Abstract
To understand the role for cytokine and growth factor receptor-mediated signaling in leukemia pathogenesis we designed a functional RNAi screen targeting 188 cytokine and growth factor receptors that we found highly expressed in primary leukemia specimens. Using this screen we identified interleukin-2 gamma receptor (IL2Rγ) as a critical growth determinant for the JAK3A572V mutation-positive AML cell line. We observed that knockdown of IL2Rγ abrogates phosphorylation of JAK3 and downstream signaling molecules, JAK1, STAT5, MAPK and pS6 ribosomal protein. Overexpression of IL2Rγ in murine cells increased the transforming potential of activating JAK3 mutations, whereas absence of IL2Rγ completely abrogated the clonogenic potential of JAK3A572V as well as the transforming potential of additional JAK3 activating mutations such as JAK3M511I. In addition, mutation at the IL2Rγ interaction site in the FERM domain of JAK3 (Y100C) completely abrogated JAK3-mediated leukemic transformation. Mechanistically, we found IL2Rγ contributes to constitutive JAK3 mutant signaling by increasing JAK3 expression and phosphorylation. Conversely, we found that mutant but not wild type JAK3 increased the expression of IL2Rγ, indicating IL2Rγ and JAK3 contribute to constitutive JAK/STAT signaling through their reciprocal regulation. Overall we demonstrate a novel role for IL2Rγ in potentiating oncogenesis in the setting of JAK3-mutation positive leukemia. Additionally, our study highlights an RNAi-based functional assay that can be used to facilitate the identification of non-kinase cytokine and growth factor receptor targets for inhibiting leukemic cell growth.
Keywords: AML, JAK3, IL2Rγ, receptors, leukemia, RNAi
INTRODUCTION
Despite the great strides that have been made in the treatment of acute myeloid leukemia (AML), current therapies are largely empirical and drug resistance is common. Prognosis is generally poor, with a five-year survival rate of less than 30% with conventional chemotherapy(1). Current therapeutic challenges stem primarily from the molecular heterogeneity found in AML, with the majority of disease-causing therapeutic targets still unknown. This underscores the need for rapid identification of candidate target genes and signaling pathways so that targeted therapy can be tailored for these patients.
Cytokines, growth factors, and their receptors are known to play important roles in cell survival, proliferation, differentiation and immune response in normal and cancer cells(2–4). Substantial evidence indicates that deregulation of growth factor and cytokine signaling contributes to leukemogenesis through aberrant activation of kinase-driven signaling pathways(2, 5). For instance, the role of interleukin-7 receptor (IL-7R) mutants in lymphoid differentiation has been demonstrated through the activation of the JAK/STAT pathway(6). Mutation in the thrombopoietin receptor MPL, a JAK2 kinase regulating receptor, is involved in myeloproliferative syndromes(5, 7). Similarly, elevated IL-3 receptor expression causes JAK/STAT activation and is associated with poor clinical outcome in AML(8). Further, pharmacological targeting of these cytokines/cytokine receptors has proven to be efficacious in treating immune and inflammatory diseases(9). Additional studies demonstrated the potential therapeutic relevance of cytokines in various hematopoietic malignancies including AML(10). For example increased CD47 expression has been demonstrated as an independent, poor prognostic factor in AML that can be targeted directly with blocking monoclonal antibodies(11). These results warrant a more rapid and systematic analysis of cytokine and growth factor receptors to identify their roles in disease pathogenesis and potential exploitation as novel targets for tailored therapeutics in leukemia.
In order to identify functional and therapeutic relevance of targeting cytokine and growth factor receptors that are critical for leukemia cell growth, we established a novel functional RNAi-based assay targeting 188 non-kinase cytokine/growth factor receptors and associated proteins. To make this assay amenable to various subtypes of leukemias we choose highly expressed cytokine/growth factor receptors and associated proteins from various hematopoietic malignancies. To show applicability of this assay we utilized an AML cell line model and identified IL2Rγ as a functional hit in JAK3-mutation positive AML.
IL2Rγ is known to regulate the signaling of the JAK3 non-receptor tyrosine kinase in normal cells in a ligand dependent manner. The IL2Rγ subunit is shared by several heteromeric cytokine receptors including IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21 and is important for the development of lymphoid cells.(12) The association of JAK3 with IL2Rγ is a functionally significant event for these cytokines to exert downstream signaling. Once the receptors are engaged by their ligands, conformational changes lead to the activation as well as auto- and trans-phosphorylation of JAK1/JAK3 followed by recruitment and JAK3-mediated phosphorylation of Signal Transducer and Activator of Transcription (STAT) factors. Phosphorylation of STAT factors allows their translocation to the nucleus to regulate the transcription of a wide variety of genes.(13) The importance of JAK3 and IL2Rγ in lymphocyte functions was underscored since inactivating mutations of either IL2Rγ or JAK3 resulted in a disease similar to severe combined immunodeficiency (SCID). (14–16)
Several studies have reported JAK3 mutations in hematopoietic malignancies such as in adult T-cell leukemia/lymphoma, cutaneous T-cell lymphoma, natural killer/T-cell lymphoma (NKTCL), T-cell acute lymphoblastic leukemia (T-ALL), T-cell prolymphocytic leukemia (T-PLL), Juvenile myelomonocytic leukemia (JMML) and AML (17–27) as well as in solid tumors such as breast and gastric carcinoma.(19) In AML, the frequency of JAK3 mutations is 3.9% (19/483) and it has been mostly observed in acute megakaryoblastic leukemia (AMKL) in either children with Down syndrome or adults without Down syndrome.(20) Previous studies have demonstrated that activating mutations in JAK3 promotes constitutive activation of STAT5.(23) However, the molecular mechanism or the specific receptors regulating mutant JAK3 activity in this context is not known.
Here, utilizing a functional siRNA screen we identified a potential regulatory role for IL2Rγ in the activation of JAK3 in JAK3 mutation-positive leukemia cells and demonstrated a novel oncogenic role of IL2Rγ in these cells. This approach will be applicable to the identification of cytokine/growth factor receptors that are crucial for cancer cell growth in a wide variety of leukemia types, and this approach will identify new target genes irrespective of the mutational status of the gene.
