Abstract
The HIV-1 envelope glycoprotein, gp120, binds the host cell receptor, CD4, in the initial step of HIV viral entry and infection. This process is an appealing target for the development of inhibitory drugs and neutralizing antibodies. To study gp120 binding and intracellular trafficking, we engineered a fluorescent fusion of the humanized gp120 JRFL HIV-1 variant and GFP. Gp120-sfGFP is glycosylated with human sugars, robustly expressed, and secreted from cultured human cells. Protein dynamics, quality control, and trafficking can be visualized in live cells. The fusion protein can be readily modified with different gp120 variants or fluorescent proteins. Finally, secreted gp120-sfGFP enables a sensitive and easy binding assay that can quantitatively screen potential inhibitors of gp120-CD4 binding on live cells via fluorescence imaging or laser scanning cytometry. This adaptable research tool should aid in studies of gp120 cell biology and the development of novel anti-HIV drugs.
Keywords: gp120, fluorescent protein, superfolder GFP, HIV-1, neutralizing antibody, laser scanning cytometry, FRAP, diffusion, CD4, envelope, inhibitory antibody
Introduction
Despite several successes in development of HIV therapeutics, an effective vaccine and cure remain elusive (Schiffner et al., 2013). The first step in viral infection is the binding of the HIV-1 envelope glycoprotein 120 (gp120) to the cellular receptor CD4, which is a major target for drug and vaccine development. Two drugs, Maraviroc and Enfuvirtide, inhibit the subsequent interactions between gp41 and the co-receptors CCR5 or CXCR4, but there are no approved drugs to inhibit gp120 binding to CD4. Broadly neutralizing antibodies (BNAbs), named for their ability to neutralize infectivity of a broad spectrum of primary HIV strains, have been identified in HIV-1 infected individuals (Simek et al., 2009) and these may aid in identification of gp120 epitopes that could be targeted for vaccine and drug development (Mascola and Haynes, 2013). Of particular interest are antibodies targeting the CD4-binding site of gp120 (Li et al., 2007). This interaction is critical for viral attachment, independent of HIV-1 virus strain. Furthermore, BNAbs potentially could be exploited to target steps of the viral life cycle that are not included in current ART (anti-retroviral therapy) drug regimens, to prevent viral rebound after cessation of ART, and to treat individuals who do not tolerate ART or develop drug resistance (Klein et al., 2013). In a related approach, potent small molecule inhibitors of viral entry that modulate binding of gp120 and CD4 have been developed. These inhibitors include BMS-378806, which binds gp120 and disrupts conformational changes required for binding to CD4 (Si et al., 2004), and the recently identified CD4-mimetic compounds, such as NBD-556 (Schon et al., 2006) and JR-II-191 (Haim et al., 2009), which can bind monomeric gp120, and block interactions with CD4. Finally it is important to note that gp120, itself, (even in the absence of virus and CD4) is pathogenic in HIV-associated neurocognitive disorders (HAND). Binding of gp120 to receptors unrelated to CD4 on neurons can trigger apoptosis (Rao et al., 2014) and emphasizes the importance of studying gp120 interactions with cells.
An easily modifiable fluorescent gp120 protein could aid in the study of HAND, the development of new therapeutics, and to evaluate inhibitors of CD4 binding. While commercial fluorescently labeled purified gp120 has been available, they have suboptimal characteristics. The products have been FITC dye-labeled gp120 expressed by insect cells, which is suboptimal because: 1) protein glycosylation patterns differ greatly between insect and human cells (Kost et al., 2005), which profoundly alters gp120 affinity for various BNAbs (Kong et al., 2010); 2) FITC is pH sensitive and highly prone to photobleaching (Swanson, 1989); 3) chemical dye-labeling, including FITC, is random and could potentially interfere with CD4 or antibody binding domains; and 4) investigators cannot modify the labeled commercial reporter with mutations or dyes for different assay needs. Here, we describe a new fluorescent protein (FP) tagged construct that can be expressed robustly in human cells and circumvents aforementioned pitfalls. The FP label enables assay development with a minimal number of manipulations. The new fluorescent reporter can be used to rapidly screen candidate drugs or antibodies using high-throughput platforms, enables visualization of trafficking in both cells that produce gp120 and cells that bind and endocytose gp120, and the reporter can be readily adapted to meet the requirements of new assays.
Results
Gp120-sfGFP construct design and characterization
In vitro production of gp120 at experimentally useful levels was initially limited by features of the native viral protein. HIV-1 envelope protein expression is significantly enhanced by a rev-responsive element in the mRNA and the small RNA-binding HIV protein rev (Felber et al., 1989). However, replacement of numerous rare human codons within the gp120 sequence with human preferred codons increased its expression and eliminated the requirement for co-expression of rev in mammalian cells (Haas et al., 1996).
Proper secretion and fluorescent tagging of a viral protein also present potential challenges. To visualize gp120 in CD4 binding assays, investigators assumed that tagging gp120 with a FP might sterically hinder interactions with CD4 (Bowley et al., 2007). It was also previously suggested that FP-tagged gp120 is not secreted because the signal sequence (SS) remains attached to the protein as a transmembrane domain (Wang and Pang, 2008). We sought to carefully evaluate a FP-tagged gp120 with the goal of improving the construct in several important ways: 1) addition of human sugars; 2) discrete and reproducible fluorescent labeling; and 3) development of a platform for easy manipulation of gp120 to create a powerful adaptable tool for investigators.
We began with a human codon optimized sequence of gp120 JRFL HIV-1 variant (Trkola et al., 1996) containing the native SS fused upstream of a FP (Figure 1A). We chose superfolder GFP (sfGFP) (Pedelacq et al., 2006) because other GFPs frequently misfold and form dark proteins in the oxidizing environment of the endoplasmic reticulum (ER) lumen (Jain et al., 2001), whereas sfGFP is resistant to formation of nonnative disulfide bonds (Aronson et al., 2011). In addition, gp120-sfGFP has the advantage of a single, genetically encoded, highly stable FP tag. SfGFP has a much greater resistance to photobleaching and lower pKa value (5.5) compared to FITC dyes (6.5), which translates to brighter sfGFP fluorescence in slightly acidic cellular compartments.
