Radial spokes are conserved macromolecular complexes that are essential for ciliary motility. Little is known about the assembly and functions of the three individual radial spokes, RS1, RS2, and RS3. In Tetrahymena, a conserved ciliary protein, FAP206, docks RS2 and dynein c to the doublet microtubule.
Abstract
Radial spokes are conserved macromolecular complexes that are essential for ciliary motility. A triplet of three radial spokes, RS1, RS2, and RS3, repeats every 96 nm along the doublet microtubules. Each spoke has a distinct base that docks to the doublet and is linked to different inner dynein arms. Little is known about the assembly and functions of individual radial spokes. A knockout of the conserved ciliary protein FAP206 in the ciliate Tetrahymena resulted in slow cell motility. Cryo–electron tomography showed that in the absence of FAP206, the 96-nm repeats lacked RS2 and dynein c. Occasionally, RS2 assembled but lacked both the front prong of its microtubule base and dynein c, whose tail is attached to the front prong. Overexpressed GFP-FAP206 decorated nonciliary microtubules in vivo. Thus FAP206 is likely part of the front prong and docks RS2 and dynein c to the microtubule.
INTRODUCTION
Radial spokes are prominent substructures of the 96-nm axonemal “motility unit” that link the outer doublet microtubules with the central pair apparatus and play a key role in regulating ciliary motility. Mutant cilia lacking radial spokes are paralyzed (Witman et al., 1978; Huang et al., 1981), and mutations in radial spoke proteins cause primary ciliary dyskinesia (Sturgess et al., 1979; Castleman et al., 2009; Zietkiewicz et al., 2012). Radial spokes are believed to act as mechanochemical bridges that transmit physical and biochemical signals from the central apparatus to dynein arms on the doublet microtubules (Warner and Satir, 1974; Huang et al., 1982; Luck et al., 1982; Diener et al., 1993; Omoto et al., 1999; Lindemann, 2003; Smith and Yang, 2004; Heuser et al., 2009). Radial spokes may regulate the phosphorylation state of dynein subunits (Smith and Sale, 1992; Howard et al., 1994; Gaillard et al., 2001, 2006; Wirschell et al., 2011).
Cilia of most studied species contain three full-length radial spokes, RS1, RS2, and RS3 (Dentler and Cunningham, 1977; Goodenough and Heuser, 1985); a well-studied exception is the unicellular algae Chlamydomonas reinhardtii, with a pair of full-length radial spokes (RS1 and RS2) and a short third spoke (RS3S; Pigino et al., 2011; Barber et al., 2012; Lin et al., 2012). Radial spokes are composed of multiple proteins that form the head and the stem, whose basal end is attached to two specific microtubule protofilaments (A2 and A3) of the A-tubule of the outer doublet (Nicastro et al., 2006). RS1 and RS2 are similar in shape and share many proteins, based on the observation that multiple single mutations destabilize RS1 and RS2 to similar extents (Huang et al., 1981; Diener et al., 1990; Curry and Rosenbaum, 1993; Yang et al., 2006). Until recently, based on classical electron microscopy, all radial spokes were believed to be identical, but cryo–electron tomography studies revealed structural differences among the three radial spokes. Each radial spoke has a microtubule base of unique shape, and the stem and head of RS3 (in the organisms that have a full-size RS3) are different in shape from the corresponding parts of RS1 and RS2 (Pigino et al., 2011, 2012; Barber et al., 2012; Heuser et al., 2012b; Lin et al., 2012). In addition to unique microtubule adapters, each radial spoke is linked to a different set of neighboring structures of the 96-nm repeat, contributing to the intricate connectivity among multiple force generators (inner and outer dynein arms) and signaling hubs (N-DRC, MIA, I1 dynein IC-LC, CSC, and OID linker; Nicastro et al., 2006; Bui et al., 2009; Heuser et al., 2009, 2012a, b; Oda et al., 2013; Yamamoto et al., 2013). It is therefore very likely that each radial spoke has a unique functional contribution. Identification of proteins that mediate the attachment of specific radial spokes to correct sites on the doublet microtubule and link the bases of radial spokes to neighboring structures is of key importance to understanding how the 96-nm axonemal repeat assembles and functions. Of importance, the calmodulin and spoke-associated complex (CSC; Dymek and Smith, 2007) localizes to the region that spans the bases of RS2 and RS3, and knockdowns of CSC destabilize RS2 and RS3 but do not affect RS1 (Dymek et al., 2011; Heuser et al., 2012b). To account for the unique organization of each radial spoke base, it is likely that in addition to the CSC, other, yet-unidentified axonemal proteins participate in docking and basal connectivity of individual radial spokes.