RESULTS
Development of a functional RNAi screen panel targeting non-kinase cytokine and growth factor receptors
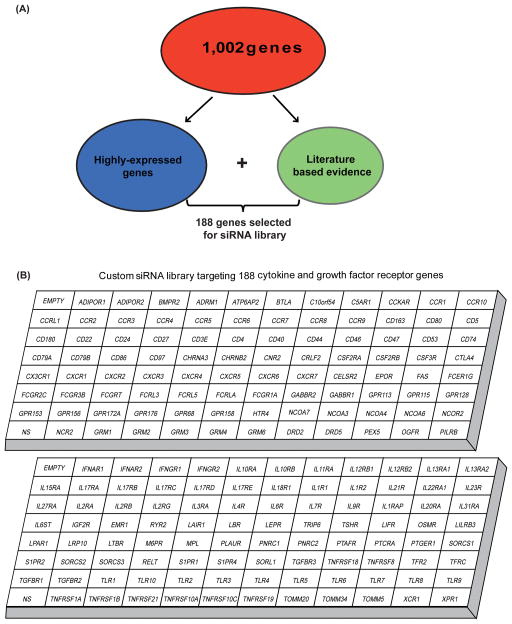
We developed an RNAi screen to assay the functional relevance of signaling mechanisms driven by non-kinase cytokine and growth factor receptors in cancer. To prioritize gene targets relevant for various hematological malignancies, we utilized microarray expression data collected from 141 leukemia patient samples including AML, B-ALL, T-ALL, CLL, and MDS. The data set was analyzed for 1,002 cytokine and growth factor receptors and associated proteins (Table S1). A total of 188 cancer related genes were selected for inclusion in our siRNA screen panel based on the filtering criteria described in Material and Methods. This final panel of 188 genes includes receptor families and associated proteins for interleukins, toll-like receptors, tumor necrosis factors, interferons, G-protein coupled receptors, growth hormone receptors and receptors associated with cell differentiation (Figure 1).
Figure 1. Functional RNAi screen panel targeting non-kinase cytokine and growth factor receptors.
(A) We performed gene expression profiling using mononuclear cells from bone marrow and peripheral blood of 141 leukemia patient samples. This data set was analyzed for 1,002 genes encoding cytokine and growth factor receptors and associated proteins. A total of 188 cancer related genes were selected for inclusion based on the following filtering criteria. First, genes consistently up-regulated compared to the average gene expression of the 100 normalization control probe sets (provided by Affymetrix) in all subjects within each subtype of disease were selected and ranked by average gene expression value. Top 15% genes from all tested subtype of diseases were selected and pooled for inclusion. These genes were designated as highly-expressed genes. Additionally, known cancer-related growth factor genes, based upon evidence found in the literature, were also included. (B) siRNA library comprising 188 genes includes non-kinase cytokine and growth factor receptor families and associated proteins for interleukins, toll-like receptors, tumor necrosis factors, interferons, G-protein coupled receptors, growth hormone receptors and receptors associated with cell differentiation.
IL2Rγ is a functional hit in JAK3A572V-positive AML cells
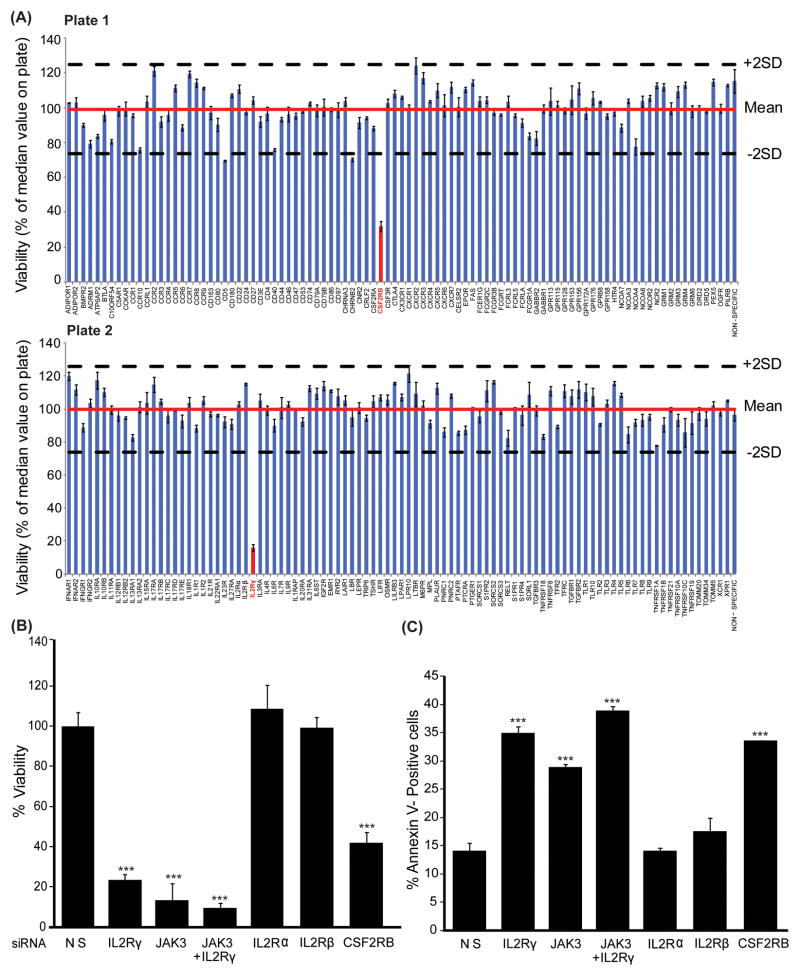
Activating JAK3 mutations have been reported in various hematopoietic malignancies(17, 19–26); however, a systematic analysis was not performed to identify the receptor regulating JAK3 kinase activity in this setting. Therefore, to demonstrate the utility of our siRNA screen and to query upstream receptors that regulate mutant JAK3 signaling, we interrogated the CMK cell line, which is derived from an acute megakaryoblastic leukemia patient (AMKL; M7 stage of AML) with Down syndrome and harbors an activating JAK3 kinase mutation, A572V(23). Utilizing our RNAi screen we identified that knockdown of IL2Rγ and colony-stimulating factor-2 receptor, beta (CSF2RB) resulted in significantly decreased viability of CMK cells (Figure 2A). These results were independently validated with individual knock down of IL2Rγ resulting in an 80% reduction of CMK cell viability and a 20% increase in apoptosis as compared to the non-silencing siRNA control (Figure 2B, C). Similar effects were observed on the growth of CMK cells when we utilized four independent IL2Rγ siRNAs (Figure S1A). Further, the effect of IL-2Rγ knockdown on the growth of AML cells is specific to JAK3 mutation positive cells as no effect on cellular growth was observed after silencing of IL2Rγ in the JAK2-mutation-positive HEL cell line (Figure S1B). This reduction in cell viability was similar to that resulting from knockdown of JAK3. Importantly, silencing of other polypeptide chains of the IL-2 receptor (IL2Rα and IL2Rβ) had no effect on cell viability or phosphorylation of JAK3 (Figure S1C), suggesting IL2Rγ is critical for the growth of leukemia cells harboring mutationally activated JAK3. CSF2RB was identified as a secondary hit (60% reduction in viability) as it had a less profound effect on cell growth as compared to IL2Rγ (Figure 2B, C). Knockdown of CSF2RB reduced phosphorylation of JAK2, STAT5, ERK1/2 kinases and S6 ribosomal protein with no effect on JAK3 phosphorylation (Figure S2). Since CMK cells have an activating JAK3 kinase mutation and IL2Rγ regulates JAK3 kinase phosphorylation we focused on the role of IL2Rγ in JAK3-mutation positive cells.
Figure 2. Functional RNAi profiling of cytokine and growth factor receptors in CMK cells.