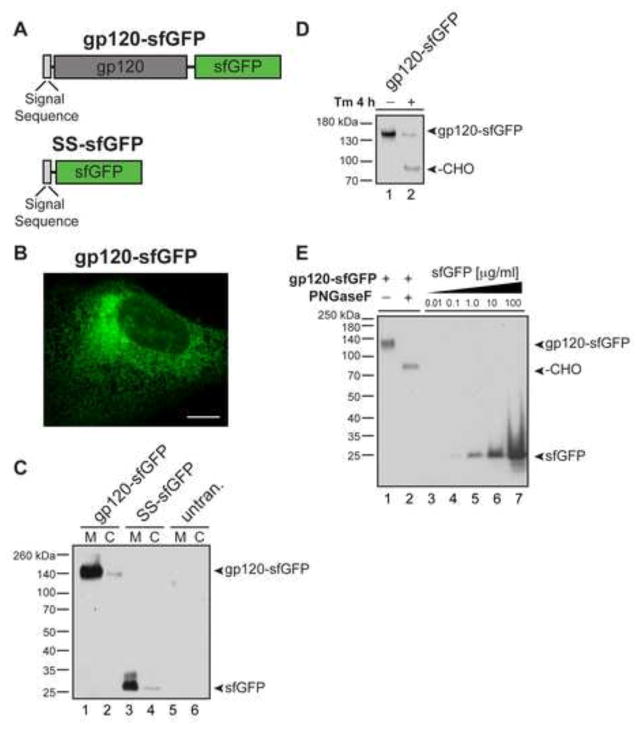
Fig. 1.
Secreted gp120-sfGFP is expressed in human cells and acquires N-linked glycans. (A) Human codon optimized gp120, including the native signal sequence, was fused upstream to sfGFP to create gp120-sfGFP. The control SS-sfGFP construct contains the bovine prolactin signal sequence (SS) followed by sfGFP. (B) Representative image of transiently transfected U-2 OS cell expressing gp120-sfGFP, scale bar = 10 μm. Media (M) and cell lysates (C) were collected and examined by immunoblotting with anti-GFP antibody from U-2 OS transiently transfected with (C) gp120-sfGFP and SS-sfGFP or (D) gp120-sfGFP with or without 5 μg/mL tunicamycin (Tm). Shift to lower molecular weight indicates non-glycosylated (-CHO) gp120-sfGFP. (E) To determine approximate concentration of gp120-sfGFP collected, serial dilutions of purified sfGFP samples of known concentration (lanes 3–7) were compared with PNGase treated gp120-sfGFP (lane 2) by immunoblotting with anti-GFP antibody.
For protein production, the human U-2 OS cell line was selected for high transfection efficiency and ability to produce robust yields of secreted gp120-sfGFP. The gp120-sfGFP plasmid and a control plasmid expressing the prolactin SS followed by sfGFP (SS-sfGFP) were transiently transfected in cells (Figure 1A). Both gp120-sfGFP and SS-sfGFP were efficiently localized to the secretory pathway and subsequently secreted from cells. Transient transfection of cells resulted in bright expression of gp120-sfGFP localized to the ER and the secretory pathway, but not to the plasma membrane (Figure 1B). We did not observe a transmembrane form of FP-tagged gp120 trapped on the cell surface.
N-linked glycosylation of gp120 is required for proper gp120 folding and for binding to CD4 (Li et al., 1993). We confirmed that gp120-sfGFP is extensively glycosylated by analyzing culture media and cell lysates by immunoblot (Figure 1C, D). Gp120-sfGFP acquired N-linked glycans, and migrated to the expected molecular weight (144 kDa). Treatment of cells expressing gp120-sfGFP with tunicamycin, a GlcNAc phosphotransferase inhibitor, blocks N-glycosylation and produced a shift to a lower molecular weight band (-CHO) corresponding to species lacking N-linked glycans (Figure 1D, Lanes 1 and 2). Finally, prevention of glycosylation with tunicamycin or removal of sugars with PNGaseF established that sugars contribute ~60 kDa to gp120-sfGFP (Figure 1E).
The large number of disulfide bonds in gp120 presents numerous opportunities for protein misfolding. It has been previously reported that secreted gp120 contains an aberrant covalent dimeric form (Finzi et al., 2010). We also observe this species (Supplementary Fig. 1A). Whereas Finzi et al. observed 20–30% dimeric form, the sfGFP fusion resulted in up to 70% covalent dimer. Importantly though, we found that the monomeric form could be readily purified using a size fractionation column (Supplementary Fig. 1B).
To determine the approximate concentration of secreted gp120-sfGFP collected, serial dilutions of sfGFP samples of known concentration (Figure 1E, Lanes 3–7) were compared with PNGase treated gp120-sfGFP by immunoblot (Figure 1E, Lane 2) and established that concentrated cell media contained ~3.4 μg/mL of secreted gp120-sfGFP. Taken together, we successfully generated a fluorescent gp120 protein decorated with human N-linked sugars.
Folding and trafficking of gp120-sfGFP in living cells
Until now, trafficking studies of gp120 have been restricted to fixed cells followed by antibody labeling or gp120 has been randomly labeled with fluorescent dyes (Hewson et al., 2001). We readily visualized trafficking of gp120 in live transiently transfected HeLa cells (Supplementary Figure 2A). In contrast to a previous report (Wang and Pang, 2008), we did not detect a plasma membrane association in expressing cells, consistent with cleavage of the gp120 SS in the ER by signal peptidase. Gp120-sfGFP was detected in two compartments, the tubular network of the ER and small highly mobile vesicles (Supplementary Fig. 2A). Future studies can exploit gp120-sfGFP to quantify rates of trafficking to and from secretory organelles in single cells with sub-second resolution.
Correct folding of proteins is essential for viral production and function. Assessment of folding can be determined with a variety of assays that typically require lysis and mixing of millions of cells followed by SDS-PAGE. While these techniques are undeniably powerful, they lack single cell resolution and do not permit interrogation of the same cell at multiple time points. For studies of gp120 protein folding events, it would be useful to have complementary live cell reporters. Gp120-sfGFP, when combined with photobleaching assays, enables quantitative detection of changes in protein folding within the ER. Fluorescence Recovery after Photobleaching (FRAP) can be used to determine effective diffusion coefficients (Deff values) of fluorescent molecules in cells (Supplementary Fig. 2B) (Lippincott-Schwartz et al., 2001). We performed FRAP on gp120-sfGFP in transiently transfected cells under homeostatic and ER stress (DTT treatment) conditions (Supplementary Fig. 2C). In the absence of stress, gp120 is fully mobile, but diffuses relatively slowly (D= 0.49 μm2/s), presumably due to its size when decorated with several 1.4 nm N-linked sugars. The much smaller reporter ER-mRFP diffuses rapidly at 8.5 μm2/s. During acute protein misfolding stress, ER-mRFP diffusion decreases only slightly (6.6 μm2/s) and this was not statistically significant (p= 0.0852). In contrast, gp120 contains nine disulfide bonds (Louwagie et al., 1995) and their formation is essential for correct gp120 folding (van Anken et al., 2008). Exposure to DTT (30 min) led to a rapid and profound 5-fold decrease in D (D = 0.07 μm2/s), consistent with an increase in size due to a less compact incompletely folded structure, aggregation, and/or binding to ER chaperones, such as BiP.
