Abstract
The Aspergillus fumigatus nonribosomal peptide synthetase FtpA is among the few of this species whose natural product has remained unknown. Both FtpA adenylation domains were characterized in vitro. Fumaric acid was identified as preferred substrate of the first and both l-tyrosine and l-phenylalanine as preferred substrates of the second adenylation domain. Genetically engineered A. fumigatus strains expressed either ftpA or the regulator gene ftpR, encoded in the same cluster of genes, under the control of the doxycycline-inducible tetracycline-induced transcriptional activation (tet-on) cassette. These strains produced fumaryl-l-tyrosine and fumaryl-l-phenylalanine which were identified by liquid chromatography and high-resolution mass spectrometry. Modeling of the first adenylation domain in silico provided insight into the structural requirements to bind fumaric acid as peptide synthetase substrate. This work adds aromatic fumaric acid amides to the secondary metabolome of the important human pathogen A. fumigatus which was previously not known as a producer of these compounds.
INTRODUCTION
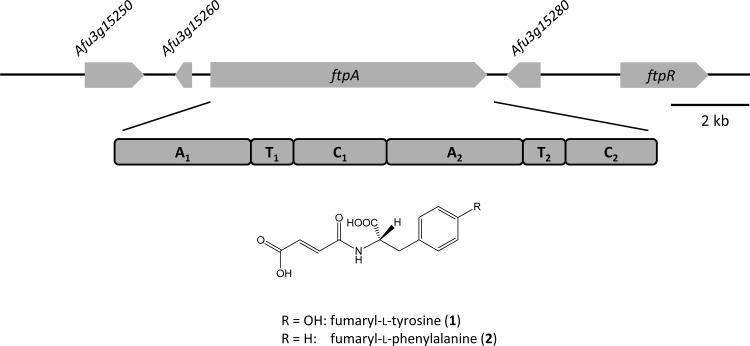
Natural products, among them nonribosomally produced peptides such as gliotoxin, fumitremorgin, and siderophores, play a critical role for fungal biology and during parasitic interactions with human, animal, or plant hosts (1). Hence, the biosynthetic capacity to produce small molecules of fungi has been the object of intensive research, as exemplified by the human pathogen Aspergillus fumigatus, which represents the most common etiological agent of invasive aspergillosis (2). The majority of its 14 nonribosomal peptide synthetases (NRPSs), among them synthetases for the mycotoxins gliotoxin (3) and fumigaclavine (4), was functionally characterized in vitro or through reverse genetics. Among the uncharacterized NRPSs of A. fumigatus is NPS7, here referred to as FtpA (fumaryl-l-tyrosine/fumaryl-l-phenylalanine synthetase, Fig. 1), which resembles the fumaryl-l-alanine synthetase SidE of the same species (5): the respective genes are paralogs, both enzymes feature two modules, each including domains for adenylation (A), thiolation (T), and condensation (C). The primary amino acid sequences of FtpA and SidE share 34% identical amino acids. While prior work demonstrated genetic induction under various physiological stress conditions for sidE (5), the function of FtpA remained obscure as its gene is not expressed under any known condition. Assumptions on the FtpA function based on phylogenetic grounds were not predictive as the SidE and FtpA A domains clearly fell in distinct phylogenetic groups. Second, besides fumaryl-l-alanine, other dipeptides have not been reported yet from A. fumigatus, and the transcriptionally inactive ftpA gene (Afu3g15270) made further investigations via analytical chemistry to identify the respective small molecular product difficult.
FIG 1.
Schematic representation of ftpA, its genetic environment in A. fumigatus, and the domain arrangement of the gene product. Introns are not shown. Gene Afu3g15250 putatively encodes a transporter of the major facilitator superfamily, gene Afu3g15260 encodes a hypothetical protein, the ftpA gene encodes the fumaryl-l-tyrosine/fumaryl-l-phenylalanine synthetase (Afu3g15280), a methyltransferase, and ftpR, a Zn2Cys6 transcription factor. Abbreviations for synthetase domains: A, adenylation; T, thiolation; C, condensation. The chemical structures of compounds 1 and 2 are shown below the enzyme.
However, analyses of other fungal metabolomes have previously identified fumaric acid as a building block in bioactive peptidic metabolites, including the sorbicillactones of Penicillium chrysogenum which are cytotoxic against leukemia cells (6), xylariamide A of a Xylaria species that showed toxicity in the brine shrimp lethality assay (7), and the thrombine inhibitor Ro09-1679 (fumaryl-l-arginyl-l-leucyl-arginal, isolated from Mortierella alpina (8). The genetic or enzymatic basis of their biosynthesis was not reported for these compounds.
Here, we report on genetically engineered A. fumigatus strains in which expression of the ftpA gene is directly or indirectly activated, along with an in vitro approach using analytical and biochemistry. The combination of these approaches allowed for assigning fumaryl-l-tyrosine (compound 1) and fumaryl-l-phenylalanine (compound 2, Fig. 1) to the repertoire of A. fumigatus small-molecule natural products. This result is consistent with biochemical assays using heterologously produced FtpA in vitro that identified l-tyrosine, l-phenylalanine, and fumaric acid as preferred substrates. We also report an alternate NRPS specificity signature for fumaric acid to aid identifying fumaric acid-incorporating fungal NRPSs more accurately and beyond the genus Aspergillus. Since such compounds occur in various taxonomic groups, as described above, we expect our data will promote efforts to predict structural elements of compounds from genomic sequences.