(A) CMK cells were electroporated with individual siRNAs targeting 188 genes from cytokine and growth factor receptor families (each siRNA is a pool of four siRNAs). Cells were re-plated in triplicate into culture media for 96 hours, and cell viability was determined by addition of a tetrazolium salt (MTS assay). All values were adjusted to a blank control within the plate and normalized to the median value for all wells on the plate. Effective hits were considered those that robustly decreased cell viability to levels below two standard deviations of the median value for the plate. Bar graph values represent mean percentage (normalized to the median value for all wells on the plate) ± SEM (n= 3). (B, C) Identified targets were confirmed using JAK3 siRNA as a positive control in CMK cells. Effect of gene knock-down on cell viability (B) and apoptosis (C) was measured by MTS assay and Annexin V+ staining, respectively. Values represent mean percentage ± SEM (n= 3). *** denotes p< 0.001.
IL2Rγ is essential for JAK3-mediated transformation
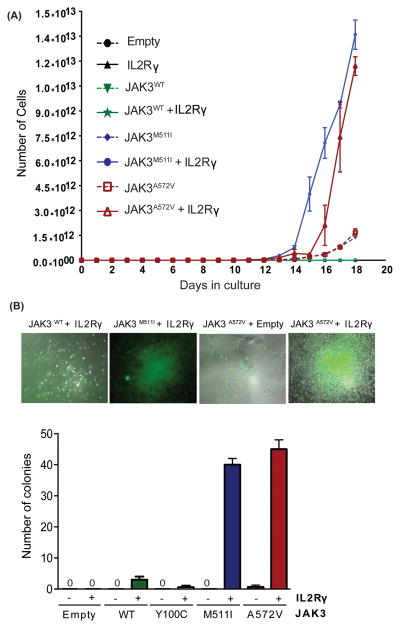
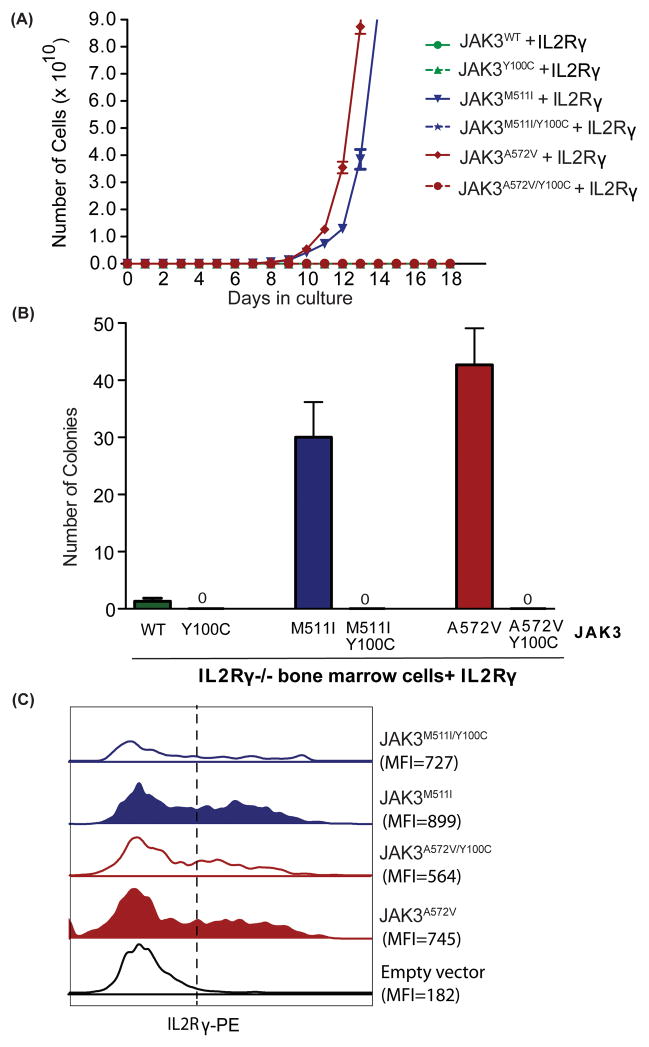
To determine the importance of IL2Rγ in JAK3-mediated cellular transformation, we overexpressed IL2Rγ in the murine pro-B cell line Ba/F3 expressing either wild-type JAK3 or the JAK3A572V mutant. Since a variety of JAK3 mutations are reported in both AML(20, 22, 23, 26) and ALL patients(19, 24, 25), we compared the effect of IL2Rγ overexpression on the growth of the cells expressing representative activating mutations of the JAK3 pseudokinase domain (A572V) and SH2 domain (M511I). We observed that overexpression of IL2Rγ imparted a significant growth advantage to cells expressing the activating JAK3A572V and JAK3M511I mutants but did not transform JAK3WT cells to IL-3 independence (Figure 3A). To establish whether IL2Rγ is essential for JAK3-mediated transformation, we transfected bone marrow cells from IL2Rγ−/− mice with retrovirus expressing JAK3WT, JAK3A572V or JAK3M511I with or without IL2Rγ. Notably, we found that the robust colony formation ability of JAK3A572V and JAK3M511I mutants that was observed in the presence of IL2Rγ was completely abrogated in the absence of IL2Rγ. In contrast, IL2Rγ expression had no effect on colony formation in cells co-expressing either JAK3WT or an inactivating JAK3 FERM domain mutation (JAK3Y100C)(13). These results suggest that IL2Rγ is necessary for JAK3 mutant-mediated transformation and support the notion that the oncogenic potential of mutant JAK3 kinases is strongly stimulated by this upstream cytokine receptor (Figure 3B).
Figure 3. IL2Rγ is necessary for transformation of JAK3 mutant-positive cells.
(A) Ba/F3 cells were retrovirally transfected with the indicated JAK3 mutants and/or IL2Rγ, and sorted using a flow cytometer. Cells were plated in medium lacking WEHI-conditioned medium (source of IL-3), and total viable cells were counted every other day for 18 days. Results shown here are representative of three independent experiments. (B) IL2Rγ−/− Lineage- mouse bone marrow cells were harvested from B6.129S4-Il2rgtm1Wjl/J mice. These cells were co-transfected with retroviruses expressing JAK3 mutants (GFP+) and IL2Rγ. Doubly positive cells (GFP+IL2Rγ+) were FACS sorted and colony formation assays were performed in cytokine-free Methocult media (M3234). Colonies were scored following 8 days of incubation at 37°C in 5% CO2. Values represent mean colony numbers ± SEM (n= 3). Representative colonies are shown.
