Abstract
Multiple yeast prions have been identified that result from the structural conversion of proteins into a self-propagating amyloid form. Amyloid-based prion activity in yeast requires a series of discrete steps. First, the prion protein must form an amyloid nucleus that can recruit and structurally convert additional soluble proteins. Subsequently, maintenance of the prion during cell division requires fragmentation of these aggregates to create new heritable propagons. For the Saccharomyces cerevisiae prion protein Sup35, these different activities are encoded by different regions of the Sup35 prion domain. An N-terminal glutamine/asparagine-rich nucleation domain is required for nucleation and fiber growth, while an adjacent oligopeptide repeat domain is largely dispensable for prion nucleation and fiber growth but is required for chaperone-dependent prion maintenance. Although prion activity of glutamine/asparagine-rich proteins is predominantly determined by amino acid composition, the nucleation and oligopeptide repeat domains of Sup35 have distinct compositional requirements. Here, we quantitatively define these compositional requirements in vivo. We show that aromatic residues strongly promote both prion formation and chaperone-dependent prion maintenance. In contrast, nonaromatic hydrophobic residues strongly promote prion formation but inhibit prion propagation. These results provide insight into why some aggregation-prone proteins are unable to propagate as prions.
INTRODUCTION
Misfolding of a wide range of proteins leads to formation of amyloid fibrils, which are ordered, β-sheet-rich protein aggregates. Many human diseases are associated with the formation of amyloid fibrils, including Alzheimer's disease, type II diabetes, and the transmissible spongiform encephalopathies (TSEs) (1). However, only a small subset of amyloids are infectious (called prions), including the causative agents of TSEs in mammals (2–4) and [URE3], [PSI+], [PIN+], and others in Saccharomyces cerevisiae (5–9).
Most of the known yeast prion proteins contain glutamine/asparagine (Q/N)-rich domains that drive amyloid formation. Q/N-rich domains are found in 1 to 4% of the proteins in most eukaryotic proteomes (10), but very few of these proteins have been shown to undergo amyloid structural conversion. Bioinformatics screens for prions in yeast have had some notable successes (reviewed in reference 11); however, despite advances in predicting which Q/N-rich domains may turn out to be bona fide prions (12, 13), predictions remain imperfect.
A well-studied model prion from yeast (S. cerevisiae) is [PSI+], the prion form of the translational terminator protein Sup35 (5). Like other yeast prion proteins, Sup35 is modular, as it contains a distinct prion-forming domain (PFD), middle domain (M), and C-terminal domain (C) (Fig. 1A) (14–17). The PFD (amino acids 1 to 114) drives the conversion of Sup35 into its amyloid form (15), the charged M domain has no known function other than its ability to stabilize [PSI+] fibers, and the C domain is an essential component responsible for translational termination (14, 17).
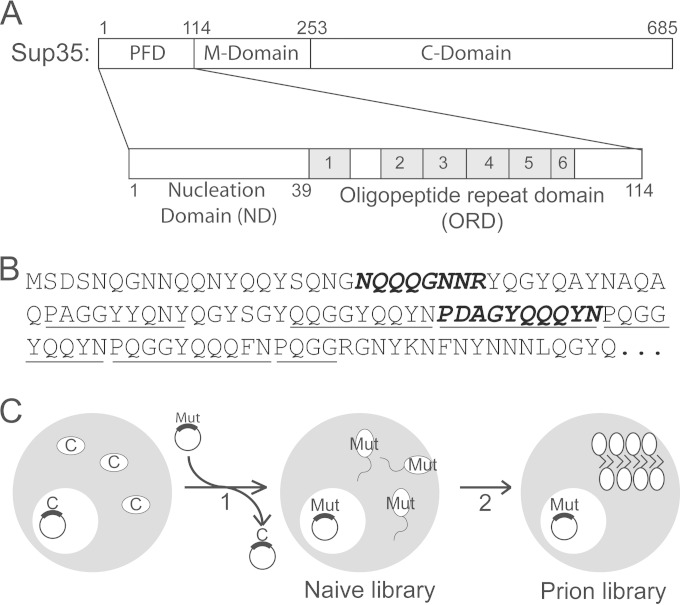
FIG 1.
Prion formation library experiment. (A) Schematic of Sup35. The PFD is enlarged below, showing the ND and ORD. (B) Sequence of Sup35. The oligopeptide repeats are underlined. The region of the ND and ORD targeted for mutagenesis are in bold italics. (C) Experimental scheme for prion formation library experiments. [psi–] cells in which Sup35C was expressed from a URA3 plasmid as the sole copy of Sup35 were transformed with a randomly mutated version of Sup35 and then selected for loss of the wild-type plasmid (step 1). Cells were screened to remove clones in which the mutant Sup35 had compromised activity, and randomly selected clones were sequenced to generate the naive library. The library was then screened for clones that could form and propagate prions (step 2).
Prion formation by Sup35 is driven primarily by the amino acid composition of the PFD (18). We previously used a quantitative mutagenesis method to determine the prion propensity of each amino acid in the context of Q/N-rich PFDs (13). Briefly, an 8-amino-acid segment in the middle of a scrambled version of the Sup35 PFD was replaced with a random sequence to generate a library of mutants. This library was then screened for the subset of mutants that maintained the ability to form and propagate prions. We then derived prion-propensity scores for each amino acid by comparing the frequency of occurrence of each amino acid among the prion-forming sequences to their frequency of occurrence in the starting library. These prion propensity values were used to develop PAPA (prion aggregation prediction algorithm), a prediction algorithm capable of accurately distinguishing between Q/N-rich domains with and without prion activity (13, 19, 20).
Although PAPA represents a significant advance in prion prediction, it is far from perfect. One likely problem is that there are multiple distinct steps required for prion activity. Specifically, prion formation requires that a protein be able to both form prion aggregates and add onto these aggregates; additionally, prion propagation to daughter cells during multiple rounds of cell division (also referred to as prion maintenance) requires that the aggregates be fragmented to create new independently segregating prion seeds to offset dilution by cell division (21). Each of these steps may have distinct amino acid sequence requirements, yet PAPA uses only a single prion propensity score for each amino acid. Making better predictions of prion propensity requires a better understanding of how amino acid composition separately affects prion formation and maintenance.
Sup35 is an ideal substrate for examining these compositional requirements. Unlike the scrambled version of Sup35 used for the initial library experiments, the wild-type Sup35 PFD has two distinct subdomains with overlapping but separate functions (Fig. 1A). The N-terminal nucleation domain (ND; amino acids 1 to 39) is highly Q/N rich and is primarily responsible for nucleation and growth of prion fibers (16, 22). The remaining portion of the PFD (amino acids 40 to 114) has been implicated in prion maintenance and contains an oligopeptide repeat domain (ORD) consisting of 5.5 imperfect repeats with the consensus sequence (P/Q)QGGYQ(Q/S)YN (16, 23–25). This separation of prion formation from prion maintenance potentially allows for dissection of how amino acid composition separately affects each activity. Importantly, the ND and the ORD have distinct compositional requirements for their respective functions (26).
The functional separation between the ND and ORD is not absolute (13, 22, 27). For example, both the ND and the first two repeats of the ORD are required for efficient de novo aggregation (22), and tyrosines in the ORD have been implicated in the early steps of prion nucleation (27). Nevertheless, significant evidence supports a role for the ORD in prion maintenance (22, 26). Removal of all or part of the ORD (14, 22, 23, 25) or replacement of the ORD with a random sequence (28) destabilizes [PSI+]. Such mutations appear to reduce prion aggregate fragmentation, resulting in larger aggregates that are frequently lost as a result of imperfect segregation of aggregates into daughter cells (29). The chaperone protein Hsp104 is essential for [PSI+] maintenance (30); Hsp104 cleaves prion fibers into smaller fragments better suited to segregate into daughter cells (21, 31, 32). The ORD repeats have been hypothesized to facilitate Hsp104-dependent aggregate cleavage; the repeats could act as Hsp104-binding sites (although recent evidence suggests that a binding site exists in the M domain [33]), conformationally modify the amyloid core to allow chaperone access, or modulate fiber fragility (24, 34). Interestingly, the mammalian prion protein PrP also contains an ORD, and PrP repeat expansion is associated with dominantly inherited prion disease (35, 36). This observation, combined with the presence of repeat elements in the PFDs of Rnq1 and New1 (22, 37), suggest a role for repeats in prion maintenance; however, other yeast PFDs, such as in Ure2, do not contain repeats, and so repeats cannot be an absolutely necessary feature for prion maintenance. Furthermore, scrambling the Sup35 ORD does not prevent prion formation or maintenance (26), indicating that the activity of the repeats is largely primary sequence independent.
The amino acid compositional requirements for ORD function have only been explored to a limited degree, mostly through targeted mutations. However, several studies have used artificial polyglutamine fragments to explore the sequence requirements for aggregate fragmentation. Targeted replacement of Gln with Tyr residues (38) or other aromatic residues (34) reduced the average aggregate size, suggesting an increase in fiber fragmentation. The elevated number and regular spacing of Tyr residues in both the Sup35 ORD and in the repeats of prion-like protein New1 likewise suggest that aromatic residues may act as recognition sites for chaperones such as Hsp104. Indeed, some chaperones are known to use exposed aromatic or hydrophobic residues as binding sites (39, 40).
To perform a more comprehensive analysis of the compositional determinants for prion formation and maintenance, we quantitatively measured how amino acid composition affects the respective activities of the Sup35 ND and ORD. We observed distinct compositional biases in these two domains. To confirm that these differences were due to distinct compositional requirements for prion formation and maintenance, we developed a new method to specifically isolate the effects of amino acid composition on prion maintenance. These studies confirm that nucleation and maintenance of prions have overlapping but nonidentical compositional requirements and highlight a divergent role for aliphatic residues in promoting prion formation while inhibiting prion maintenance.
