA molecule in a non-reactive state.
Figure 2. Major differences between AlaRS•tRNAAla/GU and AlaRS•tRNAAla/AU.
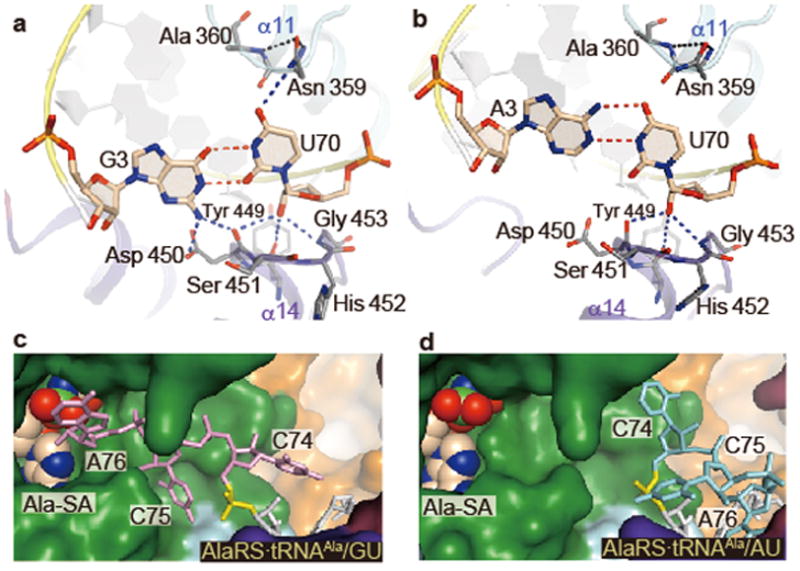
a–d, The G3•U70 (a) and A3•U70 (b) pairs and the CCA regions (c, d) of tRNAAla/GU (a, c) and tRNAAla/AU (b, d). The putative hydrogen bonds of G3•U70 and A3•U70 with AlaRS are shown with dashed lines (a, b). The CCA region of tRNAAla/GU is in the reactive route (c), while that of tRNAAla/AU is in the non-reactive route (d). The 5′-phosphate of C74 (P74) is colored yellow (c, d).
The acceptor arm•AlaRS interactions
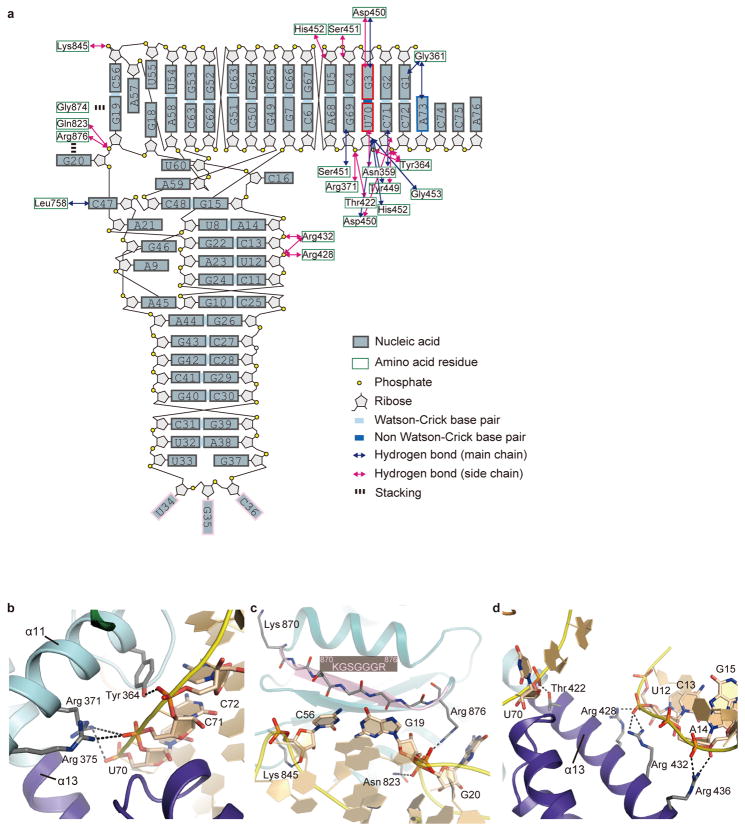
The minor-groove side of G3•U70 contacts the tip of the α14 helix from the Mid2 subdomain (Fig. 2a). The 2-amino group of G3 may hydrogen bond with the side-chain carboxyl and main-chain carbonyl groups of Asp 450 (Fig. 2a). The 2′-hydroxyl group of U70 possibly hydrogen bonds with the main-chain carbonyl groups of Asp 450 and Tyr 449 and the amino group of Gly 453. The Asp 450 side chain may also interact with the 2′-hydroxyl group of C71 (Extended Data Fig. 4d). Overall, the Tyr-Asp-Ser-His-Gly region (positions 449–453) at the α14 tip fits snugly with the unique minor groove shape of G3•U70 and its adjacent base pairs, G2•C71, C4•G69, and U5•A68 (Fig. 2a and Extended Data Fig. 4d–f). The Tyr-Asp-Ser-His-Gly sequence is conserved as Tyr-(Asp/Glu)-(Ser/Thr)-(His/Tyr)-Gly in other organisms (Extended Data Fig. 4i). The 4-carbonyl oxygen atom of U70 on the major groove side is close to and may hydrogen bond with the side-chain amino group of Asn 359 (Fig. 2a). Overall, the major-groove side from G1•C72 to G3•U70 is located near the helical loop on the N-terminal side of α11, which provides not only Asn 359 but also Gly 361 and Tyr 364 for interactions (Fig. 2a and Extended Data Fig. 4c, d). The base of the discriminator A73 is stacked on both C72 and G1, suggesting hydrogen bonding between A73 and Gly 361 (Extended Data Fig. 4g, h). A similar stacking pattern of A73 with G1•C72 is also observed for the hairpin RNA model34. The three interacting residues are almost completely conserved in the AlaRSs from bacteria to humans (Extended Data Fig. 4i).
In the AlaRS•tRNAAla/AU complex, the acceptor-stem minor groove interacts with the α14 tip, and U in A3•U70 is positioned similarly to that in G3•U70. In contrast, A3 is significantly shifted upward toward the major groove side, and is therefore much farther from Asp 450, as compared with G3 in G3•U70 (Fig. 2a, b). On the major groove side, the 4-carbonyl group of U70 is farther from the Asn 359 side chain, as the uracil base is rotated to pair with the upwardly shifted adenine base. If a G•C pair is substituted for A3•U70, then the 4-amino group of C70 might be more repulsive against Asn 359 than U70. Thus, the local geometry and the interaction manner of A3•U70 (and probably G3•C70) on both the minor and major groove sides are specifically distinct from those of G3•U70.
Geometry propagation from 3•70 to CCA
The local geometrical differences between G3•U70 and A3•U70 on both the minor and major groove sides propagate toward the 3′-CCA region. First, since A3•U70 is shifted as compared with G3•U70, G2•C71 and G1•C72 are moved together with it (Fig. 3a). These nucleotide movements reach the turning point at the 5′-phosphate group of C74, and thus result in the drastic change in the direction of the polynucleotide backbone (Fig. 3b and Extended Data Fig. 5i). In particular, A76 of tRNAAla/AU remains near A73, and the two 2′-hydroxyl groups are within hydrogen-bonding distance (< 3 Å). Thus, A76 is kept far (about 20 Å) from the catalytic site (Figs. 2d and 3a and Extended Data Fig. 5d–f), whereas A76 of tRNAAla/GU can reach it (Figs. 2c and 3a and Extended Data Fig. 5a–c).
Figure 3. Watson-Crick pair exclusion.