MATERIALS AND METHODS
General procedures.
Isolation of plasmid DNA, restrictions, and ligations were carried out according to the manufacturers' instructions (NEB, Fermentas, Zymo) and according to standard procedures (9). Isolation of chromosomal A. fumigatus DNA was performed using the MasterPure Yeast DNA purification kit (Epicentre Biotechnologies). Synthetic DNA was manufactured by GenScript and Life Technologies. Chemicals and media ingredients were from Fluka, Sigma-Aldrich, and Roth. [32P]pyrophosphate (60 Ci mmol−1) was purchased from Perkin-Elmer. Aspergillus sequence information was obtained from the Central Aspergillus Data Repository CADRE (10) and the Aspergillus Genome Database AspGD (11).
Construction of recombinant A. fumigatus strains ftpR-Oex and ftpA-Oex.
For overexpression of the transcription factor gene ftpR, the tetracycline-induced transcriptional activation (tet-on) system using plasmid pCH008 (12) was applied. The ftpR gene was amplified with primers npsR_PmeI_for and npsR_PmeI_rev (see Table S1 in the supplemental material) introducing PmeI restriction sites at both ends, using genomic A. fumigatus DNA as the template and Phusion Flash master mix (Thermo Fisher Scientific) according to the manufacturer's instructions. The resulting 2.2-kb fragment was cloned into plasmid pJet1.2/blunt (Thermo Fisher Scientific) and was then transferred via PmeI to plasmid pCH008. The final plasmid pteton-ftpR was used to transform protoplasts of A. fumigatus wild-type strain CBS144.89 leading to strain ftpR-Oex. The identical procedure was performed for the ftpA gene but using primers nps7_PmeI_for and nps7_PmeI_rev (see Table S1 in the supplemental material) instead, resulting in a 7.1-kb fragment. The integration of the teton-ftpA and teton-ftpR constructs into the genome of A. fumigatus was verified by Southern blot and PCR analysis using genomic A. fumigatus DNA of wild type (control) and the respective transformants (see Fig. S1 in the supplemental material). Doxycycline-mediated inducibility of ftpA and ftpR was verified by reverse transcription-PCR (see Fig. S1 in the supplemental material).
Cloning of gene expression constructs.
The full-length A. fumigatus ftpA gene was assembled from four fragments (I-IV; see Fig. S2 in the supplemental material). The portions encoding fragment I and III (2,561 and 1,811 bp, respectively) were supplied in vector pUC57 as synthetic DNA, optimized for Escherichia coli codon usage. Fragments II and IV (1,424 and 1,435 bp, respectively) were amplified by PCR, using genomic A. fumigatus DNA as the template. For both fragments, the respective reaction (50 μl) consisted of 2 mM MgSO4, 0.1 mM (each) deoxynucleoside triphosphate, 20 pmol (each primer) of oligonucleotide primer pairs NPS7_F2_For1 and NPS7_F2_Rev1 (for fragment II) or NPS7_F4_For1 and NPS7_F4_Rev1 (for fragment IV), and 2.5 U of Pfu DNA polymerase. Primer sequences are shown in Table S1 in the supplemental material. Thermal cycling conditions were: 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 3 min, followed by a terminal hold at 72°C for 10 min. The PCR products were ligated into pBluescript SK which had been linearized with EcoRV and treated with calf intestinal phosphatase to yield plasmids pDK11 (fragment II) and pDK12 (fragment IV). Fragment I was cloned into pET28a via the NdeI and HindIII sites to yield plasmid pDK9, followed by insertion of fragment II (released from pDK11 by restriction with SacII and HindIII) between the SacII and HindIII sites of pDK9 to yield plasmid pDK13. Fragment III was cloned into pET28a also via the NdeI and HindIII sites to give plasmid pDK10, into which fragment IV (released from pDK12) was inserted between the SacII and HindIII sites to yield pDK14. The latter plasmid was used to produce the A2-T2-C2 tridomain protein. The combined fragments I and II were excised from pDK13 by restriction with NdeI and NheI and inserted into pDK14 via the same sites to yield expression plasmid pDK15 that includes the reconstituted full-length ftpA gene to produce N-terminally hexahistidine-tagged fusion protein. DNA sequencing confirmed the correct assembly of the respective constructs.
The expression plasmid to produce the FtpA A1-domain heterologously as N-terminal polyhistidine fusion (pDK17) was made by amplifying the respective gene portion by PCR using pDK15 as the template. The PCR (50 μl) was set up as described above but using primers NPS7_A1_f1 and NPS7_A1_r1 (see Table S1 in the supplemental material). The thermal cycling conditions were as follows: 10 cycles of 95°C for 30 s, 49°C for 30 s, and 72°C for 3 min, followed by 25 cycles with increased annealing temperature (58°C) and a terminal hold at 72°C for 10 min. The PCR product was cloned into pBluescript SK (linearized by EcoRV restriction) to yield plasmid pDK16. The insert was then recloned between the NheI and NotI sites of pET28a to yield plasmid pDK17.