MATERIALS AND METHODS
Yeast strains and media.
Standard yeast media and methods were as previously described (41), except that yeast extract-peptone-dextrose (YPD) medium contained 0.5% yeast extract in place of the standard amount (1%). In all experiments, yeast were grown at 30°C.
A complete strain list can be found in Table 1. To build YER709/pER589, the HIS3 gene was amplified from pRS313 using primers EDR1314 and EDR1315 (see Table S1 in the supplemental material for primer sequences). The resulting product was transformed into YER632/pJ533 (42); pJ533 expresses SUP35 from a URA3 plasmid as the sole copy of SUP35 in the cell (43) (see Table S2 in the supplemental material for a complete plasmid list). Successful knockout of ppq1 was confirmed by PCR and sequencing. Two rounds of plasmid shuffling were then used to replace pJ533 with pER589 (a URA3 plasmid expressing Sup35MC from the SUP35 promoter).
TABLE 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| YER709/pER589 | MATα kar1-1 SUQ5 ade2-1 his3 leu2 trp1 ura3 ppq1::HIS3 sup35::KanMx [psi−] [PIN+] pER589 (URA3 SUP35MC) | This study |
| YER632/pJ533 | MATα kar1-1 SUQ5 ade2-1 his3 leu2 trp1 ura3 sup35::KanMx [psi−] [PIN+] pJ533 (URA3 SUP35) | 42 |
| YER282/pER1112 | MATa kar1-1 SWQ5 ade2-1 his3 leu2 trp1 ura3 arg1::HIS3 sup35::KanMx [psi−] [PIN+] pER1112 (URA3 SUP35-27) | 18 |
| 780-1D/pJ533 | MATα kar1-1 SUQ5 ade2-1 his3 leu2 trp1 ura3 sup35::KanMx [PSI+] [PIN+] pJ533 (URA3 SUP35) | 43 |
Building the libraries.
To randomly mutate regions of the SUP35 PFD, first the C-terminal portion of Sup35 was amplified with EDR304 paired with either EDR1388 or EDR1384 for the ND and ORD libraries, respectively. These products were then reamplified with EDR304 paired with either EDR1380 [GCAAAACTACCAGCAATACAGCCAGAACGGT(NNB)8TACCAAGGCTACCAGGCTTACAATGC] or EDR1377 [CTGGGTACCAACAAGGTGGCTATCAACAGTACAAT(NNB)10CCTCAAGGAGGCTACCAGCAATACAAC]. These oligonucleotides, made by Invitrogen, contain degenerate segments encoding a 25% mix of each nucleotide at positions 1 and 2 of each mutated codon and a 33.3% mix of C, G, and T at the third position. In a second PCR, a primer complementary to the nondegenerate 5′ region of EDR1380 or EDR1377 (EDR1389 or EDR1385, respectively) was paired with EDR302 to amplify the N-terminal portion of Sup35. The N- and C-terminal PCR products were combined and reamplified with EDR301 and EDR262. The final PCR products were cotransformed with BamHI/HindIII-cut pJ526 (cen LEU2; from Dan Masison, National Institutes of Health [18]) into yeast strain YER709/pER589 for the prion formation experiments and YER282/pER1112 for the prion maintenance experiments. Transformants were selected on synthetic complete (SC)-Leu medium.
Prion formation library experiments.
Transformants were spotted onto 5-fluoroorotic acid (5-FOA)-containing medium to select for loss of pER589. Library mutants that grew on 5-FOA were stamped onto SC-Ade, YPD, and yeast extract-peptone-adenine-dextrose (YPAD) and grown for 3 to 5 days at 30°C. Isolates that grew red colonies on YPD and did not grow on SC-Ade were pooled into minilibraries containing ∼80 clones. Random isolates were sequenced to generate the naive data set. Minilibraries were plated onto SC-Ade at concentrations of 106 and 105 cells per plate and grown for 5 days at 30°C. To test curability, Ade+ colonies were grown on YPD and on YPD plus 4 mM guanidine-HCl (GdHCl) and then restreaked on YPD to test for loss of the Ade+ phenotype. Clones in which the Ade+ phenotype was stable and curable were sequenced. The odds ratio (OR) for each amino acid or group of amino acids was calculated as follows:
| (1) |
where ƒpf is the per-residue frequency of the amino acid in the mutated region of prion-forming isolates and ƒn is the per-residue frequency of the amino acid in the mutated region of the naive library (44, 45). Prion propensity scores for each amino acid (PPaa) are then calculated as follows:
| (2) |
Prion maintenance library experiments.
Transformants were replica plated onto 5-FOA-containing medium to select for loss of pER1112. Cells were pooled and mated with 780-1D/pJ533 for 24 h on YPAD. Diploids were selected by replica plating on SD-Ade-Trp-Ura medium and then replica plating onto 5-FOA-containing medium to select for loss of pJ533. Cells were then plated for single colonies on YPD medium to allow color selection. Ade+ colonies were streaked on YPD and YPD plus 4 mM GdHCl to test for curability. Clones in which the Ade+ phenotype was stable and curable were defined as propagators and sequenced. Clones with a strong Ade− phenotype were defined as nonpropagators and sequenced.
The prion maintenance odds ratio (ORm) for each amino acid or group of amino acids was calculated as follows:
| (3) |
where ƒp is the per-residue frequency of the amino acid in the mutated region of prion-positive clones and ƒnp is the per-residue frequency of the amino acid in the mutated region of nonprion clones. Prion maintenance propensity scores for each amino acid (PMPaa) were then calculated as follows:
| (4) |
To test whether library mutants that failed to maintain [PSI+] could add onto wild-type aggregates when coexpressed with wild-type Sup35, plasmids expressing nonpropagating mutants were isolated and transformed into 780-1D/pJ533. Cells were then spread on SD-Trp-Ura medium supplemented with limiting adenine (10 μg/ml) to allow color selection. To confirm an inability to propagate [PSI+], cells were spotted on 5-FOA-containing medium to select for loss of pJ533 and then spread on YPD to test for prion loss.
Prion maintenance library experiments, preselecting for the ability to add to existing aggregates.
To preselect against any mutants that were unable to add onto wild-type Sup35 aggregates, the library experiments were performed as described above, except that selection for diploids was performed on medium lacking adenine (SD-Trp-Ura).
Leave-one-out analysis.
To calculate the predicted prion maintenance propensity (PMP) for each isolate in the prion maintenance library data set, PMPaa scores were first calculated based on the other 151 isolates in the data set (i.e., “leaving out” the one sequence to be scored), as shown in equation 4. The PMP score for the left-out isolate was then calculated as the sum of the PMPaa scores for the 10 amino acids in the mutagenized region (the third repeat). This process was iteratively repeated for all 152 isolates in the data set. Four isolates were excluded from the analysis because they contained amino acids for which PMPaa scores could not be calculated. The three lysine-containing red sequences were excluded because the absence of lysine among the propagating clones made lysine's PMPaa score indeterminate; likewise, the only methionine-containing prion-propagating clone could not be scored, because when it was left out of the PMPaa calculation the methionine PMPaa score became indeterminate. The accuracies of the leave-one-out PMP scores were assessed using a receiver operator characteristic (ROC) plot.
Creation of de novo mutants in the ORD.
A random proteome of 65,386 residues was generated using the random number function of the Microsoft Excel software program, with an equal chance of selecting any of the 20 natural amino acids at each position. Windows of 10 amino acid were scored using the calculated PMP values (from the full library data set). A total of 3,628 sequences did not contain any of the low-abundance residues (E, K, M, Q, and W) and were chosen for further evaluation. Sequences with PMP scores at the 95th and 5th percentiles were chosen. Sequences were constructed using the same protocol as that used to build the ORD library, except that EDR1471 to EDR1473 and EDR1490 to EDR1492 were used in place of EDR1377 for the 95th percentile mutants and EDR1480 to EDR1482 and EDR1493 to EDR1495 were used for the 5th percentile mutants.
Tyrosine substitutions in the ORD.
To make the tyrosine substitution mutations, first the portion of SUP35 immediately after the site of mutation was PCR amplified with EDR304 and EDR1890. This product was reamplified with EDR304 paired with EDR1892 to EDR1895 or EDR2156 to EDR2158. In a separate reaction, the portion of SUP35 immediately before the site of mutation was amplified with EDR302 and EDR1891. These two reaction products were then combined and reamplified with EDR301 and EDR262. The final PCR products were cotransformed with BamHI/HindIII-cut pJ526 into YER632/pJ533. 5-FOA was used to select for loss of pJ526. Plasmids for transient overexpression of each PFD from the GAL1 promoter were constructed, and prion formation assays were performed as previously described (42).
Plasmids expressing PFD-green fluorescent protein (GFP) fusions were constructed as previously described (42). To test for focus formation, these plasmids were transformed into 780-1D/pJ533 and YER632/pJ533. Cells were grown for 2 h in galactose/raffinose dropout medium and visualized by fluorescence microscopy.
In silico reanalysis of the Alberti et al. data set.
Amino acid compositions were compared by calculating the percentage of each amino acid out of the total number of amino acids in each predicted PFD (12). The 18 proteins that passed all four tests in the assays of Alberti et al. (12) were the following: Ure2, Sup35, Rnq1, New1, Puf2, Nrp1, Swi1, Ybr016w, Cbk1, Lsm1, Ybl081w, Pub1, Ksp1, Asm4, Nsp1, Gln3, Ypr022c, and Rlm1. The 12 proteins that failed only in the Sup35 fusion protein expression assay were the following: Snf5, Gts1, Scd5, Sgf73, Sok2, Mot3, Ngr1, Jsn1, Pdr1, Cyc8, Pan1, and Ybr108w.
