ABSTRACT
Biofilms are surface-attached multicellular communities. Using single-cell tracking microscopy, we showed that a pilY1 mutant of Pseudomonas aeruginosa is defective in early biofilm formation. We leveraged the observation that PilY1 protein levels increase on a surface to perform a genetic screen to identify mutants altered in surface-grown expression of this protein. Based on our genetic studies, we found that soon after initiating surface growth, cyclic AMP (cAMP) levels increase, dependent on PilJ, a chemoreceptor-like protein of the Pil-Chp complex, and the type IV pilus (TFP). cAMP and its receptor protein Vfr, together with the FimS-AlgR two-component system (TCS), upregulate the expression of PilY1 upon surface growth. FimS and PilJ interact, suggesting a mechanism by which Pil-Chp can regulate FimS function. The subsequent secretion of PilY1 is dependent on the TFP assembly system; thus, PilY1 is not deployed until the pilus is assembled, allowing an ordered signaling cascade. Cell surface-associated PilY1 in turn signals through the TFP alignment complex PilMNOP and the diguanylate cyclase SadC to activate downstream cyclic di-GMP (c-di-GMP) production, thereby repressing swarming motility. Overall, our data support a model whereby P. aeruginosa senses the surface through the Pil-Chp chemotaxis-like complex, TFP, and PilY1 to regulate cAMP and c-di-GMP production, thereby employing a hierarchical regulatory cascade of second messengers to coordinate its program of surface behaviors.
IMPORTANCE
Biofilms are surface-attached multicellular communities. Here, we show that a stepwise regulatory circuit, involving ordered signaling via two different second messengers, is required for Pseudomonas aeruginosa to control early events in cell-surface interactions. We propose that our studies have uncovered a multilayered “surface-sensing” system that allows P. aeruginosa to effectively coordinate its surface-associated behaviors. Understanding how cells transition into the biofilm state on a surface may provide new approaches to prevent formation of these communities.
INTRODUCTION
Bacteria are able to live as members of surface-attached microbial communities called biofilms. The formation of a biofilm in pseudomonads begins with cells “reversibly” attaching to a surface and progresses to more stable “irreversible” attachment. The transition from reversible to irreversible attachment is the first committed step in forming a biofilm (1). Alternatively, once cells contact a surface, they also have the ability to move across the surface using either type IV pilus (TFP)-mediated twitching motility or flagellator-mediated swarming motility (2–4). A comprehensive understanding of these early events remains elusive.
The intracellular second messenger cyclic di-GMP (c-di-GMP) is now appreciated as a master regulator for the transition between motile and sessile lifestyles in many bacterial species (5). In the case of Pseudomonas aeruginosa, studies to date have linked c-di-GMP and its effector proteins primarily to the control of flagellar motility and EPS production; that is, elevated levels of c-di-GMP stimulate biofilm formation by increasing EPS production and inhibiting flagellator-mediated swarming motility (4, 6–9). The levels of c-di-GMP are modulated by two classes of enzymes: diguanylate cyclases (DGCs) and phosphodiesterases (PDEs). DGCs synthesize c-di-GMP from two molecules of GTP, while PDEs degrade c-di-GMP to pGpG or GMP (10–12).
In studying the mechanisms of the early events in biofilm formation, we reported that c-di-GMP levels were elevated 3- to 5-fold in P. aeruginosa PA14 when cells were grown on an agar surface compared to this microbe grown in a liquid broth (13). Concomitant with this increase in c-di-GMP is the upregulation of the cell surface-associated protein PilY1, which shares sequence similarity with the pilus-associated adhesin PilC of Neisseria. PilY1 also contains a von Willibrand A mechanosensory domain (14, 15). Genetic analyses showed that PilY1, as well as the DGC SadC, is required for the increased c-di-GMP in surface-grown cells and, furthermore, that PilY1 functions upstream of SadC in boosting c-di-GMP levels (16). These data point toward a central role for PilY1 in controlling surface behaviors by this microbe (17).
In this study, we dissected the earliest event in biofilm initiation and explored the regulatory network of surface-induced PilY1 expression. Together, our data revealed a complex regulatory system, whereby P. aeruginosa senses the surface through the Pil-Chp chemotaxis-like complex, TFP, and the FimS-AlgR two-component system (TCS) and employs a hierarchical regulatory cascade of two different second messengers (cyclic AMP [cAMP] and c-di-GMP) to initiate its program of surface behaviors.
RESULTS
The pilY1 mutant is defective for irreversible attachment.
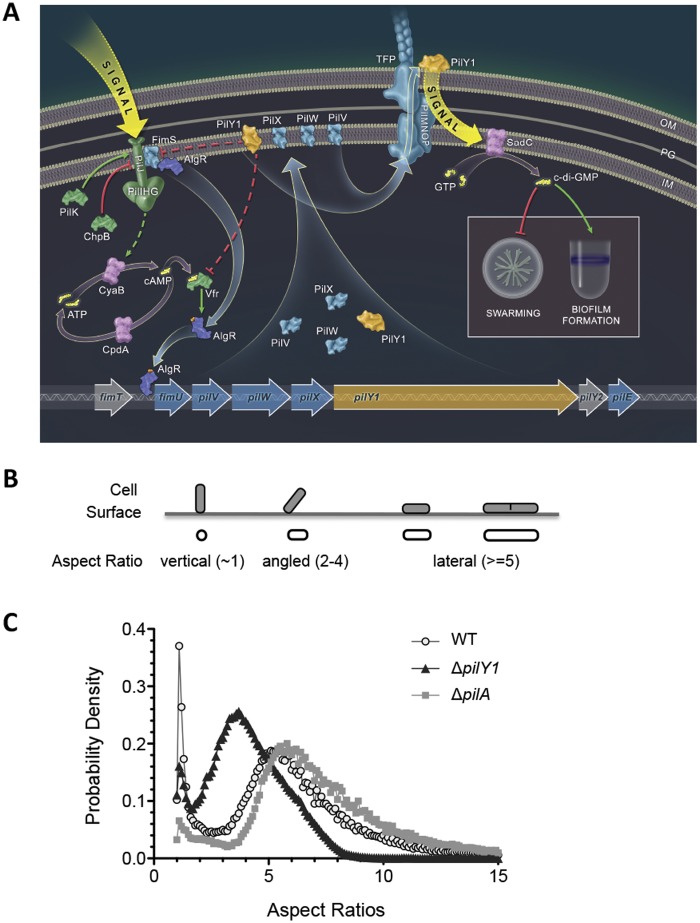
To begin to address the question of how P. aeruginosa adapts to a surface lifestyle, we wanted to identify a target gene/protein whose expression was specifically upregulated on a surface and then search for mutants that were altered for expression of this target. Based on the criteria outlined below, we selected the pilY1 gene as this target. The pilY1 gene is expressed as part of a polycistronic operon fimU-pilVWXY1Y2E (here, pilY1 operon) (Fig. 1A). The PilY1 protein can be detected in the cytosol, inner membrane (IM), and on the cell surface (13, 14, 18–20), is required for biogenesis and function of TFP (18, 21), and has been proposed to be a pilus-associated adhesin (14, 15). We showed previously that PilY1 is required to repress swarming when levels of c-di-GMP are elevated and is upregulated when bacteria are grown on an agar plate compared to in liquid medium. We also showed that c-di-GMP levels increase for agar-grown bacteria, and the increase in levels of this dinucleotide requires PilY1 (13, 16). Here, we demonstrate that a strain lacking PilY1 is defective in irreversible attachment during biofilm formation (Fig. 1B and C). We monitored ~500,000 cells of the wild type (WT) and a pilY1 mutant strain over a period of 20 to 40 h in flow cells, as reported (3), and determined the orientation of each cell relative to the surface using the apparent aspect ratio, defined as the ratio of the length to the width of an object (Fig. 1B). We found that WT cells centered at aspect ratios of 1 and 5.5 (Fig. 1C), which represent the reversible and irreversible attachment status, respectively. Compared to the WT, the pilY1 mutant also showed a “reversibly attached” peak at 1, but the second peak was left-shifted to 3.5 (Fig. 1C). Since the pilY1 and WT strains have a similar growth rate (see Fig. S1 in the supplemental material), this shift to a lower value is not due to differences in cell length but instead due to the increased population of cells at an angle relative to the surface. These data indicate that the pilY1 mutant initiates surface adhesion but cannot stabilize irreversible attachment, a finding consistent with the observation that the pilY1 mutant shows increased surface-associated swarming motility (13, 16).