IL2Rγ regulates mutant JAK3 kinase activity and is reciprocally regulated by JAK3
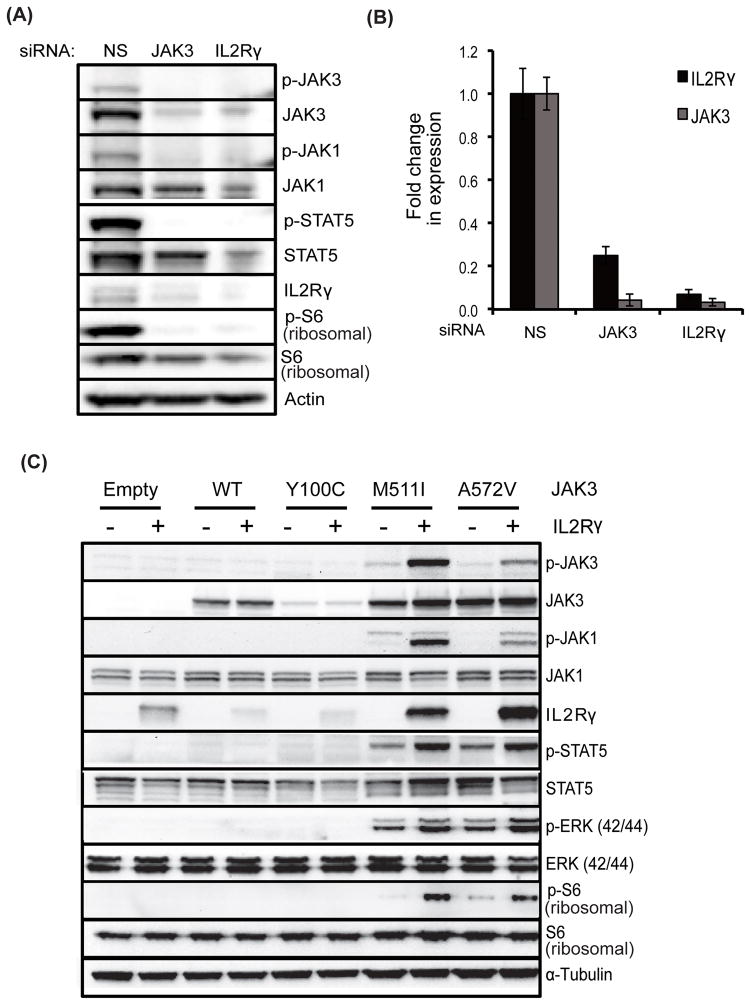
Given the critical role of IL2Rγ in JAK3-mediated cellular transformation, we next tested whether IL2Rγ regulates the activity of mutationally activated JAK3. IL2Rγ knockdown in CMK cells reduced phosphorylation of JAK3, JAK1, STAT5 and S6 ribosomal protein, and JAK3 knockdown reduced JAK1, STAT5 and S6 ribosomal protein phosphorylation, suggesting that IL2Rγ regulates JAK3 kinase (Figure 4A). Co-expression of JAK3 mutants in Ba/F3 cells with IL2Rγ revealed increased phosphorylation of JAK3, JAK1, STAT5, ERK1/2 and S6 ribosomal protein in cells expressing activating JAK3 mutants but had no effect on cells expressing wild-type JAK3 or the inactivating FERM domain mutant JAK3Y100C (Figure 4C). These results suggest that IL2Rγ is a critical regulator of JAK3 kinase signaling in leukemia cells harboring activating JAK3 mutations. We also observed that knock down of either IL2Rγ or JAK3 led to reciprocal reduction of protein levels in CMK cells (Figure 4A). These results were also validated at the transcript expression level by quantitative PCR (Figure 4B). Additionally, co-expression of activating JAK3 mutants and IL2Rγ in murine Ba/F3 cells led to elevated levels of both JAK3 and IL2Rγ compared to those in JAK3 mutant only cells and in cells co-expressing wild-type or kinase-inactive JAK3 with IL2Rγ (Figure 4C), suggesting reciprocal regulation of their expression. Interestingly, cells expressing the JAK3Y100C mutant, which is incapable of interacting with IL2Rγ, exhibited reduced expression of both JAK3 and IL2Rγ, implying that interaction between JAK3 and IL2Rγ is necessary for JAK3-mediated leukemic transformation.
Figure 4. IL2Rγ regulates JAK3 kinase signaling and reciprocally regulate each other.
(A, B) CMK cells were electroporated with JAK3 and IL2Rγ siRNA and cells were cultured for 48 hours. Whole cell lysates were subjected to immunoblotting with the indicated antibodies and actin was used as a loading control (A). Quantitative PCR analysis was performed using specific primers and GAPDH as a reference control (B). (C) Stable Ba/F3 cells were prepared by co-expressing IL2Rγ and JAK3 mutants. The effect of IL2Rγ overexpression on downstream signaling was analyzed by immunobloting with the indicated antibodies and α-tubulin was used as a loading control.
Interaction of IL2Rγ with JAK3 is required for transformation of leukemic cells harboring activating JAK3 mutations
Previous studies have shown that IL2Rγ interacts with the JAK3 FERM domain at tyrosine 100 residue of JAK3(13). To understand whether the interaction between JAK3 and IL2Rγ is necessary for the transformation of activating JAK3 mutation-positive cells we created double mutants harboring both Y100C and the activating mutations in the same allele of JAK3 (JAK3A572V/Y100C and JAK3M511I/Y100C). We observed that co-expression of JAK3A572V/Y100C or JAK3M511I/Y100C with IL2Rγ in the murine pro-B cell line Ba/F3 (Figure 5A) and in IL2Rγ−/− murine bone marrow cells (Figure 5B) completely abrogated the JAK3A572V or JAK3M511I -mediated Ba/F3 transformation to growth factor independence and clonogenic potential in murine bone marrow cells. These results suggest that direct physical interaction between IL2Rγ and JAK3 mutants is necessary for JAK3-mediated leukemic transformation. Further, the FACS analysis of murine bone marrow cells co-expressing JAK3 mutants and IL2Rγ demonstrated decreased surface expression of IL2Rγ in the cells expressing JAK3 inactivating mutations including JAK3A572V/Y100C and JAK3M511I/Y100C as compared to the cells expressing activating mutations JAK3A572V or JAK3M511I, respectively (Figure 5C). Overall these results suggest that IL2Rγ regulates JAK3 mediated transformation by its direct interaction with JAK3.
Figure 5. Direct interaction of IL2Rγ and JAK3 is necessary for the transformation of JAK3 mutant-positive cells.
(A) Ba/F3 cells were retrovirally transfected with the indicated JAK3 mutants and IL2Rγ, sorted using a flow cytometer. Cells were plated in medium lacking WEHI-conditioned medium (source of IL-3), and total viable cells were counted every other day for 18 days. Results shown here are representative of three independent experiments. (B) IL2Rγ−/− Lineage- mouse bone marrow cells were co-transfected with retroviruses expressing JAK3 mutants (GFP+) and IL2Rγ. Doubly positive cells (GFP+IL2Rγ+) were FACS sorted and colony formation was performed in cytokine-free Methocult media (M3234). Colonies were scored following 8 days of incubation at 37°C in 5% CO2. Values represent mean colony numbers ± SEM (n= 3). (C) Surface expression of IL2Rγ (anti-PE) was analyzed using FACS in JAK3 mutant positive murine bone marrow cells (GFP+) after 48 hrs of co-transfection with retroviruses. Representative histograms showing mean fluorescent intensity is shown.