Gp120-sfGFP binds CD4-expressing cells
To determine whether the gp120-sfGFP was functionally capable of binding CD4, TZMbl cells (HeLa cells stably expressing CD4 and CCR5), were exposed to gp120-sfGFP, SSsfGFP or control media for 1 h. Incubation with gp120-sfGFP resulted in bright fluorescence labeling of the exterior edge of cells (Figure 2). The fluorescence intensity of sfGFP corresponded to a concentration gradient of gp120-sfGP added to the cells (Figure 2A). The largest two volumes of 20 and 40 μl, which corresponds to ~0.2 and 0.45 μg/mL of gp120-sfGFP, were readily detected (Figure 2A). Similar results were observed with purified monomeric gp120-sfGFP (Supplementary Fig. 1D). Subsequent experiments were conducted utilizing ~0.45 μg/mL of gp120-sfGFP for maximal signal to noise. Incubation with SS-sfGFP and control media did not label cells with fluorescence (Figure 2B). Additionally, the parental cell line HeLa was exposed to 40 μl of gp120-sfGFP; no fluorescence labeling was observed (Figure 2C).
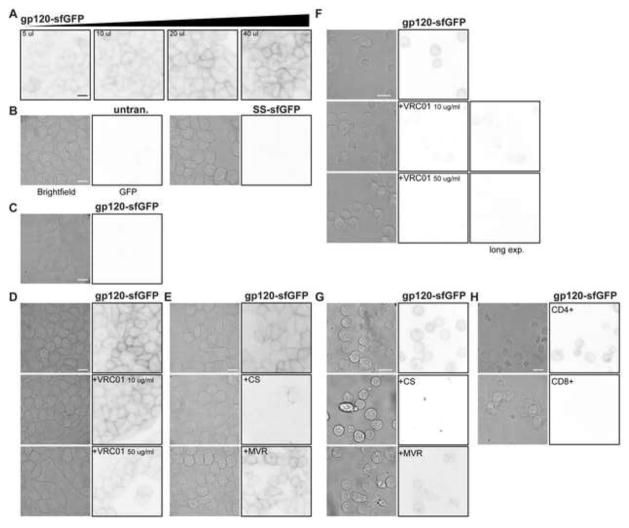
Fig. 2.
gp120-sfGFP binds CD4/CCR5 expressing TZM-bl cells. TZM-bl or HeLa cells were plated in chamber slides and incubated overnight. TZM-bl cells were then exposed to (A) a range of concentrations of gp120-sfGFP, 5–40 μL (~0.2–0.45 μg/mL) and (B) SS-sfGFP and control media. (C) Parental HeLa cells were exposed to 40 μl gp120-sfGFP and no binding was apparent. TZM-bl cells were co-incubated with 40 μL (~0.45 μg/mL) gp120-sfGFP and (D) 10 or 50 μg/ml VRC01 antibody, (E) 100 μg/ml cellulose sulfate (CS) or 10 μM Maraviroc (MVR). Brightfield and inverted fluorescence images were used to determine the fluorescence binding pattern; images are representative of cells from at least two independent experiments, scale bar = 10 μm. (E–G) gp120-sfGFP binds primary lymphocytes. After three days of stimulation with PHA, PBMCs were incubated with 40 μL (~0.45 μg/mL) gp120-sfGFP or controls in the presence or absence of the inhibitors (E) VRC01 antibody, (F) cellulose sulfate (CS) or Maraviroc (MVR). After incubation, PBMCs were transferred to lysine-coated coverglass chamber slides to promote PBMC attachment to the glass for fluorescence microscopy. (G) After 3 days of PHA-stimulation, CD8-negative (labeled as CD4-positive) and CD8-positive (labeled as CD8-positive) PBMC were incubated with gp120-sfGFP for 1 h. Brightfield paired with inverted fluorescence images were used to determine the fluorescence binding pattern. Incubation of PBMCs with SS-sfGFP or control media did not result in fluorescence (data not shown). Images are representative of cells from at least two different donors, scale bar = 10 μm.
To determine the effect of an inhibitory antibody on gp120-sfGFP binding, TZM-bl cells were co-incubated with gp120-sfGFP and the anti-gp120 BNAb VRC01. Fluorescent images acquired using equivalent incubation conditions revealed that VRC01 decreased gp120-sfGFP binding in a dose dependent manner (Figure 2D). These results were further confirmed with experiments conducted with purified monomeric gp120-sfGFP (Supplementary Fig. 3). Similarly, to determine the effect of an antimicrobial compound on gp120-sfGFP binding, 100 μg/ml of cellulose sulfate (CS), a sulfonated polymer which competitively blocks binding of gp120 to CD4 (Moulard et al., 2000; Scordi-Bello et al., 2005), also markedly reduced gp120-sfGFP binding (Figure 2E). In contrast, binding was unaffected by the CCR5 antagonist Maraviroc (10 μM) as expected (Figure 2E).
To evaluate gp120-sfGFP binding to primary cells, PHA-stimulated human PBMC were incubated with gp120-sfGFP, SS-sfGFP or control media. As previously demonstrated with TZM-bl cells, incubation of PBMC with gp120-sfGFP resulted in intense GFP fluorescence (Figure 2F). The addition of VRC01 (10 and 50 μg/ml) decreased gp120-sfGFP binding, even when viewing fluorescence intensity with longer exposure time (Figure 2F). Moreover, 100 μg/mL cellulose sulfate, but not 10 μM Maraviroc, also inhibited gp120-sfGFP binding to PBMC (Figure 2G). To assess binding specificity, PBMC were subdivided into CD8-positive and CD8-negative populations by positive selection with nanoparticles and PHA-stimulated, before incubation with gp120-sfGFP and controls. Intense GFP fluorescence was observed on CD8-negative cells (primarily CD4 T cells) whereas CD8-positive cells did not exhibit GFP fluorescence (Figure 2H).