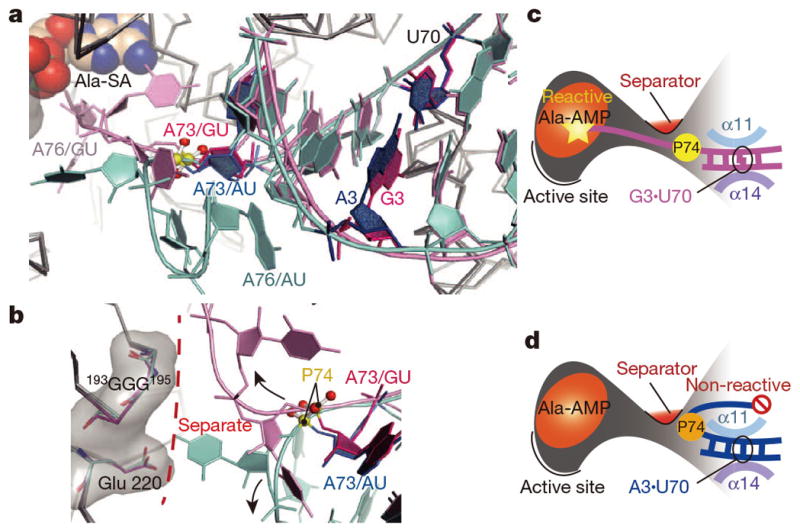
a, The acceptor arms of tRNAAla/GU (pink) and tRNAAla/AU (cyan), with A73 (red) and G3•U70/A3•U70 (magenta/blue) highlighted. b, Route separation for the CCA region, with the turning point at P74, by the separator 193GGG195 and Glu220 (sticks with translucent surface). c, d, The reactive state of tRNAAla/GU (c) and the non-reactive state of tRNAAla/AU (d).
The reactive and non-reactive routes for the CCA region are separated by the 193GGG195 region and Glu 220, and the CCA region is sterically hindered from shuttling between the two routes (Fig. 3b–d). Consequently, the structure formed by 193GGG195 and Glu 220 serves as “the route separator”. This mechanism of tRNAAla selection would apply to tRNAAla variants with Watson-Crick pairs replacing G3•U70 (tRNAAla/WC). In this context, Asp 174 in E. coli AlaRS is spatially near the site corresponding to the 193GGG195 region and Glu 220 in A. fulgidus AlaRS (Extended Fig. 5j, k), and its Gly substitution reportedly causes mis-aminoacylation of the G3•C70 variant of a model RNA36.
Figure 5. Alanylation assays of tRNAAla with AlaRS.
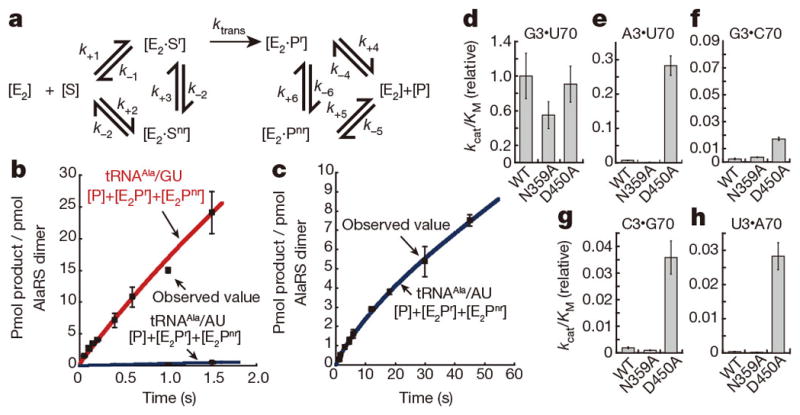
a, Reaction scheme including the non-reactive state. b, c, Alanylation of tRNAAla/GU (red) and tRNAAla/AU (blue), with curves of [E2·Pr]+[E2·Pnr]+[P] simulated according to a. d–h, The kcat/KM values of the wild-type, N359A, and D450A AlaRSs for tRNAAla/GU (d) and tRNAAla/WC (e–h). Error bars: standard deviations of the mean of three replicates (b–h).
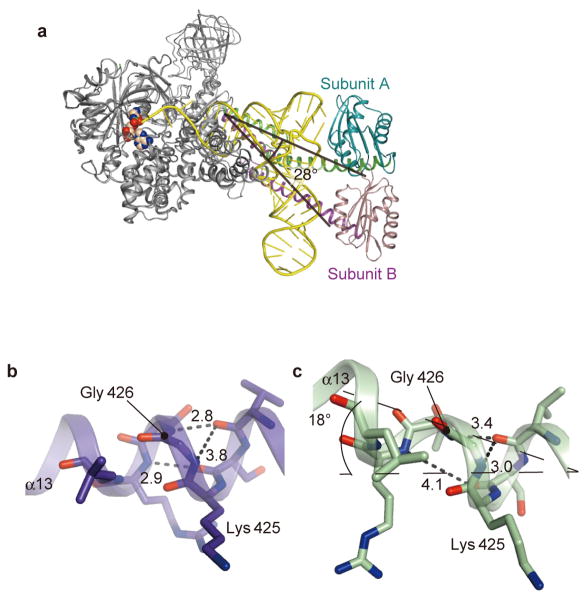
The geometrical difference between G3•U70 and A3•U70 is smaller than the range of the positional flexibility of the single-stranded CCA region. Consequently, the tRNA selection mechanism directing the CCA region into either the reactive or non-reactive route requires precise positioning of the acceptor stem. The main mechanism is the firm clamping of the major and minor grooves of the acceptor stem by the Mid1 and Mid2 subdomains of AlaRS. First, both the major and minor grooves of the G1•C72, G2•C71, G3•U70, and C4•G69 pairs interact with α11 of Mid1 and α14 of Mid2, respectively (Fig. 4a), while more interactions are formed with the 3′-strand (residues 69–73) than the 5′-strand of the acceptor stem (Extended Data Fig. 3a, b). In the tRNA-free subunit B, α13 connecting Mid1 and Mid2 is kinked by ~18°, due to the 310-like conformation around the conserved Gly 426 near the subdomain boundary (Fig. 4c and Extended Data Fig. 1b, c). In contrast, α13 is straight in the tRNA-bound subunit A, due to the change around Gly 426 from a 310-like to α-helical conformation (Fig. 4c and Extended Data Fig. 1b, c). Accordingly, α14 and α15 of Mid2 are reoriented upon tRNA binding. This Mid2 reorientation enables Mid1 and Mid2 to snugly clamp the acceptor-stem base pairs (Fig. 4a). In contrast, Mid1 α11 and Mid2 α14 directly contact each other in the tRNA-free subunit B (Fig. 4b).
Figure 4. The tRNAAla acceptor stem is widened by the clamp.
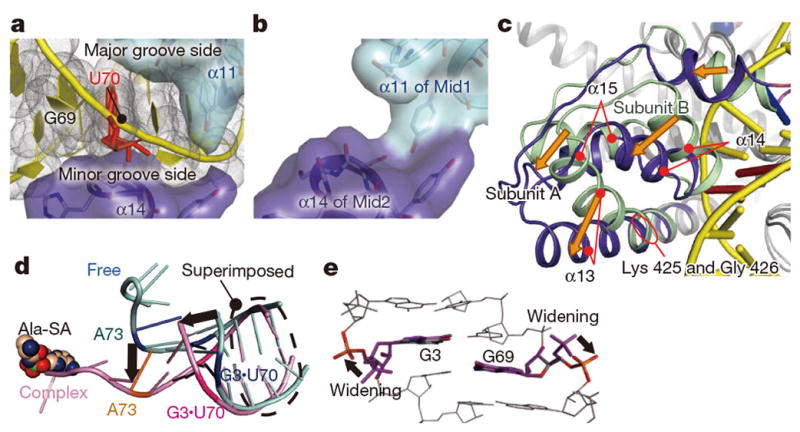
a, The tRNAAla acceptor arm trapped by the clamp (α11 and α14). b, The clamp without tRNAAla. c, Mid2 reorientation. Superimposition on Mid1. d, The AlaRS-induced widening of the tRNAAla acceptor stem (pink), as compared to the model RNA (PDB ID: 1IKD) (cyan). e, Conformational changes of G3 and G69 relative to the A-form RNAs.