Here we identify FAP206, a conserved ciliary protein, as an RS2-specific microtubule-docking factor. FAP206 was discovered in the ciliary proteome of C. reinhardtii (Pazour et al., 2005) and has been linked to the 96-nm outer doublet repeat by multiple studies; FAP206 is physically associated with the radial spoke protein RSP3, a major component of the spoke stem of RS1 and RS2 (Gupta et al., 2012), and phosphorylation of FAP206 is affected by mutations in subunits of the N-DRC (Heuser et al., 2009; Lin et al., 2011). However, the localization, function, and significance of FAP206 remain unknown. We show that FAP206 acts as an adapter required for stable attachment of only one of the three radial spokes, RS2, and an inner dynein arm, dynein c, which is closely associated with RS2.
RESULTS
FAP206 is an axoneme protein required for normal ciliary motility in Tetrahymena
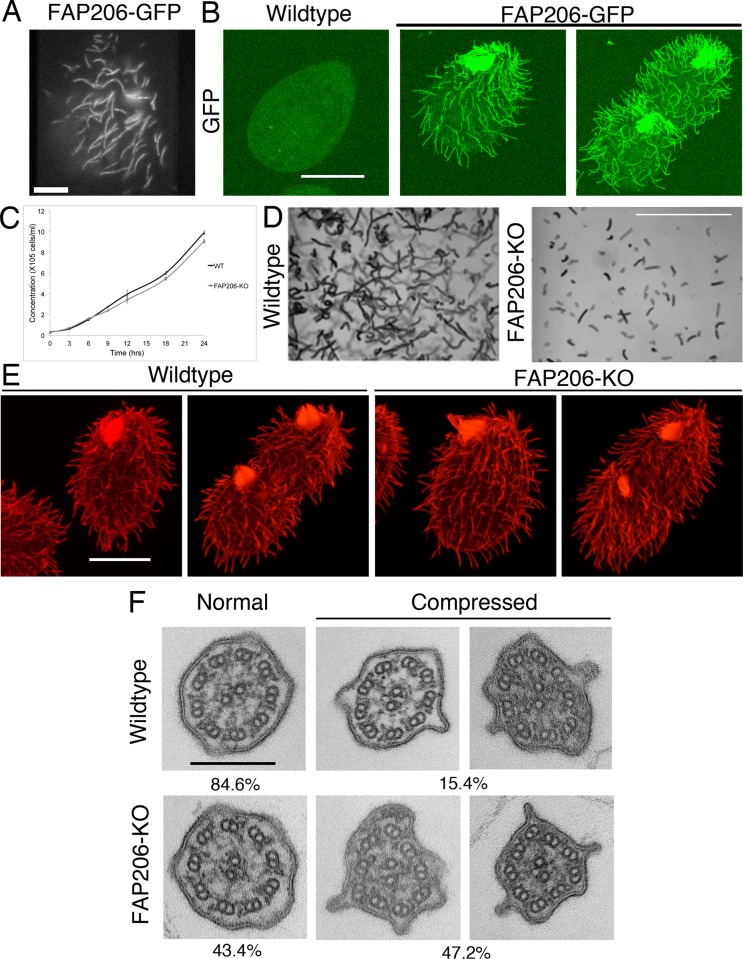
To localize FAP206 under native conditions of expression, we tagged the FAP206 gene with a sequence encoding a C-terminal green fluorescent protein (GFP). The gross phenotype of the FAP206-GFP strain appeared normal. In both live (Figure 1A and Supplemental Movie S1) and detergent-treated (Triton X-100) cells (Figure 1B), FAP206-GFP was detected exclusively in cilia, where it was distributed uniformly. The detergent resistance indicates that FAP206 is stably associated with the axoneme, in agreement with published biochemical studies (Pazour et al., 2005; Lin et al., 2011; Gupta et al., 2012).