Endocytic trafficking of gp120-sfGFP
In our studies of gp120-sfGFP binding to TZM-bl cells, we observed that the protein localized to both the cell surface and an internal perinuclear compartment, most likely endosomes (Figure 3). No internal labeling of parent HeLa cells was observed (Figure 3). Visualization of binding to and endocytosis of gp120-sfGFP into live cells may prove useful in studies of HAND pathogenesis and gp120-mediated loss of CD4 (Hewson et al., 2001). In Figure 3, gp120-sfGFP was visualized in various endocytic compartments after an initial binding step, in which no internalized gp120-sfGFP is detectable (0 min). Within minutes, gp120-sfGFP localizes to early endosomes (rab5 and then rab7) and later in lysosomes (identified by LAMP-1 association). Gp120 trafficking can be followed out of and into cells. This reagent or an even brighter variant using a brighter FP, such as mNeonGreen (Shaner et al., 2013), may enable researchers, for the first time, to follow gp120 production and uptake in a closed system.
Fig. 3.
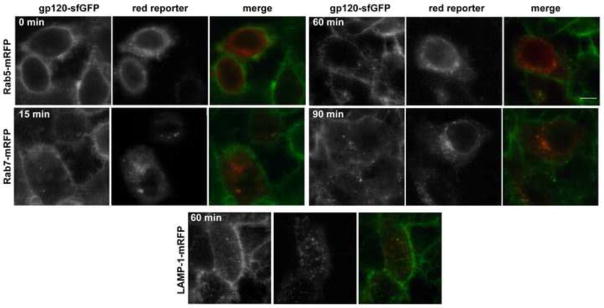
gp120-sfGFP uptake and trafficking in living cells. Endocytosis and colocalization of gp120-sfGFP in TZM-bl cells transiently transfected with Rab5-mRFP, Rab7-mRFP or LAMP-1-mRFP. Confocal images of cells at different times during incubation reveals progressive trafficking of gp120-sfGFP from early endosomes to lysosomes. Scale bars = 10 μm.
Antibody screening with gp120-sfGFP
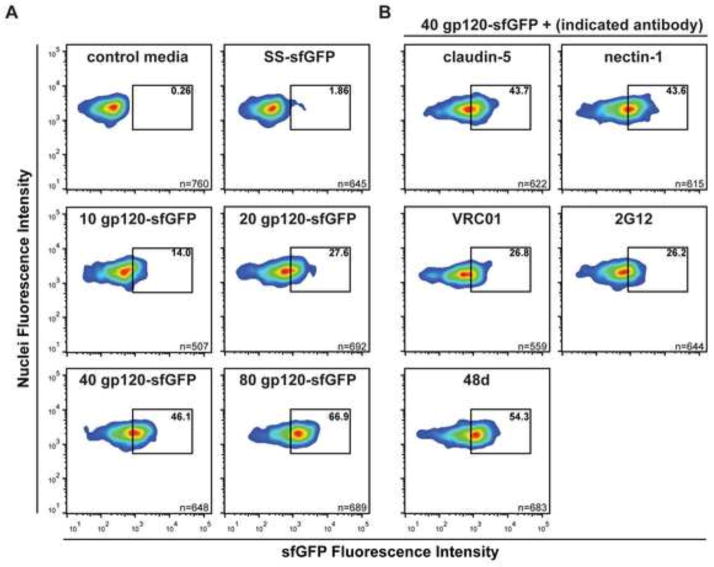
To evaluate gp120-sfGFP for high throughput screening of drugs or antibodies, we performed Laser Scanning Cytometry (LSC), a powerful high throughput fluorescence microscopy technology that generates large quantitative data sets for adherent cells with subcellular resolution. With several similarities to flow cytometry (Mach et al., 2010; Pozarowski et al., 2013), the data generated by LCS can be presented as dot-plots and histograms. TZM-bl cells were plated 18 h prior to incubation with gfp120-sfGFP, controls, and inhibitors for 1 h. As observed with fluorescence microscopy, SS-sfGFP and control media did not significantly label cells (Figure 4A, top). Incubation of TZM-bl cells with increasing amounts of gp120-sfGFP resulted in a corresponding quantitative increase in the number of GFP positive cells (Figure 4A, middle and bottom). Gp120-sfGFP labeling of cells (40 μl) was reduced by co-incubation with established anti-gp120 BNAbs: VRC01, which binds to the CD4 binding site of gp120 (Zhou et al., 2010), reduced binding of gp120-sfGFP to 26.8% (Fig. 4B). 2G12, which binds to the outer domain glycans of gp120 at a site distinct from the CD4 binding site (Sanders et al., 2002; Scanlan et al., 2002), also decreased the binding to 26.2% (Figure 4B). Possibly, this reflects distal effects of 2G12 on CD4 binding to gp120, such as by the introduction of allosteric constraints by the binding of 2G12 to target gp120 N-glycans, which inhibits residue movements required for the optimal binding of CD4 to gp120 (Platt et al., 2012). Note that 2G12 can not bind the sugars that decorate insect cell glycoproteins, emphasizing the importance of using recombinant gp120 from human cells. As expected, binding was unaffected by mAb 48d, which inhibits gp120 binding to the co-receptor CCR5 (Trkola et al., 1996) (Figure 4B). Importantly, isotype-matched control antibodies (claudin-5 and nectin-1) did not affect gp120-sfGFP binding to TZM-bl cells (Figure 4B).
Fig. 4.
gp120-sfGFP can be used to screen gp120 inhibitors by LSC. TZM-bl cells grown overnight in optical imaging microplates were exposed to (A) a range of concentrations of gp120-sfGFP in the presence or absence of (B) 10 μg/mL anti-HIV antibodies (VRC01, 2G12, and 48d) or 10 μg/mL isotype-matched control antibodies (claudin-5 and nectin-1). After 1 h of incubation, media was replaced and nuclei were stained using Hoechst; LSC was then used to identify nuclei and GFP intensities in 10 fields per well with triplicate wells. The SS-sfGFP control was used to set the negative gate to enable detection of specific GFP cell labeling. Results are representative of two independent experiments.