FIG 1 .
A role for PilY1 in regulation of surface behaviors. (A) Model for surface behavior regulation in P. aeruginosa. From left to right, a surface-associated signal activates the Pil-Chp complex and induces the production of cAMP through CyaB. CyaB, a putative membrane-associated protein, was illustrated as a cytoplasmic protein due to space restriction. cAMP, together with its receptor Vfr, regulates transcription at the fimS-algR locus. AlgR-P (the phosphate indicated by the red dot) directly binds the pilY1 operon promoter region and activates its transcription. FimS and PilJ physically interact, and we propose that PilJ may act via the FimS-AlgR two-component system, along with cAMP-Vfr, to stimulate production of and autoregulate pilY1 gene expression. PilY1 and minor pilins PilVWX are localized to the inner membrane (IM), where they feedback inhibit their own expression via PilJ, Vfr, and AlgR-FimS. PilY1 is also secreted to the cell surface through the TFP apparatus and remains associated with the outer membrane (OM). The external PilY1 can signal through components of TFP alignment complex PilMNOP and induce SadC activity. SadC synthesizes c-di-GMP, leading to promotion of biofilm formation and repression of swarming motility. Straight arrows indicate activation, and T arrows are inhibition. Solid arrows indicate direct regulation, and dashed arrows indicate indirect regulation. Illustration courtesy of William Scavone, Kestrel Studio, reprinted with permission. (B) Schematic illustration of the aspect ratio. The aspect ratio is calculated as cell length (as it is projected on the surface) divided by the projected cell width. The cell orientation and relative aspect ratios were defined based on the measurements with WT cells. Since cells were visualized with a camera that was oriented 90° to the attachment surface, cells show “stadium-shaped” contour (aspect ratio of ≥5, as in the WT) when they are perfectly horizontal and irreversibly attached and circular contour (aspect ratio of ~1) when they are perfectly vertical and reversibly attached. There are a large number of cells that exhibit behavior between these two limiting cases and have different tilt angles (between 0 and 90°) with respect to the surface; therefore, intermediate aspect ratios are observed. Besides cell attachment angles, the aspect ratio can also be affected by cell growth. For example, a dividing cell has longer cell length than a nondividing cell and hence a larger aspect ratio. (C) Shown are the aspect ratio histograms plotted as probability density versus aspect ratio of the indicated strain. Bin size = 0.1.
Because the pilY1 mutant is also defective for TFP biogenesis (14, 18), we also assessed the phenotype of the pilA mutant in this assay, which codes for the major pilin in TFP, and thus the pilA mutant is also defective in TFP biogenesis. In contrast to the pilY1 mutant and the WT, the attachment profile of the pilA mutant showed that most cells had an aspect ratio of ~5.5, a finding consistent with earlier work showing that a mutant defective in TFP biogenesis was unable to “stand” (3), and thus few cells had an aspect ratio near 1. Taken together, our data suggest that the impact of PilY1 on attachment in this assay is not simply due to the loss of the TFP fiber but also the loss of another PilY1-mediated function, a conclusion consistent with the data presented below.
Identification of genes that affect surface-specific pilY1 transcription.
To better understand how the pilY1 gene is regulated when grown on a surface, we sought to identify candidate regulators of the pilY1 operon using transposon mutagenesis screening. To facilitate high-throughput screens, we constructed both pilY1 transcriptional and translational lacZ reporter fusions. We observed a 2-fold increase in PpilY1-lacZ transcription for cells grown on an agar surface (147 ± 9 Miller units) compared to those in liquid broth (72 ± 2 Miller units). A similar fold change was also observed when the pilY1 translational lacZ reporter was inserted at the pilY1 site (Fig. 2A) and when the PilY1 protein level was monitored by Western blotting (13). These results indicated that the surface-associated expression of pilY1 is regulated mainly at the transcriptional level and that the PpilY1-lacZ transcriptional reporter is suitable for genetic screening.
FIG 2 .
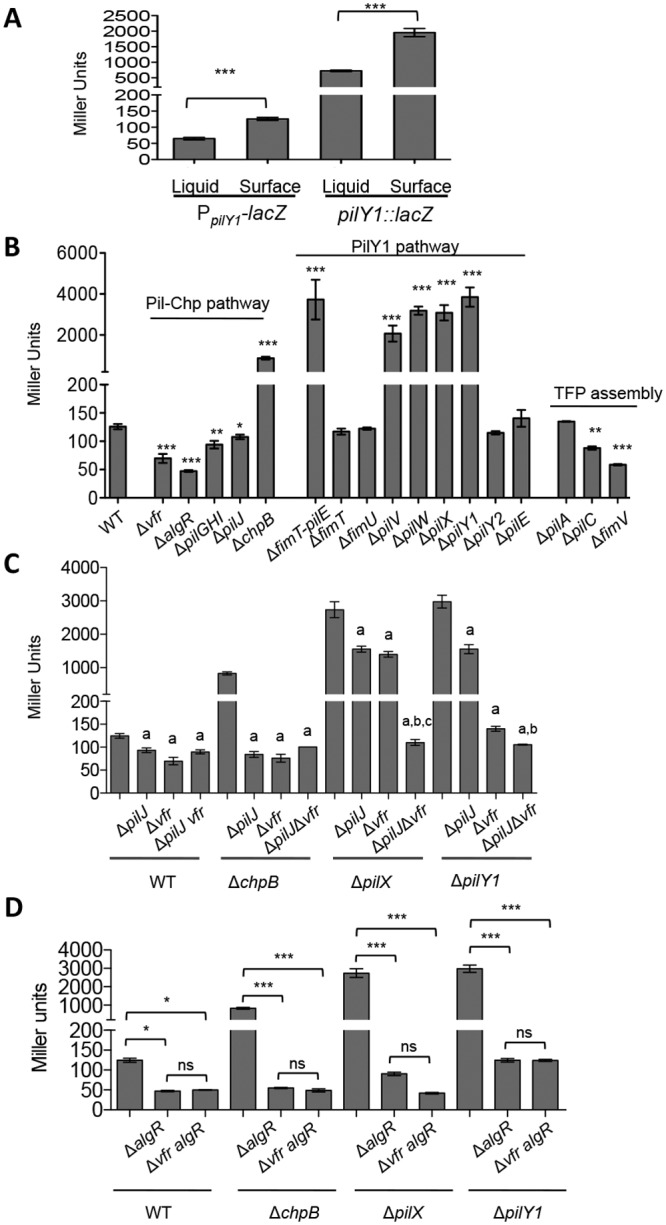
β-Galactosidase activities of PpilY1-lacZ transcriptional reporter in various strain backgrounds. All strains were grown overnight on an M8 agar plate (1% agar); M8 medium is the standard swarm medium used and thus was used throughout our studies. The data represent a minimum of three independent experiments with two biological replicates each, and values are reported as means ± standard errors of the mean (SEM). (A) β-Galactosidase activities of the transcriptional reporter PpilY1-lacZ and the translational reporter pilY1::lacZ. Cells were grown in liquid M8 minimal medium or on the surface of M8 minimal medium supplemented with 1% agar. The data represent a minimal of three independent experiments with two biological replicates each, and values are reported as means ± SEM. Significance was determined by one-way analysis of variance (ANOVA) followed by a Tukey posttest comparison. ***, P < 0.001. (B) Pathways involved in pilY1 expression. Expression of the pilY1-lacZ fusion is presented as Miller units, for mutants in the Pil-Chp and PilY1 pathways and TFP assembly. Significance was determined by one-way ANOVA followed by a Dunnett posttest comparison for the difference between WT and individual mutants. *, P < 0.05; ***, P < 0.001. (C) Epistasis analysis with the vfr mutant. Expression of the pilY1-lacZ fusion is presented as Miller units for the mutants indicated. Statistical analyses were performed using one-way ANOVA followed by a Tukey posttest comparison, P < 0.05. a, significant difference between the ΔpilJ, Δvfr, or ΔpilJ vfr mutant and the parental strain; b, significant difference between the ΔpilJ and ΔpilJ vfr mutants in the same strain background; c, significant difference between the Δvfr and ΔpilJ vfr mutants in the same strain background. (D) Epistasis analysis with the algR mutant. Expression of the pilY1-lacZ fusion is presented as Miller units for the mutants indicated. Statistical analyses were performed by one-way ANOVA followed by a Tukey posttest comparison. *, P < 0.05; ***, P < 0.001; ns, no significant difference.