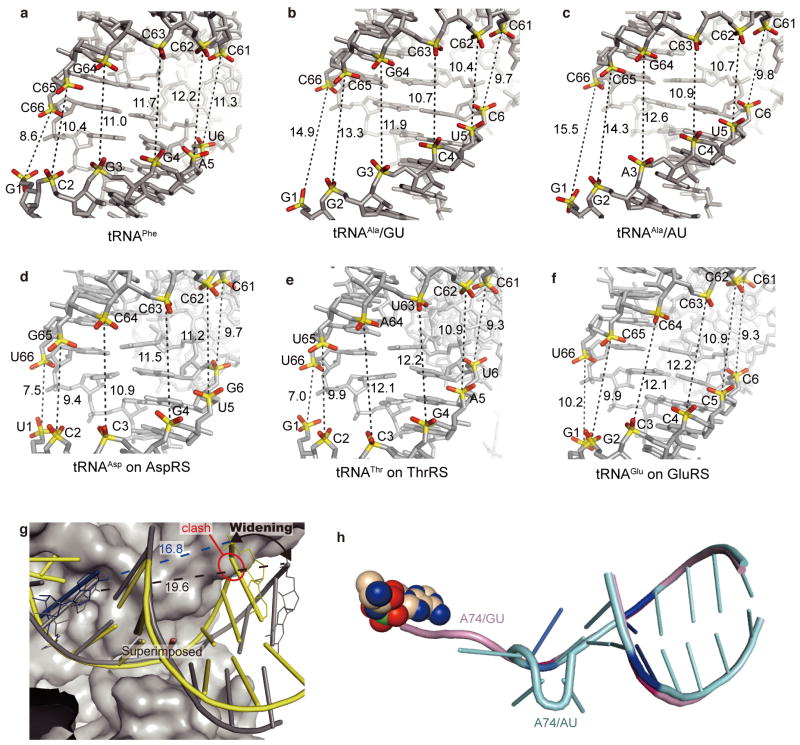
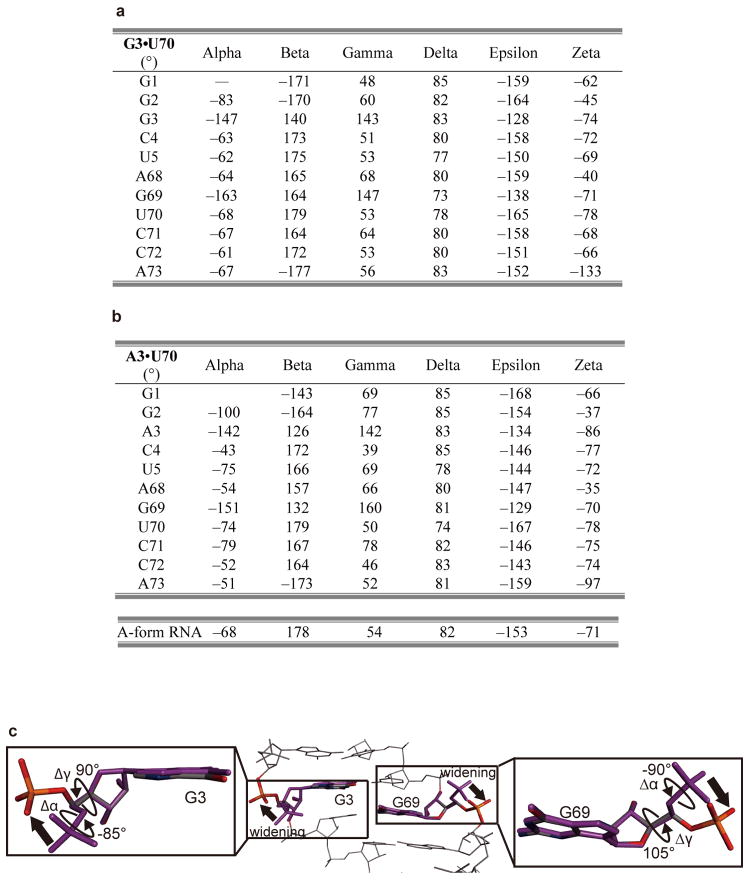
Concomitantly, the major groove of the acceptor stem is widened by the clamp, as compared with the model RNA hairpin structures34,35 (Fig. 4d) and the canonical tRNA structures (Extended Data Fig. 6a–f). If the major groove widening did not occur, then the major groove recognition would cause a serious clash of the G68 phosphate group with the editing core subdomain (Extended Data Fig. 6g), and the CCA region could not enter the reactive (or non-reactive) route selectively (Fig. 4d). Specifically, the conformations of the ribose-phosphate backbones of G3 and G69 are both changed from the gauche−/gauche+ form, typical of the A-form RNA duplex, to the trans/trans form (Fig. 4e and Extended Data Fig. 7). The trans/trans conformation has been reported for G3, but not G69, in the NMR structure of the model RNA34, whereas the crystal structure of a similar model RNA showed the gauche−/gauche+ form for G335. On the other hand, in the non-reactive AlaRS•tRNAAla/AU complex, the major-groove of the tRNAAla/AU acceptor stem is widened in a similar manner to that of tRNAAla/GU (Extended Data Figs. 6h, 7a–c), with almost the same AlaRS conformation as in the wild-type AlaRS•tRNAAla/GU complex.
In addition to the Mid2 subdomain, the C-terminal domain is remarkably reoriented between the tRNA-bound and -free subunits (Fig. 4c and Extended Data Fig. 1). The globular subdomain of the C-terminal domain thereby interacts with the outer corner of the L-shaped tRNA molecule. Actually, the conserved Gly-rich segment (870KGSGGGR876)28,37 stacks on the G19•C56 tertiary base pair connecting the D and T loops (Extended Data Fig. 3c). Thus, the C-terminal globular subdomain (“backrest”) snugly holds the back of tRNAAla, while the tRNAAla acceptor stem sits on the tRNA recognition domain (“seat”). Furthermore, the long linker (residues 485–500) connecting the Mid2 and β-barrel subdomains (“safety belt”) runs over the tRNAAla acceptor stem, near the minor groove side of G2•C71 (Fig. 1e). These features of the tRNA-embedding mechanism distinguish AlaRS from the other aaRSs, which bind tRNA on their convex surfaces (Fig. 1e and Extended Data Fig. 2).
In the non-reactive conformation of the CCA region of tRNAAla/AU, A76 is likely to interact with A73 and the linker, which might stabilize the non-reactive orientation. On the other hand, tRNAAla/AU in the non-reactive conformation hardly seems to dissociate from AlaRS, since the bulky CCA region should prevent the acceptor stem from going through the tunnel between the tRNA recognition domain and the linker (Fig. 2d and Extended Data Fig. 5e, f).
These unique mechanisms for holding the tRNA seem to be required to place the base pair at position 3•70 in the precise recognition site.
Selection of tRNAAla on the kcat level
By pre-steady state kinetics analyses using a quench flow apparatus, the kcat values of the A. fulgidus AlaRS dimer for tRNAAla/GU and tRNAAla/AU were determined to be 14.4 ± 0.2 and 0.14 ± 0.02 s−1, respectively. Therefore, the strict tRNA discrimination is achieved by the 100-fold difference in the kcat value. Next, we performed single turn-over analyses of the alanylation of tRNAAla/GU and tRNAAla/AU. Here, kchem is the rate of the overall product formation through two steps, and is almost equal to the rate of the aminoacyl transfer to tRNA, ktrans, in the case of E. coli AlaRS38,39. The kchem (s−1) and Kd (μM) values of the A. fulgidus AlaRS dimer for tRNAAla/AU (24 ± 6 s−1 and 0.59 ± 0.40 μM, respectively) were only about two-fold smaller and larger, respectively, than those of tRNAAla/GU (52 ± 1 s−1 and 0.34 ± 0.08 μM, respectively), and thus the differences in the kchem and Kd values between the two tRNAs are much smaller than the two-orders of magnitude difference in the kcat values (s−1).
To interpret these observations, a simplified numerical simulation of the pre-steady state reactions of AlaRS for tRNAAla/GU and tRNAAla/AU was performed, on the assumption of a reaction scheme involving the non-reactive state in addition to the reactive state (Fig. 5a). The simulated curves fit the observed data well (Fig. 5b, c), when the reactive state was the major state for tRNAAla/GU, but the amount of the reactive state was much smaller for tRNAAla/AU. Here, product release is what limits Ala-tRNAAla/AU, as it is trapped mostly in the non-reactive state (see Methods). The non-reactive state is most likely to correspond to the present structure of tRNAAla/AU on AlaRS. Therefore, the strict tRNA selection dependent on kcat can be accomplished by this unprecedented mechanism with the reactive and non-reactive states.