Conclusions
In summary, we have described a powerful new fluorescent tool for studies of gp120 in live cells. The gp120-sfGFP fusion protein can be readily expressed and visualized in single cells to enable studies of the endocytic and exocytic trafficking of gp120. Additionally, by combining gp120-sfGFP with photobleaching techniques in live cells, we found that, unlike another viral envelope protein, VSV G-GFP, misfolded gp120 is effectively immobilized in the ER lumen. This difference could arise for at least two significant and not mutually exclusive reasons. First, diffusion in a membrane is much less sensitive to size changes of a molecule and this may mask the effects of chaperone binding on an unfolded protein. However, this seems unlikely to fully account for the dramatic decrease in misfolded gp120 mobility. Another explanation is that by uncoupling the protein from the ER membrane, all of the protein can be completely accessed and bound by ER chaperones. Regions close to the membrane are likely to be sterically unavailable to chaperones. Thus, the soluble protein could potentially bind more chaperones, creating a larger and less mobile complex. More fundamentally, our findings and those of others (Finzi et al., 2010; Trkola et al., 1996), suggest that artificially untethering viral proteins from membranes, inadvertently increases potential folding and misfolding outcomes including novel dimers or more extensive protein misfolding. This is not to suggest that soluble membrane proteins lack utility. Rather, as with any reagent, characterization of reporters can help avoid pitfalls and aid interpretation of results.
Curiously, gp120-sfGFP behaved quite differently from another viral envelope protein, vesicular stomatitis virus G protein (VSV G). DTT and other inhibitors of post-translational folding modifications in the ER disrupt VSV G folding and can induce formation of very large complexes, which can be detected by ultracentrifuge gradient separation (de Silva et al., 1993). In a live cell photobleaching study by Nehls et al., VSV G-GFP mobility did not decrease in cells treated with DTT and remained highly mobile (Nehls et al., 2000). Our data with the inert ER-mRFP reporter established that gp120-sfGFP misfolding did not grossly alter the luminal environment. One explanation is that integration of VSV G into the ER membrane could dampen the effects of size changes on diffusion (i.e. a 10-fold increase in size decreases diffusion by half) (Saffman and Delbruck, 1975). In contrast, the mobility of soluble proteins is inversely proportional to changes in molecular size (a 2-fold increase in size decreases mobility by half) (Einstein, 1905). It is also possible that the extensively glycosylated gp120 would be effectively much larger in an unfolded state. To date, the organization and dynamics of few misfolded secretory proteins have been studied in live cells. Gp120-sfGFP represents a useful new reagent for basic research and mutants/variants of gp120-sfGFP can now be assayed for aspects of folding and quality control in their native environment.
The fluorescent gp120 binding assays described here provide a robust and rapid quantitative readout of gp120-CD4 binding. We have outlined the minimal steps to purify and assay gp120-sfGFP and provided examples of qualitative and quantitative methods to evaluate gp120-CD4 binding events. More sensitive analysis of binding inhibitors can be performed using live cell fluorescence microscopy with a high numerical aperture objective. For high-throughput approaches and large scale screens, LSC and similar technologies can assess effects with greater speed than possible with manual imaging.
An important feature of the gp120-sfGFP reagent is its adaptability for new assays. For example, even sfGFP is not ideal for acidic (pH ~4–5.5) endocytic cellular compartments. Thus, one can easily replace sfGFP with a low pKa FP, i.e. mCherry (pKa<4). For testing other Env variants, the JRFL sequence can be mutated as needed (Wu et al., 2010). More disparate Env sequences can be synthesized with human codon optimization. If desired, the gp120-sfGFP could be modified for purification, i.e. a Hisx6 tag or a Fc tag (Binley et al., 2006) following the sfGFP protein. Alternatively, the gp120-sfGFP construct could be affinity-purified by binding to an anti-gp120 or anti-GFP antibody. Given that there are at least two forms of gp120-sfGFP (monomers and covalent dimers), purification on a molecular sizing column is likely to be useful for some applications. We have demonstrated that the material can be readily fractionated and it is likely that a larger sizing column will increase yield of the monomer. Equally importantly, we note that the ratio of dimer to monomer with our fusion appears higher than that reported for gp120 alone (Finzi et al., 2010). This raises the possibility that sfGFP may contribute to dimer formation and we hypothesize the cysteines in sfGFP may be the cause. In future studies, we seek to identify monomeric bright cysteine-less fluorescent proteins for fusions to potentially further improve yield. In conclusion, the gp120-sfGFP provides a powerful and improved fluorescent tool with consistent labeling and human sugar modifications to examine CD4-binding, cellular uptake, and trafficking within live cells.
Materials and methods
Plasmids
The codon optimized gp120-sfGFP was generated by PCR amplification of JRFL gp120 (Trkola et al., 1996) with forward-GATCGCTAGCGCCACATGGATGCAATGAAGAGAGGG and reverse-GATCACCGGTCCCTTCTCCCTCTGCAC primers, the fragment was inserted into sfGFP N1 Clontech vector via NheI/AgeI restriction sites. The codon-optimized gp120-sfGFP was further modified to include the native Hxb2 gp120 signal sequence. PCR amplification of the codon optimized gp120-sfGFP fusion (described above) with forward-GATCGGTACCTGTGTGGAAG and reverse-GGTTCAGGGGGAGGTGT was inserted downstream of Hxb2 gp120 signal sequence with KpnI/NotI restriction sites. The parental vector HXB2 gp120 signal sequence-containing vector was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HXB2-env from Dr. Kathleen Page and Dr. Dan Littman (Page et al., 1990). SS-sfGFP was previously described (Aronson et al., 2011). Rab5-mRFP and Rab7-mRFP were produced by the laboratory of Dr. Ari Helenius (Vonderheit and Helenius, 2005) and obtained from Addgene (Cambridge, MA). LAMP-1-mRFP1 was created by the laboratory of Dr. Walter Mothes (Sherer et al., 2003). ER-mRFP was previously described (Snapp et al., 2006).