DISCUSSION
The success of targeted therapies utilizing monoclonal antibodies and small molecule inhibitors in numerous malignancies has highlighted the potential of these agents for the treatment of cancer. However, precise implementation of these strategies requires a detailed understanding of the principal genetic targets involved in cancer pathogenesis in an individual patient. Cytokines and growth factor receptors signal through their specific ligands and regulate a number of cellular processes in normal and cancer cells. To establish the functional relevance of these receptors in cancer pathogenesis we designed a novel functional siRNA screen targeting highly expressed cytokine and growth factor receptors in leukemia. When we designed the siRNA screen, our goal was to create a broad application for this assay across various malignancies for the identification of functionally important cytokine and growth factor receptors. Therefore we selected those genes that were highly expressed in mononuclear cells of AML, B-ALL, T-ALL, CLL, and MDS primary samples by gene expression analysis. Here we provide a proof-of-principle example by utilizing AML cell line to show the applicability of this functional siRNA screen to identify critical cytokine and growth factor receptors required for leukemia cell growth.
The activating mutations in JAK3 have been reported in various hematopoietic malignancies including AML, ALL and JMML (17, 19–26) and in solid tumors including breast carcinoma and gastric carcinoma(19). However, the molecular mechanism leading to the constitutive activation of JAK3 kinase is not well defined. Therefore to demonstrate the applicability of siRNA screening in identifying functionally relevant cytokine and growth factor receptors we tested the CMK cell line derived from an AMKL patient harboring an activating JAK3 kinase mutation (JAK3A572V)(23). Utilizing this siRNA screen we identified that CMK cell growth is dependent on IL2Rγ and showed that IL2Rγ regulates JAK3 kinase signaling in these cells. Additionally, we found that CMK cell growth is partially dependent on CSF2RB. CSF2RB is a common beta-receptor for GM-CSF, IL-3, and IL-5 signaling(28) and is known to regulate JAK2 and SRC signaling. High expression of CSF2RB has been previously implicated in AML pathogenesis(29). Our data showed that CSF2RB regulates JAK2 but not mutant JAK3 kinase in the CMK cells. Therefore we focused this study on the role of IL2Rγ. Interestingly, overexpression of IL2Rγ potentiated an oncogenic role of JAK3 by increasing the transforming potential of activating JAK3 kinase mutations. More importantly, the ablation of IL2Rγ completely abrogated the transforming potential of JAK3 activating mutations. These results suggested that IL2Rγ is necessary for JAK3-mediated transformation. Our results are consistent with the role of EPOR or TPOR in activation of JAK2V617F mutant proteins in myeloproliferative neoplasms, which transduce hyperproliferative signals only in the presence of the JAK2-binding receptors EPOR or TPOR(30, 31). Although it should be noted that activating mutations in JAK3 were able to transform parental BaF3 cells to IL3-independence (Figure 3A), this is likely due to the use of the endogenous IL2Rγ for scaffolding.
JAK3 is known to be activated by various cytokines including IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21 as their specific cytokine receptors contain the IL2Rγ subunit as a heterodimerization partner(12). Our results suggested that IL2Rγ-mediated JAK3 signaling in JAK3A572V -positive cell lines is ligand-independent as activating JAK3 mutations induced colony formation with IL2Rγ in the absence of cytokines (Figure 3B). Further, none of the specific α/β cytokine receptor chains for IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21 were identified as ‘potential hits’ in our RNAi screen. However, we cannot completely exclude the possibility of having trace amounts of cytokine in the experimental system, which might be sufficient for ligand-dependent constitutive activation of JAK3 kinase signaling. We propose that IL2Rγ may be serving as a scaffold for direct interaction of JAK3 which enables the auto-and trans-phosphorylation by JAK1 as has been shown previously(23). Further, we demonstrated that the constitutive activation of mutant JAK3 kinase by IL2Rγ not only induced canonical JAK3/STAT5 signaling but also induced additional signaling cascades of ERK and S6 signaling pathways. These results provide additional insights into our understanding of IL2Rγ-mediated JAK3/STAT5 signaling mechanisms.
In normal cells, JAK3 is thought to bind specifically and exclusively to IL2Rγ which is primarily involved in lymphopoiesis and immune responses (12). It has been shown that the membrane-proximal region of IL2Rγ is essential for its association with JAK3. Specifically, the Y100 residue within the FERM domain of JAK3 is necessary and sufficient for its binding to IL2Rγ and to deliver downstream signals(13). Our result suggested that although IL2Rγ-mediated regulation of JAK3 activation was ligand independent, the oncogenic potential of this complex still required JAK3 interaction with IL2Rγ as a mutation in the FERM domain of JAK3 (Y100C) completely abrogated JAK3-mediated leukemic transformation. In addition our data suggested that IL2Rγ and JAK3 reciprocally regulated their expression both at the RNA and protein levels. Interestingly, we found that association between JAK3 and IL2Rγ was critical for their expression as mutation in JAK3 at Y100C reduced expression of JAK3 and IL2Rγ as compared to wild-type control. Furthermore, activating mutations in JAK3 increased both JAK3 and IL2Rγ levels. However, it is not clear whether the increased expression of activated JAK3 mutant and IL2Rγ is a cause of the mutations or an effect of JAK3 activation. It is possible that activation induces an allosteric change that stabilized JAK3. These results are consistent with previous studies where JAK kinase family members have been reported to regulate plasma membrane expression of their cognate receptors. (30, 32)
Previous studies have reported activating JAK3 kinase mutations in various hematopoietic malignancies (19, 20, 22–26). However, targeted treatment options are limited for patients with activating JAK3 mutations. Several small molecule kinases inhibitors such as (CP-690, 550, R348, and VX509) targeting the JAK3 kinase at the level of the ATP binding pocket have been reported(33). Some of these inhibitors have been tested in hematopoietic malignancies(34) and inflammatory diseases(35); however, none have been clinically approved (reviewed in (36)). We think that the finding of constitutive activation of multiple signaling pathways and reciprocal regulation of IL2Rγ and JAK3 by direct interaction is clinically relevant for rational design of therapeutic strategies. Our results support further investigation into therapeutic strategies that directly block the interaction between JAK3 and IL2Rγ, similar to how the mutation in Y100C inhibits constitutive activation of JAK3 mediated signaling. Therefore a small molecule inhibitor targeting interaction of the FERM domain of JAK3 with IL2Rγ would be able to block constitutive activation of JAK3 kinases irrespective of kinase domain mutations. This approach might be applicable to other JAK kinases, for example, JAK2 activity is also regulated by the FERM domain through direct interaction with its cognate receptors (30, 37).
Overall, these results demonstrate that IL2Rγ plays an onogenic role in JAK3 mutation positive leukemias and provides an excellent proof-of-principle example whereby a functional screening methodology facilitated identification of a receptor regulating mutant JAK3 kinase activity. We established that functional screening with an siRNA library targeting cytokine and growth factor receptor is an efficient and useful approach that can identify known and unknown therapeutic targets in malignant cells. These targets can then in turn provide additional insights into the identification of signaling mechanisms or can serve as an alternative targets for therapeutic intervention. Using this functional profiling approach we also demonstrate that oncogenes are not limited to mutated genes in malignant cells. Taken together, we have shown that an RNAi-based functional screen for non-kinase cytokine and growth factor receptors can rapidly identify novel oncogenic pathophysiological mechanisms, underscoring the importance of targeting cellular growth factor receptors in leukemia.