To identify potential regulators of the pilY1 operon, we performed a mariner transposon (Tn) mutagenesis screen in a strain carrying the PpilY1-lacZ transcriptional reporter described above. We screened over 60,000 random Tn insertion mutants (~10-fold coverage of the genome), and the identified 32 candidate genes whose expression was altered (up or down) compared to the WT when grown on an agar plate are listed in Table S1 in the supplemental material. A number of the genes identified in this screen were previously implicated in pilY1 regulation, and new genes involved in this process were also identified. We found transposon insertions in the gene coding for FimS-AlgR TCS, known positive regulators of the pilY1 operon (22). We also recovered multiple insertions in the cyclic-AMP (cAMP)-responsive transcription factor Vfr and in the pilGHIJK-chpABC gene cluster, which encodes a chemotaxis-like chemosensory signal transduction system. Finally, we identified mutations in TFP biogenesis genes, namely, pilC and fimV. The genes identified in the screen allowed us to develop the model that serves as a framework for this report (Fig. 1A).
pilY1 transcription on a surface is regulated by two pathways.
To validate the results from the Tn screen, we constructed nonpolar, unmarked deletion mutations of select genes or gene clusters listed in Table S1 in the supplemental material and quantified PpilY1-lacZ reporter activities of agar-grown cells in these backgrounds using β-galactosidase assays. Based upon previously established relationships (22–24) and our findings here, we organized these genes into two pathways, namely, the Pil-Chp pathway and the PilY1 pathway (Fig. 2B).
The Pil-Chp pathway includes the PilGHIJK-ChpABC complex (here, Pil-Chp complex) and two transcriptional regulators, Vfr and AlgR (Fig. 1). The Pil-Chp complex is a chemotaxis-like system with components that share sequence similarity with the flagellar chemotaxis system of Escherichia coli (25–28). PilJ is a methyl-accepting chemotaxis protein (MCP), and its activity, by analogy with chemotaxis, is likely regulated by the opposing activities of the methyltransferase PilK, which stimulates signaling through PilJ, and the methylesterase ChpB, which dampens PilJ signaling. The activated form of PilJ is thought to stimulate histidine kinase ChpA via its adaptors PilI and ChpC, which in turn activate the response regulators PilH and PilG, leading to cAMP synthesis by CyaB, the major cAMP cyclase of P. aeruginosa (23). cAMP, in turn, binds to its receptor Vfr, regulating transcription of the fimS-algR genes (24, 29). The response regulator AlgR protein directly binds the pilY1 operon promoter region and activates its transcription; AlgR activity is regulated by its cognate sensor kinase FimS, also known as AlgZ (22, 30, 31).
As shown in Fig. 2B, when the positive-acting components of this regulatory cascade (pilGHI, pilJ, vfr, or algR) were deleted, PpilY1-lacZ expression was reduced in surface-grown cells. When the negative-acting component (chpB) was deleted, significantly higher β-galactosidase activity was observed. These results support the involvement of the Pil-Chp, cAMP-Vfr, and FimS-AlgR regulatory factors in pilY1 expression.
The second pathway identified implicates PilY1 in its own regulation. We isolated two Tn insertions in pilY1 that increased PpilY1-lacZ expression (see Table S1). To dissect the role of pilY1 operon in its own expression, we generated nonpolar deletion mutants of the entire operon as well as the individual genes (Fig. 1). Compared to the WT strain, the deletion mutant of the pilY1 operon (ΔfimT-pilE mutant) showed significantly increased β-galactosidase activity. A similar significant increase was also observed for single mutants of the pilV, pilW, pilX, or pilY1 genes but not the other genes tested (using complemented mutants from a previous study [13, 16]), suggesting these four genes participate in autoregulation of their own transcription (Fig. 2B). This finding is consistent with previous work from Bohn et al. (21). Together, these data show that PilY1 and the minor pilins PilVWX regulate their own transcription; we suggest a model below to explain this regulation.
Finally, we identified mutations in TFP assembly complex components pilC and fimV, which significantly reduced PpilY1-lacZ expression. Interestingly, mutating the major TFP pilin gene, pilA, had minimal impact on PpilY1-lacZ activity (Fig. 2B). This result indicates that loss of the pilus is not sufficient to explain the observed impact on surface-mediated PpilY1-lacZ expression in strains lacking pilV, pilX, pilW, pilY1, pilC, or fimV, all of which are also defective in TFP assembly (18, 19, 32–34).
The PilY1 pathway is epistatic to the Pil-Chp pathway.
We noticed that there are two transcriptional regulators (Vfr and AlgR) in the Pil-Chp pathway and hypothesized that PilY1 and PilVWX are epistatic to the Pil-Chp pathway, and, furthermore, we propose that the pilY1 operon is regulated by Pil-Chp, cAMP-Vfr, and FimS-AlgR. To test this idea, we performed an epistasis test with mutations in the pilX and pilY1 genes and with mutations in components of the Pil-Chp pathway, namely, pilJ, vfr, and algR. A chpB mutant was included as a positive control.
As shown in Fig. 2C, the increased PpilY1-lacZ activity in the chpB mutant (compared to the WT) was eliminated by introducing mutations in the pilJ or vfr genes, illustrating an epistatic relationship among chpB, pilJ, and vfr. In the pilX mutant background, introducing mutations in the pilJ or vfr genes reduced PpilY1-lacZ activity of surface-grown cells to about half of that of the parental pilX mutant strain. The PpilY1-lacZ activity, however, is significantly lower in the triple mutant of pilX pilJ vfr than that in each double pilX pilJ and pilX vfr mutants, suggesting that PilJ and Vfr both contribute to PpilY1-lacZ expression; that is, PilJ acts via a cAMP-dependent as well as a cAMP-independent pathway. Finally, mutating algR eliminated PpilY1-lacZ activity in all the strain backgrounds tested (WT, ΔchpB, ΔpilX, and ΔpilY1 strains) (Fig. 2D), possibly masking the effects of Vfr, which functions upstream of AlgR (Fig. 1). This result shows that AlgR acts with cAMP-Vfr in the transcriptional regulation of the pilY1 operon. We found similar regulation in the P. aeruginosa PAK strain (see Fig. S2 in the supplemental material).
Together, these genetic analyses suggest that Pil-Chp regulates PilY1/PilVWX via both cAMP-Vfr as well as FimS-AlgR (Fig. 1), a model consistent with that previously proposed for planktonically grown bacteria (22, 23, 29). The fact that we found the same pathway in surface-grown cells indicates the previously unrecognized surface induction of this pathway. Furthermore, we show that the increased expression of the pilY1 operon in a pilX or pilY1 mutant strain can be eliminated by mutating the pilJ, vfr, and algR genes, indicating that PilY1 and PilX feedback inhibit their own expression through the Pil-Chp, cAMP-Vfr, and FimS-AlgR signal transduction pathways.