Statistics.
Both a two-sided Student's t test and Fisher's exact test were performed using the GraphPad QuickCalcs website. Standard errors (SE) for log odds ratios are estimated as follows:
| (5) |
where np and nnp are the numbers of times that the amino acid is found in the prion and naive (or nonprion for the prion maintenance experiments) libraries, respectively, and tp and tnp are the total numbers of amino acids in the prion and naive (or nonprion) libraries, respectively (44). To determine if the difference between two log odds ratios was statistically significant, z scores were calculated, using a two-sample z-test:
| (6) |
where OR1 and OR2 are the two odds ratios and SE1 and SE2 are the standard errors for the respective log odds ratios.
RESULTS
Prion formation library experiments with the SUP35 ND and ORD.
To define the distinct compositional requirements of the Sup35 ND and ORD, libraries of Sup35 mutants were created in which segments of the ND or ORD were replaced with a segment of random amino acids (Fig. 1B, bold italics). The ND segment (amino acids 21 to 28) was selected because it overlaps the portion of the ND that was previously shown to be critical for aggregate growth (16) and because it contains a mixture of predicted prion-promoting and -inhibiting residues. In the ORD, the third repeat (amino acids 65 to 74) was targeted because this repeat is important for efficient prion maintenance but dispensable for prion nucleation or fiber growth (22).
We utilized an oligonucleotide-based mutagenesis method to build each library (13). Oligonucleotides were designed to anneal to the regions flanking the site of mutagenesis, but with the target codons replaced with the sequence (NNB)n, where N is any of the four nucleotides, B is any of the nucleotides except adenine, and n is the number of targeted codons (8 for the ND library and 10 for the ORD library). Disallowing adenine at the final position eliminated two of the three stop codons while still allowing all 20 amino acids to be incorporated in the mutated region. PCR with these oligonucleotides was used to create libraries of randomly mutated versions of SUP35, which were then transformed into yeast cells in which SUP35C was expressed from a plasmid as the sole copy of SUP35 in the cell. Through plasmid shuffling, the SUP35C-expressing plasmid was replaced with the random library (Fig. 1C). Prion formation by Sup35 is extremely rare without PFD overexpression, and only a small fraction of library mutants was expected to form prions. Therefore, to enhance prion detection, a ppq1 strain was used; this mutation enhances [PSI+] formation by approximately 10-fold (46).
The prion-forming libraries were screened as previously described (13). Briefly, to remove any clones that might have compromised Sup35 activity, each clone was first screened for Sup35 activity by monitoring nonsense suppression of the ade2-1 allele (47). ade2-1 mutants are unable to grow in the absence of adenine and turn red in the presence of limiting adenine. [PSI+] causes stop codon read-through, allowing for growth without adenine and white or pink colony formation in the presence of limiting adenine. Colonies that grew red on limiting adenine and did not grow without adenine were pooled into minilibraries consisting of ∼80 mutants. Sup35 was sequenced from randomly selected clones to generate a naive library data set (see Table S3 in the supplemental material for the full set of sequences).
The minilibraries were plated onto SC-Ade medium to select for prion formation. Ade+ colonies can result from either DNA mutation or prion formation. To distinguish between these, Ade+ cells were grown on YPD with and without guanidine-HCl and then restreaked onto YPD to test for loss of the Ade+ phenotype. Guanidine-HCl cures [PSI+] (48) by inhibiting Hsp104 activity (49, 50). Cells that lost the Ade+ phenotype after growth on guanidine-HCl but that maintained the Ade+ phenotype after growth on YPD were considered prion positive and were sequenced (see Table S3 in the supplemental material).
Compositional biases among the ND and ORD prion-forming isolates.
For each amino acid, an odds ratio was determined, which represented the degree of over- or underrepresentation of that amino acid among the [PSI+] isolates (13) (Table 2). In many cases, the odds ratios for individual amino acids carried large confidence intervals due to limitations of the library sample sizes. This was particularly true for Met, Trp, Lys, Gln, and Glu; because adenine was excluded at the third position of each codon, each was only encoded by a single codon, and thus each was quite rare among the libraries (Table 2). Nevertheless, there was a strong correlation (P = 0.016 by Spearman rank analysis) between the odds ratios for the ND library and our previously determined odds ratios based on mutagenesis of Sup35-27 (13), a version of Sup35 with a scrambled PFD (Table 3). Excluding the five single-codon amino acids further strengthened this correlation (P = 0.0064).
TABLE 2.
Amino acid representation within the libraries
| Amino acid(s) | ND prion formation librarya |
ORD prion formation libraryb |
Prion maintenance libraryc |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Frequency of amino acid in: |
Odds ratiod | Frequency of amino acid in: |
Odds ratiod | Frequency of amino acid in: |
Odds ratioe | ||||
| Selected [PSI+] library | Unselected naive library | Selected [PSI+] library | Unselected naive library | White [PSI+] colonies | Red [psi−] colonies | ||||
| Individual amino acids | |||||||||
| Alanine | 0.053 | 0.055 | 0.96 | 0.095 | 0.068 | 1.40 | 0.089 | 0.069 | 1.32 |
| Arginine | 0.024 | 0.095 | 0.26*** | 0.044 | 0.107 | 0.41** | 0.054 | 0.121 | 0.41**** |
| Asparagine | 0.061 | 0.031 | 1.95 | 0.047 | 0.021 | 2.24 | 0.038 | 0.032 | 1.20 |
| Aspartic acid | 0.028 | 0.046 | 0.60 | 0.033 | 0.058 | 0.57 | 0.054 | 0.064 | 0.83 |
| Cysteine | 0.065 | 0.071 | 0.91 | 0.084 | 0.062 | 1.35 | 0.072 | 0.047 | 1.58* |
| Glutamic acid | 0.003 | 0.008 | 0.42 | 0.006 | 0.012 | 0.53 | 0.015 | 0.006 | 2.70 |
| Glutamine | 0.007 | 0.016 | 0.41 | 0.003 | 0.003 | 0.94 | 0.008 | 0.007 | 1.12 |
| Glycine | 0.13 | 0.213 | 0.61* | 0.144 | 0.151 | 0.95 | 0.114 | 0.109 | 1.05 |
| Histidine | 0.031 | 0.029 | 1.08 | 0.047 | 0.038 | 1.26 | 0.04 | 0.043 | 0.94 |
| Isoleucine | 0.088 | 0.055 | 1.60 | 0.04 | 0.058 | 0.69 | 0.025 | 0.043 | 0.57 |
| Leucine | 0.065 | 0.071 | 0.91 | 0.058 | 0.06 | 0.96 | 0.04 | 0.063 | 0.62* |
| Lysine | 0 | 0 | N/A | 0.016 | 0.009 | 0.31 | 0 | 0.003 | 0.00 |
| Methionine | 0.014 | 0.014 | 0.96 | 0.088 | 0.06 | 1.89 | 0.002 | 0.009 | 0.17 |
| Phenylalanine | 0.121 | 0.042 | 2.88*** | 0.037 | 0.053 | 1.45 | 0.068 | 0.049 | 1.40 |
| Proline | 0.017 | 0.035 | 0.48 | 0.123 | 0.113 | 0.70 | 0.048 | 0.045 | 1.07 |
| Serine | 0.109 | 0.125 | 0.87 | 0.04 | 0.038 | 1.09 | 0.12 | 0.102 | 1.20 |
| Threonine | 0.061 | 0.042 | 1.45 | 0.016 | 0.016 | 1.07 | 0.037 | 0.039 | 0.94 |
| Tryptophan | 0.014 | 0.021 | 0.67 | 0.076 | 0.055 | 1.04 | 0.018 | 0.005 | 4.07* |
| Tyrosine | 0.069 | 0.042 | 1.63 | 0.076 | 0.086 | 1.40 | 0.074 | 0.028 | 2.81**** |
| Valine | 0.125 | 0.078 | 1.60 | 0.095 | 0.068 | 0.89 | 0.085 | 0.116 | 0.70* |
| Groups of amino acids | |||||||||
| Charged (D, E, K, R) | 0.054 | 0.139 | 0.35*** | 0.129 | 0.210 | 0.43*** | 0.123 | 0.194 | 0.58*** |
| Nonaromatic hydrophobic (I, L, M, V) | 0.267 | 0.206 | 1.41 | 0.181 | 0.200 | 0.88 | 0.151 | 0.231 | 0.59**** |
| Prion-promoting nonaromatic hydrophobic (I, M, V) | 0.206 | 0.139 | 1.61* | 0.126 | 0.143 | 0.86 | 0.111 | 0.168 | 0.62** |
| Aromatic (F, W, Y) | 0.186 | 0.101 | 2.04** | 0.168 | 0.124 | 1.42 | 0.160 | 0.082 | 2.14*** |
| Polar (N, Q, S, T) | 0.220 | 0.198 | 1.14 | 0.197 | 0.162 | 1.27 | 0.203 | 0.180 | 1.16 |
The ND library consists of 37 selected [PSI+] sequences (296 amino acids) and 62 unselected sequences (496 amino acids).
The ORD prion formation library consists of 31 selected [PSI+] sequences (310 amino acids) and 58 unselected sequences (580 amino acids).
The prion maintenance library consists of 65 white [PSI+] sequences (650 amino acids) and 87 red [psi−] sequences (870 amino acids).