Gene expression and protein purification.
E. coli KRX was transformed with plasmids pDK14 and pDK17 to produce the A1 domain and the A2-T2-C2 tridomain, respectively. E. coli transformants were cultivated overnight in Luria-Bertani (LB) medium as 5-ml shake cultures which were used to inoculate 400 ml of LB medium. Cultures were incubated at 180 rpm and 37°C on an orbital shaker until an optical density at 600 nm (OD600) of 0.35 was reached. Subsequently, the temperature was decreased to 16°C for 30 min before gene expression was induced by adding 0.1% (wt/vol) l-rhamnose. The media were amended with kanamycin (50 mg liter−1), wherever appropriate.
Full-length apo-FtpA was produced in E. coli BL21(DE3) transformed with pDK15, which was grown as described above, but to an OD600 of 0.6, followed by induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Incubation of transformed E. coli cultures proceeded for 24 h before the biomass was collected by centrifugation (4°C, 1,200 × g, 15 min). The pellet was resuspended in lysis buffer (50 mM NaH2PO4·H2O, 300 mM NaCl, 10 mM imidazole [pH 8.0]) and sonicated using a Bandelin sonopulse sonifier. Cell debris was removed by subsequent centrifugation at 4°C and 14,000 × g for 15 min. The proteins were purified by immobilized metal affinity chromatography on Ni2+-NTA resin (Macherey & Nagel). The A1 domain and the A2-T2-C2 tridomain were purified on gravity flow columns, whereas full-length FtpA was purified on an Äkta Pure 25 FPLC instrument equipped with a His Trap FF crude column (GE Healthcare). Buffers used to equilibrate, wash, and elute FtpA were buffers A (50 mM NaH2PO4·H2O, 300 mM NaCl [pH 7.4]) and buffer B (500 mM imidazole in buffer A). Equilibration was at 5% buffer B; the column was washed with 12% buffer B. FtpA elution was accomplished at 100% buffer B. The flow rate was set to 1 ml min−1. The eluted protein was desalted on a PD-10 column (GE Healthcare), equilibrated with Tris buffer (80 mM Tris-HCl, 5 mM MgCl2·6H2O, 100 μM EDTA). Purification was verified with polyacrylamide gels (12% Laemmli gels). The protein concentrations were determined by Bradford method (13).
In vitro characterization of adenylation domains.
ATP-[32P]pyrophosphate exchange assays were carried out in triplicate. The reactions (100 μl) were incubated at 37°C (varied between 4 and 45°C to determine the temperature optimum) and consisted of Tris buffer (pH 7.5 [varied from pH 5.0 to 8.0 to determine the optimum pH]), 5 mM MgCl2, 125 nM EDTA, 5 mM ATP, 100 nM purified protein, 0.1 μM [32P]pyrophosphate, and 1 mM substrate (fumaric acid, l-phenylalanine, l-tyrosine). The reactions proceeded for 1 h before they were stopped and further processed as described previously (14). Exchange of pyrophosphate was quantified using a Perkin-Elmer TriCarb 2910TR scintillation counter. Substrates used to determine A domain specificities are listed in Table S2 in the supplemental material.
In vitro product formation.
FtpA was converted in vitro in its holo-form by incubation with 50 nM A. nidulans phosphopantetheinyl transferase NpgA (15) and 250 μM coenzyme A in Tris buffer (pH 7.5) for 30 min at 37°C. Subsequently, 5-ml reactions were set up, containing 80 mM Tris buffer, 5 mM MgCl2, 125 nM EDTA, 5 mM ATP, 100 nM FtpA, and 1 mM substrates (l-phenylalanine or l-tyrosine and fumaric acid). The incubation proceeded for 24 h. The reaction was then acidified with HCl to pH 1 and extracted three times with ethyl acetate. The organic phases were combined, concentrated under reduced pressure in a rotary evaporator, and subsequently lyophilized. The dried sample was then solved in methanol and filtered, and a portion was injected into the liquid chromatography/high-resolution mass spectrometry (LC-HRMS) instrument.
In vivo product formation.
For the detection of fumaryl-l-tyrosine and fumaryl-l-phenylalanine in A. fumigatus, the wild-type and overexpression strains ftpR-Oex and ftpA-Oex were used (see above). Aspergilli were grown in 1 liter of minimal medium (AMM) as described previously (16). The medium was inoculated with 6 × 105 conidia per ml and incubated for 48 h at 30°C before inducing with doxycycline (10 μg ml−1) for further 72 h. The mycelia were removed by filtration. The broth was then acidified and further processed, as described above.
Synthesis of standards.