Physical interaction between PilJ and FimS.
To further explore the relationship between the Pil-Chp and PilY1 pathways, we examined whether the inner membrane-localized components of these two pathways physically interact using the bacterial adenylate cyclase two-hybrid assay (BACTH). Of all the pairwise tests between the proteins in the Pil-Chp and PilY1 pathways tested in the BACTH assay (listed in the supplemental data), we detected an interaction only between PilJ-FimS (see Fig. S3A and B in the supplemental material). FimS is a sensor histidine kinase whose cognate response regulator is AlgR. FimS, like PilJ, is a predicted IM protein, consistent with the ability of these proteins to interact. Furthermore, as shown above, AlgR plays a key role in the regulation of the pilY1 operon. Together, these data suggest that PilJ and FimS activities may be coordinated through protein-protein interactions in the IM (Fig. 1), thus allowing coordination between the input signal(s) of the Pil-Chp and FimS-AlgR pathways and the downstream expression of the pilY1 gene.
cAMP levels are upregulated when cells are grown on a solid surface.
We next asked how cells upregulate pilY1 expression when they grow on a solid surface. We hypothesized that surface growth might induce PilJ activity, which would then in turn stimulate downstream cAMP production and, consequently, pilY1 transcription. To test this idea, we utilized a cAMP-dependent promoter fusion, P1-lacZ, whose activity has been shown to reflect cellular cAMP levels (23). We deduced cellular cAMP levels based on the fold change of P1-lacZ activity over a vector control, as reported (23), and tracked these levels over time.
When cells were grown in liquid culture, both the WT and the ΔpilJ mutant showed constant, low levels of cAMP production over time (Fig. 3A). The cAMP level in the ΔpilJ mutant strain was slightly lower than that in the WT strain, which is consistent with the positive role of PilJ in cAMP synthesis (23). When the WT was grown on an agar surface, cAMP levels increased sharply during the first 7 h, with a detectable increase observed as soon as 2 h postinoculation. This surface-specific increase was abolished in the cells lacking the pilJ gene, where cAMP levels remained at low levels, similar to levels measured for the liquid-grown cells. As expected, we also found that the cAMP levels in a chpB mutant remained constantly high when cells were grown in liquid or on a surface (Fig. 3B). This result suggests that cells increase cAMP production when growing on an agar surface and that this increase is dependent on signaling through the PilJ chemotaxis-like receptor.
FIG 3 .
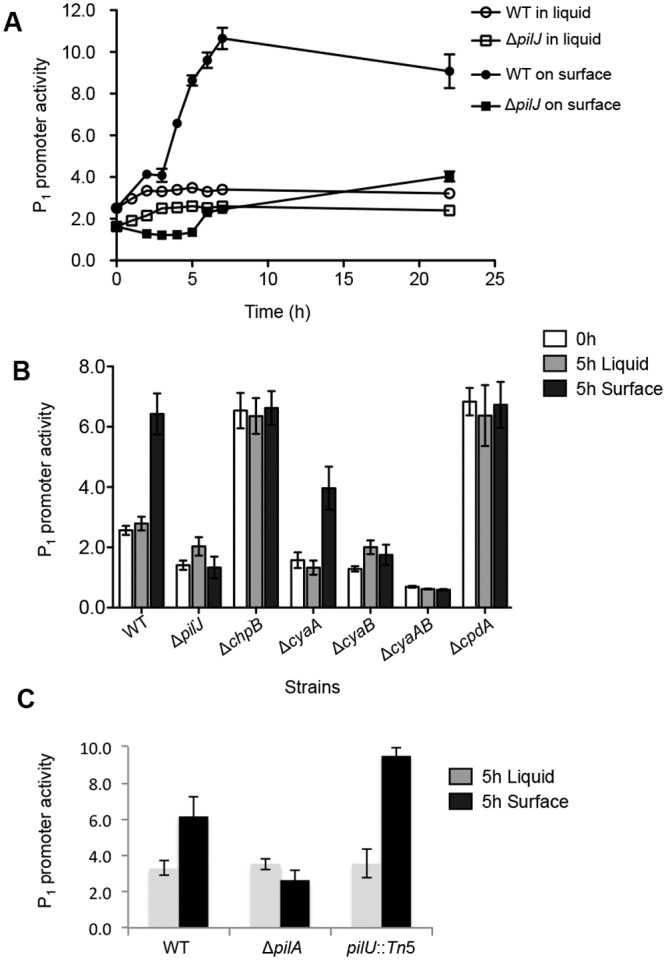
The cellular cAMP levels in cells grown in liquid broth and on agar surface. (A) Representative time lapse plot of cAMP levels in cells grown in liquid broth or on an agar surface with the same base medium (M8). The cellular cAMP level is expressed as P1-lacZ activity divided by the vector control. The means ± SEM from three biological replicates were shown. (B) cAMP levels in the WT and mutant strains grown in M8 liquid broth or on agar surface. Each strain was grown to early log phase in M8 broth. At time = 0 h, the cultures were split and either continued to be incubated in M8 broth or spread on M8 plate (1% agar) for an additional 5 h. The data represent a minimum of three independent experiments with two biological replicates each, and values are reported as means ± SEM. (C) cAMP levels in the WT and mutant strains grown in M8 liquid broth or on agar surface, performed as outlined in panel B.
To confirm that the cAMP level is altered in surface-grown cells, we mutated the genes that are known to be involved in cAMP synthesis (cyaA and cyaB) and degradation (cpdA) and determined their effects on cAMP levels in both liquid- and surface-grown cells (Fig. 3B). When cells were grown in liquid broth, cAMP levels were reduced in single mutants lacking either cyaA or cyaB and further reduced in the cyaAB double mutant. Upon surface growth, the cAMP levels increased only in the cyaA mutant but remained unchanged in the cyaB and cyaAB mutants. These results confirm that cAMP level is upregulated in surface-grown cells and further suggest that this change is largely dependent on the presence of CyaB. Consistent with this observation, CyaB has been shown to be involved in the PilJ-induced cAMP synthesis in planktonic cultures (23, 29).
Our findings suggested that several factors regulating pilus biogenesis impacted cAMP levels. We directly addressed whether pilus biogenesis impacts cAMP levels by testing the levels of this signaling molecule in a strain lacking TFP (ΔpilA mutant) or a hyperpiliated strain defective for pilus retraction (a pilU::Tn mutant) (35). Consistent with a role for TFP in regulating cAMP levels, the ΔpilA mutant showed no surface induction of cAMP; however, fully functional pili capable of extension and retraction are not required for cAMP production, as the hyperpiliated pilU mutant shows increased cAMP signaling (Fig. 3C), indicating that the TFP may act as a surface-sensing appendage independent of active retraction/extension.
Based on these findings, we conclude that both the Pil-Chp and TFP are required for the surface-specific increase in cAMP levels, implying the surface-deployed TFP might signal via the IM-localized Pil-Chp system. Second, given that the loss of TFP does not significantly impact the downstream surface-dependent expression of PilY1 (Fig. 2B), we also conclude that TFP and cAMP signaling are not sufficient to drive pilY1 expression, and as described in the following sections, we show that the FimS-AlgR TCS is also required for surface-specific upregulation of the pilY1 gene.
cAMP functions upstream of c-di-GMP in the regulation of surface behaviors.
