Abstract
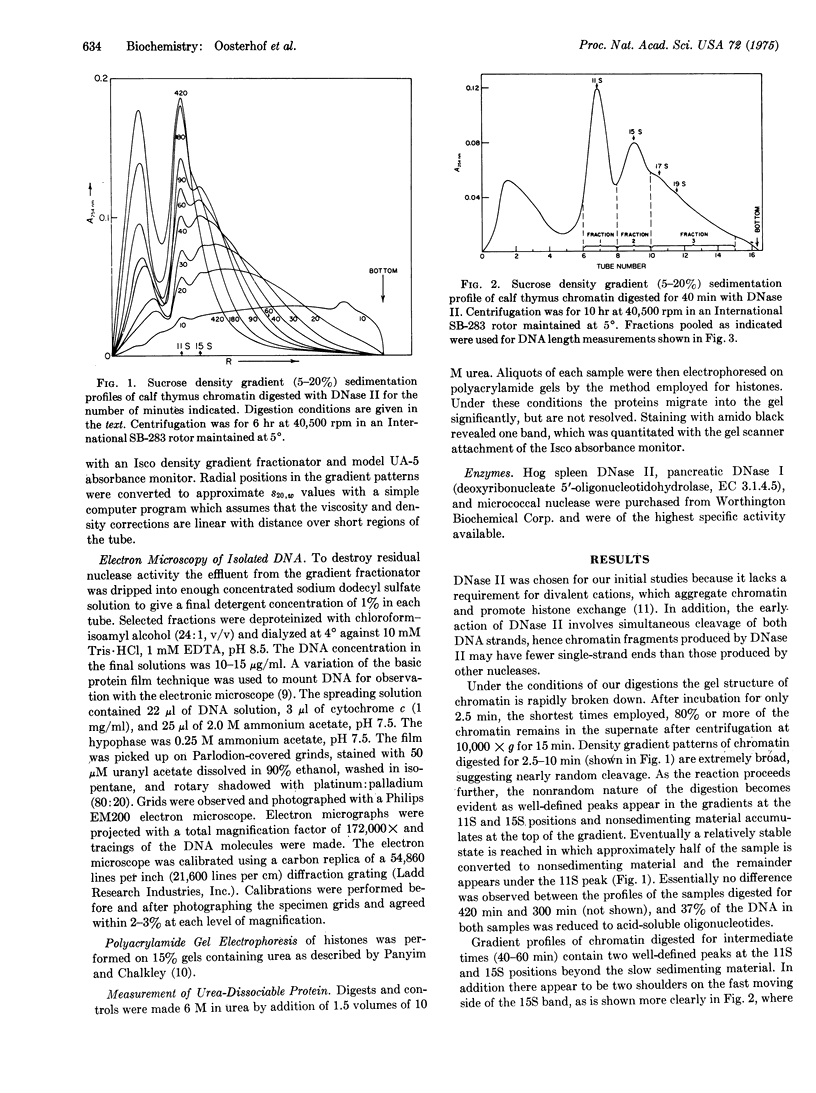
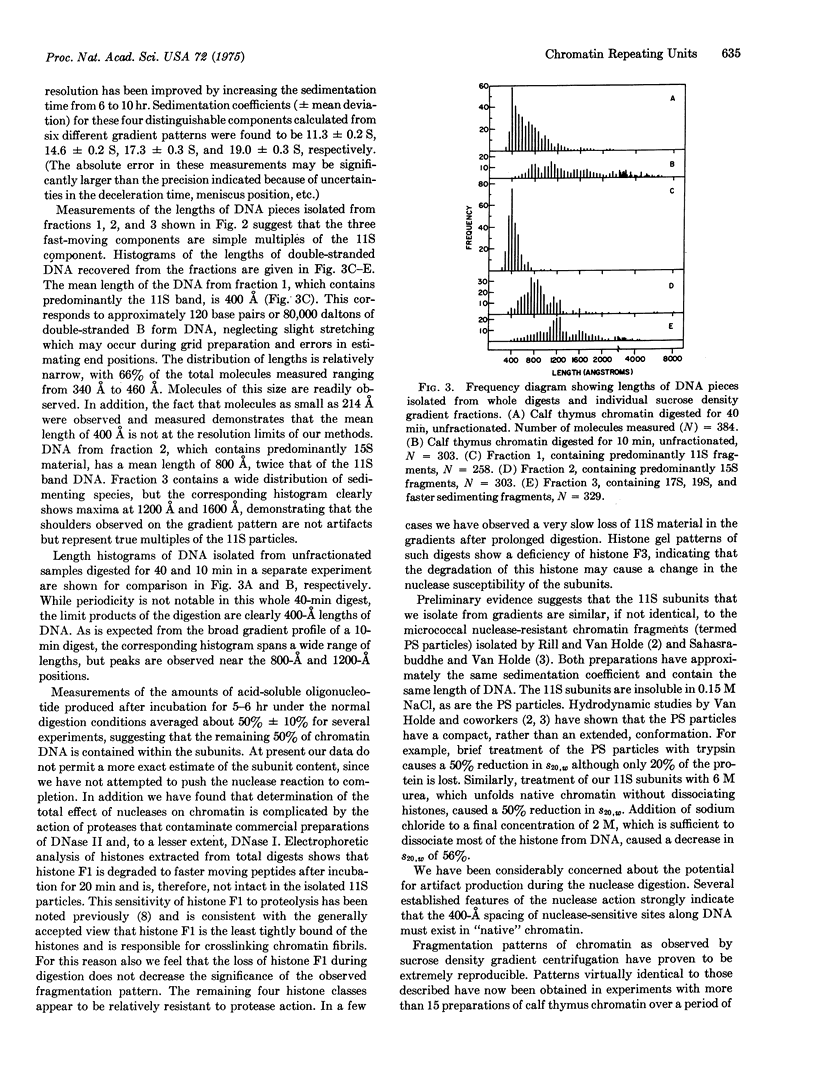
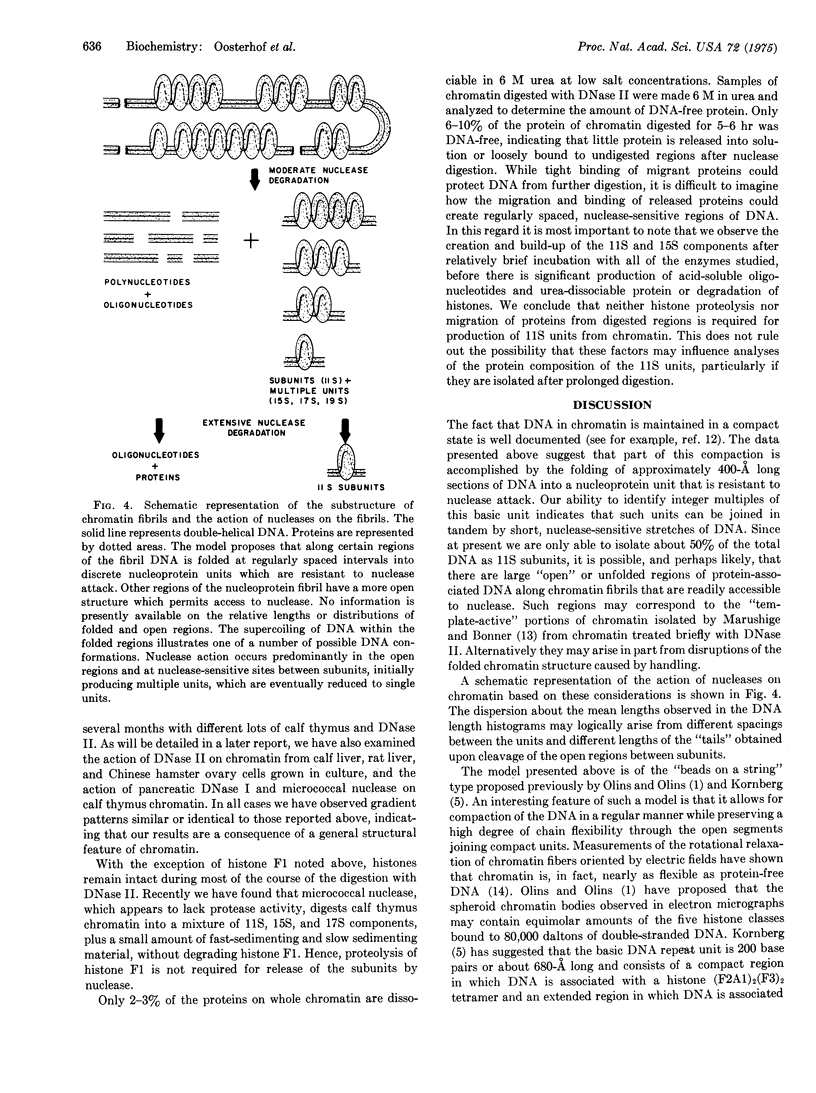
The time course of the fragmentation of calf thymus chromatin by DNase II (deoxyribonucleate 3'-oligonucleotidohydrolase, EC 3.1.4.6) has been examined by sedimentation of chromatin digests through linear (5-20%) sucrose gradients. The action of nuclease is decidedly nonrandom, ultimately producing roughly equal amounts of acid-soluble oligonucleotides and 11S nucleoprotein particles. The 11S particles contain double-stranded DNA that is approximately 400 A or 120 base-pairs long, as measured by electron microscopic examination of deproteinized samples, and is maintained in a compact conformation within the intact particles. In addition, 15S nucleoprotein particles containing predominantly 800-A lengths of DNA have been isolated from less extensively digested chromatin. Evidence is presented which indicates that the 11S particles are fundamental structural units that are arranged in tandem along certain regions of chromatin fibrils. Preliminary experiments with different nucleases and with chromatin from different mammalian species indicate that these results are a natural consequence of the arrangement of DNA and proteins in mammalian chromatin and are not peculiar to the system described in detail.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartley J., Chalkley R. Further studies of a thymus nucleohistone-associated protease. J Biol Chem. 1970 Sep 10;245(17):4286–4292. [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Association of arginine-rich histones with G-C-rich regions of DNA in chromatin. Nat New Biol. 1972 Dec 20;240(103):226–229. doi: 10.1038/newbio240226a0. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Chemical probes of chromatin structure. Biochemistry. 1974 Aug 13;13(17):3622–3628. doi: 10.1021/bi00714a034. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Huberman J. A. Structure of chromosome fibers and chromosomes. Annu Rev Biochem. 1973;42:355–378. doi: 10.1146/annurev.bi.42.070173.002035. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Marushige K., Bonner J. Fractionation of liver chromatin. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2941–2944. doi: 10.1073/pnas.68.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Rill R., Van Holde K. E. Electric dichroism of chromatin. J Mol Biol. 1974 Mar 15;83(4):459–471. doi: 10.1016/0022-2836(74)90507-5. [DOI] [PubMed] [Google Scholar]
- Rill R., Van Holde K. E. Properties of nuclease-resistant fragments of calf thymus chromatin. J Biol Chem. 1973 Feb 10;248(3):1080–1083. [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]