The procedure resembled the synthesis of fumaryl-l-alanine described previously (17). A total of 6.5 mmol of l-phenylalanine or l-tyrosine was dissolved in 6.5 ml of sodium hydroxide solution (1 N). Fumaric acid chloride monoethyl ester (6.5 mmol) was added dropwise, while the reaction was stirred on ice, for 1 h. During the reaction, the pH was kept constant at pH 10.2 by addition of NaOH (2 N). Afterward, the solution was stirred at pH 12 at room temperature for an additional 5 h. After washing with diethyl ether, the aqueous phase was acidified with HCl to pH 1.5, and the product was extracted four times with ethyl acetate. The combined extracts were dried over Na2SO4, evaporated to dryness, and solved in methanol. Final purification was performed by preparative high-pressure liquid chromatography (HPLC) on an Eclipse XDB-C8 column (21.2 by 250 mm, 7-μm particle size) using gradient elution from 5% solvent B to 100% solvent B within 18 min. Solvent A was 0.1% trifluoroacetic acid in H2O; solvent B was 83% acetonitrile. The flow rate was 15 ml min−1. 1H nuclear magnetic resonance (NMR) spectra were recorded in CD3OD on a Bruker Avance III spectrometer at 600 MHz.
Chemical analysis.
Chemical analysis of the FtpA products fumaryl-l-tyrosine and fumaryl-l-phenylalanine was performed on a Thermo Accela liquid chromatograph equipped with a C18 column (Grom-Sil 100 ODS-0 AB, 250 by 4.6 mm, 3-μm particle size) and coupled to an Exactive OrbiTrap spectrometer operated in positive and negative mode using electrospray ionization. Chromatography solvents were 0.1% formic acid in water (solvent A) and acetonitrile (solvent B). After an initial hold at 5% solvent B for 1 min, a linear gradient up to 100% solvent B within 15 min was applied. The flow rate was 1.0 ml min−1. Synthetic fumaryl-l-tyrosine and fumaryl-l-phenylalanine were used as standards.
Protein modeling.
A homology model of the FtpA A1 domain was constructed by YASARA software version 12.11.25 (18) using PheA (PDB:1AMU) as a structural template. The phenylalanine ligand was replaced by fumarate, preserving the original orientation of the carboxyl group. Afterward, a molecular dynamics refinement simulation was run for ∼50 ps in a water-filled cell using the YASARA2 force field.
RESULTS AND DISCUSSION
Genetic and biochemical characterization of FtpA.
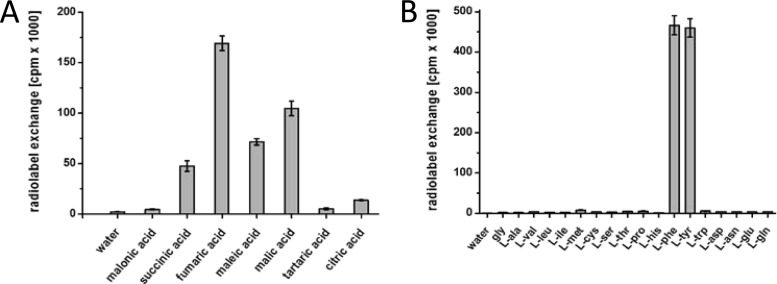
The A. fumigatus ftpA gene (nps7, Afu3g15270) is disrupted by a single 54-bp intron and encodes a 2,353-amino-acid (aa) bimodular NRPS enzyme (domain setup: A1-T1-C1-A2-T2-C2) with a calculated molecular mass of 258.5 kDa. It is located in a presumed cluster of genes (Afu3g15250-15290) that putatively code for a transporter, a hypothetical protein, a methyltransferase, and the GAL4-like Zn2Cys6-transcription factor FtpR (Fig. 1). None of the transcriptomic data, taken from various growth conditions, indicated any transcription of ftpA or of other ftp cluster genes (19–21). Therefore, we could not a priori exclude a pseudogene scenario. To verify enzymatic functionality and for a first insight into a possible product of FtpA, we relied on a biochemical in vitro approach, rather than reverse genetics. The A1 domain (556 aa, including a hexahistidine tag, 62 kDa) and the entire second module (i.e., the A2-T2-C2 tri-domain, 1,083 aa, a including hexahistidine tag, 119 kDa) were heterologously produced as N-terminal hexahistidine fusion proteins in E. coli KRX. These partial enzymes were separately investigated for their respective substrate preferences by assaying the substrate-dependent ATP-[32P]pyrophosphate radiolabel exchange. This method is used to detect the A domain-catalyzed formation of an aminoacyl adenylate intermediate. As the reverse reaction takes place, the aminoacyl adenylate is converted back to the free amino acid while 32P-labeled ATP is formed. To this end, a total of 38 substrates (see Table S2 in the supplemental material) was tested that included glycine and all proteinogenic l-amino acids, six α-keto acids, and four aryl acids, as well as eight di- and tricarboxylic acids. The assay was first carried out with pools of substrates, followed by a second round with individual substrates of positive pools.