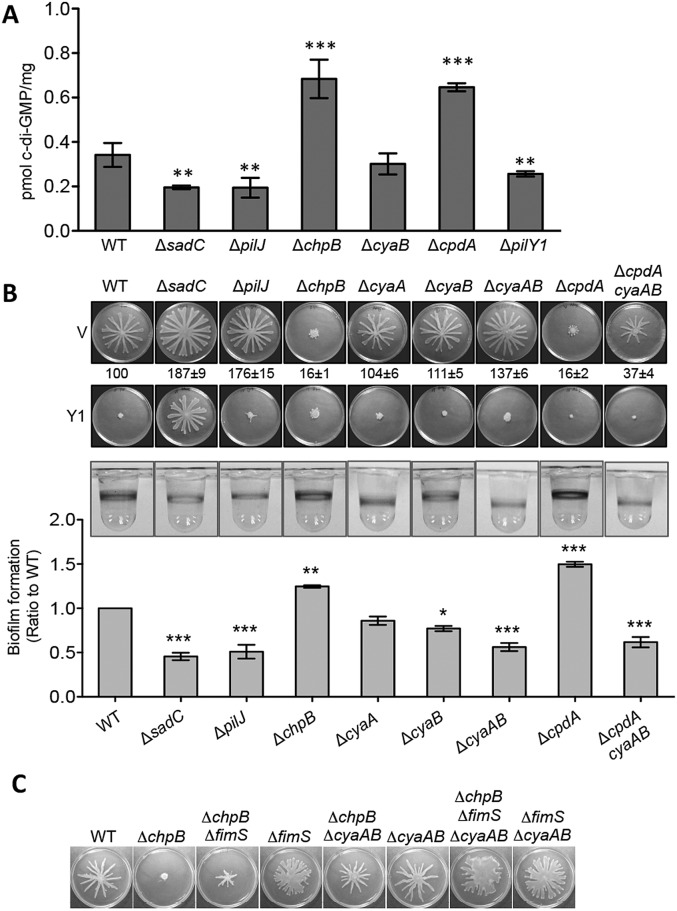
The observation that cAMP is upregulated soon after cells are inoculated on an agar surface led us to hypothesize that cAMP may act upstream of c-di-GMP in the regulation of surface behaviors. To test this idea, we measured cellular c-di-GMP levels in strains with altered levels of cAMP (Fig. 4A). The strain lacking the pilJ (low in cAMP) gene showed significantly reduced c-di-GMP levels compared to those of the WT strain, as did the control strains carrying mutations in the sadC or pilY1 genes. In contrast, the c-di-GMP levels were increased in the ΔchpB and ΔcpdA mutant strains, both of which are known to have high levels of cAMP. Surprisingly, the level of c-di-GMP in the cyaB mutant (low cAMP) was not different from the WT, suggesting that signaling via cAMP may be necessary but is not sufficient for increased c-di-GMP signaling, a point that we address in the next section.
FIG 4 .
cAMP functions upstream of c-di-GMP in surface behavior regulation. (A) Quantification of cellular c-di-GMP levels by liquid chromatography-mass spectrometry (LC-MS) for the indicated strains grown on swarm plates. Data are expressed as picomoles of c-di-GMP per milligram (dry weight) of the cell pellets from which the nucleotides were extracted. The data represent six independent experiments, and values are reported as means ± SEM. Significance was determined by one-way ANOVA followed by a Dunnett posttest comparison for the difference between WT and individual mutants. **, P < 0.01; ***, P < 0.001. (B) Surface behaviors of mutants in the cAMP-Vfr pathway. Top and middle panels are representative swarm plates of strains carrying either empty vector (V) or PilY1-expressing plasmid (Y1). Plates contained 0.2% arabinose. The ratio numbers below the top panel indicate the percentage (means ± SEM, from 20 plates) of the plate surface coverage of the mutant strains (harboring empty vectors) relative to that of the WT strain (set at 100%). The bottom panel shows representative wells of a 96-well biofilm assay for each strain. The biofilm ratios (mutant/WT) were calculated using the WT and mutant strains grown in the same 96-well plate. The means and SEM from three independent experiments are reported. Significance was determined by one-way ANOVA followed by a Dunnett posttest comparison for the difference between WT and individual mutants. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Swarms of the indicated strains grown on M8 agar plate containing 0.5% agar.
We also tested the effect of mutations analyzed in Fig. 4A on surface behaviors, including swarming motility and biofilm formation (Fig. 4B). Swarming motility was increased in the ΔpilJ mutant and repressed in the ΔchpB mutant. A small but reproducible increase in swarming motility was observed with the ΔcyaB or ΔcyaAB mutant but not to the degree observed for the sadC mutant (which is defective in upregulating c-di-GMP on a surface [Fig. 4A]). As expected, loss of the cAMP phosphodiesterase CpdA and overstimulation of cAMP signaling results in stimulated c-di-GMP levels (Fig. 4A), as well as a complete loss of swarming motility, which is restored by introduction of the ΔcyaAB mutations (Fig. 4B). However, if cAMP signaling were sufficient to regulate downstream c-di-GMP signaling, we would have expected the ΔcyaAB mutant to show both reduced c-di-GMP levels (Fig. 4A) and a hyperswarming phenotype similar to the sadC mutant (Fig. 4B); we did not observe either of these phenotypes. Again, these data suggest that a surface-dependent increase in cAMP levels alone is not sufficient to drive downstream c-di-GMP-mediated behaviors, a point we address in the next section.
Finally, when PilY1 was overexpressed in the mutants with low cAMP levels (i.e., ΔpilJ, ΔcyaB, and ΔcyaAB strains), swarming motility was repressed, but no such repression was observed in the strain defective for c-di-GMP production (i.e., the sadC mutant [Fig. 4B]). Together, these results are consistent with the idea that PilY1 functions downstream of cAMP in swarming regulation and argue for both a cAMP-dependent and cAMP-independent pathway for downstream regulation of pilY1 gene expression and subsequent PilY1-mediated c-di-GMP levels.
We also observed reduced biofilms in the ΔpilJ, ΔcyaB, and ΔcyaAB mutants and increased biofilm in the ΔchpB and ΔcpdA mutant strains (Fig. 4B, bottom). Taken together, our data suggest that cAMP signaling functions upstream of c-di-GMP-mediated regulation, consistent with the model presented in Fig. 1.
Regulation of pilY1 gene expression, and thus increased c-di-GMP, is also mediated by the FimS-AlgR TCS.
The data given above indicated that the Pil-Chp system signaled via both a cAMP-dependent and cAMP-independent mechanism to regulate pilY1 gene expression, downstream stimulation of c-di-GMP levels on a surface, and c-di-GMP-mediated control of surface behaviors. A candidate for this alternative pathway was the FimS-AlgR TCS given that FimS and PilJ interact and that we identified mutations in the fimS and algR genes in our genetic screen. To test this possibility, we started with a ΔchpB mutant that we showed in Fig. 4B is nonswarming, likely due to increased signaling through hypermethylated PilJ (Fig. 1), and introduced a mutation in fimS as well as mutations in the cyaAB genes, coding for the two adenylate cyclases. As shown in Fig. 4C, mutating the fimS gene partially restored swarming to the ΔchpB mutant. Because the ΔfimS mutant is defective in pili biogenesis, the swarm patterns observed typically lack the defined tendrils observed in the WT strain (13, 16). Similarly, the ΔchpB ΔcyaAB mutant also showed partial rescue of the swarming defect observed in the ΔchpB mutant, while the ΔchpB ΔfimS ΔcyaAB strain shows swarm plate coverage comparable to or greater than the WT. Together, these data show that signaling through the Pil-Chp system to modulate surface behaviors requires both stimulation of cAMP signaling and activation of the FimS-AlgR system to fully induce PilY1 production, as well as PilY1 surface localization, downstream c-di-GMP signaling, and the associated modulation of surface behaviors.
PilY1 secretion is dependent mainly on the TFP assembly complex.