Steady-state kinetics experiments (Fig. 5d–h) also revealed that the discrimination of tRNAAla/GU from tRNAAla/WC variants (tRNAAla/GC, tRNAAla/AU, tRNAAla/CG, and tRNAAla/UA) was mainly due to the two orders of magnitude differences in the kcat values (Extended Data Table 2). These drastic distinctions may correspond to the differences in the probability of adopting the reactive orientation of the CCA region. In addition, we tested kcat-level tRNAAla/AU discrimination with human AlaRS. Strikingly, human tRNAAla/AU binds to human AlaRS and competes with tRNAAla/GU. Moreover, even in the presence of excess amounts of tRNAAla/AU, the discrimination against human tRNAAla/AU is ultimately via kcat (Extended Data Fig. 8).
In contrast, the tRNAAla-binding affinity of AlaRS is essentially the same between tRNAAla/GU and tRNAAla/WC, as it is mostly governed by various extensive interactions, such as firmly embedding the tRNA in the huge cavity and snugly clamping the acceptor stem. To examine how the snug clamping contributes to the tRNA selection, we mutated Asn 359 and Asp 450, which directly interact with G3•U70 (Fig. 2a). The kcat/KM value of the N359A mutant for tRNAAla/GU was 1.7-fold lower than that of the wild-type AlaRS. By contrast, the D450A mutation affected neither kcat nor KM (Extended Data Table 2), but remarkably increased the alanylation activity for tRNAAla/WC (Fig. 5d–h and Extended Data Table 2). These enhanced activities of the D450A mutant are ascribed mostly to an increase in the kcat value (Extended Data Table 2). It is likely that the D450A mutant loosened the clamping of the 3•70 pair, and promoted the misorientation of the CCA region of tRNAAla/WC toward the reactive route (Extended Data Fig. 5l).
On the other hand, several mismatch base pairs in place of G3•U70 in an alanine suppressor tRNA reportedly cause aminoacylation by AlaRS in E. coli cells (G•A, C•A, and C•C are more active and U•U, C•U, and A•C are less active), even though their activities are not as high as that of tRNAAla/GU40–42. The effects of these mismatch base pairs may be analogous to those of the D450A mutation, which incorrectly guides the CCA region into the reactive route.
This unprecedented mechanism now provides an answer to the long-standing question of how a small number of nucleotides about 30 Å away from the catalytic site can predominantly determine the tRNA identity, particularly on the kcat level.
METHODS
Preparation and characterization of AlaRS and tRNAAla
The AlaRS from Archaeoglobus fulgidus consists of 906 amino-acid residues (103 kDa), and is composed of the aminoacylation, tRNA-recognition, editing, and dimerization domains. The wild-type, full-length A. fulgidus AlaRS was prepared according to the procedure reported for the AlaRS-ΔC preparation22,28. The expression plasmids for the mutant AlaRS proteins were prepared by site-directed mutagenesis, from the vector bearing the wild-type AlaRS. The proteins were overexpressed in the E. coli strain Rosetta 2 (DE3), and were purified by the same procedure as for the wild-type AlaRS. The yields of the purified proteins obtained from 1 l of cell culture were about 2 mg. The in vitro transcript corresponding to A. fulgidus tRNAAla with the UGC anticodon was prepared by using T7 RNA polymerase. The plasmids for the tRNAAla variants were prepared by site-directed mutagenesis, and the in vitro transcripts were prepared in the same manner as the wild-type tRNAAla transcript. To characterize the purified A. fulgidus AlaRS and tRNAAla, two methods were employed. First, the sedimentation equilibrium analyses gave a molecular mass of 174 kDa, confirming that A. fulgidus AlaRS forms a homodimer. This experiment was performed as reported previously43 with minor modifications, such as the use of 10 mM Tris–HCl buffer (pH 8.0), containing 200 μM zinc acetate and 5 mM β-mercaptoethanol. Second, isothermal titration calorimetry revealed that one AlaRS dimer binds one tRNAAla, with Kd values of 0.12±0.2 and 0.14±0.3 μM at 10 and 20 °C, respectively. The ITC experiments were performed essentially as reported44, except that the solution was 20 mM Tris–HCl buffer (pH 7.5), containing 200 μM zinc acetate, 10 mM MgCl2, and 5 mM β-mercaptoethanol.
Crystallization and data collection
For co-crystallization, the wild-type A. fulgidus AlaRS and the A. fulgidus tRNAAla transcript (either the wild type or the AU variant) were mixed to final concentrations of 90 μM and 120 μM, respectively (molar ratio of 1:1.3), and the solution was incubated at 65 °C for 5 minutes. A 1 μl aliquot of the solution was mixed with 1 μl of reservoir solution, and the mixture was equilibrated by the sitting-drop vapor diffusion technique against 200 μl of the reservoir solution. Crystallization screenings were performed with Crystal Screen, Crystal Screen 2, PEG/Ion Screen, Natrix, MembFac (Hampton Research), and Wizard I and II (Emerald Biosystems), at 20 °C. Polymorphic crystals were obtained with the Wizard I condition No. 6 (20% polyethylene glycol (PEG) 3000 and 100 mM citrate buffer (pH 5.5)). The optimized conditions were 100 mM citrate buffer (pH 6.0), containing 10% PEG 6000, 1 mM CdCl2, and 3% MPD, at 20 °C. Co-crystals were obtained in a few days. The crystals belong to the space group P212121, with unit cell dimensions of a = 99.7 Å, b = 169.8 Å, c = 175.3 Å. For cryo-cooling, the crystals were transferred to a cryo-protectant solution, containing 17% PEG 6000, acetate buffer (pH 4.6), and 20% glycerol. The crystals were then picked up with a nylon loop (Hampton Research), and were flash-cooled with liquid nitrogen. Diffraction datasets of the wild-type and tRNAAla/AU-variant complexes were collected at a 1 Å wavelength at −173 °C, at the beamline NW12A in the Photon Factory (Tsukuba, Japan) and the beamlines BL32XU and BL41XU in SPring-8 (Harima, Japan), respectively. The data were processed by the CCP4 suite45, and were indexed, integrated, and scaled with the HKL-2000 program suite46.
Structure determination and refinement
The structure of the full-length A. fulgidus AlaRS in complex with the wild-type A. fulgidus tRNAAla transcript was solved by molecular replacement with the PHASER program47, by using the coordinates of the N- and C- terminal fragments of A. fulgidus AlaRS [AlaRS-ΔC (2ZTG) and AlaRS-C (2ZVF), respectively]28 and S. cerevisiae tRNAAsp (1IL2)32. Initially, two AlaRS-ΔC molecules were searched. This solution was composed of a dimeric AlaRS-ΔC molecule in the asymmetric unit. Subsequently, a tRNA molecule was searched, and this solution was composed of one tRNA molecule. Finally, a dimeric AlaRS-C molecule was searched, and the initial phase of the wild-type AlaRS•tRNAAla complex structure was determined. The structure was refined by iterative cycles of manual model fitting with the Coot program48,49, followed by positional and group B-factor refinements with the CNS and Phenix programs50–52. For the refinement, 5% of the data were randomly chosen and set aside for cross validation. The refinement converged to R and Rfree factors of 25.0% and 28.9%, respectively (Extended Data Table 1). In the Ramachandran plot, 95.2%, 4.4%, and 0.4% of the residues fell in the favored, allowed, and outlier regions, respectively. This is not only the first AlaRS•tRNA complex structure but also the first full-length AlaRS structure. Next, the structure of A. fulgidus AlaRS in complex with the variant tRNAAla/AU was solved by molecular replacement with the PHASER program47, by using the coordinates of the wild-type AlaRS•tRNAAla complex. The structure was refined by iterative cycles of manual model fitting with the Coot program48,49, followed by positional and group B-factor refinements with the CNS and Phenix programs50–52. For the refinement, 5% of the data were randomly chosen and set aside for cross validation. The refinement converged to R and Rfree factors of 24.1% and 27.7%, respectively (Extended Data Table 1). In the Ramachandran plot, 95.7%, 4.2%, and 0.17% of the residues fell in the favored, allowed, and outlier regions, respectively.