The FtpA A1 domain accepted four-carbon dicarboxylic acids (Fig. 2). The preferred substrate was fumaric acid (169,200 cpm). Malic and maleic acid were turned over at appreciable levels (104,700 and 71,500 cpm, respectively). Low turnover was found for succinic and l-aspartic acid (47,500 and 40,200 cpm, respectively). Optimum ATP-pyrophosphate radiolabel exchange was observed at 37°C and pH 7.5 (see Table S3 in the supplemental material). The FtpA A2 domain was tested with the same set of 38 substrates and showed a virtually equal preference for l-phenylalanine and l-tyrosine (466,500 and 460,000 cpm). All other tested compounds resulted in negligible amounts of pyrophosphate exchange (<5%, Fig. 2). Additional evidence for these substrates stems from in vitro product formation. Full-length N-terminally hexahistidine-tagged FtpA (see Fig. S3 in the supplemental material) was produced in E. coli BL21(DE3) and converted into its holo-form by the Aspergillus nidulans phosphopantetheinyl transferase NpgA (15, 22) prior to the reaction. Subsequently, FtpA was incubated in two separate reactions, i.e., with fumaric acid and either l-tyrosine or l-phenylalanine at 1 mM (each) final concentrations. Control experiments using heat-inactivated enzyme were run in parallel.
FIG 2.
Substrate preference of FtpA A domains determined using the ATP-[32P]pyrophosphate exchange assay. (A) Results for the A1 domain; (B) results for the A2 domain. Error bars indicate the standard deviations.
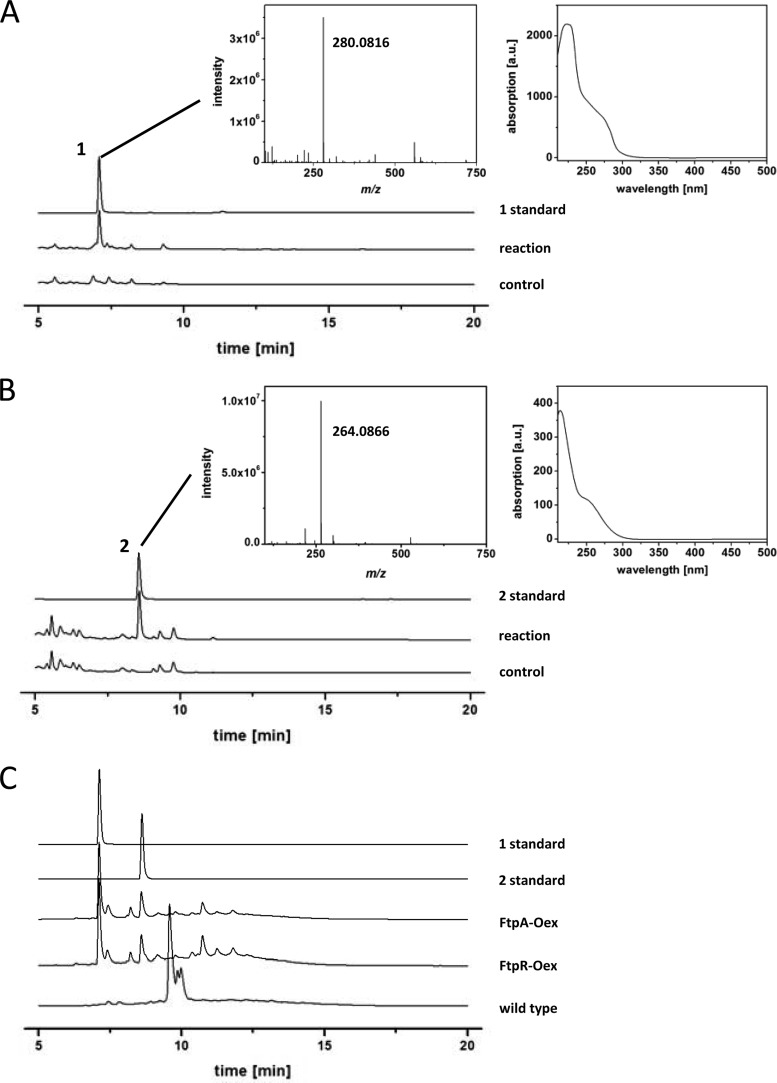
For the reaction with fumaric acid and l-tyrosine as the substrates, LC-HRMS demonstrated formation of a product that was identical to synthetic fumaryl-l-tyrosine (compound 1) (Fig. 1 and Fig. 3A; see also Fig. S4 in the supplemental material) with regard to the UV/Vis spectrum (local maximum at λ = 224 nm), the retention time (tR = 7.1 min), and the mass (m/z 280.0816 [M+H]+, calculated for C13H14NO6 as m/z 280.0821). When fumaric acid and l-phenylalanine were used as the substrates, a compound was detected chromatographically and by HR-MS (Fig. 3B) that turned out to be identical to synthetic fumaryl-l-phenylalanine (compound 2) (Fig. 1; see also Fig. S4 in the supplemental material) regarding its UV/Vis spectrum (local maximum at λ = 214 nm), its retention time (tR = 8.6 min), and its molecular mass (m/z 264.0866 [M+H]+, calculated for C13H14NO5 as m/z 264.0872). Neither of the above aromatic fumaric acid amides was detectable in the respective control reactions (Fig. 3A and B).
FIG 3.