MATERIALS AND METHODS
Patient samples and microarray data
All samples were obtained from patients evaluated at the Oregon Health & Science University (OHSU) Center for Hematologic Malignancies following informed consent as approved by the OHSU Institutional Review Board. Gene expression profiling on mononuclear cells from bone marrow and peripheral blood of 141 leukemia patient samples was performed, including AML (N=38), precursor B-cell acute lymphoblastic leukemia (B-ALL; N=17), T-cell acute lymphoblastic leukemia (T-ALL; N=13), chronic lymphocytic leukemia (CLL; N=41), chronic myeloid leukemia (CML; N=22), and myelodysplastic syndrome (MDS; N=10). Gene expression analysis was conducted using a GeneChip® Affymetrix Human Genome U133 (HG–U133A & B) array, NCBI Gene Expression Omnibus accessions GPL96 and GPL97, with more than 39,000 transcripts. After signal quantification, the resulting data was pre-processed and normalized within each disease subtype using Robust Multi-array Average (RMA) using R Bioconductor (http://www.bioconductor.org/)(38). This gene expression data has been deposited to NCBI’s Gene Expression Omnibus and is accessible through GEO series accession number GSE51082 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=edmdqoourhebtyz&acc=GSE51082).
Microarray data filtering criteria for designing siRNA screen
For individual patient samples, expression of each gene was compared to the average gene expression of the 100 normalization control probe sets (provided by Affymetrix). Genes with expression level higher than the average expression of normalization control probe sets were identified for each sample. We conventionally designated these genes as well-expressed in these samples. If the individual gene was identified as well-expressed for all samples in a subtype of disease, this gene passed the first filtering criteria. In our hand this is a very stringent criteria. For example, the probability of falsely identifying a gene expressed in AML (N=38) is 1.30E−6 even if the true probability of it being well-expressed is only 0.3. In other words, the probability of passing the first criteria is 1.30E−6 when the genes with higher expression than the average expression level for control probe sets have only a 30% chance of being a true well-expressed gene. Additionally, the probability of false identification for MDS, which is the smallest subtype of disease (N=10) is 0.0282. The probability of false identification is defined by the probability of a gene passing the filtering criteria when a gene is not well-expressed. Those genes that passed the first criteria were ranked by average expression of each subtype of disease, and the top 15% genes from all tested disease subtypes were selected and pooled for inclusion. These genes were designated as highly-expressed genes. This list was manually curated and also included 5% known cancer-related genes. (2, 3, 39, 40)
siRNA knockdown
CMK cells were obtained from the German National Resource Center for Biological Material. Cells were maintained in RPMI 1640 medium supplemented with 2 mM L-glutamine, 10% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin at 37°C in 5% CO2. Cell lines used in this study were not cultured for longer than 6 months from initial purchase or characterization. For siRNA knockdown, cells (1×105/well) were electroporated at 200V, with 2 pulses of 1.5ms in two separate 96-well electroporation plates (Bio-Rad Laboratories, Hercules, CA) containing 0.1 nM of individual gene siRNAs per well in 100 μl of siPORT buffer (120mM Trehalose, 20mM HEPES, 1mM Myo-Inositol, 1mM KCl, 1mM MgCl2, 1mM K2HPO4, 0.4mM KH2PO4, 2.14 mM KOH, and 1mM Glutathione) as described previously (41). siRNA utilized for the study are pool of four individual siRNAs (siGENOME SMARTpool format from Thermo Scientific, Pittsburgh, PA). Cells were cultured in triplicate for 96 hours and subjected to a cell viability assay (CellTiter; Promega, Madison, WI). All values from the MTS assay were adjusted to a blank control and normalized to the median value for all wells on the plate. Effective hits were considered those that robustly decreased cell viability to levels below two standard deviations of the median value for the plate. Apoptosis was measured by AnnexinV+ staining (Guava Technology, EMD Millipore Corp, Billerica, MA).
Plasmid constructs
JAK3 was cloned into the MSCV-IRES-GFP vector and IL2Rγ was cloned into a Gateway compatible pMXs-IRES-Puromycin vector. Site directed mutagenesis was performed using the QuikChange®IIXL kit (Agilent Technologies, Santa Clara, CA).
Retroviral infection and IL3 withdrawal assay
Ba/F3 cells were cultured in standard media containing RPMI 1640 media, 10% fetal bovine serum (FBS), L-glutamine, penicillin-streptomycin (Life Technologies, Grand Island, NY) containing IL-3 from WEHI conditioned media. JAK3-GFP mutant and IL2Rγ retrovirus was generated by transfecting 293T/17 cells with EcoPac helper plasmid (a kind gift of Dr. Rick Van Etten, Tufts University, Boston, MA) and MSCV retroviral constructs using Fugene (Roche Diagnostics Corp., Indianapolis, IN). For infection, 1 mL of viral supernatant was used to infect 1×106 cell using two rounds of spinoculation in the presence of 5ug/mL polybrene and HEPES at 2,500 rpm for 90 mins. Cells were cultured for several days, followed by sorting of double positive cells using FACSAriaII (BD Bioscience, San Jose, CA) for JAK3 and IL2Rγ expression gated on GFP and anti-human IL2Rγ-PE (BioLegend, San Diego, CA), respectively. Equal density of stable cell lines plated in medium lacking WEHI-conditioned medium (source of IL-3), and total viable cells were counted every other day for three weeks.
Colony assays
Bone marrow cells were harvested from 6 week old B6.129S4-Il2rgtm1Wjl/J mice (Jackson Laboratory, Bar Harbor, Maine). Cells were subjected to red cell lysis in NH4Cl solution (0.8% NH4Cl with 0.1 mM EDTA). Lineage-cells were enriched using a Lineage Cell Depletion Kit according to the manufacturer’s protocol (Miltenyi Biotech, Germany). Cells were pre-stimulated at 37°C in DMEM with 15% FCS, 15% WEHI-3B, murine IL-3 (7 ng/mL), IL-6 (12 ng/mL), and SCF (56 ng/mL) (Stem Cell Technologies, Canada). JAK3 mutants and IL2Rγ retroviruses were generated as described above. After 24 hrs, the cells were spinoculated into 6-well plates with viral supernatant in pre-stimulation medium in the presence of 5 μg/mL polybrene and HEPES at 2,500 rpm for 90 mins. This was repeated at 48 hrs. Cells expressing JAK3 and IL2Rγ were doubly sorted by FACS, gated on GFP (JAK3) and anti-human IL2Rγ-PE (BioLegend). 1×104 cells per 35 mm dish were plated in triplicate in methylcellulose medium in the absence of cytokines (MethoCult M3234, Stem Cell Technologies). Colonies were scored following 8 days of incubation at 37°C in 5% CO2.