Alanylation assay
Alanylation reactions of tRNAAla (the wild-type tRNAAla/GU and the variant tRNAAla/WC) were performed at 65 °C in 60 mM Tris–HCl buffer (pH 7.5), containing 10 mM MgCl2, 3 mM ATP, 40 μM alanine, 2 μM [3H]-alanine, and various concentrations of the AlaRS protein and tRNAAla. Aliquots were removed at 30 seconds, 1 minute, and 1.5 minutes, and were quenched on filter papers (Whatman, 3 mm) equilibrated with 5% trichloroacetic acid (TCA). The filters were washed three times with 5% ice-cold TCA and twice with 80% ethanol. The radioactivities of the precipitates were quantitated by scintillation counting. Under the same conditions used for tRNAAla/GU and tRNAAla/WC, alanylation reactions of tRNAVal and its variant with G3•U70 transplantation (tRNAVal/GU) were performed (the kcat values of 0.004 and 0.338 s−1, respectively), which confirmed that the identity determinant of A. fulgidus tRNAAla is G3•U70.
Rapid chemical-quench kinetics of Ala-tRNAAla-formation
Pre-steady state kinetics
Ala-tRNAAla/GU and Ala-tRNAAla/AU formation by the wild-type and D450A AlaRSs was analyzed under pre-steady state conditions at 60 °C in 100 mM Tris–HCl buffer (pH 7.5), containing 10 mM MgCl2, 3 mM ATP, 50 μM alanine, 3–5 μM [3H]-alanine, 0.2 μM AlaRS protein and 10 μM tRNAAla, by using a rapid chemical-quench flow apparatus (RQF-3, KinTek Instruments). Solutions of AlaRSs and tRNAs (15 μl each) in the sample loops were mixed rapidly in a single reaction loop. The reactions were quenched in the rapid quench instrument with 55 μL of 0.5 M NaOAc (pH 5.0), and precipitated on the filters (Whatman, 3 mm) with 5% ice-cold TCA. The filters were washed four times with 5% ice-cold TCA and twice with 80% ethanol. The radioactivities of the precipitates were quantitated by scintillation counting. Data were fitted to the equation , where k1 is the apparent rate constant of the exponential phase, and k2 is the rate constant of the steady-state phase.
Single turnover kinetics
Single turnover reactions were performed under the same conditions as those for the pre-steady state kinetics, except for the AlaRS and tRNAAla concentrations. The AlaRS concentrations were maintained in at least two-fold molar excess over tRNAAla, in the reactions where the tRNAAla concentrations were less than 2 μM. The amounts of Ala-tRNA were plotted versus time and were fitted to the first-order exponential equation , where y0 is the y intercept, A is a scaling constant, kapp is the apparent rate constant, and t is time in seconds. The concentration-dependence of single turnover reactions for AlaRS was determined by titrating the AlaRS concentration. To determine the Kd for tRNAAla, the reactions included 2–8 μM concentrations of the AlaRS protein. The rate constants (kapp) versus the substrate concentrations were fitted to the hyperbolic binding curve to calculate the Kd and kchem values, using the KaleidaGraph software. By using three independently obtained sets of Kd and kchem values, their standard deviations were estimated.
Simulation of the aminoacylation reactions
The aminoacylation reactions were simulated based on the assumption of a reaction scheme involving the reactive and non-reactive states (Fig. 5a), by numerically solving the differential equations. Here, the ktrans values were taken from the kchem values determined by the single turnover kinetics experiments, described above. All of the other rate constants (from k+1 to k−6) were fitted as adjustable parameters, where k+4, k−4, k+5, k−5, k+6, and k−6 were postulated to be equal to k−1, k+1, k−2, k+2, k+3, and k−3, respectively, to minimize the difference between the simulated and observed amounts of produced aminoacyl-tRNA (Fig. 5b, c) and between the value following the equation and the observed value. Finally, the k+1, k−1, k+2, k−2, k+3, k−3, k+4, k−4, k+5, k−5, k+6, k−6, and ktrans values for tRNAAla/GU were determined to be 85, 30, 0.1, 0.074, 0.693, 0.01, 30, 85, 0.074, 0.1, 0.577, 0.01, and 52, respectively, and those for tRNAAla/AU were 1.0, 10, 20.0, 0.074, 0.693, 12.73, 10.0, 1.0, 0.074, 20.0, 0.693, 12.729, and 25, respectively. The simulated curves for aminoacylation are shown in Fig. 5b, c, together with the observed values.
Gly-tRNAAla-deacylation assay
[14C]-Gly-tRNAAla was prepared by incubating reaction mixtures in 30 mM Tris–HCl buffer (pH 7.5), containing 10 mM MgCl2, 3 mM ATP, 5 μM A. fulgidus tRNAAla, 80 μM [14C]-glycine (300 cpm pmol−1) and 5 μM editing-deficient C703A mutant of A. fulgidus AlaRS, at 65 °C for 10 min. The [14C]-Gly-tRNAs thus produced were purified by the acidic phenol chloroform method and ethanol precipitation. The [14C]-Gly-tRNAAla deacylation reactions were performed at 55 °C in 30 mM Tris–HCl buffer (pH 7.5), containing 150 mM KCl, 10 mM MgCl2, 4 μM [14C]-aminoacyl-tRNA, and 3 μM AlaRS enzyme. Aliquots were removed at 2, 4, and 8 minutes. The reactions were quenched and washed according to the same procedure used for the analysis of the Ala-tRNAAla-formation reactions.
Extended Data
Extended Data Figure 1. Conformational differences between the tRNA-bound and -free subunits of A. fulgidus AlaRS.
a, Rotation of the C-terminal domain. The C-terminal domain of subunit A is inclined ~28° toward the outer corner of the L-shaped tRNA, relative to subunit B, and thus the globular subdomain stacks with the outer corner of the tRNA. b, c, Mid2 reorientation. α13 is straight with tRNAAla (b) and kinked without tRNAAla (c).
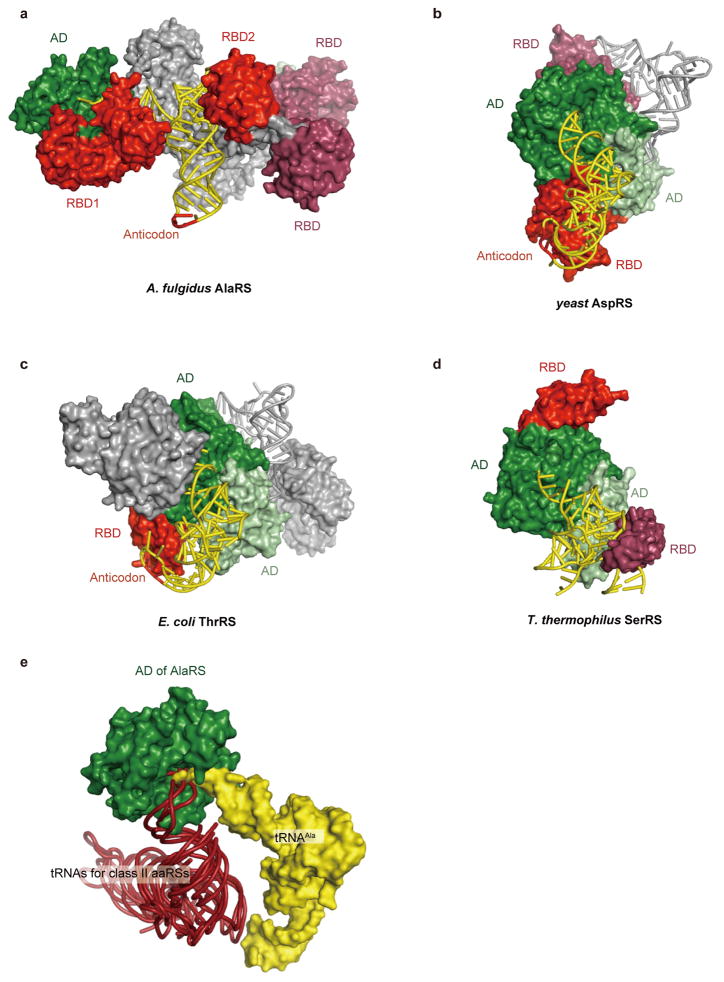
Extended Data Figure 2. The positions of tRNAs relative to the aminoacylation domains of AlaRS and other class II aaRSs depend on the specific spatial arrangements of their tRNA-binding domain(s).
a–d, Class II aaRS dimers bound to tRNAs. A. fulgidus AlaRS, yeast AspRS, E. coli ThrRS, and T. thermophilus SerRS are shown as surface models, with their cognate tRNAs displayed as tube models. For each aaRS, one of the two tRNA-binding aminoacylation domains (ADs) and its bound tRNA are colored green and yellow, respectively, while the other AD and its bound tRNA are colored light green and grey, respectively. In each of the aaRSs, except for A. fulgidus AlaRS, the tRNA-binding domain (RBD) corresponding to the green AD is colored red, and that of the other subunit is colored raspberry. A. fulgidus AlaRS has two RBDs (RBD1 and RBD2), colored in the same manner as the RBDs of the other aaRSs. e, The position of tRNAAla relative to the AD of AlaRS (displayed as yellow and green surface models, respectively) is completely different from the well conserved positions of tRNAAsp, tRNAThr, tRNAPro, tRNASer, tRNAPhe, tRNAPyl, and tRNACys (brown tube models) shown in the same positions relative to the ADs of AspRS32, ThrRS53, ProRS54, SerRS33, PheRS55, PylRS56, and SepRS57, respectively (PDB IDs: 1ASY, 1QF6, 1H4Q, 1SER, 1EIY, 2ZNI and 2DU3, respectively).