HPLC-analysis of compound 1 and 2 production in vitro and by A. fumigatus. Chromatograms for λ = 254 nm are shown. (A) Chromatograms represent (upper to lower) synthetic compound 1 standard, FtpA-catalyzed in vitro production of compound 1, and negative control with heat-treated FtpA. (B) Chromatograms represent (upper to lower) synthetic compound 2 standard, FtpA-catalyzed in vitro production of compound 2, and negative control with heat-treated FtpA. The insets display HRMS- and UV/Vis spectra extracted at tR = 7.1 min (A) and 8.6 min (B). (C) Compound 1 and 2 standards and HPLC analysis of A. fumigatus overexpression strains of ftpA (ftpA-Oex) and the transcription regulator ftpR (ftpR-Oex). The lower chromatogram shows A. fumigatus wild type. The signal at tR = 9.6 min represents pseurotin A, as identified by its UV-Vis-spectrum and LC-MS. Two other pseurotins eluted at tR = 9.85 and 10.0 min.
Activating fumaric acid amide biosynthesis in A. fumigatus.
Given that FtpA catalyzed the formation of two structurally related enzymatic products in vitro, ambiguity remained whether solely one, or both, would reflect the biosynthetic situation in vivo. To clarify this situation, A. fumigatus was genetically engineered so as to activate the ftpA gene. Strain ftpA-Oex was created in which the ftpA gene was placed under the control of a tet-on cassette that allows for doxycycline-induced transcriptional activation (12). The gene ftpR (= Afu3g15290), located downstream of ftpA (Fig. 1), encodes a Zn2Cys6 GAL4-type transcriptional regulator. We hypothesized that it may govern the expression of ftpA. Hence, a second manipulated strain, ftpR-Oex, was created in which the promoter region of ftpR was engineered to replace the native promoter by the tet-on cassette. The engineered strains A. fumigatus ftpR-Oex and ftpA-Oex, and wild-type for control, were grown in liquid minimal medium under inducing conditions, that is, in the presence of 10 μg of doxycycline ml−1. Acidified ethyl acetate extracts of the culture supernatants were subjected to LC-HRMS. The extracts of the ftpA- and ftpR-overexpressing strains showed virtually identical metabolic profiles (Fig. 3C): both strains produced compound 1 as a major metabolite, while compound 2 was the second most abundant compound (in a 2.6:1 ratio, according to the areas under the curve). Synthetic standards served as controls and showed identical retention times, UV/Vis profiles, and masses. In the A. fumigatus wild-type control extract, LC-HRMS analyses identified the signals appearing between 9.5 and 9.9 min (Fig. 3C) as pseurotins, which are known products of A. fumigatus (23). Neither compound 1 nor compound 2 was detectable, which corroborates the transcriptional inactivity of ftpA that was evident by transcriptomic data. Our study therefore suggests that ftpA is the regulatory target of FtpR.
FtpA substrate specificity signatures.
The term “nonribosomal code” (24, 25) refers to a set of 10 mostly nonconsecutive amino acid residues within NRPS A domains and represents a specificity signature for substrate preference. Hence, it allows for the prediction of structural features of the enzymatic product. Extraction of this code of the FtpA A1 domain by the software NRPSpredictor (26) yielded S-A-R-D-V-G-S-Q-L-K as a signature indicative for fumaric acid that is deviating from the fumaric acid code previously identified for SidE (S-A-R-G-T-V-S-Q-L-K) (5). Phylogenetic analyses placed the A1 domains of SidE and FtpA in dissimilar evolutionary clades (5, 27, 28), and this phylogenetic divergence may account for the deviating NRPS codes in these A domains. The identification of FtpA as a fumaric acid-adenylating NPRS is intriguing since this dicarboxylic acid is a building block that occurs in various natural products. However, current algorithms to predict NRPS substrate preferences, such as NRPSpredictor and NRPSsp (29), do not include this substrate and, consequently, fail to decipher this particular nonribosomal code. Together with our previous work on SidE, the present study provides the basis to further refine the predictive capacity of such software.
In silico modeling of the FtpA A1 domain.
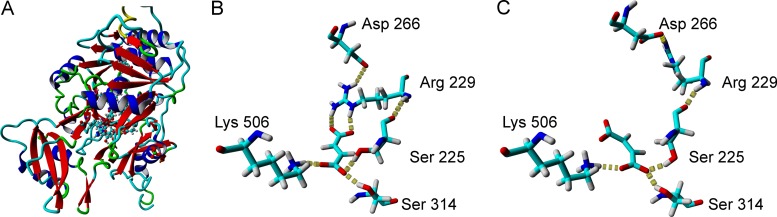
For a more profound insight into the structural requirements of fumaric acid binding by FtpA, the A1 domain was modeled in silico and constructed a homology model using the PheA crystal structure (30) as a template. The fumarate substrate was placed in a way that the conserved lysine K506 (PheA-K517) served as an anchor for the carboxy group (Fig. 4). Typically, amino acid-binding A domains share a conserved aspartate (PheA-D235), which is known to interact with the amino group of the substrate (24). In the case of the fumarate-accepting A1 domain of FtpA, aspartate is replaced by serine (S225). Intriguingly, a molecular dynamics simulation suggests that both S225 and S314 hydrogen bond with the C-1 carboxy group of fumarate. Furthermore, the model suggests that the C-4 carboxy group of fumarate is held in place by the guanidinium group of R229 via two hydrogen bonds. Thus, R229 might represent a key residue to support the selectivity for dicarboxylates. When maleic acid was chosen as a substrate, it became apparent that it does not come into contact with the R229 residue. Hence, this in silico result is consistent with the low turnover of maleic acid in our biochemical assays. The nonribosomal code of the l-tyrosine- and l-phenylalanine-accepting FtpA A2 domain was recognized as D-G-Y-N-A-G-S-I-C-K, by both NRPSpredictor and NRPSsp. This code is unprecedented but distantly reminiscent of that of the PsoA A domain (D-A-Y-T-M-A-A-I-C-K) that adenylates l-phenylalanine for pseurotin biosynthesis in A. fumigatus (23).