Cells
Peripheral blood mononuclear cells (PBMC) were isolated from human leukopacks (New York Blood Center) by gradient centrifugation using FicollPaque Plus (Sigma-Aldrich, St. Louis, MO). For isolation of T cell subpopulations, EasySep Human CD8 Positive Selection Kit (STEMCELL Technologies, Vancouver, BC) was used as recommended. Total PBMC or CD8-negaitve and CD8-positive populations were cultured for 3 days (1 × 106 cells/mL) in phenol red free RPMI 1640 (Gibco, Life Technologies, Grand Island, NY) containing 2 mM glutamine, 10% fetal bovine serum (FBS) (HyClone, Life Technologies), 100 U of penicillin/mL, 100 U of streptomycin/mL (Life Technologies), and 5 μg/mL phytohaemagglutinin (PHA) (Sigma-Aldrich) prior to use. TZM-bl (HeLa cells stably expressing CD4 and CCR5 as well as luciferase and β-galactosidase under the HIV-1 promoter) cells were obtained from the NIH AIDS Research and Reference Reagent Program and cultured as recommended with the exception of using phenol red free DMEM (Kappes and Wu, 2005). Human bone epithelial cells (U-2 OS; ATCC) and HeLa cells (ATCC) were cultured in RPMI medium supplemented with 5 mM glutamine, penicillin/streptomycin, and 10% heat inactivated FBS.
gp120-sfGFP expression and concentration
U-2 OS cells were cultured in RPMI phenol red free media containing 2% FBS, supplemented with 5 mM glutamine and penicillin/streptomycin. Gp120-sfGFP or SS-sfGFP plasmids were transiently transfected with Lipofectamine 2000 (Life Technologies) according to manufacturer instructions. Two days post-transfection, culture media was collected and centrifuged at 3,000 × g to remove cellular debris. Media was then concentrated using centrifugal 15 ml 10 kDa filter units (Millipore, Billerica, MA). Media was concentrated 20x, for gp120-sfGFP 40 mL of media was concentrated to 2 mL and SS-sfGFP and untransfected media, 10 mL of media was concentrated to 500 μL. Samples were stored at 4°C.
Immunoblot
U-2 OS were transfected with gp120-sfGFP in the presence (6 h) or absence of tunicamycin-treatment (Tm; Calbiochem, La Jolla, CA) and were lysed in 1% SDS, 0.1 M Tris, pH 8.0 with 100 mM DTT. Proteins were separated using a 6% Tris-tricine gel, transferred to nitrocellulose, probed with an anti-GFP antibody (a generous gift from Ramanujan S. Hegde, Laboratory of Molecular Biology, Cambridge, UK) and horseradish peroxidase-labeled anti-rabbit secondary antibody (Jackson Immunoresearch Laboratories, West Grove, PA), and developed using enhanced chemiluminescent reagents (Pierce, Rockford, IL), and exposed to X-ray film. The relative concentration of gp120-sfGFP was approximated by comparison with purified sfGFP of a known concentration. For gp120-sfGFP, the concentrated media was determined to contain ~3.4 μg/mL. Experiments were conducted with a dynamic range of dilutions corresponding to ~0.2–0.45 μg/mL.
Live cell fluorescence microscopy
PBMCs were plated and exposed to gp120-sfGFP in round-bottom 96-well tissue culture plates for 1 h at 37°C before being moved to 0.1% poly-L-lysine coated 8-well Nunc LabTek #1.0 borosilicate coverglass chambers (Nunc, Life Technologies, Rochester, NY). TZM-bl cells were grown overnight in 8-well Nunc LabTek #1.0 borosilicate coverglass chambers, cells were exposed to gp120-sfGFP or controls in the presence or absence of the indicated inhibitor for 1 h at 37°C and finally, phenol red free media was replaced for microscopy. Gp120-sfGFP images were acquired using an Axiovert 200 widefield fluorescence microscope (Carl Zeiss Microimaging, Inc., Thornwood, NY) with a 63x oil immersion 1.4 NA objective, and 470/40 excitation, 525/50 emission bandpass filter for sfGFP. Transiently transfected HeLa cells expressing gp120-sfGFP were imaged in a 37°C environmentally controlled chamber of a confocal microscope system (Zeiss LSM 5 LIVE microscope with Duoscan attachment; Carl Zeiss MicroImaging, Inc.) with a 63X/1.4 NA oil objective and a 489 nm 100mW diode laser with a 495–555 and 520–555 nm bandpass filter. Image analysis and composite figures were prepared using ImageJ (National Institutes of Health), Photoshop CS4 and Illustrator CS4 software (Adobe Systems, San Jose, CA).
FRAP was performed on the Zeiss LIVE with Duoscan. Data were analyzed using an in homogenous diffusion simulation (Siggia et al., 2000).
Antibodies and drugs
HIV-1 gp120 monoclonal antibody 2G12, 48D, and VRC01 were obtained from the NIH AIDS Reagent and Reference Program. Anti-claudin-5 and anti-nectin-1 antibodies (Santa Cruz Biotechnology, Dallas, TX) were used as isotype-matched control antibodies. Cellulose sulfate (Acros Organics, Life Technologies, Rockford, IL) was rehydrated in 1 x PBS. Maraviroc was provided by ViiV Healthcare.
Laser Scanning Cytometry
TZM-bl cells were plated in glass-bottom 96-well optical imagining microplates (Brooks Life Science, Chelmsford, MA) and incubated at 37°C O/N. Cells were exposed to constructs or control media in the presence or absence of the indicated inhibitors or controls for 1 h at 37°C. Cells were stained with 0.1 μg/mL Hoechst for 30 min, washed, and phenol red free media supplemented with 10 mM HEPES was added. Cells were then imaged with the iCys Research Imaging Cytometer (CompuCyte, Thorlabs Inc., Newton, NJ) using a 60X (Figure 4) or 40X (Supplementary Fig. 3) 0.9 NA objective. sfGFP was excited using a laser at 488 nm and fluorescence was collected with a 515–545 nm emission filter. Ten fields were imaged with biological triplicates for each condition specified. Using the iCyte (CompuCyte) imaging software individual cells were identified and analyzed for green fluorescence. Data were exported to FloJo (Treestar, Ashland, OR) for analysis of cytometric data.
Supplementary Material
Secreted gp120-sfGFP includes correct monomeric and aberrant dimeric species. Secreted media was concentrated and denatured with or without 100 mM DTT and then separated by SDS-PAGE and immunoblotted with anti-GFP. In the reducing lane, a single species is observed and runs at ~150 kDa. In the nonreducing lane three species are observed that likely correspond to monomeric, disulfide-bonded dimeric, and a potentially trimeric or tetrameric form. (B) Fractions of concentrated gp120sfGFP media (Start) separated over a Superdex 200 prep grade (GE Healthcare) column in 1X PBS and then immunoblotted with SDS-PAGE and anti-GFP antibody. Fractions 19–23 were pooled and further concentrated with a 10 kDa MWCO spin column. (C) The concentrated pooled material from B was diluted, run on SDS-PAGE, and immunoblotted with anti-GFP. (D) Indicated volumes of concentrated unpurified media or purified concentrated monomer were incubated in 150 ul volumes with TZM-bl cells for 1 h, washed and imaged with a Zeiss Axiovert, 63x NA 1.4 oil objective. Scale bar = 10 μm.