Odds ratios reflect the degree of overrepresentation or underrepresentation of each amino acid among the prion-forming isolates, as calculated according to equation 1. Values above 1 indicate overrepresentation among prion-forming isolates. Statistical significance of the over- or underrepresentation is indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Odds ratios reflect the degree of overrepresentation or underrepresentation of each amino acid among the white (prion-propagating) isolates, as calculated using equation 3.
TABLE 3.
Prion propensity and prion maintenance propensity scores from each library
| Amino acid(s)a | Prion propensity score for prion formation library ofb: |
Prion maintenance propensity scorec | ||
|---|---|---|---|---|
| Sup35-27d | Sup35 ND | Sup35 ORD | ||
| Individual amino acids | ||||
| Phenylalanine | 0.84 | 1.06 | 0.37 | 0.33 |
| Isoleucine | 0.81 | 0.47 | −0.37* | −0.57* |
| Valine | 0.81 | 0.47 | −0.12* | −0.35* |
| Tyrosine | 0.78 | 0.49 | 0.34 | 1.03 |
| Methionine | 0.67 | −0.04 | 0.63 | −1.80* |
| Tryptophan | 0.67 | −0.41 | 0.039 | 1.40 |
| Cysteine | 0.42 | −0.10 | 0.30 | 0.45 |
| Serine | 0.13 | −0.14 | 0.084 | 0.18 |
| Asparagine | 0.08 | 0.67 | 0.81 | 0.18 |
| Glutamine | 0.069 | −0.88 | −0.067 | 0.11 |
| Glycine | −0.039 | −0.49 | −0.047 | 0.047 |
| Leucine | −0.04 | −0.10 | −0.039 | −0.48 |
| Threonine | −0.12 | 0.37 | 0.069 | −0.059 |
| Histidine | −0.28 | 0.077 | 0.23 | −0.064 |
| Alanine | −0.40 | −0.036 | 0.34 | 0.28 |
| Arginine | −0.41 | −1.37 | −0.89 | −0.88 |
| Glutamic acid | −0.61 | −0.87 | −0.63 | 0.99 |
| Proline | −1.20 | −0.73 | −0.36 | 0.065* |
| Aspartic acid | −1.28 | −0.51 | −0.56 | −0.19 |
| Lysine | −1.58 | NAe | −1.17 | NAe |
| Groups of amino acids | ||||
| Charged (D, E, K, R) | −0.90 | −1.04 | −0.83 | −0.54 |
| Hydrophobic (I, L, M, V) | 0.68 | 0.34 | −0.13** | −0.53**** |
| Prion-promoting nonaromatic hydrophobic (I, M, V) | 0.88 | 0.47 | −0.15*** | −0.48**** |
| Aromatic (F, W, Y) | 0.84 | 0.71 | 0.35 | 0.76 |
| Polar (N, Q, S, T) | 0.064 | 0.13 | 0.24 | 0.15 |
Amino acids are listed in the order of their prion propensity according to PAPA.
Prion formation libraries were compiled as illustrated in Fig. 1C. Prion propensity scores were calculated as the natural log of the odds ratios, according to equation 2. Statistically significant differences relative to the Sup35-27 library (13) are indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
The prion maintenance library experiment was performed as illustrated in Fig. 1D. Prion maintenance propensity scores were calculated according to equation 4. Statistically significant differences relative to the Sup35-27 library (13) are indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
From Toombs et al. (13).
NA, not applicable; lysine was not found in any of the prion-forming sequences in the ND library or in any of the white colonies in the prion propagation library.
Grouping similar amino acids can effectively increase sample sizes, thereby improving statistical significance. Doing so confirmed that the same broad trends observed for Sup35-27 were seen in the ND library, with both aromatic and nonaromatic hydrophobic residues promoting prion activity and charged residues strongly inhibiting prion activity (Fig. 2A).
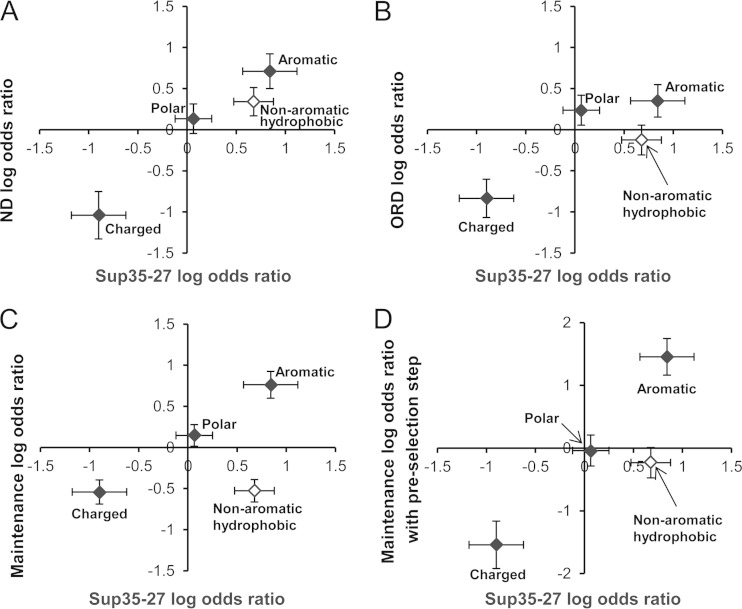
FIG 2.
Nonaromatic hydrophobic residues show different prion formation and maintenance propensities. Comparisons of the previously determined log odds ratios based on mutagenesis of Sup35-27 (13) were undertaken to the log odds ratios from the ND (A) or ORD (B) prion formation library experiments, the ORD prion maintenance library experiment (C), or the prion propagation library experiment in which an additional step was added to remove mutants that were not efficiently recruited into wild-type prion aggregates (D). While the odds ratios for charged, aromatic, polar residues (filled diamonds) showed similar trends in each library, nonaromatic hydrophobic residues (open diamonds) scored substantially worse in the ORD prion formation and maintenance libraries. Charged residues are Asp, Glu, Lys, and Arg. Polar residues are Ser, Thr, Asn, and Gln. Aromatic residues are Trp, Tyr, and Phe. Nonaromatic hydrophobic residues are Leu, Ile, Val, and Met. Error bars indicate standard errors, calculated according to equation 5.
There was also a statistically significant correlation between our previous Sup35-27 odds ratios and those for the ORD library (P = 0.017). As in the ND and Sup35-27 libraries, there was a statistically significant bias against charged residues among prion-forming sequences in the ORD library (P = 0.0002) (Table 2) and an overrepresentation of aromatic residues, albeit below the threshold for statistical significance (P = 0.084) (Table 2). However, there was one striking difference. With the exception of leucine (which is known to have a low β-sheet propensity [51]), the nonaromatic hydrophobic residues (Ile, Val, and Met) were highly enriched among prion-forming sequences in both the Sup35-27 library (P = 0.0001) and the ND library (P = 0.017), yet they were actually modestly underrepresented among prion-forming sequences in the ORD library (Table 3). The differences between the ORD library and both the Sup35-27 and ND libraries were both statistically significant (P = 0.0031 and 0.029, respectively) (Table 3), demonstrating that these residues have significantly different effects in these locations.
Prion maintenance library experiments with the SUP35 ORD.
The simplest explanation for the different biases observed in the ND versus ORD is that these differences reflect the distinct functions of the two regions (16, 23–25) and, thus, that nonaromatic hydrophobic residues promote prion formation but not prion maintenance. However, the functional separation between the two regions is not absolute, so it is possible that some of the ORD biases that we observed were due to effects on prion formation.
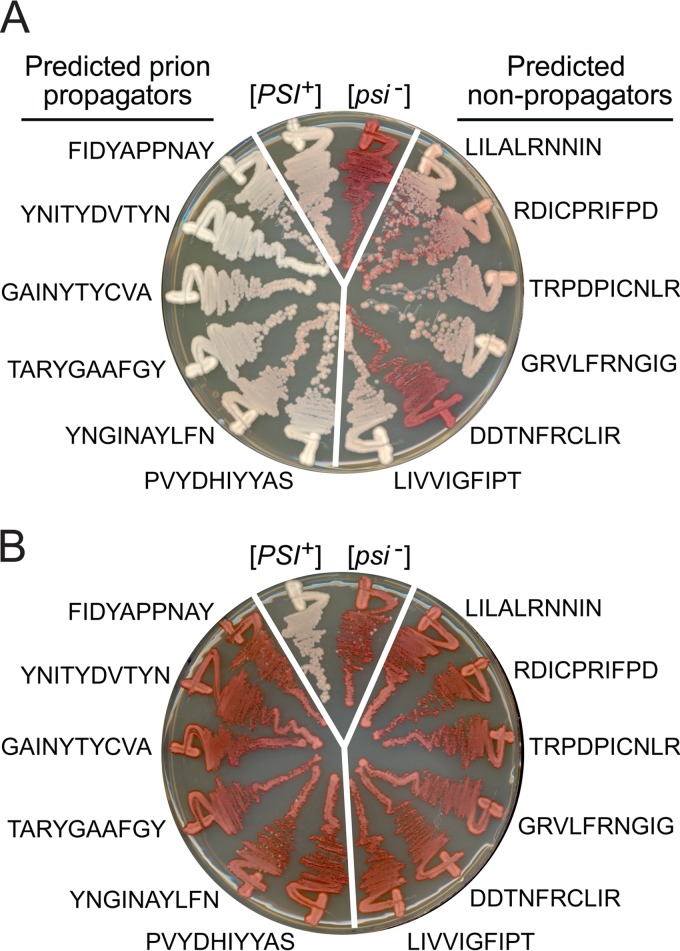
Therefore, we adapted our library screening method to specifically isolate the effects of composition on prion maintenance (Fig. 3A). We constructed a second ORD library as described above, but this time we assessed the abilities of mutants to maintain an existing prion. To accomplish this, we utilized a two-step process. Because our plasmid libraries were constructed directly in yeast by using homologous recombination (by cotransforming a mutagenized PCR product and a linearized vector), the libraries were inevitably contaminated with products of recombination between the initial Sup35 maintenance plasmid that was present in the cell and the linearized vector. Thus, when the libraries were built directly in a [PSI+] cell, because prion-propagating mutants are relatively rare, a large fraction of the prion-propagating clones turned out to contain wild-type Sup35 (data not shown). To avoid this problem, the libraries were constructed in a [psi−] strain expressing scrambled Sup35 (Sup35-27) from a URA3 plasmid as the sole copy of Sup35 in the cell. After transformation, we selected for loss of the URA3 plasmid, so that the library mutants were the sole copy of Sup35. These cells were then mated with wild-type [PSI+] cells in which the sole copy of Sup35 was again expressed from a URA3 plasmid. After selection for diploids and for loss of the URA3 plasmid, clones were plated for single colonies and screened for Sup35 activity by using the ade2-1 allele. Because Sup35-27 is unable to propagate wild-type [PSI+], any spurious recombination events between the Sup35-27 maintenance plasmid and the linearized vector during the cloning step would result in [psi−] cells. These Sup35-27 clones were excluded from our analysis.