FIG 4.
Structural model of the FtpA A1 domain. (A) Homology model of NPS7 using the crystal structure of PheA as the template molecule. (B) Close-up view of fumarate in the binding pocket. While PheA-D222 binds the amino group of the phenylalanine substrate in case of PheA, S225 forms an additional hydrogen bond with the C-1 carboxylate of fumarate. R229 positions the C-4 carboxylate and most likely defines a key residue for dicarboxylate specificity. (C) Due to the (E)-configuration, maleate does not hydrogen bond with R229. Hydrogen bonds are illustrated by dotted lines. Labels indicate residue type and number. For clarity, only key residues are shown.
Concluding remarks.
Compounds 1 and 2 were not antimicrobially and antifungally active at concentrations up to 100 μg ml−1 and did not show antiproliferative or cytotoxic effects (data not shown). Modest in vitro bioactivity (50% inhibitory concentration [IC50] = 2.3 mM) as angiotensin-converting enzyme inhibitor was previously reported for compound 2 (31). The relevance of FtpA-produced aromatic fumaric acid amides for the producer A. fumigatus remains elusive and warrants further investigation. We cannot discount a highly specific function for the identified aromatic fumaric acid amides under conditions that were not covered during transcriptomic sampling. Alternatively, the FtpA products may have been functionally replaced by the SidE product, the aliphatic fumaric acid amide fumaryl-l-alanine, which may explain the silent ftpA gene.
In summary, in vitro work combined with gene activation identified FtpA as fumaryl-l-phenylalanine and fumaryl-l-tyrosine synthetase and expanded the known natural product repertoire of A. fumigatus by aromatic fumaric acid amides. We verified an alternative A domain substrate signature for fumaric acid that may help predict fungal metabolites more accurately. Therefore, our results are relevant beyond this human pathogen and generally contribute to genomics-based natural product chemistry.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the following members of the Leibniz Institute for Natural Product Research and Infection Biology/Hans Knöll Institute, Jena, Germany: Carmen Schult, Heike Heinecke, Andrea Perner, and Christiane Weigel for technical assistance, recording NMR spectra, mass spectra, and for antimicrobial tests, respectively.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03268-14.
REFERENCES
- 1.Scharf DH, Heinekamp T, Brakhage AA. 2014. Human and plant fungal pathogens: the role of secondary metabolites. PLoS Pathog 10:e1003859. doi: 10.1371/journal.ppat.1003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brakhage AA, Langfelder K. 2002. Menacing mold: the molecular biology of Aspergillus fumigatus. Annu Rev Microbiol 56:433–455. doi: 10.1146/annurev.micro.56.012302.160625. [DOI] [PubMed] [Google Scholar]
- 3.Balibar CJ, Walsh CT. 2006. GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry 45:15029–15038. doi: 10.1021/bi061845b. [DOI] [PubMed] [Google Scholar]
- 4.O'Hanlon KA, Gallagher L, Schrettl M, Jochl C, Kavanagh K, Larsen TO, Doyle S. 2012. Nonribosomal peptide synthetase genes pesL and pes1 are essential for fumigaclavine C production in Aspergillus fumigatus. Appl Environ Microbiol 78:3166–3176. doi: 10.1128/AEM.07249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinchen W, Lackner G, Yasmin S, Schrettl M, Dahse HM, Haas H, Hoffmeister D. 2013. Bimodular peptide synthetase SidE produces fumarylalanine in the human pathogen Aspergillus fumigatus. Appl Environ Microbiol 79:6670–6676. doi: 10.1128/AEM.02642-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bringmann G, Lang G, Gulder TAM, Tsuruta H, Mühlbacher J, Maksimenka K, Steffens S, Schaumann K, Stöhr R, Wiese J, Imhoff JF, Perovic-Ottstadt S, Boreiko O, Müller WEG. 2005. The first sorbicillinoid alkaloids, the antileukemic sorbicillactones A and B, from a sponge-derived Penicillium chrysogenum strain. Tetrahedron 61:7252–7265. doi: 10.1016/j.tet.2005.05.026. [DOI] [Google Scholar]
- 7.Davis RA. 2005. Isolation and structure elucidation of the new fungal metabolite (−)-xylariamide A. J Nat Prod 68:769–772. doi: 10.1021/np050025h. [DOI] [PubMed] [Google Scholar]