Dynamics of gp120-sfGFP in living cells. (A) Trafficking of gp120-sfGFP in HeLa cell transiently transfected with gp120-sfGFP and imaged with confocal microscopy. Secretory vesicles (yellow arrows) are highly dynamic throughout the cell. Thumbnail insets enlarge vesicle images 2X for ease of visualization. (B) FRAP series of U-2 OS cell co-expressing ER-mRFP and gp120-sfGFP. The unbleached proteins diffuse into the bleach regions (white boxes) at different rates, consistent with significant differences in size. (C) Quantitation of effective diffusion coefficients (Deff) in untreated cells and cells treated with 5 mM DTT for 30–60 min. Each circle represents a value for one cell. Scale bars = 10 μm.
Characterization of purified monomeric gp120-sfGFP. (A) iCys histograms of fluorescence intensities of populations of TZM-bl cells labeled with different amounts of purified concentrated monomeric gp120-sfGFP (see Supplementary Figure 1C). Increasing concentrations of gp120-sfGFP lead to increased GFP intensity. (B) Representative images (merged gp120-sfGFP and Hoechst 33342 top row, gp120-sfGFP staining alone in grayscale in lower row of panels) acquired by iCys of TZM-bl cells stained with monomeric gp120-sfGFP, and in the final two panels with two concentrations (10 and 50 μg/ml) of VRC01 antibody. Scale bar = 40 μm. (C) Plots of iCys data, analyzed as in Figure 4. Note the addition of 50 μg/ml VRC01 reduces monomeric gp120-sfGFP labeling of TZM-bl cells to background fluorescence levels.
Highlights.
Development of fluorescent protein labeled HIV-1 envelope gp120
Imaging of gp120 dynamics and trafficking in live cells
Quantitative visual assay of antibody-mediated inhibition of gp120 binding to CD4 on live cells
Acknowledgments
This work was supported in part by the Center for AIDS Research at the Albert Einstein College of Medicine and Montefiore Medical Center funded by the National Institutes of Health (NIH AI-051519 to ELS, T32A1007501 to SCI, and R01AI065309 to BCH). ELS was a recipient of a CFAR pilot award that helped support this research.
List of Abbreviations
- ART
anti-retroviral therapy
- BNAbs
broadly neutralizing antibodies
- CS
cellulose sulfate
- ER
endoplasmic reticulum
- FP
fluorescent protein
- FRAP
fluorescence recovery after photobleaching
- gp120
HIV-1 envelope glycoprotein
- MR
maraviroc
- PHA
phytohaemagglutinin
- sfGFP
superfolder GFP
- SS
signal sequence
- Tm
tunicamycin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aronson DE, Costantini LM, Snapp EL. Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic. 2011;12:543–548. doi: 10.1111/j.1600-0854.2011.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Ngo-Abdalla S, Moore P, Bobardt M, Chatterji U, Gallay P, Burton DR, Wilson IA, Elder JH, de Parseval A. Inhibition of HIV Env binding to cellular receptors by monoclonal antibody 2G12 as probed by Fc-tagged gp120. Retrovirology. 2006;3:39. doi: 10.1186/1742-4690-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowley DR, Labrijn AF, Zwick MB, Burton DR. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng Des Sel. 2007;20:81–90. doi: 10.1093/protein/gzl057. [DOI] [PubMed] [Google Scholar]
- de Silva A, Braakman I, Helenius A. Posttranslational folding of vesicular stomatitis virus G protein in the ER: involvement of noncovalent and covalent complexes. J Cell Biol. 1993;120:647–655. doi: 10.1083/jcb.120.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einstein A. Über die von der molekularkinetischen Theorie der Wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Ann Phys. 1905;17:549–560. [Google Scholar]
- Felber BK, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis GN. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci U S A. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi A, Pacheco B, Zeng X, Kwon YD, Kwong PD, Sodroski J. Conformational characterization of aberrant disulfide-linked HIV-1 gp120 dimers secreted from overexpressing cells. J Virol Methods. 2010;168:155–161. doi: 10.1016/j.jviromet.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB, 3rd, Sodroski J. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 2009;5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson TJ, Logie JJ, Simmonds P, Howie SE. A CCR5-dependent novel mechanism for type 1 HIV gp120 induced loss of macrophage cell surface CD4. J Immunol. 2001;166:4835–4842. doi: 10.4049/jimmunol.166.8.4835. [DOI] [PubMed] [Google Scholar]
- Jain RK, Joyce PB, Molinete M, Halban PA, Gorr SU. Oligomerization of green fluorescent protein in the secretory pathway of endocrine cells. Biochem J. 2001;360:645–649. doi: 10.1042/0264-6021:3600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes J, Wu X. Cell-based method and assay for measuring the infectivity and drug sensitivity of immunodeficiency virus. Google Patents 2005 [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Sheppard NC, Stewart-Jones GB, Robson CL, Chen H, Xu X, Krashias G, Bonomelli C, Scanlan CN, Kwong PD, Jeffs SA, Jones IM, Sattentau QJ. Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J Mol Biol. 2010;403:131–147. doi: 10.1016/j.jmb.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost TA, Condreay JP, Jarvis DL. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol. 2005;23:567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Luo L, Rasool N, Kang CY. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J Virol. 1993;67:584–588. doi: 10.1128/jvi.67.1.584-588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, Mascola JR. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- Louwagie J, Janssens W, Mascola J, Heyndrickx L, Hegerich P, van der Groen G, McCutchan FE, Burke DS. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J Virol. 1995;69:263–271. doi: 10.1128/jvi.69.1.263-271.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach WJ, Thimmesch AR, Orr JA, Slusser JG, Pierce JD. Flow cytometry and laser scanning cytometry, a comparison of techniques. J Clin Monit Comput. 2010;24:251–259. doi: 10.1007/s10877-010-9242-4. [DOI] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunological reviews. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulard M, Lortat-Jacob H, Mondor I, Roca G, Wyatt R, Sodroski J, Zhao L, Olson W, Kwong PD, Sattentau QJ. Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120. J Virol. 2000;74:1948–1960. doi: 10.1128/jvi.74.4.1948-1960.