Previous work has shown that the cell surface-associated PilY1 can stimulate c-di-GMP synthesis via the IM-localized DGC SadC (16); however, it is not clear how PilY1 is localized to the cell surface. In the genetic analysis described above, we found that mutations in pilC or fimV, but not in pilA (pilin), impact pilY1 expression, suggesting that the TFP secretion machinery has a role in regulating expression of the pilY1 gene. Based on this finding, we hypothesized that the TFP complex may be involved in PilY1 secretion. To test this idea, we assessed localization of PilY1 in the WT and TFP assembly mutants with antiserum specific to PilY1 (Fig. 5). The absence of IM platform protein (PilC), OM secretin (PilQ), or members of the alignment complex (PilMNOP and FimV) has strong negative impacts on PilY1 accumulation at the cell surface (Fig. 5A). Consistent with the transcriptional analysis described above, the ΔpilX mutant showed increased PilY1 levels compared to those of the WT (see figure legend). The FliC (flagellin) protein served as a loading control and surface protein marker. Antibody to the cytoplasmic Cas3 protein served to show that there was no cell lysis in our cell surface protein preparations (Fig. 5B). The total protein levels of PilY1 were reduced in the pilC and fimV mutant but largely unchanged in the other strains, in contrast to the severe reduction of PilY1 level observed at the cell surface (Fig. 5A compared to C). These results suggest that PilY1 secretion is dependent mainly on the TFP assembly system.
FIG 5 .
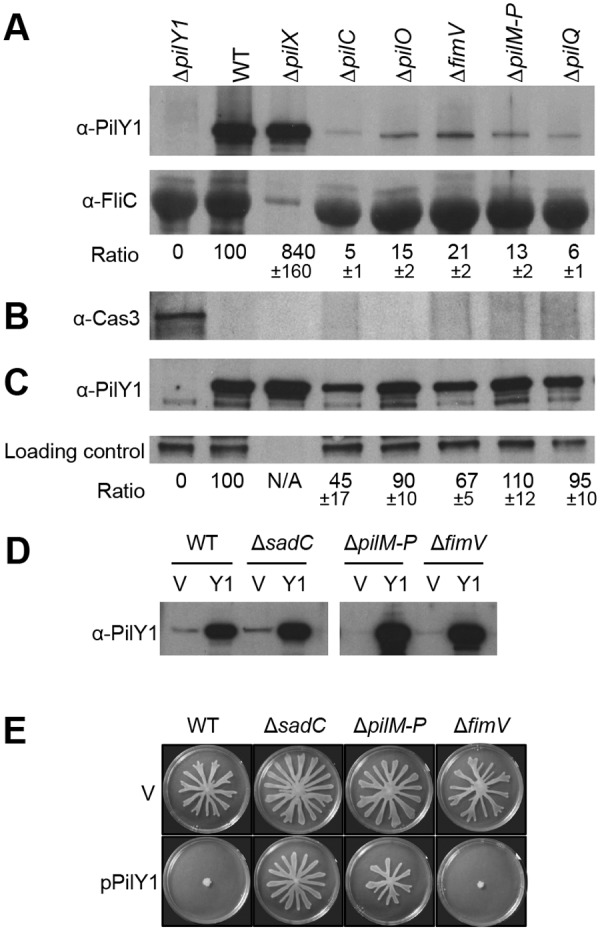
The TFP complex is involved in PilY1 secretion and signaling. (A) Detection of PilY1 in cell surface (CS) fractions. The CS protein samples were concentrated using TCA precipitation before loading. Western blots were probed with anti-PilY1 antibody (top) or anti-FliC antibody (middle). Strains tested in all blots are indicated at the top of panel A. FliC (flagellin) served as a cell surface protein marker and a protein loading control. The intensity of the PilY1 protein band is normalized to FliC. The ratio numbers below each band represent the percentage (means ± standard deviations [SD], from three experiments) of the normalized PilY1 in mutants relative to that in the WT strain (lane 2). (B) Western blot probed with anti-Cas3 antibody (bottom) was included as a cytoplasmic protein control. Lane 1, whole-cell lysate from WT cells; lanes 2 to 8, the same cell surface fraction samples used in lanes 2 to 8 in the top and middle panels. (C) Detection of PilY1 in whole-cell lysate. An unknown protein cross-reacting with anti-PilY1 antibody served as a protein-loading control. The strains analyzed here are indicated at the top of panel A. The experiment was repeated three times, and one representative gel was shown. The ratio is calculated as the percentage of normalized PilY1 intensity relative to that in the WT strain and is reported as means ± SD from three experiments. Note: in panels A and C, the protein extracts from the ΔpilX strain (lane 3) were diluted 10-fold before loading. (D) Assessment of PilY1 protein levels in the unconcentrated CS fraction. Strains carrying either empty vector (V) or the pilY1-expressing plasmid pPilY1 (Y1) were grown on an M8 agar plate containing 1% agar and 0.2% arabinose. (E) Swarms of the indicated strains grown on an M8 agar plate containing 0.5% agar and 0.2% arabinose.
We also noted that there were small amounts of PilY1 (5 to 20% of the WT level) detected in the cell surface fraction in the TFP assembly mutants (Fig. 5A). When overexpressed in trans, a large quantity of PilY1 can be found at the cell surface in strains lacking the TFP complex (Fig. 5D). These results suggest that there are additional pathway(s) involved in PilY1 secretion. We speculated that PilY1 might be secreted through the T2SS, an evolutionarily related pathway to the TFP system (36). However, additional mutations in T2SS secretin genes xcpQ and hxcQ (37, 38) did not affect PilY1 secretion compared to the ΔpilQ single mutant (see Fig. S4 in the supplemental material). We concluded that PilY1 secretion is dependent primarily on the TFP assembly complex, and small amounts of PilY1 can be secreted through an unknown alternative pathway.
Importantly, taken together with our genetic studies described above, these findings suggest that PilY1 deployment occurs downstream of signaling via Pil-Chp, cAMP-Vfr, and FimS-AlgR. In other words, in response to cell growth on surfaces, Pil-Chp-mediated cAMP synthesis and signaling via FimS-AlgR first promotes assembly of the TFP biogenesis machinery, which is in turn required for robust PilY1 secretion. Thus, PilY1 is not deployed on the cell surface until after the TFP system is assembled.
PilY1-mediated stimulation of c-di-GMP requires components of the TFP alignment complex.
Given the extracellular localization of PilY1, we hypothesized that PilY1 likely communicates with the IM-localized SadC through intermediary membrane proteins. In the search for components connecting PilY1 to SadC, we took advantage of the PilY1-mediated swarming phenotypes. Overexpressing PilY1 in the WT strain can repress swarming motility (Fig. 5E); however, swarming cannot be repressed in a strain lacking a functional sadC gene (16). Similarly, one could expect that if the intermediate signaling components are missing, PilY1, when overexpressed, would not be able to repress swarming.
We speculated that components of the TFP complex might participate in signal transduction between PilY1 and SadC. This analysis was complicated by the fact that PilY1 requires a functional TFP complex for maximal secretion, but as mentioned above, there was residual PilY1 secretion in a strain lacking TFP components. In addition, overexpressing PilY1 from a plasmid could bypass the requirement for TFP in its secretion, allowing PilY1 to accumulate at the cell surface (Fig. 5D).
We tested the impact of overexpression of PilY1 on repression of swarming motility in TFP mutants. Overexpression of PilY1 completely repressed swarming motility of the ΔfimV mutant but failed to do so with the ΔpilM-P mutant (Fig. 5E) or with the single nonpolar mutations of pilM, pilN, pilO, or pilP genes (see Fig. S5 in the supplemental material). PilMNOP and FimV are components of the TFP alignment complex, a nonessential component of the TFP assembly system that is required for optimal TFP assembly (32, 33). In contrast, overexpression of PilY1 can repress swarming motility of strains lacking pilC (see Fig. S5) or pilA (16), again indicating that TFP play only a partial role in controlling biofilm formation and other surface behaviors. Taken together, we concluded that PilMNOP are involved in the PilY1-SadC signaling pathway and thus involved in affecting c-di-GMP levels of surface-grown cells.