Extended Data Figure 3. Summary of tRNA-protein interactions.
a, Schematics of tRNA-protein interactions. The tRNA backbones and bases are colored as indicated. The G3•U70 base pair and the anticodon bases are highlighted by red and pink squares, respectively. The hydrogen bonds between the nucleotides and the AlaRS main chain are indicated with blue arrows. Those between the nucleotides and the AlaRS side chains are depicted with magenta arrows. The stacking interactions are indicated with black dashed lines. b, The tRNAAla interaction with the _11 helix. The α11 helix interacts with the backbone phosphate groups of the 3′ half of the acceptor stem. The side chains of Tyr 364 and Arg 371 hydrogen bond with C72 and C71, respectively. The side chain of Arg 375 hydrogen bonds with U70. c. Interactions between the tRNAAla outer corner region and the globular subdomain. The conserved Gly-rich segment 870KGSGGGR878 stacks with the hinge region of the T- and D-loops. The side chain of Arg 876 stacks and hydrogen bonds with the base and the backbone phosphate group, respectively, of G20. The side chain of Asn 823 hydrogen bonds with the backbone phosphate group of G20. d, The tRNAAla interactions with the _13 helix. The α13 helix interacts with the backbone phosphate groups of the D arm (U12-C13-A14-G15) and that of U70. The side chain of Thr 422 in α13 hydrogen bonds with U70. The side chains of Arg 428 and Arg 432 hydrogen bond with C13. The side chain of Arg 436 hydrogen bonds with A14 and G15.
Extended Data Figure 4. Nucleotide interactions with the conserved residues of AlaRS.
a, b, |Fo–Fc| omit electron density maps contoured at 3σ, superimposed on the refined models of G3•U70 (a) and A3•U70 (b). c–g, The interactions of G1•C72 (c), G2•C71 (d), C4•G69 (e), U5•A68 (f), and A73 (g) with AlaRS. The amino-acid side chains and the nucleotides are shown as gray and beige stick models, respectively. Hydrogen bonds are indicated with dashed lines. h, A73 stacking with G1 and C72. i, Alignments of the AlaRS sequences. A total of 249 AlaRS sequences were initially aligned with the ClustalW program58, and then were manually adjusted based on the structural information. The sequences of the A. fulgidus, P. horikoshii, Homo sapiens, E. coli, and A. aeolicus AlaRSs are shown. The highly-conserved residues are boxed in red. The residues that are involved the acceptor stem interactions in A. fulgidus AlaRS are boxed in blue. Secondary structure information is shown above the A. fulgidus sequence. The full-sequence alignments were reported previously28.
Extended Data Figure 5. The routes of the CCA regions of tRNAAla/GU and tRNAAla/AU.
a–c, tRNAAla/GU. d–f, tRNAAla/AU. tRNA is shown in stick models (a, d), cartoon models (b, e), and surface models (c, f). The |Fo–Fc| omit electron density maps contoured at 3σ are superimposed on the refined models of the CCA regions of tRNAAla/GU (a) and tRNAAla/AU (d). The CCA region of tRNAAla/GU is in the reactive route (b, c), while that of tRNAAla/AU is in the non-reactive route (e, f). The CCA region runs under the linker (b, c, e, f). g, h, Heterologous superimpositions of the |2Fo–Fc| electron density maps contoured at 1σ of tRNAAla/AU (cyan, g) and tRNAAla/GU (pink, h) on the stick models of the structures of the acceptor stems and the CCA regions of tRNAAla/GU (pink, g) and tRNAAla/AU (cyan, h), respectively, which clearly demonstrate the conformational difference between the two tRNAs. i, The |Fo–Fc| omit electron density maps for tRNAAla/AU, contoured at 2.5σ, superimposed on the refined models of the C74s of tRNAAla/GU (pink) and tRNAAla/AU (cyan). j, k, The similarity in the spatial positions between D220/GGG of A. fulgidus AlaRS (j) and E174 of E. coli AlaRS (k), marked in red on the surface models of the aminoacylation and tRNA-recognition domains with the tube models of tRNAAla (the crystal structure and the docking model, respectively). l, Misorientation of the CCA region of tRNAAla/WC on the D450A mutant of AlaRS.
Extended Data Figure 6. Major-groove widening of the tRNAAla acceptor stem.
a, The phosphate–phosphate distances of G1–C66, C2–C65, G3–G64, G4–C63, A5–C62, and U6–C61 in tRNAPhe (refs. 59,60). b, The phosphate–phosphate distances of G1–C66, G2–C65, G3–G64, C4–C63, U5–C62, and C6–C61 in the wild-type tRNAAla (tRNAAla/GU) complexed with AlaRS. The phosphate–phosphate distances between G1–C66, G2–C65, and G3–G64 across the major groove of tRNAAla/GU are extended by 6.3, 2.9, and 0.9 Å, respectively, as compared to those of tRNAPhe. Thus, AlaRS clearly widens the major groove of the acceptor stem of tRNAAla/GU. c, The phosphate–phosphate distances of G1–C66, G2–C65, A3–G64, C4–C63, U5–C62, and C6–C61 in the variant tRNAAla/AU complexed with AlaRS. The major-groove of the tRNAAla/AU acceptor stem is slightly wider than that in the wild-type complex structure. Therefore, the replacement of G3•U70 by A3•U70 does not seem to impair the major-groove widening by AlaRS. d, The phosphate–phosphate distances of U1–U66, C2–G65, C3–C64, G4–C63, U5–C62, and G6–C61 in tRNAAsp bound to AspRS32 (PDB ID: 1ASY). e, The phosphate–phosphate distances of G1–U66, C2–U65, C3–A64, G4–U63, A5–C62, and U6–C61 in tRNAThr bound to ThrRS53 (PDB ID: 1QF6). f, The phosphate–phosphate distances of G1–U66, G2–C65, C3–C64, C4–C63, C5–C62, and C6–C61 in tRNAGlu bound to GluRS61 (PDB ID: 1G59). g, The AlaRS-induced widening of the tRNAAla acceptor stem. Without widening, AlaRS clashes with A67-G68. h, Widening of the acceptor stems of tRNAAla/GU (pink tube models) and tRNAAla/AU (cyan tube models).
Extended Data Figure 7. Torsion angles of nucleotides in the acceptor stems of tRNAAla/GU and tRNAAla/AU.
a, b, Tables for torsion angles of nucleotides in the acceptor stems of tRNAAla/GU (a) and tRNAAla/AU (b). c, Conformational changes of G3 and G69 relative to the A-form RNAs.
Extended Data Figure 8. tRNAAla/AU inhibits aminoacylation of human tRNAAla/GU by human AlaRS.