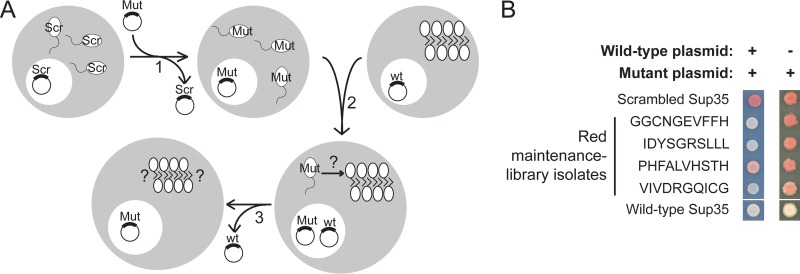
FIG 3.
Prion maintenance library experiments. (A) Experimental scheme. [psi–] cells in which Sup35C was expressed from a URA3 plasmid as the sole copy of Sup35 were transformed with a randomly mutated version of Sup35 and then selected for loss of the wild-type plasmid (step 1). These cells were mated with wild-type [PSI+] cells in which the sole copy of Sup35 was expressed from a URA3 plasmid (step 2). After selection for loss of the URA3 plasmid (step 3), red and white clones were sequenced. In the modified protocol to select against mutants with a defect in adding onto wild-type aggregates, selection for diploid cells in step 2 was done in the absence of adenine. (B) Plasmids expressing mutant Sup35s from individual red prion maintenance library isolates were transformed into [PSI+] cells in which the sole copy of Sup35 was expressed from a URA3 plasmid. To test whether the mutant Sup35s were inactivated in the presence of wild-type [PSI+], cells were plated on limiting adenine medium, selecting for both the wild-type and mutant Sup35-expressing plasmids (left). Cells were then retested on YPD after selection for loss of the wild-type plasmid (right). Representative examples are shown, with the sequences of the mutagenized regions indicated. Sup35-27, a scrambled version of Sup35 that is not incorporated into wild-type [PSI+] aggregates, and wild-type Sup35 are shown as controls.
Red colonies were deemed to have lost [PSI+]. In contrast, colonies were considered capable of efficiently maintaining [PSI+] if they were white on YPD and stably maintained this white phenotype upon restreaking on YPD but grew red on YPD after guanidine treatment. Clones of intermediate phenotype (which were substantially pink or sectored on YPD, or that were initially white on YPD but showed any loss of the prion upon restreaking on nonselective medium) were excluded from the study. A total of 65 distinct white mutants and 87 red mutants were sequenced (see Table S3 in the supplemental material).
Compositional biases among the propagating prion isolates.
For each of the amino acids, an odds ratio was calculated according to equation 3. Six amino acids showed statistically significant differences between the prion-maintaining (white) and prion-losing (red) isolates: Trp, Tyr, and Cys were significantly overrepresented among the prion-maintaining isolates, while Val, Leu, and Arg were significantly underrepresented (Table 2). When chemically similar amino acids were grouped together, aromatic amino acids were overrepresented among the [PSI+] isolates (P < 10−4), and charged residues were underrepresented (P = 0.0002), as in each of the previous libraries; however, nonaromatic hydrophobic residues were substantially underrepresented (P = 10−4). Thus, focusing specifically on prion maintenance amplified the previously observed differences seen between the ND and ORD libraries for nonaromatic hydrophobic residues (Fig. 2B and C). The differences observed for nonaromatic hydrophobic residues between the ORD prion-propagating library experiments and both Sup35-27 and the ND were highly statistically significant (P < 10−4). Interestingly, even the difference between the ORD prion-propagating experiments and the original ORD library experiments approached statistical significance (P = 0.077), suggesting that in the original ORD experiments, nonaromatic hydrophobic residues may have had partially offsetting effects, promoting prion formation while inhibiting prion maintenance.
Failures of ORD mutants to maintain [PSI+] were not due to failure to add onto existing wild-type aggregates.
In the prion maintenance library experiments, a protein could fail to maintain [PSI+] for one of two reasons: a mutant could fail to add onto the preexisting wild-type aggregates (Fig. 3A, after step 2), or the mutant could successfully add onto preexisting aggregates but have a defect in the subsequent prion maintenance steps (Fig. 3A, after step 3). To distinguish between these two possibilities, plasmids expressing mutant SUP35 from individual nonpropagating clones were isolated and retransformed into wild-type [PSI+] cells. The phenotype of transformants was examined before and after selection for the loss of wild-type plasmid (analogous to before and after step 3 in Fig. 3A). If a mutant is unable to add onto the preexisting wild-type aggregates, then it should remain soluble (active), even in the presence of wild-type [PSI+], resulting in a red phenotype (Fig. 3B). Of the 14 clones examined (indicated with an asterisk in Table S3 in the supplemental material), none was fully red when the wild-type and mutant proteins were coexpressed, although three (FYSVSILDRR, GCPRVVIHVD, and PHFALVHSTH) showed a mild pink phenotype, suggesting a slightly reduced efficiency of adding onto wild-type aggregates or a partially dominant defect in prion aggregate fragmentation; by contrast, all 14 were red or highly sectored after loss of the wild-type plasmid (Fig. 3B and data not shown).
These results indicate that the majority of library sequences that failed to maintain [PSI+] were competent for adding onto the preexisting wild-type aggregates but had a defect in the subsequent maintenance steps. However, it remained possible that rare mutants with a defect in addition to preexisting aggregates could skew the results of the library screen. To more comprehensively examine this issue, the library experiment was repeated with an additional selection step to remove such mutants. After mating the mutant library strains with wild-type [PSI+]-containing cells (Fig. 3A, step 2), the selection step to select for diploid cells was undertaken in the absence of adenine; this selected against mutants that remained functional in the presence of wild-type [PSI+] (i.e., that were not efficiently incorporated into [PSI+] aggregates). Then, after selecting for loss of the wild-type plasmid, each clone was examined as before for its ability to propagate [PSI+] when expressed as the sole copy in the cell. This method has the substantial downside that it adds an additional prion selection step; nevertheless, it allowed us to confirm that selecting against mutants with a defect in adding to preexisting aggregates did not substantially change the outcome. With a smaller set of 19 prion-maintaining mutants and 26 nonpropagators (see Table S3 in the supplemental material), the broad trends from the original maintenance library held in this altered experimental system (Fig. 2C and D).
Predicting propagating versus nonpropagating sequences.
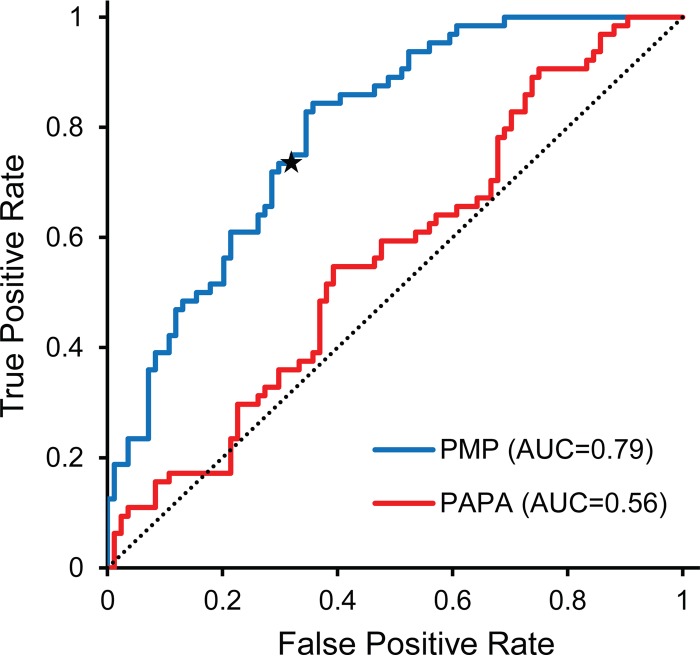
To assess whether the biases seen in the prion maintenance library experiments were sufficient to predict the behavior of individual library isolates, we used the standard leave-one-out method of cross-validation. Briefly, there were 152 sequences in the prion maintenance library data set (65 white and 87 red). To calculate the PMP score for each sequence, the sequence was excluded (i.e., left out) from the data set, and the remaining 151 sequences were used to calculate prion maintenance propensity scores for each amino (PMPaa) acid according to equation 4. The PMP score for the excluded sequence was then calculated as the sum of the PMPaa values for each of the 10 amino acids in the mutated region. This process was iteratively repeated for all 152 sequences. White clones had significantly higher PMP scores (P < 0.0001 by two-sided t test) than red clones, although there was significant overlap between the two sets (Fig. 4). For example, 73.4% of the white clones had positive PMP scores, while only 32% of the red clones did, and the 26 lowest-scoring sequences were all red. In contrast, PAPA showed almost no ability to distinguish between the red and white clones (Fig. 4), consistent with the idea that PAPA is better correlated with prion formation propensity than prion maintenance propensity (Fig. 2).