Immunoblotting
Whole cell lysates from cells were prepared in 1x lysis buffer (Cell Signaling, Boston, MA) and 1% protease and phosphatase inhibitors (Sigma Chemicals, St. Louis, MO). Equal amounts of protein (50–100 μg) were fractionated on 4–15% Tris-glycine gels (Bio-Rad, Hercules, CA), transferred to PVDF membranes, and probed with the indicated antibodies. Antibodies used for immunoblotting were: JAK3, pJAK3, IL2Rγ, CSF2RB (Santa Cruz Biotechnology, Dallas, TX), pJAK1, JAK1, STAT5, pSTAT5, AKT, pAKT, ERK1/2, pERK1/2 (Cell Signaling, Beverly, MA), and α-tubulin (Sigma Chemicals).
Quantitative RT-PCR analysis
CMK cells were transfected with the indicated siRNA. Total RNA was extracted using an RNeasy Mini Kit (QIAGEN, Valencia, CA). cDNA was synthesized using Oligo(dT)-primers with SuperScript-First-Strand Synthesis kit (Invitrogen, Carlsbad, CA). cDNA was amplified with Opticon-3 thermal cycler (MJ Research, Waltham, MA) using 2xSYBR Green (Invitrogen). Primers used were as follows: JAK3 Forward: 5′-GGCCCCATCACTCTGGACT-3′, JAK3 Reverse: 5′-GCCCTTATAATCAGGACCAAGG-3′, IL2Rγ Forward: 5′-CCACAGCTGATTTCTTCCTGA-3′; IL2Rγ Reverse: 5′-GCAGAGTGAGGTTGGTAGGC-3′, GAPDH Forward: 5′-TCCTGCACCACCAACTGCTTAG-3′, and GAPDH Reverse: 5′-GGCATGGACTGTGGTCATGAG-3′.
Statistical methods
For siRNA knockdown experiments, a 2-tailed Student’s t-test was carried out for each well in comparison to pooled nonspecific siRNA controls. For proliferation and apoptosis assays continuous variables were compared by pairwise Student’s t-test for 2 independent samples. A p value less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
Financial Support
A.A. is supported by a National Cancer Institute Career Development Award (5 K99 CA151670-02), Collins Foundation, Knight Pilot Project, and Friends of Doernbecher grants. J.W.T. is supported by grants from the V-Foundation, Gabrielle’s Angel Foundation, and the National Cancer Institute (5R00CA151457-04; 1R01CA183947-01). B.J.D. is a Howard Hughes Medical Institute investigator. This work is also supported by The Leukemia & Lymphoma Society.
Footnotes
CONFLICT OF INTEREST
OHSU and B.J.D. have a financial interest in Molecular MD (5% equity or less). This potential conflict of interest has been reviewed and managed by the OHSU Conflict of Interest in Research Committee and the Integrity Program Oversight Council. OHSU has clinical trial contracts with Novartis, and Bristol Myers Squibb to pay for patient costs, nurse and data manager salaries, and institutional overhead. B.J.D. does not derive salary, nor do their laboratories receive funds, from these contracts. B.J.D serves as a consultant for Roche and Nodality (Consulting income $10,000 or over).
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
AUTHORSHIP CONTRIBUTION
We thank Sarah Bowden for administrative support. A.A., R.J.M., K.W., R.B. and M.A.D. designed the research, performed experiments, and wrote the paper. S.M. and B.P. helped with microarray gene expression analysis. J.W.T., C.E.T., C.A.E., and B.J.D. provided critical feedback and helped with manuscript preparation.
References
- 1.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006 Nov 25;368(9550):1894–907. doi: 10.1016/S0140-6736(06)69780-8. eng. [DOI] [PubMed] [Google Scholar]
- 2.Graf M, Hecht K, Reif S, Pelka-Fleischer R, Pfister K, Schmetzer H. Expression and prognostic value of hemopoietic cytokine receptors in acute myeloid leukemia (AML): implications for future therapeutical strategies. Eur J Haematol. 2004 Feb;72(2):89–106. doi: 10.1046/j.0902-4441.2003.00184.x. eng. [DOI] [PubMed] [Google Scholar]
- 3.Van Etten RA. Aberrant cytokine signaling in leukemia. Oncogene. 2007 Oct 15;26(47):6738–49. doi: 10.1038/sj.onc.1210758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2013 May 23;32(21):2601–13. doi: 10.1038/onc.2012.347. [DOI] [PubMed] [Google Scholar]
- 5.Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2010 Jun;24(6):1128–38. doi: 10.1038/leu.2010.69. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009 Sep 24;114(13):2688–98. doi: 10.1182/blood-2009-03-208397. eng. [DOI] [PubMed] [Google Scholar]
- 7.Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS medicine. 2006 Jul;3(7):e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin L, Lee EM, Ramshaw HS, Busfield SJ, Peoppl AG, Wilkinson L, et al. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 2009 Jul 2;5(1):31–42. doi: 10.1016/j.stem.2009.04.018. eng. [DOI] [PubMed] [Google Scholar]
- 9.O’Shea JJ, Park H, Pesu M, Borie D, Changelian P. New strategies for immunosuppression: interfering with cytokines by targeting the Jak/Stat pathway. Current opinion in rheumatology. 2005 May;17(3):305–11. doi: 10.1097/01.bor.0000160781.07174.db. [DOI] [PubMed] [Google Scholar]
- 10.Ferretti E, Cocco C, Airoldi I, Pistoia V. Targeting acute myeloid leukemia cells with cytokines. Journal of leukocyte biology. 2012 Sep;92(3):567–75. doi: 10.1189/jlb.0112036. [DOI] [PubMed] [Google Scholar]
- 11.Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, Jr, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009 Jul 23;138(2):286–99. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Shea JJ, Pesu M, Borie DC, Changelian PS. A new modality for immunosuppression: targeting the JAK/STAT pathway. Nature reviews Drug discovery. 2004 Jul;3(7):555–64. doi: 10.1038/nrd1441. [DOI] [PubMed] [Google Scholar]
- 13.Tang W, Huo H, Zhu J, Ji H, Zou W, Xu L, et al. Critical sites for the interaction between IL-2Rgamma and JAK3 and the following signaling. Biochemical and bio research communications. 2001 May 11;283(3):598–605. doi: 10.1006/bbrc.2001.4824. eng. [DOI] [PubMed] [Google Scholar]
- 14.Cacalano NA, Migone TS, Bazan F, Hanson EP, Chen M, Candotti F, et al. Autosomal SCID caused by a point mutation in the N-terminus of Jak3: mapping of the Jak3-receptor interaction domain. The EMBO journal. 1999 Mar 15;18(6):1549–58. doi: 10.1093/emboj/18.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baird AM, Thomis DC, Berg LJ. T cell development and activation in Jak3-deficient mice. Journal of leukocyte biology. 1998 Jun;63(6):669–77. doi: 10.1002/jlb.63.6.669. [DOI] [PubMed] [Google Scholar]