- 8.Kamiyama T, Umino T, Nakayama N, Itezono Y, Satoh T, Yamashita Y, Yamaguchi A, Yokose K. 1992. Ro 09-1679, a novel thrombin inhibitor. J Antibiot 45:424–427. doi: 10.7164/antibiotics.45.424. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Russell DW. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 10.Mabey JE, Anderson MJ, Giles PF, Miller CJ, Attwood TK, Paton NW, Bornberg-Bauer E, Robson GD, Oliver SG, Denning DW. 2004. CADRE: the Central Aspergillus Data REpository. Nucleic Acids Res 32:D401–D405. doi: 10.1093/nar/gkh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnaud MB, Chibucos MC, Costanzo MC, Crabtree J, Inglis DO, Lotia A, Orvis J, Shah P, Skrzypek MS, Binkley G, Miyasato SR, Wortman JR, Sherlock G. 2010. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res 38:D420–D427. doi: 10.1093/nar/gkp751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmschrott C, Sasse A, Samantaray S, Krappmann S, Wagener J. 2013. Upgrading fungal gene expression on demand: improved systems for doxycycline-dependent silencing in Aspergillus fumigatus. Appl Environ Microbiol 79:1751–1754. doi: 10.1128/AEM.03626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 14.Schneider P, Weber M, Rosenberger K, Hoffmeister D. 2007. A one-pot chemoenzymatic synthesis for the universal precursor of antidiabetes and antiviral bis-indolylquinones. Chem Biol 14:635–644. doi: 10.1016/j.chembiol.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Keszenman-Pereyra D, Lawrence S, Twfieg ME, Price J, Turner G. 2003. The npgA/cfwA gene encodes a putative 4′-phosphopantetheinyl transferase which is essential for penicillin biosynthesis in Aspergillus nidulans. Curr Genet 43:186–190. [DOI] [PubMed] [Google Scholar]
- 16.Weidner G, d'Enfert C, Koch A, Mol PC, Brakhage AA. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr Genet 33:378–385. doi: 10.1007/s002940050350. [DOI] [PubMed] [Google Scholar]
- 17.Rossi D, Calcagni A, Romeo A. 1979. Approach to the use of benzylpenicillinacylase for configurational correlations of amino-compounds. 2. Hydrolysis of N-(para-aminophenylacetyl) derivatives of some chiral primary amines. J Org Chem 44:2222–2225. [Google Scholar]
- 18.Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. 2009. Improving physical realism, stereochemistry, and side chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77(Suppl 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillmann F, Linde J, Beckmann N, Cyrulies M, Strassburger M, Heinekamp T, Haas H, Guthke R, Kniemeyer O, Brakhage AA. 2014. The novel globin protein fungoglobin is involved in low oxygen adaptation of Aspergillus fumigatus. Mol Microbiol 93:539–553. doi: 10.1111/mmi.12679. [DOI] [PubMed] [Google Scholar]
- 20.Gibbons JG, Beauvais A, Beau R, McGary KL, Latgé J, Rokas A. 2012. Global transcriptome changes underlying colony growth in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot Cell 11:68–78. doi: 10.1128/EC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonagh A, Fedorova ND, Crabtree J, Yu Y, Kim S, Chen D, Loss O, Cairns T, Goldman G, Armstrong-James D, Haynes K, Haas H, Schrettl M, May G, Nierman WC, Bignell E. 2008. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog 4:e1000154. doi: 10.1371/journal.ppat.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider P, Bouhired S, Hoffmeister D. 2008. Characterization of the atromentin biosynthesis genes and enzymes in the homobasidiomycete Tapinella panuoides. Fungal Genet Biol 45:1487–1496. doi: 10.1016/j.fgb.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Maiya S, Grundmann A, Li X, Li SM, Turner G. 2007. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. Chembiochem 8:1736–1743. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- 24.Stachelhaus T, Mootz HD, Marahiel MA. 1999. The specificity conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 25.Kalb D, Lackner G, Hoffmeister D. 2013. Fungal peptide synthetases: an update on functions and specificity signatures. Fungal Biol Rev 27:43–50. doi: 10.1016/j.fbr.2013.05.002. [DOI] [Google Scholar]
- 26.Röttig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O. 2011. NRPSpredictor2: a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res 39:W362–W367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bushley KE, Ripoll DR, Turgeon BG. 2008. Module evolution and substrate specificity of fungal nonribosomal peptide synthetases involved in siderophore biosynthesis. BMC Evol Biol 8:328. doi: 10.1186/1471-2148-8-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushley KE, Turgeon BG. 2010. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol Biol 10:26. doi: 10.1186/1471-2148-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prieto C, Garcia-Estrada C, Lorenzana D, Martin JF. 2012. NRPSsp: non-ribosomal peptide synthase substrate predictor. Bioinformatics 28:426–427. doi: 10.1093/bioinformatics/btr659. [DOI] [PubMed] [Google Scholar]
- 30.Conti E, Stachelhaus T, Marahiel MA, Brick P. 1997. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S EMBO J 16:4174–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park Choo H-Y, Peak K-H, Park J, Kim DH, Chung HS. 2000. Design and synthesis of α,β-unsaturated carbonyl compounds as potential ACE inhibitors. Eur J Med Chem 35:643–648. doi: 10.1016/S0223-5234(00)00158-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.