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls S, Snapp EL, Cole NB, Zaal KJ, Kenworthy AK, Roberts TH, Ellenberg J, Presley JF, Siggia E, Lippincott-Schwartz J. Dynamics and retention of misfolded proteins in native ER membranes. Nat Cell Biol. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- Page KA, Landau NR, Littman DR. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nature Biotech. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Platt EJ, Gomes MM, Kabat D. Kinetic mechanism for HIV-1 neutralization by antibody 2G12 entails reversible glycan binding that slows cell entry. Proc Natl Acad Sci U S A. 2012;109:7829–7834. doi: 10.1073/pnas.1109728109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozarowski P, Holden E, Darzynkiewicz Z. Laser scanning cytometry: principles and applications-an update. Methods Mol Biol. 2013;931:187–212. doi: 10.1007/978-1-62703-056-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VR, Ruiz AP, Prasad VR. Viral and cellular factors underlying neuropathogenesis in HIV associated neurocognitive disorders (HAND) AIDS Res Ther. 2014;11:13. doi: 10.1186/1742-6405-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman PG, Delbruck M. Brownian motion in biological membranes. Proc Natl Acad Sci U S A. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing antihuman immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffner T, Sattentau QJ, Dorrell L. Development of prophylactic vaccines against HIV-1. Retrovirology. 2013;10:72. doi: 10.1186/1742-4690-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB, 3rd, Sodroski J, Freire E. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry. 2006;45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scordi-Bello IA, Mosoian A, He C, Chen Y, Cheng Y, Jarvis GA, Keller MJ, Hogarty K, Waller DP, Profy AT, Herold BC, Klotman ME. Candidate sulfonated and sulfated topical microbicides: comparison of anti-human immunodeficiency virus activities and mechanisms of action. Antimicrob Agents Chemother. 2005;49:3607–3615. doi: 10.1128/AAC.49.9.3607-3615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang J. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer NM, Lehmann MJ, Jimenez-Soto LF, Ingmundson A, Horner SM, Cicchetti G, Allen PG, Pypaert M, Cunningham JM, Mothes W. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 2003;4:785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB, 3rd, Sodroski JG. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci U S A. 2004;101:5036–5041. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggia ED, Lippincott-Schwartz J, Bekiranov S. Diffusion in inhomogeneous media: theory and simulations applied to whole cell photobleach recovery. Biophysical journal. 2000;79:1761–1770. doi: 10.1016/S0006-3495(00)76428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp EL, Sharma A, Lippincott-Schwartz J, Hegde RS. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc Natl Acad Sci U S A. 2006;103:6536–6541. doi: 10.1073/pnas.0510657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. Fluorescent labeling of endocytic compartments. Methods Cell Biol. 1989;29:137–151. doi: 10.1016/s0091-679x(08)60192-2. [DOI] [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- van Anken E, Sanders RW, Liscaljet IM, Land A, Bontjer I, Tillemans S, Nabatov AA, Paxton WA, Berkhout B, Braakman I. Only five of 10 strictly conserved disulfide bonds are essential for folding and eight for function of the HIV-1 envelope glycoprotein. Mol Biol Cell. 2008;19:4298–4309. doi: 10.1091/mbc.E07-12-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderheit A, Helenius A. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 2005;3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Pang S. An intercellular adhesion molecule-3 (ICAM-3) -grabbing nonintegrin (DC-SIGN) efficiently blocks HIV viral budding. FASEB J. 2008;22:1055–1064. doi: 10.1096/fj.07-9443com. [DOI] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Secreted gp120-sfGFP includes correct monomeric and aberrant dimeric species. Secreted media was concentrated and denatured with or without 100 mM DTT and then separated by SDS-PAGE and immunoblotted with anti-GFP. In the reducing lane, a single species is observed and runs at ~150 kDa. In the nonreducing lane three species are observed that likely correspond to monomeric, disulfide-bonded dimeric, and a potentially trimeric or tetrameric form. (B) Fractions of concentrated gp120sfGFP media (Start) separated over a Superdex 200 prep grade (GE Healthcare) column in 1X PBS and then immunoblotted with SDS-PAGE and anti-GFP antibody. Fractions 19–23 were pooled and further concentrated with a 10 kDa MWCO spin column. (C) The concentrated pooled material from B was diluted, run on SDS-PAGE, and immunoblotted with anti-GFP. (D) Indicated volumes of concentrated unpurified media or purified concentrated monomer were incubated in 150 ul volumes with TZM-bl cells for 1 h, washed and imaged with a Zeiss Axiovert, 63x NA 1.4 oil objective. Scale bar = 10 μm.
Dynamics of gp120-sfGFP in living cells. (A) Trafficking of gp120-sfGFP in HeLa cell transiently transfected with gp120-sfGFP and imaged with confocal microscopy. Secretory vesicles (yellow arrows) are highly dynamic throughout the cell. Thumbnail insets enlarge vesicle images 2X for ease of visualization. (B) FRAP series of U-2 OS cell co-expressing ER-mRFP and gp120-sfGFP. The unbleached proteins diffuse into the bleach regions (white boxes) at different rates, consistent with significant differences in size. (C) Quantitation of effective diffusion coefficients (Deff) in untreated cells and cells treated with 5 mM DTT for 30–60 min. Each circle represents a value for one cell. Scale bars = 10 μm.
Characterization of purified monomeric gp120-sfGFP. (A) iCys histograms of fluorescence intensities of populations of TZM-bl cells labeled with different amounts of purified concentrated monomeric gp120-sfGFP (see Supplementary Figure 1C). Increasing concentrations of gp120-sfGFP lead to increased GFP intensity. (B) Representative images (merged gp120-sfGFP and Hoechst 33342 top row, gp120-sfGFP staining alone in grayscale in lower row of panels) acquired by iCys of TZM-bl cells stained with monomeric gp120-sfGFP, and in the final two panels with two concentrations (10 and 50 μg/ml) of VRC01 antibody. Scale bar = 40 μm. (C) Plots of iCys data, analyzed as in Figure 4. Note the addition of 50 μg/ml VRC01 reduces monomeric gp120-sfGFP labeling of TZM-bl cells to background fluorescence levels.