FIG 4.
Receiver operator characteristic (ROC) plot (57) assessing the ability of PMP scores and PAPA to predict the prion-propagating library mutants. A leave-one-out method of cross-validation was used to assess whether PMP scores from the prion maintenance library experiment were sufficient to predict which library members would successfully propagate [PSI+]. PMP scores showed reasonable prediction accuracy (area under the curve [AUC], 0.79); the star indicates the point on the ROC plot for a PMP score of zero. PAPA showed virtually no ability to distinguish between red and white isolates (AUC, 0.56), with prediction accuracy barely above what would be expected by random chance (dotted line). False positive rate = (number of red isolates scored as prion propagating)/(total number of red isolates). True positive rate = (number of white isolates scored as prion propagating)/(total number of red isolates).
We then tested whether the observed PMPaa values from the full library data set (Table 3) were sufficient to rationally design sequences that could substitute for the third repeat of the ORD (the region mutagenized in the ORD library experiments) in supporting prion propagation. We constructed a random library of 10-amino-acid segments in silico and then used the PMP scores to identify segments with predicted high or low prion maintenance propensities. Six of the randomly designed sequences that were predicted to be very good at maintaining [PSI+] (in the 95th percentile among the in silico library) and six versions predicted to maintain [PSI+] poorly (5th percentile) were inserted in the place of the third repeat. Plasmids expressing these mutants were transformed into wild-type [PSI+] cells in which the sole copy of Sup35 was expressed from a plasmid. After selection for loss of wild-type plasmid, cells were examined for [PSI+] loss (Fig. 5A). While all six predicted prion-maintaining mutants were uniformly white when plated on YPD (Fig. 5A, left side), the predicted nonpropagators were more variable. Three clones showed a mixture of red and white colonies, reflecting a high degree of prion loss, while the others showed only very modest pink phenotypes (Fig. 5A, right side). All 12 mutants were red after treatment with guanidine-HCl (Fig. 5B). Collectively, these results suggest that our PMP values are sufficient to identify broad trends but not sufficient to predict whether a given sequence will support prion maintenance.
FIG 5.
Successful design of prion-propagating sequences. A library of random 10-amino-acid sequences was built in silico. The library was screened using the PMP scores from the ORD prion propagation library experiment. Six high-scoring sequences (left side of each panel) and six low-scoring sequences (right side) were selected and inserted into Sup35 in place of the third repeat of the ORD. Mutants were introduced to wild-type [PSI+] cells. Transformants were spotted onto 5-FOA to select for loss of the plasmid expressing wild-type Sup35, and either streaked onto YPD medium to test for loss of [PSI+] (A) or streaked onto SC medium plus 4 mM guanidine-HCl and then streaked onto YPD medium to test for loss of [PSI+] (B). Untreated wild-type [PSI+] and [psi−] cells are shown as controls.
Essential role for aromatic residues in prion maintenance.
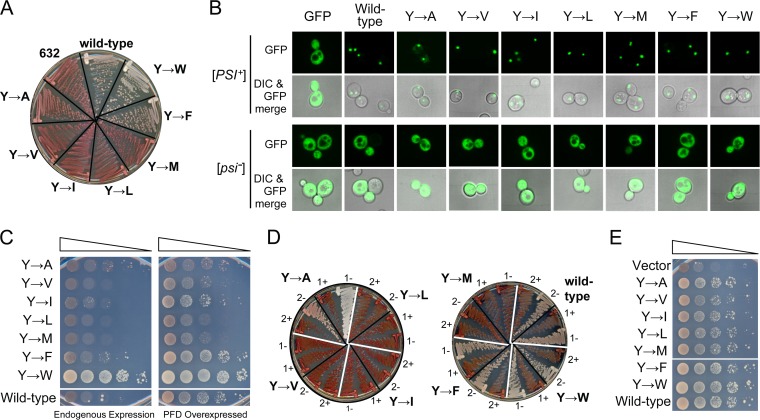
We designed targeted mutations to further examine the differences between the effects of aromatic and nonaromatic residues on [PSI+] maintenance. The Sup35 PFD contains 20 Tyr residues, one Phe, and no Trp, Ile, or Val. Five of the Tyr residues are located in the ND, and nonaromatic hydrophobic residues can replace these ND tyrosines in supporting prion activity (42). Here, we examined the effects of replacing different residues for Tyr in the ORD.
We replaced the five tyrosine residues in the third, fourth, and fifth repeat of the ORD with Ala, Val, Ile, Leu, Met, Phe, or Trp. Each mutant was transformed into a wild-type [PSI+] cell in which the sole copy of Sup35 was expressed from a plasmid. After selection for loss of the wild-type plasmid, only the two constructs with aromatic substitutions were able to stably maintain [PSI+] (Fig. 6A). Prion loss was not due to an inability to be recruited to preexisting Sup35 aggregates. When GFP fusions of each mutant PFD were transiently expressed for 2 h in wild-type [psi−] cells, each remained diffuse (Fig. 6B); however, when the GFP fusions were transiently expressed in [PSI+] cells, each rapidly coalesced into foci (Fig. 6B), indicating that the mutants were efficiently recruited into wild-type [PSI+] aggregates.
FIG 6.
Aromatic residues in the ORD are critical for prion propagation. (A) Prion maintenance by tyrosine substitution mutants. The five tyrosines in repeats 3 to 5 of the Sup35 ORD were replaced with Ala, Val, Ile, Leu, Met, Phe, or Trp. These mutants were introduced into wild-type [PSI+] cells expressing wild-type Sup35 from a plasmid. After selection for loss of the wild-type plasmid, cells were streaked onto YPD medium to test for the ability to maintain [PSI+]. Strain YER632/pJ533 was included as a [psi−] control (632). (B) Tyrosine substitution mutants were efficiently incorporated into wild-type [PSI+] aggregates. Plasmids expressing GFP fusions of each tyrosine substitution mutant PFD under the control of the GAL1 promoter were transformed into wild-type [PSI+] and [psi−] strains. Cells were grown in galactose/raffinose dropout medium for 2 h and visualized by confocal microscopy. Foci were observed for each fusion in [PSI+] cells but not [psi−] cells. (C) Prion formation by tyrosine substitution mutants. [psi−] strains expressing each mutant as the sole copy of Sup35 were transformed either with an empty vector (left) or with a plasmid expressing the matching Sup35 mutant under the control of the GAL1 promoter (right). All strains were cultured for 3 days in galactose/raffinose dropout medium, and then 10-fold serial dilutions were plated onto medium lacking adenine to select for [PSI+]. (D) Tryptophan, alanine, and phenylalanine substitution mutants form stable, curable prions. Ade+ isolates from panel B were streaked onto either SC medium (−) or SC plus 3 mM guanidine-HCl (+) and then restreaked onto YPD to test for prion loss. Two representative Ade+ isolates are shown for each mutant. (E) Overexpression of the tyrosine substitution mutants induced wild-type [PSI+] formation. Yeast expressing wild-type Sup35 were transformed with either an empty vector (vector) or the vector modified to express either the wild-type Sup35 NM domain (wild-type) or the NM domain of the ORD tyrosine substitution mutants under the control of the GAL1 promoter. Cells were then tested for [PSI+] formation.
Although these results suggest that replacement of ORD tyrosines with nonaromatic hydrophobic residues results in a defect in [PSI+] maintenance, it remained possible that these constructs are able to maintain some variant of [PSI+], but just not the specific [PSI+] variant present in these cells. Therefore, each of the mutants was tested for the ability to form [PSI+] de novo when expressed as the sole copy of Sup35 in the cell. Cells were grown either with or without overexpression of the matching PFD and then plated onto SC-Ade medium to test for prion formation. PFD overexpression increases prion formation by increasing the probability of the initial prion-forming nucleation events (5). All of the constructs were able to form Ade+ colonies upon PFD overexpression (Fig. 6C), albeit at various frequencies; in fact, the Trp substitutions actually substantially increased prion formation. However, the Ade+ colonies formed by the Phe and Trp substitution constructs, and to a lesser extent by the Ile construct, were substantially bigger than those formed by the Ala, Val, Leu, or Met constructs. Furthermore, as in the plasmid shuffling experiments (Fig. 6A), Phe and Trp constructs were able to consistently maintain a white [PSI+] phenotype when passaged on YPD medium, while the Val, Ile, Leu, and Met Ade+ isolates all reverted to a red phenotype after growth on nonselective medium (Fig. 6D). The only mutant that behaved differently from the results of the shuffling experiment was the Ala substitution mutant, which was able to form rare stable, curable prions (Fig. 6D).