DISCUSSION
How cells sense and respond to a surface is a fundamental question in biofilm research. Our data show that the Pil-Chp complex, a chemotaxis-like complex located at the IM, is critical in surface sensing. Mutating the pilJ gene, which codes for an MCP-like protein, leads to a reduction in PilY1 expression, a loss in biofilm formation, and a hyperswarming phenotype. Mutation of ChpB, a methylesterase, presumably results in hypermethylation of PilJ and increased basal signaling through this MCP. In contrast to the pilJ mutant, loss of ChpB results in increased biofilm formation and a nonswarming phenotype, as well as increased PilY1 expression. Given the close regulation between the Pil-Chp complex and TFP synthesis (25–27), and the observation that PilY1 is not deployed until the TFP is assembled, it is tempting to speculate that the TFP or TFP-associated factor(s) may serve as a mechanosensor for a surface, a finding consistent with our observation that loss of pili results in reduced cAMP signaling. The observation that the hyperpiliated pilU mutant, defective in retraction, shows increased cAMP signaling suggests that active extension/retraction of the TFP is not required for cAMP signaling.
Our data show that signaling through the Pil-Chp system to stimulate PilY1 production and downstream c-di-GMP-mediated regulation requires both stimulation of cAMP levels via CyaA/B and signaling through the FimS-AlgR two-component system, likely through direct physical interaction of PilJ and FimS. Our findings are in good agreement with previous reports that cAMP-Vfr upregulates transcription at the fimS-algR operon (24) and that FimS-AlgR is required for pilY1 operon expression, as the phosphorylated AlgR directly binds to the pilY1 operon promoter and activates its transcription (22, 30).
Our data also show that the TFP assembly complex is intimately involved in PilY1 production, secretion, and signal transduction. The TFP assembly complex is comprised of four interacting subcomplexes, including (i) the IM motor subcomplex (PilC, PilB, PilT, and PilU), (ii) the outer membrane (OM) secretin pore subcomplex (PilQ and PilF), (iii) the alignment subcomplex (PilM, PilN, PilO, PilP, and FimV) that bridges the IM and OM subcomplexes but is not required for pili assembly (31, 39), and finally (iv) the pilus itself (PilA) plus the minor pilins FimU, PilV, PilW, PilX, and PilE (40). We showed that PilY1 is secreted through the TFP apparatus. Furthermore, our published studies show that cell surface-associated PilY1 can signal through SadC to control swarming motility (16), and this report shows that the PilMNOP alignment complex is also required for PilY1 to stimulate c-di-GMP production via SadC. Taken together, our studies show that PilY1 plays a central role in coordinating the surface behaviors in P. aeruginosa, and this function is exerted via two discrete regulatory pathways: (i) PilY1 in the IM represses its own transcription via modulation of cAMP signaling through Pil-Chp, cAMP-Vfr, and FimS-AlgR, and (ii) PilY1 on the cell surface is required to regulate c-di-GMP levels via the TFP alignment complex and the DGC SadC (Fig. 1).
In a broader context, we believe we are now at the beginning stage of answering the question of how cells sense a surface. Here, we propose that P. aeruginosa utilizes a stepwise regulatory circuit to detect and respond to surface growth; this circuit employs a hierarchical regulatory cascade of two second messengers (cAMP and c-di-GMP) to initiate this microbe’s program of surface behaviors. It appears that TFP assembly and the pilus are required for robust cAMP signaling, indicating that the low levels of pili found on planktonic cells participate in initial surface engagement. We propose that the FimS-AlgR system, perhaps with input from a direct physical interaction with PilJ, also is required for sensing surface engagement. Subsequent c-di-GMP signaling does not require the pilus per se but instead requires PilY1, whose expression requires Pil-Chp, cAMP-Vfr, and the FimS-AlgR TCS; and furthermore, PilY1 is secreted in a TFP-dependent manner. Thus, we can propose a hierarchical model of signaling because downstream c-di-GMP-mediated events via PilY1 cannot occur until PilY1 is synthesized, secreted, and localized to the cell surface. Furthermore, previous studies by Harwood and colleagues implicated surface-dependent clustering of WspR and phosphorylation of this protein, likely by a chemotaxis-like system, are required for surface-dependent stimulation of WspR activity (41, 42). Given the timing of WspR clustering upon surface contact (~6 to 8 h) and the observation that cAMP signaling is stimulated as early as 2 h, we believe the system we describe here occurs upstream of WspR signaling.
A key implication of our findings is that there may not be a single “surface signal” to trigger the surface program in P. aeruginosa. Instead, we suggest that there may be multiple reinforcing signals mediated via numerous sensing proteins (i.e., PilJ, TFP, FimS, PilY1, Wsp), and likely additional downstream factors, to continually reassure the bacterium that it has engaged and remains in contact with a solid surface. Blocking any of these inputs may result in a microbe that is not fully committed to life on a surface and thus offers a number of potential targets for antibiofilm therapeutics.
MATERIALS AND METHODS
Additional details are provided in the supplemental material.
Bacterial strains and plasmids.
Bacterial strains and plasmids used here are listed in Table S2 in the supplemental material. Mariner transposon mutagenesis and mapping mutations were performed as reported (16, 43, 44).
β-Galactosidase activity assays.
The β-galactosidase activity assay of strains harboring lacZ fusions was performed as previously described (45), and results were presented as Miller units, except that the reactions were performed at room temperature and the absorbance was measured in 96-well flat-bottom plates using a SpectraMax M2 microplate reader (Molecular Devices). In most studies, we performed β-galactosidase activity assays with both planktonic- and surface-grown bacteria, but typically only the results of assays from surface-grown cells are included to simplify presentation.
Swarming and biofilm assays.
Swarming and biofilm formation assays were performed as previously described (46–48). M8 medium was used, as this is the standard medium for swarming assays.
PilY1 cellular localization, SDS-PAGE, and Western blotting.
Fractionation assays and Western blot analysis was performed as reported previously (16).
Bacterial adenylate cyclase two-hybrid (BATCH) assay.
The E. coli strain bth101 was cotransformed with relevant recombinant T18 and T25 plasmids as previously described (49).
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
The ΔpilY1 mutant has no growth defect. Download
β-Galactosidase activities of transcriptional reporters PpilY1-lacZ and PfimS-lacZ in the P. aeruginosa PAK strain background. Download
Detection of in vivo interaction between PilJ and FimS by a BACTH analysis. Download
T2SS is not involved in PilY1 secretion. Download
Swarms of the WT and the ΔsadC, ΔfimV, ΔpilC, ΔpilM, ΔpilN, ΔpilO, and ΔpilP mutants carrying either empty vector or pPilY1 plasmid. Download
Transposon insertion mutants with altered PpilY1-lacZ reporter activity.
Strains, plasmids, and oligonucleotide primers used in this study.
ACKNOWLEDGMENTS
This work was supported by NIH grant R37-AI83256-06 to G.A.O., the Human Frontiers in Science Program to G.C.L.W., NIH grant AI069116 to M.C.W., and NSF Graduate Research Fellowship to A.E.B.
Footnotes
Citation Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GCL, O’Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6(1):e02456-14. doi:10.1128/mBio.02456-14.