- 16.Candotti F, Oakes SA, Johnston JA, Notarangelo LD, O’Shea JJ, Blaese RM. In vitro correction of JAK3-deficient severe combined immunodeficiency by retroviral-mediated gene transduction. The Journal of experimental medicine. 1996 Jun 1;183(6):2687–92. doi: 10.1084/jem.183.6.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellanger D, Jacquemin V, Chopin M, Pierron G, Bernard OA, Ghysdael J, et al. Recurrent JAK1 and JAK3 somatic mutations in T-cell prolymphocytic leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2014 Feb;28(2):417–9. doi: 10.1038/leu.2013.271. [DOI] [PubMed] [Google Scholar]
- 18.De Vita S, Mulligan C, McElwaine S, Dagna-Bricarelli F, Spinelli M, Basso G, et al. Loss-of-function JAK3 mutations in TMD and AMKL of Down syndrome. British journal of haematology. 2007 May;137(4):337–41. doi: 10.1111/j.1365-2141.2007.06574.x. eng. [DOI] [PubMed] [Google Scholar]
- 19.Jeong EG, Kim MS, Nam HK, Min CK, Lee S, Chung YJ, et al. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008 Jun 15;14(12):3716–21. doi: 10.1158/1078-0432.CCR-07-4839. [DOI] [PubMed] [Google Scholar]
- 20.Kiyoi H, Yamaji S, Kojima S, Naoe T. JAK3 mutations occur in acute megakaryoblastic leukemia both in Down syndrome children and non-Down syndrome adults. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2007 Mar;21(3):574–6. doi: 10.1038/sj.leu.2404527. eng. [DOI] [PubMed] [Google Scholar]
- 21.Sakaguchi H, Okuno Y, Muramatsu H, Yoshida K, Shiraishi Y, Takahashi M, et al. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nature genetics. 2013 Jul 7; doi: 10.1038/ng.2698. [DOI] [PubMed] [Google Scholar]
- 22.Sato T, Toki T, Kanezaki R, Xu G, Terui K, Kanegane H, et al. Functional analysis of JAK3 mutations in transient myeloproliferative disorder and acute megakaryoblastic leukaemia accompanying Down syndrome. British journal of haematology. 2008 May;141(5):681–8. doi: 10.1111/j.1365-2141.2008.07081.x. [DOI] [PubMed] [Google Scholar]
- 23.Walters DK, Mercher T, Gu TL, O’Hare T, Tyner JW, Loriaux M, et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer cell. 2006 Jul;10(1):65–75. doi: 10.1016/j.ccr.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Bains T, Heinrich MC, Loriaux MM, Beadling C, Nelson D, Warrick A, et al. Newly described activating JAK3 mutations in T-cell acute lymphoblastic leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2012 Mar 19; doi: 10.1038/leu.2012.74. Eng. [DOI] [PubMed] [Google Scholar]
- 25.Blink M, Buitenkamp TD, van den Heuvel-Eibrink MM, Danen-van Oorschot AA, de Haas V, Reinhardt D, et al. Frequency and prognostic implications of JAK 1–3 aberrations in Down syndrome acute lymphoblastic and myeloid leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2011 Aug;25(8):1365–8. doi: 10.1038/leu.2011.86. [DOI] [PubMed] [Google Scholar]
- 26.Tefferi A. JAK and MPL mutations in myeloid malignancies. Leukemia & lymphoma. 2008 Mar;49(3):388–97. doi: 10.1080/10428190801895360. [DOI] [PubMed] [Google Scholar]
- 27.Constantinescu SN, Girardot M, Pecquet C. Mining for JAK-STAT mutations in cancer. Trends in biochemical sciences. 2008 Mar;33(3):122–31. doi: 10.1016/j.tibs.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Murphy JM, Young IG. IL-3, IL-5, and GM-CSF signaling: crystal structure of the human beta-common receptor. Vitamins and hormones. 2006;74:1–30. doi: 10.1016/S0083-6729(06)74001-8. [DOI] [PubMed] [Google Scholar]
- 29.Hercus TR, Dhagat U, Kan WL, Broughton SE, Nero TL, Perugini M, et al. Signalling by the betac family of cytokines. Cytokine & growth factor reviews. 2013 Jun;24(3):189–201. doi: 10.1016/j.cytogfr.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Funakoshi-Tago M, Pelletier S, Moritake H, Parganas E, Ihle JN. Jak2 FERM domain interaction with the erythropoietin receptor regulates Jak2 kinase activity. Molecular and cellular biology. 2008 Mar;28(5):1792–801. doi: 10.1128/MCB.01447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spivak JL, Merchant A, Williams DM, Ophelia Rogers O, Zhao W, Moliterno AR, et al. A Functional Thrombopoietin Receptor Is Required for Full Expression of Phenotype in a JAK2 V617F Transgenic Mouse Model of Polycythemia Vera. American Society of Hematology: Blood. 2012 [Google Scholar]
- 32.Hofmann SR, Lam AQ, Frank S, Zhou YJ, Ramos HL, Kanno Y, et al. Jak3-independent trafficking of the common gamma chain receptor subunit: chaperone function of Jaks revisited. Molecular and cellular biology. 2004 Jun;24(11):5039–49. doi: 10.1128/MCB.24.11.5039-5049.2004. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornejo MG, Boggon TJ, Mercher T. JAK3: a two-faced player in hematological disorders. The international journal of biochemistry & cell biology. 2009 Dec;41(12):2376–9. doi: 10.1016/j.biocel.2009.09.004. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ju W, Zhang M, Jiang JK, Thomas CJ, Oh U, Bryant BR, et al. CP-690,550, a therapeutic agent, inhibits cytokine-mediated Jak3 activation and proliferation of T cells from patients with ATL and HAM/TSP. Blood. 2011 Feb 10;117(6):1938–46. doi: 10.1182/blood-2010-09-305425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijayakrishnan L, Venkataramanan R, Gulati P. Treating inflammation with the Janus kinase inhibitor CP-690,550. Trends in pharmacological sciences. 2011 Jan;32(1):25–34. doi: 10.1016/j.tips.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Kontzias A, Kotlyar A, Laurence A, Changelian P, O’Shea JJ. Jakinibs: a new class of kinase inhibitors in cancer and autoimmune disease. Current opinion in pharmacology. 2012 Aug;12(4):464–70. doi: 10.1016/j.coph.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wernig G, Gonneville JR, Crowley BJ, Rodrigues MS, Reddy MM, Hudon HE, et al. The Jak2V617F oncogene associated with myeloproliferative diseases requires a functional FERM domain for transformation and for expression of the Myc and Pim proto-oncogenes. Blood. 2008 Apr 1;111(7):3751–9. doi: 10.1182/blood-2007-07-102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003 Apr;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 39.Rubin JB. Chemokine signaling in cancer: one hump or two? Semin Cancer Biol. 2009 Apr;19(2):116–22. doi: 10.1016/j.semcancer.2008.10.001. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiss C, Benko I, Kovacs P. Leukemic cells and the cytokine patchwork. Pediatric blood & cancer. 2004 Feb;42(2):113–21. doi: 10.1002/pbc.10436. [DOI] [PubMed] [Google Scholar]
- 41.Tyner JW, Walters DK, Willis SG, Luttropp M, Oost J, Loriaux M, et al. RNAi screening of the tyrosine kinome identifies therapeutic targets in acute myeloid leukemia. Blood. 2008 Feb 15;111(4):2238–45. doi: 10.1182/blood-2007-06-097253. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.