The low frequency of Ade+ colonies seen for some of the mutants (Fig. 6C) could have been due to either a defect in prion nucleation or in maintenance of prion aggregates. However, overexpression of each of the mutants efficiently stimulated wild-type Sup35 to form prions, suggesting that these mutants do not have a nucleation defect (Fig. 6E). Collectively, these results indicated that the mutants containing nonaromatic hydrophobic replacements for tyrosine are able to efficiently aggregate but are unable to stably propagate these aggregates as prions.
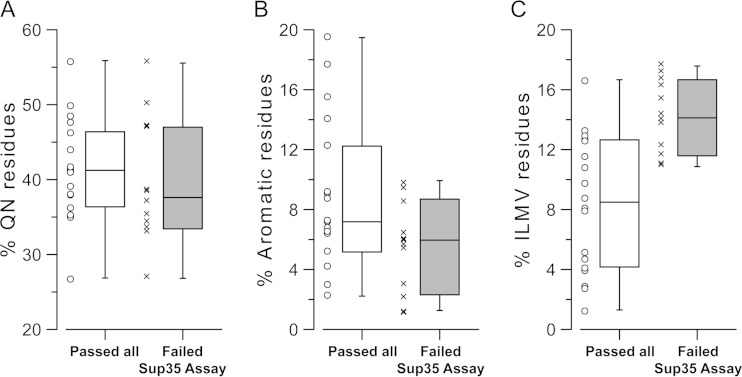
Yeast PFDs that successfully propagate show similar compositional biases.
Alberti et al. previously generated a large data set in which the 100 yeast protein fragments (averaging about 160 amino acids in length) with the greatest compositional similarity to the Sup35, Ure2, Rnq1, and New1 PFDs were tested in four distinct assays of prion-like activity (12). The four assays used in this study included three measures of aggregation (formation of fluorescent foci when expressed as an enhanced yellow fluorescent protein [EYFP] fusion, formation of SDS-resistant aggregates in an SDD-AGE assay, and in vitro aggregation of purified recombinant proteins, as monitored by thioflavin-T fluorescence) and one assay (replacement of the PFD of Sup35 with each fragment) that was tested for the ability to support true prion activity (12).
Eighteen of the fragments in the data set passed all four assays (12). Another 12 of the fragments passed all three of the aggregation assays but failed the Sup35 fusion assay; this indicated that these domains have an ability to form aggregates but may have a defect in prion maintenance, although it is important to note that proteins can fail the Sup35 fusion assay for a variety of reasons and that even some known PFDs fail in this assay (see Discussion). These two sets had very similar Q/N contents (Fig. 7A) and predicted aggregation propensities according to PAPA (data not shown). However, consistent with the results of our library screens, aromatic residues were overrepresented (Fig. 7B) and nonaromatic hydrophobic residues were underrepresented (Fig. 7C) among the proteins that passed all four assays. Strikingly, each of the most hydrophobic nonaromatic residues (Ile, Met, Val, and Leu) was more common among the proteins that passed the three aggregation assays but failed the Sup35 assay, although this bias was only statistically significant for Leu and Val (P = 0.008 and 0.0002, respectively). While neither the overrepresentation of aromatics nor the underrepresentation of nonaromatic hydrophobics was absolute, both were statistically significant (P = 0.0003 for nonaromatic hydrophobics and P = 0.05 for aromatics), suggesting that the trends identified in our library experiments may extend to other prion-like domains.
FIG 7.
Aromatic residues are overrepresented and nonaromatic hydrophobics are underrepresented among domains with prion activity. Alberti et al. (12) tested 100 prion-like domains in four assays for prion-like activity. Three of the assays tested aggregation activity, while a fourth tested the ability of the domains to support prion activity when inserted in place of the Sup35 PFD. Box-and-whisker plots show the frequency of Q/N residues (A), aromatic residues (B), and nonaromatic hydrophobic residues (C; Ile, Leu, Met, and Val) among each of the Alberti proteins that passed all tests (white bars) or that passed all tests except the Sup35-fusion protein assay (gray bars).
DISCUSSION
We previously showed that the ND and ORD have distinct compositional requirements (26). Here, we made the first steps toward quantitatively defining these requirements. Most significantly, we found that aliphatic residues promote prion activity in the ND while inhibiting prion activity in the ORD. It appears that this difference is due to the distinct functions of the two regions in supporting prion activity. Consistent with earlier work suggesting that the ORD is largely dispensable for prion formation (22), replacement of aromatic residues in the ORD with aliphatic residues did not significantly affect the ability of the PFD to nucleate prion formation, but it did disrupt maintenance of prion aggregates.
These experiments nicely complement previous work that used polyglutamine to study the effects of amino acid composition on fiber fragmentation (34, 38). Alexandrov et al. inserted different residues into polyglutamine stretches and found that aromatic residues reduced average aggregate size (34). However, there are challenges in interpreting these experiments. While poly-Q forms aggregates, it does not form prions per se, and it is not clear how similar the structures of poly-Q aggregates are to prion aggregates; Alexandrov et al. suggested that the uniform sequence of poly-Q likely results in “staggered” aggregates, rather than the ordered, in-register parallel β-sheet aggregates formed by Sup35 (34). Additionally, while a smaller aggregate size is consistent with an increase in fiber fragmentation, average aggregate size would also be expected to be a function of the frequency of spontaneous aggregate nucleation and the rate of fiber growth rates. For example, spontaneous nucleation, like fragmentation, creates new independently segregating aggregates, so if nucleation rates were exceptionally high, such that many nucleation events happen per-cell division, this would increase the number of independent aggregates and thus decrease average aggregate size. The current experiments expand on this previous work by beginning to parse the specific steps in prion activity affected by each amino acid.
The observation that specific amino acids can have different effects at different positions is itself not surprising or unprecedented. For example, Bondarev et al. recently showed that insertion of lysine residues into the first or second repeat of the Sup35 ORD resulted in [PSI+] loss, but similar insertions in the other repeats did not (52). This result makes sense; the ND and first two repeats of the ORD are required for efficient nucleation of prion formation and for addition to preexisting [PSI+] aggregates (22), suggesting that this region forms critical contacts that mediate fiber growth. In contrast, the third through fifth repeats are dispensable for these activities. Therefore, it is not surprising that mutations in the first two repeats might have stronger effects. Indeed, we saw what may be a similar effect: proline and glycine, both of which have low β-sheet propensities, were better tolerated in the ORD than the ND. However, our results also showed something more unexpected—that amino acids that promote prion activity at one region in a PFD can actually inhibit prion activity in other regions.
Although the differences between the amino acid compositions of red and white clones in our prion maintenance library experiments were highly statistically significant, they were not sufficient to predict with 100% accuracy whether a given mutant could propagate prions (Fig. 4 and 5). Part of this could be due to the large confidence intervals associated with each amino acid's PMP score. Also, factors other than just simple amino acid composition may affect prion maintenance. For example, there may be certain positions where specific amino acids are favored or disfavored. We did not observe any strong positional biases in any of our libraries, but this in part could have been due to limitations of our sample sizes. We also examined whether a number of other factors might, in conjunction with PMP score, improve discrimination between the sets. These include presence/absence of groups of amino acids, total number of charges, net charges of each 10-mer, distribution of charges within each 10-mer, hydrophobicity, predicted β-sheet propensity, and disorder propensity. However, none of these improved the discrimination between the propagating and nonpropagating sequences compared to PMP scores alone (data not shown).
Similarly, the biases for aromatic residues and against aliphatic residues among domains that can substitute for the Sup35 PFD in supporting prion activity were also not absolute. The prion protein Ure2 is a good example of this. The Ure2 PFD has only two aromatic residues (both F) and 12 nonaromatic hydrophobic residues (I, L, V, and M) (53). It is possible that other residues that modestly promote prion maintenance can substitute for aromatic residues when present at a high enough density; for example, Ure2 has very high Ser and Asn content, both of which scored as modestly promoting prion maintenance in our assays. Alternatively, different prions have different chaperone requirements (54), so the trends that we observed might be specific for the constellation of chaperones that propagate [PSI+]. Consistent with this, Crist et al. (28) identified repeat sequences lacking aromatic residues that could substitute for the Sup35 ORD in supporting prion activity, but the resulting prions were Hsp104 independent. Thus, more detailed comparison of the amino acid compositions and chaperone requirements of different PFDs may provide insight into the mechanism by which specific compositional features promote prion maintenance.
Because of the distinct chaperone requirements for different prions, it may prove difficult to develop a simple method to predict whether a given sequence will be able support prion maintenance. The prion prediction algorithm PAPA is able to effectively discriminate between Q/N-rich proteins that have high versus low aggregation propensity (19), but for proteins that show high aggregation propensity it is ineffective at predicting which will be able to support full-fledged prion activity. The current experiments explain why: the prion propensity scores that make up PAPA match the ND library scores more closely than the ORD library scores (Fig. 2), suggesting that PAPA predominantly scores aggregation propensity.
There are some important caveats to consider when analyzing constructs in the Sup35 fusion assay (as in Fig. 7). First, Sup35 is an essential gene, so any [PSI+] prion that too effectively sequesters and inactivates Sup35 will be lethal; indeed, many spontaneously formed [PSI+] variants are lethal (55). Thus, a fragment could fail the Sup35 assay because it forms too strong a prion variant. Additionally, context does affect prion activity. Some known PFDs fail to support prion activity when fused to Sup35, and conversely, many of the fragments that support prion activity when fused to Sup35 have not yet been shown to form prions in their native context. Therefore, while our analysis may help explain why some prion-like fragments fail in the Sup35 fusion assay, additional experiments will be needed to determine whether similar effects would be seen in other sequence contexts.