REFERENCES
- 1.Monds RD, O’Toole GA. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol 17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Conrad JC, Gibiansky ML, Jin F, Gordon VD, Motto DA, Mathewson MA, Stopka WG, Zelasko DC, Shrout JD, Wong GC. 2011. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophys J 100:1608–1616. doi: 10.1016/j.bpj.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibiansky ML, Conrad JC, Jin F, Gordon VD, Motto DA, Mathewson MA, Stopka WG, Zelasko DC, Shrout JD, Wong GC. 2010. Bacteria use type IV pili to walk upright and detach from surfaces. Science 330:197. doi: 10.1126/science.1194238. [DOI] [PubMed] [Google Scholar]
- 4.Merritt JH, Ha DG, Cowles KN, Lu W, Morales DK, Rabinowitz J, Gitai Z, O’Toole GA. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1(4):e00183-10. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baraquet C, Harwood CS. 2013. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci U S A 110:18478–18483. doi: 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lory S, Merighi M, Hyodo M. 2009. Multiple activities of c-di-GMP in Pseudomonas aeruginosa. Nucleic Acids Symp Ser (Oxf) 53:51–52. doi: 10.1093/nass/nrp026. [DOI] [PubMed] [Google Scholar]
- 8.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. 2007. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 11.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 13.Kuchma SL, Griffin EF, O’Toole GA. 2012. Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J Bacteriol 194:5388–5403. doi: 10.1128/JB.00899-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heiniger RW, Winther-Larsen HC, Pickles RJ, Koomey M, Wolfgang MC. 2010. Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and a novel pilus-associated adhesin. Cell Microbiol 12:1158–1173. doi: 10.1111/j.1462-5822.2010.01461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson MD, Garrett CK, Bond JE, Coggan KA, Wolfgang MC, Redinbo MR. 2011. Pseudomonas aeruginosa PilY1 binds integrin in an RGD- and calcium-dependent manner. PLoS One 6:e29629. doi: 10.1371/journal.pone.0029629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuchma SL, Ballok AE, Merritt JH, Hammond JH, Lu W, Rabinowitz JD, O’Toole GA. 2010. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J Bacteriol 192:2950–2964. doi: 10.1128/JB.01642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siryaporn A, Kuchma SL, O’Toole GA, Gitai Z. 2014. Surface attachment induces Pseudomonas aeruginosa virulence. Proc Natl Acad Sci U S A 111:16860–16865. doi: 10.1073/pnas.1415712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alm RA, Hallinan JP, Watson AA, Mattick JS. 1996. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol 22:161–173. doi: 10.1111/j.1365-2958.1996.tb02665.x. [DOI] [PubMed] [Google Scholar]
- 19.Giltner CL, Habash M, Burrows LL. 2010. Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J Mol Biol 398:444–461. doi: 10.1016/j.jmb.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 20.Orans J, Johnson MD, Coggan KA, Sperlazza JR, Heiniger RW, Wolfgang MC, Redinbo MR. 2010. Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc Natl Acad Sci U S A 107:1065–1070. doi: 10.1073/pnas.0911616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohn YS, Brandes G, Rakhimova E, Horatzek S, Salunkhe P, Munder A, van Barneveld A, Jordan D, Bredenbruch F, Häussler S, Riedel K, Eberl L, Jensen PØ, Bjarnsholt T, Moser C, Hoiby N, Tümmler B, Wiehlmann L. 2009. Multiple roles of Pseudomonas aeruginosa TBCF10839 PilY1 in motility, transport and infection. Mol Microbiol 71:730–747. doi: 10.1111/j.1365-2958.2008.06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belete B, Lu H, Wozniak DJ. 2008. Pseudomonas aeruginosa AlgR regulates type IV pilus biosynthesis by activating transcription of the fimU-pilVWXY1Y2E operon. J Bacteriol 190:2023–2030. doi: 10.1128/JB.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. 2010. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanack KJ, Runyen-Janecky LJ, Ferrell EP, Suh SJ, West SE. 2006. Characterization of DNA-binding specificity and analysis of binding sites of the Pseudomonas aeruginosa global regulator, Vfr, a homologue of the Escherichia coli cAMP receptor protein. Microbiology 152:3485–3496. doi: 10.1099/mic.0.29008-0. [DOI] [PubMed] [Google Scholar]
- 25.Darzins A. 1994. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol 11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 26.Whitchurch CB, Leech AJ, Young MD KD, Sargent JL, Bertrand JJ, Semmler AB, Mellick AS, Martin PR, Alm RA, Hobbs M, Beatson SA, Huang B, Nguyen L, Commolli JC, Engel JN, Darzins A, Mattick JS. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 27.Bertrand JJ, West JT, Engel JN. 2010. Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. J Bacteriol 192:994–1010. doi: 10.1128/JB.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sampedro I, Parales RE, Krell T, Hill JE 6 August 2014. Pseudomonas chemotaxis. FEMS Microbiol Rev. doi: 10.1111/1574-6976.12081. [DOI] [PubMed] [Google Scholar]
- 29.Wolfgang MC, Lee VT, Gilmore ME, Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell 4:253–263. doi: 10.1016/S1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 30.Whitchurch CB, Alm RA, Mattick JS. 1996. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 93:9839–9843. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okkotsu Y, Tieku P, Fitzsimmons LF, Churchill ME, Schurr MJ. 2013. Pseudomonas aeruginosa AlgR phosphorylation modulates rhamnolipid production and motility. J Bacteriol 195:5499–5515. doi: 10.1128/JB.00726-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takhar HK, Kemp K, Kim M, Howell PL, Burrows LL. 2013. The platform protein is essential for type IV pilus biogenesis. J Biol Chem 288:9721–9728. doi: 10.1074/jbc.M113.453506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wehbi H, Portillo E, Harvey H, Shimkoff AE, Scheurwater EM, Howell PL, Burrows LL. 2011. The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin. J Bacteriol 193:540–550. doi: 10.1128/JB.01048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunn D, Bergman S, Lory S. 1990. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol 172:2911–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitchurch CB, Mattick JS. 1994. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol Microbiol 13:1079–1091. doi: 10.1111/j.1365-2958.1994.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 36.Ayers M, Howell PL, Burrows LL. 2010. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol 5:1203–1218. doi: 10.2217/fmb.10.76. [DOI] [PubMed] [Google Scholar]
- 37.Ball G, Durand E, Lazdunski A, Filloux A. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol Microbiol 43:475–485. doi: 10.1046/j.1365-2958.2002.02759.x. [DOI] [PubMed] [Google Scholar]
- 38.Filloux A, Michel G, Bally M. 1998. GSP-dependent protein secretion in Gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev 22:177–198. [DOI] [PubMed] [Google Scholar]
- 39.Ayers M, Sampaleanu LM, Tammam S, Koo J, Harvey H, Howell PL, Burrows LL. 2009. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J Mol Biol 394:128–142. doi: 10.1016/j.jmb.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 40.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 41.Güvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huangyutitham V, Güvener ZT, Harwood CS. 2013. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. mBio 4(3):e00242-13. doi: 10.1128/mBio.00242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 44.Caetano-Anollés G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl 3:85–94. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 45.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 46.Ha DG, Kuchma SL, O’Toole GA. 2014. Plate-based assay for swarming motility in Pseudomonas aeruginosa, p [bull4][bull4] In Filloux A, Ramos J-L (ed), Pseudomonas methods and protocols, vol 1149. Humana Press, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 48.O’Toole GA. 2011. Microtiter dish biofilm formation assay. J Vis Exp 47:p = 2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Battesti A, Bouveret E. 2012. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58:325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
The ΔpilY1 mutant has no growth defect. Download
β-Galactosidase activities of transcriptional reporters PpilY1-lacZ and PfimS-lacZ in the P. aeruginosa PAK strain background. Download
Detection of in vivo interaction between PilJ and FimS by a BACTH analysis. Download
T2SS is not involved in PilY1 secretion. Download
Swarms of the WT and the ΔsadC, ΔfimV, ΔpilC, ΔpilM, ΔpilN, ΔpilO, and ΔpilP mutants carrying either empty vector or pPilY1 plasmid. Download
Transposon insertion mutants with altered PpilY1-lacZ reporter activity.
Strains, plasmids, and oligonucleotide primers used in this study.