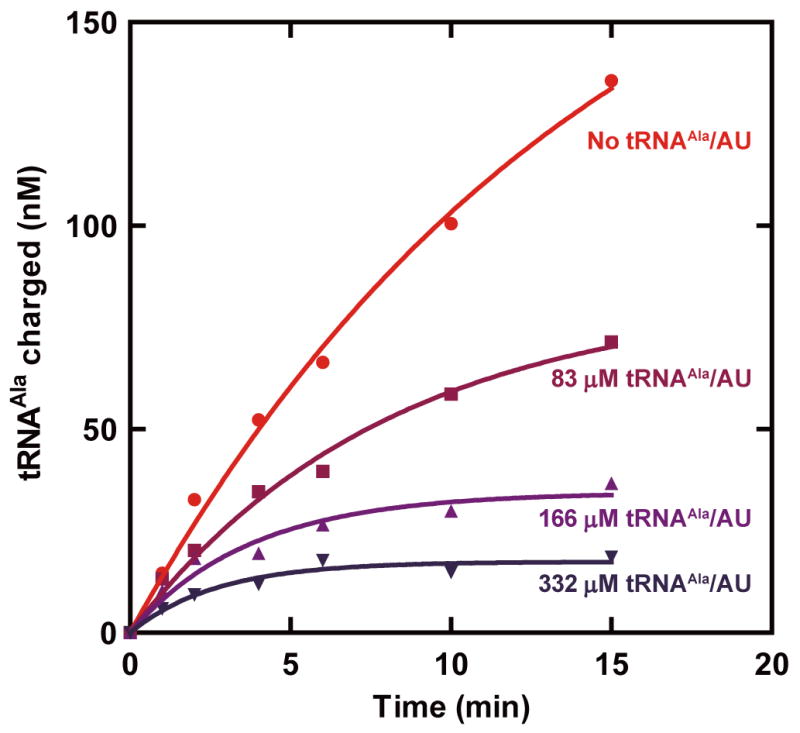
In vitro transcribed human tRNAAla/AU was added to aminoacylation reactions containing 2 _M human tRNAAla and 50 nM human AlaRS, in pH 6.0 reaction buffer at room temperature. The dose-dependent inhibition of aminoacylation of tRNAAla/GU by excess tRNAAla/AU shows that the ultimate discrimination against A3•U70 is via kcat.
Extended Data Table 1.
Data collection and refinement statistics (Molecular replacement).
| AlaRS•tRNAAla/GU | AlaRS•tRNAAla/AU | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 99.7, 169.8, 175.3 | 99.7, 169.9, 176.7 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–3.3 (3.42–3.30)* | 50–3.5 (3.63–3.5) |
| Rsym or Rmerge | 0.141 (0.654) | 0.120 (0.689) |
| I/σI | 11.7 (2.2) | 17.1 (2.5) |
| Completeness (%) | 99.6 (99.3) | 99.9 (100) |
| Redundancy | 5.4 (4.2) | 6.8 (6.3) |
| Refinement | ||
| Resolution (Å) | 50–3.3 | 50–3.5 |
| No. reflections | 45304 | 38766 |
| Rwork/Rfree | 0.250/0.289 | 0.241/0.277 |
| No. atoms | ||
| Protein | 14421 | 14412 |
| RNA | 1609 | 1608 |
| Water | 7 | – |
| Zn ion | – | 2 |
| Mg ion | 3 | – |
| B-factors | ||
| Protein | 100.4 | 122.5 |
| RNA | 66.5 | 103.4 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.004 |
| Bond angles (°) | 0.886 | 0.762 |
Highest resolution shell is shown in parentheses.
Extended Data Table 2.
Kinetics of wild-type and mutant AlaRSs.
| G3•U70 | ||||
|---|---|---|---|---|
| kcat(1/s) | KM (μM) | kcat/KM (1/Ms) | kcat/KM (relative) | |
| WT | 2.6±0.38 | 1.2±0.14 | 2200000±560000 | 1±0.25 |
| N359A | 1.9±0.30 | 1.5±0.05 | 1300000±330000 | 0.59±0.15 |
| D450A | 2.2±0.52 | 1.2±0.37 | 1800000±440000 | 0.81±0.21 |
| SC* | 0.046±0.012 | 1.5±0.31 | 31000±2100 | 0.014±0.001 |
| A3•U70 | ||||
|---|---|---|---|---|
| kcat (1/s) | KM (μM) | kcat/KM (1/Ms) | kcat/KM (relative) | |
| WT | 0.06±0.0030 | 4.1±0.6 | 15000±1600 | 0.0068±0.00073 |
| N359A | 0.010±0.0034 | 3.8±1.4 | 2600±390 | 0.0012±0.00018 |
| D450A | 1.0±0.064 | 1.7±0.11 | 590000±60000 | 0.27±0.027 |
| G3•C70 | ||||
|---|---|---|---|---|
| kcat (1/s) | KM (μM) | kcat/KM (1/Ms) | kcat/KM (relative) | |
| WT | 0.020±0.0044 | 4.3±1.4 | 4700±1000 | 0.0021±0.00045 |
| N359A | 0.036±0.0015 | 4.7±0.22 | 7700±300 | 0.0035±0.00014 |
| D450A | 0.12±0.028 | 3.2±0.75 | 38000±2500 | 0.017±0.0011 |
| U3•A70 | ||||
| kcat (1/s) | KM (μM) | kcat/KM (1/Ms) | kcat/KM (relative) | |
| WT | 0.0050±0.00038 | 6.5±1.6 | 770±170 | 0.00035±0.000077 |
| N359A | 0.0013±0.00036 | 3.5±1.2 | 370±170 | 0.00017±0.000077 |
| D450A | 0.15±0.0082 | 2.6±0.46 | 58000±8400 | 0.026±0.0040 |
| C3•G70 | ||||
|---|---|---|---|---|
| kcat (1/s) | KM (μM) | kcat/KM (1/Ms) | kcat/KM (relative) | |
| WT | 0.017±0.00082 | 4.0±0.45 | 4300±720 | 0.0020±0.00033 |
| N359A | 0.006±0.00066 | 3.1±0.7 | 1900±320 | 0.00086±0.00015 |
| D450A | 0.020±0.019 | 2.7±0.5 | 74000±13000 | 0.034±0.0060 |
The SC or shortcut mutant of AlaRS has the substitution of 450GGG453 for 450DSHG453, which was designed to drastically deform the α14 tip structure.
Acknowledgments
We are grateful to the staffs of the beam line AR-NW12A at the Photon Factory and the beam lines BL32XU and BL41XU at SPring-8 for their assistance during data collection. This work was supported by the Targeted Proteins Research Program, and the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. This work was also supported by a Grant-in-Aid for Scientific Research (No. 20247008) from the Japan Society for the Promotion of Science and MEXT, and the Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from MEXT.
Footnotes
Author contributions M. N., S. S. and S. Y. designed the research. M. N. and S. S. performed the structural analysis. M. N., Y. E. C., M. G., X.-L. Y., H. G., and Y.-M. H. performed the enzyme kinetics analyses. M. N., S. S., P. S., and S. Y. wrote the paper. All authors discussed the results and commented on the manuscript.
The coordinates and structure factors for AlaRS complexed with its cognate tRNAAla with G3•U70 and a variant with A3•U70 have been deposited in the Protein Data Bank (PDB) under accession codes 3WQY and 3WQZ, respectively.
The authors declare no competing financial interests.