Finally, it should be noted that the prion maintenance library experiments were done with a single strong [PSI+] strain. We chose to use a strong [PSI+] variant for two reasons. First, it increased the chances that any red clones were due to a prion maintenance defect, as opposed to the spontaneous prion loss that would be common with a weak strain. Second, various evidence suggests that the amyloid core extends further into the ORD in weak prion variants (24, 56), increasing the chances that red isolates could be due to an inability to add onto preexisting aggregates rather than a defect in the subsequent maintenance steps. The difference in prion maintenance ability between aromatic and aliphatic residues appears to be prion variant independent, as the ORD mutants in which tyrosines were replaced with aliphatic residues not only failed to propagate an existing strong prion variant (Fig. 6A) but also were unable to form their own stable prion variants (Fig. 6D). Nevertheless, it remains possible that some of the other observed biases are prion variant dependent.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (MCB-1023771) and National Institutes of Health (GM105991).
We thank the laboratories of P. Shing Ho and Olve Peersen for helpful comments. We thank the undergraduate researchers who assisted in library screening, including Robert Newell, Jr., Lauren Gonzales, Taylor Beairsto, Stephen Gross, and Alexander Queen. We also thank Connor Hendrich and the rest of the Ross lab, as well as Emily Davis and James Knox in the MacLea lab, for comments and technical support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01020-14.
REFERENCES
- 1.Chiti F, Dobson CM. 2006. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner SB. 1991. Molecular biology of prion diseases. Science 252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 4.Weissmann C. 1991. Spongiform encephalopathies. The prion's progress. Nature 349:569–571. doi: 10.1038/349569a0. [DOI] [PubMed] [Google Scholar]
- 5.Wickner RB. 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264:566–569. [DOI] [PubMed] [Google Scholar]
- 6.Wickner RB, Masison DC, Edskes HK. 1995. [PSI] and [URE3] as yeast prions. Yeast 11:1671–1685. [DOI] [PubMed] [Google Scholar]
- 7.Lindquist S. 1997. Mad cows meet psi-chotic yeast: the expansion of the prion hypothesis. Cell 89:495–498. [DOI] [PubMed] [Google Scholar]
- 8.Sondheimer N, Lindquist S. 2000. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell 5:163–172. doi: 10.1016/S1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 9.Derkatch IL, Bradley ME, Hong JY, Liebman SW. 2001. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106:171–182. doi: 10.1016/S0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 10.Michelitsch MD, Weissman JS. 2000. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A 97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacLea KS, Ross ED. 2011. Strategies for identifying new prions in yeast. Prion 5:1–6. doi: 10.4161/pri.17918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toombs JA, McCarty BR, Ross ED. 2010. Compositional determinants of prion formation in yeast. Mol Cell Biol 30:319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. 1993. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol 7:683–692. [DOI] [PubMed] [Google Scholar]
- 15.Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. 1994. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137:671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePace AH, Santoso A, Hillner P, Weissman JS. 1998. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93:1241–1252. [DOI] [PubMed] [Google Scholar]
- 17.Liu J-J, Sondheimer N, Lindquist SL. 2002. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. Proc Natl Acad Sci U S A 99(Suppl 4):S16446–S16453. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross ED, Edskes HK, Terry MJ, Wickner RB. 2005. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A 102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toombs JA, Petri M, Paul KR, Kan GY, Ben-Hur A, Ross ED. 2012. De novo design of synthetic prion domains. Proc Natl Acad Sci U S A 109:6519–6524. doi: 10.1073/pnas.1119366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross ED, MacLea KS, Anderson C, Ben-Hur A. 2013. A bioinformatics method for identifying Q/N-rich prion-like domains in proteins. Methods Mol Biol 1017:219–228. doi: 10.1007/978-1-62703-433-8_16. [DOI] [PubMed] [Google Scholar]
- 21.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 22.Osherovich LZ, Cox BS, Tuite MF, Weissman JS. 2004. Dissection and design of yeast prions. PLoS Biol 2:e86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu JJ, Lindquist S. 1999. Oligopeptide-repeat expansions modulate “protein-only” inheritance in yeast. Nature 400:573–576. [DOI] [PubMed] [Google Scholar]
- 24.Shkundina IS, Kushnirov VV, Tuite MF, Ter-Avanesyan MD. 2006. The role of the N-terminal oligopeptide repeats of the yeast Sup35 prion protein in propagation and transmission of prion variants. Genetics 172:827–835. doi: 10.1534/genetics.105.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parham SN, Resende CG, Tuite MF. 2001. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J 20:2111–2119. doi: 10.1093/emboj/20.9.2111-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toombs JA, Liss NM, Cobble KR, Ben-Musa Z, Ross ED. 2011. [PSI+] maintenance is dependent on the composition, not primary sequence, of the oligopeptide repeat domain. PLoS One 6:e21953. doi: 10.1371/journal.pone.0021953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohhashi Y, Ito K, Toyama BH, Weissman JS, Tanaka M. 2010. Differences in prion strain conformations result from non-native interactions in a nucleus. Nat Chem Biol 6:225–230. doi: 10.1038/nchembio.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crist CG, Nakayashiki T, Kurahashi H, Nakamura Y. 2003. [PHI+], a novel Sup35-prion variant propagated with non-Gln/Asn oligopeptide repeats in the absence of the chaperone protein Hsp104. Genes Cells 8:603–618. doi: 10.1046/j.1365-2443.2003.00661.x. [DOI] [PubMed] [Google Scholar]
- 29.Derdowski A, Sindi SS, Klaips CL, DiSalvo S, Serio TR. 2010. A size threshold limits prion transmission and establishes phenotypic diversity. Science 330:680–683. doi: 10.1126/science.1197785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268:880–884. [DOI] [PubMed] [Google Scholar]
- 31.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. 2001. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol 21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ness F, Ferreira P, Cox BS, Tuite MF. 2002. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol 22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helsen CW, Glover JR. 2012. Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J Biol Chem 287:542–556. doi: 10.1074/jbc.M111.302869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandrov AI, Polyanskaya AB, Serpionov GV, Ter-Avanesyan MD, Kushnirov VV. 2012. The effects of amino acid composition of glutamine-rich domains on amyloid formation and fragmentation. PLoS One 7:e46458. doi: 10.1371/journal.pone.0046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wadsworth JDF, Hill AF, Beck JA, Collinge J. 2003. Molecular and clinical classification of human prion disease. Br Med Bull 66:241–254. doi: 10.1093/bmb/66.1.241. [DOI] [PubMed] [Google Scholar]
- 36.Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. 1998. Prion protein biology. Cell 93:337–348. [DOI] [PubMed] [Google Scholar]
- 37.Vitrenko YA, Pavon ME, Stone SI, Liebman SW. 2007. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr Genet 51:309–319. doi: 10.1007/s00294-007-0127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexandrov IM, Vishnevskaya AB, Ter-Avanesyan MD, Kushnirov VV. 2008. Appearance and propagation of polyglutamine-based amyloids in yeast: tyrosine residues enable polymer fragmentation. J Biol Chem 283:15185–15192. doi: 10.1074/jbc.M02071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. 1997. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J 16:1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rüdiger S, Schneider-Mergener J, Bukau B. 2001. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J 20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman F. 1991. Getting started with yeast. Methods Enzymol 194:3–21. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez Nelson AC, Paul KR, Petri M, Flores N, Rogge RA, Cascarina SM, Ross ED. 2014. Increasing prion propensity by hydrophobic insertion. PLoS One 9:e89286. doi: 10.1371/journal.pone.0089286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song Y, Wu Y-X, Jung G, Tutar Y, Eisenberg E, Greene LE, Masison DC. 2005. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot Cell 4:289–297. doi: 10.1128/EC.4.2.289-297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris JA, Gardner MJ. 1988. Statistics in medicine: calculating confidence intervals for relative risks (odds ratios) and standardised ratios and rates. BMJ 296:1313–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agresti A. 2007. An introduction to categorical data analysis, 2nd ed. Wiley, Hoboken, NJ. [Google Scholar]
- 46.Tyedmers J, Madariaga ML, Lindquist S. 2008. Prion switching in response to environmental stress. PLoS Biol 6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox BS. 1965. Ψ, a cytoplasmic suppressor of super-suppressor in yeast. Heredity (Edinb) 20:505–521. [Google Scholar]
- 48.Tuite MF, Mundy CR, Cox BS. 1981. Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics 98:691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung G, Masison DC. 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol 43:7–10. doi: 10.1007/S002840010251. [DOI] [PubMed] [Google Scholar]
- 50.Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol 40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- 51.Street AG, Mayo SL. 1999. Intrinsic beta-sheet propensities result from van der Waals interactions between side chains and the local backbone. Proc Natl Acad Sci U S A 96:9074–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bondarev SA, Shchepachev VV, Kajava AV, Zhouravleva GA. 2013. Effect of charged residues in the N-domain of Sup35 protein on prion [PSI+] stability and propagation. J Biol Chem 288:28503–28513. doi: 10.1074/jbc.M113.471805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du Z. 2011. The complexity and implications of yeast prion domains. Prion 5:311–316. doi: 10.4161/pri.18304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reidy M, Masison DC. 2011. Modulation and elimination of yeast prions by protein chaperones and co-chaperones. Prion 5:245–249. doi: 10.4161/pri.17749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGlinchey RP, Kryndushkin D, Wickner RB. 2011. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci U S A 108:5337–5341. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toyama BH, Kelly MJS, Gross JD, Weissman JS. 2007. The structural basis of yeast prion strain variants. Nature 449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 57.Fawcett T. 2006. An introduction to ROC analysis. Pattern Recognit Lett 27:861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.