References
- 1.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–35. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beuning PJ, Musier-Forsyth K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999;52:1–28. doi: 10.1002/(SICI)1097-0282(1999)52:1<1::AID-BIP1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 3.Pütz J, Puglisi JD, Florentz C, Giegé R. Additive, cooperative and anti-cooperative effects between identity nucleotides of a tRNA. EMBO J. 1993;12:2949–57. doi: 10.1002/j.1460-2075.1993.tb05957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weygand-Duraševic I, Rogers MJ, Söll D. Connecting anticodon recognition with the active site of Escherichia coli glutaminyl-tRNA synthetase. J Mol Biol. 1994;240:111–8. doi: 10.1006/jmbi.1994.1425. [DOI] [PubMed] [Google Scholar]
- 5.Rogers MJ, Adachi T, Inokuchi H, Söll D. Functional communication in the recognition of tRNA by Escherichia coli glutaminyl-tRNA synthetase. Proc Natl Acad Sci USA. 1994;91:291–5. doi: 10.1073/pnas.91.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uter NT, Perona JJ. Long-range intramolecular signaling in a tRNA synthetase complex revealed by pre-steady-state kinetics. Proc Natl Acad Sci USA. 2004;101:14396–401. doi: 10.1073/pnas.0404017101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou YM, Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–5. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 8.McClain WH, Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science. 1988;240:793–6. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 9.de Duve C. Transfer RNAs: the second genetic code. Nature. 1988;333:117–8. doi: 10.1038/333117a0. [DOI] [PubMed] [Google Scholar]
- 10.Park SJ, Hou YM, Schimmel P. A single base pair affects binding and catalytic parameters in the molecular recognition of a transfer RNA. Biochemistry. 1989;28:2740–6. doi: 10.1021/bi00432a056. [DOI] [PubMed] [Google Scholar]
- 11.Hou YM, Schimmel P. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry. 1989;28:6800–4. doi: 10.1021/bi00443a003. [DOI] [PubMed] [Google Scholar]
- 12.Shi JP, Francklyn C, Hill K, Schimmel P. A nucleotide that enhances the charging of RNA minihelix sequence variants with alanine. Biochemistry. 1990;29:3621–6. doi: 10.1021/bi00467a005. [DOI] [PubMed] [Google Scholar]
- 13.Beuning PJ, Gulotta M, Musier-Forsyth K. Atomic Group “Mutagenesis” Reveals Major Groove Fine Interactions of a tRNA Synthetase with an RNA Helix. J Am Chem Soc. 1997;119:8397–8402. [Google Scholar]
- 14.Musier-Forsyth K, Schimmel P. Functional contacts of a transfer RNA synthetase with 2′-hydroxyl groups in the RNA minor groove. Nature. 1992;357:513–5. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- 15.Beuning PJ, et al. Efficient aminoacylation of the tRNAAla acceptor stem: dependence on the 2:71 base pair. RNA. 2002;8:659–70. doi: 10.1017/s1355838202020277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francklyn C, Shi JP, Schimmel P. Overlapping nucleotide determinants for specific aminoacylation of RNA microhelices. Science. 1992;255:1121–5. doi: 10.1126/science.1546312. [DOI] [PubMed] [Google Scholar]
- 17.Francklyn C, Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989;337:478–81. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 18.Varani G, McClain WH. The G•U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musier-Forsyth K, et al. Specificity for aminoacylation of an RNA helix: an unpaired, exocyclic amino group in the minor groove. Science. 1991;253:784–6. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- 20.Chang KY, Varani G, Bhattacharya S, Choi H, McClain WH. Correlation of deformability at a tRNA recognition site and aminoacylation specificity. Proc Natl Acad Sci U S A. 1999;96:11764–9. doi: 10.1073/pnas.96.21.11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueda H, et al. X-ray crystallographic conformational study of 5′-O-[N-(L-alanyl)-sulfamoyl]adenosine, a substrate analogue for alanyl-tRNA synthetase. Biochim Biophys Acta. 1991;1080:126–34. doi: 10.1016/0167-4838(91)90138-p. [DOI] [PubMed] [Google Scholar]
- 22.Fukunaga R, Yokoyama S. Crystallization and preliminary X-ray crystallographic study of alanyl-tRNA synthetase from the archaeon Archaeoglobus fulgidus. Acta Crystallogr F. 2007;63:224–8. doi: 10.1107/S1744309107006264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dignam JD, et al. Allosteric interaction of nucleotides and tRNAala with E. coli alanyl-tRNA synthetase. Biochemistry. 2011;50:9886–900. doi: 10.1021/bi2012004. [DOI] [PubMed] [Google Scholar]
- 24.Jasin M, Regan L, Schimmel P. Modular arrangement of functional domains along the sequence of an aminoacyl tRNA synthetase. Nature. 1983;306:441–7. doi: 10.1038/306441a0. [DOI] [PubMed] [Google Scholar]
- 25.Beebe K, Ribas De Pouplana L, Schimmel P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 2003;22:668–75. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swairjo MA, et al. Alanyl-tRNA synthetase crystal structure and design for acceptor-stem recognition. Mol Cell. 2004;13:829–41. doi: 10.1016/s1097-2765(04)00126-1. [DOI] [PubMed] [Google Scholar]
- 27.Guo M, et al. Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma. Nature. 2009;462:808–12. doi: 10.1038/nature08612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naganuma M, Sekine S, Fukunaga R, Yokoyama S. Unique protein architecture of alanyl-tRNA synthetase for aminoacylation, editing, and dimerization. Proc Natl Acad Sci USA. 2009;106:8489–94. doi: 10.1073/pnas.0901572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokabe M, et al. The structure of alanyl-tRNA synthetase with editing domain. Proc Natl Acad Sci USA. 2009;106:11028–33. doi: 10.1073/pnas.0904645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–6. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 31.Cusack S. Eleven down and nine to go. Nature Struct Biol. 1995;2:824–31. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 32.Ruff M, et al. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science. 1991;252:1682–9. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- 33.Biou V, Yaremchuk A, Tukalo M, Cusack S. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science. 1994;263:1404–10. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 34.Ramos A, Varani G. Structure of the acceptor stem of Escherichia coli tRNAAla: role of the G3·U70 base pair in synthetase recognition. Nucleic Acids Res. 1997;25:2083–90. doi: 10.1093/nar/25.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller U, Schübel H, Sprinzl M, Heinemann U. Crystal structure of acceptor stem of tRNAAla from Escherichia coli shows unique G•U wobble base pair at 1.16 Å resolution. RNA. 1999;5:670–7. doi: 10.1017/s1355838299982304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller WT, Hou YM, Schimmel P. Mutant aminoacyl-tRNA synthetase that compensates for a mutation in the major identity determinant of its tRNA. Biochemistry. 1991;30:2635–41. doi: 10.1021/bi00224a011. [DOI] [PubMed] [Google Scholar]
- 37.Guo M, et al. The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science. 2009;325:744–7. doi: 10.1126/science.1174343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang CM, Perona JJ, Ryu K, Francklyn C, Hou YM. Distinct kinetic mechanisms of the two classes of Aminoacyl-tRNA synthetases. J Mol Biol. 2006;361:300–11. doi: 10.1016/j.jmb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 39.Guth E, Connolly SH, Bovee M, Francklyn CS. A substrate-assisted concerted mechanism for aminoacylation by a class II aminoacyl-tRNA synthetase. Biochemistry. 2005;44:3785–94. doi: 10.1021/bi047923h. [DOI] [PubMed] [Google Scholar]
- 40.McClain WH, Chen YM, Foss K, Schneider J. Association of transfer RNA acceptor identity with a helical irregularity. Science. 1988;242:1681–4. doi: 10.1126/science.2462282. [DOI] [PubMed] [Google Scholar]
- 41.Gabriel K, Schneider J, McClain WH. Functional evidence for indirect recognition of G•U in tRNAAla by alanyl-tRNA synthetase. Science. 1996;271:195–7. doi: 10.1126/science.271.5246.195. [DOI] [PubMed] [Google Scholar]
- 42.McClain WH, Jou YY, Bhattacharya S, Gabriel K, Schneider J. The reliability of in vivo structure-function analysis of tRNA aminoacylation. J Mol Biol. 1999;290:391–409. doi: 10.1006/jmbi.1999.2884. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa H, et al. Crystallographic and mutational studies on the tRNA thiouridine synthetase TtuA. Proteins: Structure, Function, and Bioinformatics. 2013;81:1232–1244. doi: 10.1002/prot.24273. [DOI] [PubMed] [Google Scholar]
- 44.Yamashita S, et al. Structures of the first and second double-stranded RNA-binding domains of human TAR RNA-binding protein. Protein Science. 2011;20:118–130. doi: 10.1002/pro.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collaborative Computational Project No. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 51.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–33. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 53.Sankaranarayanan R, et al. The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–81. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 54.Yaremchuk A, Tukalo M, Grøtli M, Cusack S. A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: comparison with histidyl-tRNA synthetase. J Mol Biol. 2001;309:989–1002. doi: 10.1006/jmbi.2001.4712. [DOI] [PubMed] [Google Scholar]
- 55.Goldgur Y, et al. The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNAPhe. Structure. 1997;5:59–68. doi: 10.1016/s0969-2126(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 56.Nozawa K, et al. Pyrrolysyl-tRNA synthetase-tRNAPyl structure reveals the molecular basis of orthogonality. Nature. 2009;457:1163–7. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukunaga R, Yokoyama S. Structural insights into the first step of RNA-dependent cysteine biosynthesis in archaea. Nat Struct Mol Biol. 2007;14:272–9. doi: 10.1038/nsmb1219. [DOI] [PubMed] [Google Scholar]
- 58.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robertus JD, et al. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature. 1974;250:546–51. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 60.Kim SH, et al. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–40. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- 61.Sekine S, et al. ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. EMBO J. 2003;22:676–88. doi: 10.1093/emboj/cdg053. [DOI] [PMC free article] [PubMed] [Google Scholar]