FIGURE 1:
FAP206 localizes to the ciliary axoneme, and knockout of FAP206 results in cilia-related defects. (A) TIRF image of a live cell expressing FAP206-GFP under the native promoter. (B) A wild-type (negative control) cell (left) and a cell expressing FAP206-GFP under the native promoter (right) extracted with Triton X-100, fixed with paraformaldehyde, and imaged for GFP using a confocal microscope. (C) Culture growth rates for a wild-type CU428 and an FAP206-KO strain. Each data point represents an average for three independent experiments. (D) Paths of swimming wild-type and FAP206-KO cells recorded for 1 s. The average swim velocities were 170 μm/s for the wild type and 50 μm/s for FAP206-KO. (E) Immunofluorescence images of tubulin for wild-type and FAP206-KO cells. For each genotype, an interphase (left) and a dividing cell (right) are shown. (F) Classical TEM images of cilia cross-sections that are either circular (left) or compressed (right). The percentages represent fractions of either circular or compressed axonemes (n = 52 for wild type, n = 48 for FAP206-KO). Scale bars, 20 μm (A, B, and E), 1 mm (D), 0.2 μm (F).
To determine the importance of FAP206, we used homologous DNA recombination to obtain a Tetrahymena strain lacking the FAP206 gene. The resulting FAP206-knockout (FAP206-KO) cells grew at a nearly normal rate (Figure 1C) but swam with a rate of ∼30% of the wild type (Figure 1D). The FAP206-KO cells were covered with a normal number of cilia (Figure 1E) that were slightly longer than in wild type (5.27 ± 0.06 μm, n = 337 for wild type, and 5.44 ± 0.06 μm for FAP206-KO cilia, n = 307; p = 0.044). Based on classical transmission electron microscopy (TEM) of chemically fixed cells, the cross-sections of the FAP206-KO cilia showed a normal 9 + 2 organization of microtubules, except that the mutant axonemes were more frequently compressed, and some had a nearly triangular shape (Figure 1F). High-speed video recording showed that FAP206-KO cilia had an abnormal waveform characterized by decreased bend amplitude and decreased metachronal coordination (compare Supplemental Movies S2 and S3). Typically, an abnormal waveform is observed in mutants affected in the inner dynein arms (IDAs) or components of the radial spokes or the central apparatus, whereas a reduction in the beat frequency is attributed to the function of outer dynein arms (ODAs; Brokaw and Kamiya, 1987; Kamiya, 2002; Yokoyama et al., 2004; Lechtreck et al., 2008; Yang et al., 2008). The uncoordinated motility of FAP206-KO cilia made the measurement of the beat frequency difficult. Thus, to gain an insight into the functionality of the ODAs, we measured the velocity of doublet microtubule sliding in isolated axonemes reactivated with ATP. In this assay, the rate of microtubule sliding is determined primarily by the activity of ODAs that override the force contribution by IDAs (reviewed in Kamiya, 2002). The rate of microtubule sliding in FAP206-KO axonemes was similar to that in wild-type axonemes (6.97 ± 0.83 μm/s, n = 79, for FAP206-KO and 6.99 ± 0.89 μm/s, n = 75, for wild type), indicating that the net activity of ODAs is not affected by the absence of FAP206. Overall the slow-swimming phenotype of the FAP206-KO cells is consistent with a defect in the IDAs, the radial spokes, or the central apparatus.
FAP206 is needed for assembly of radial spoke RS2
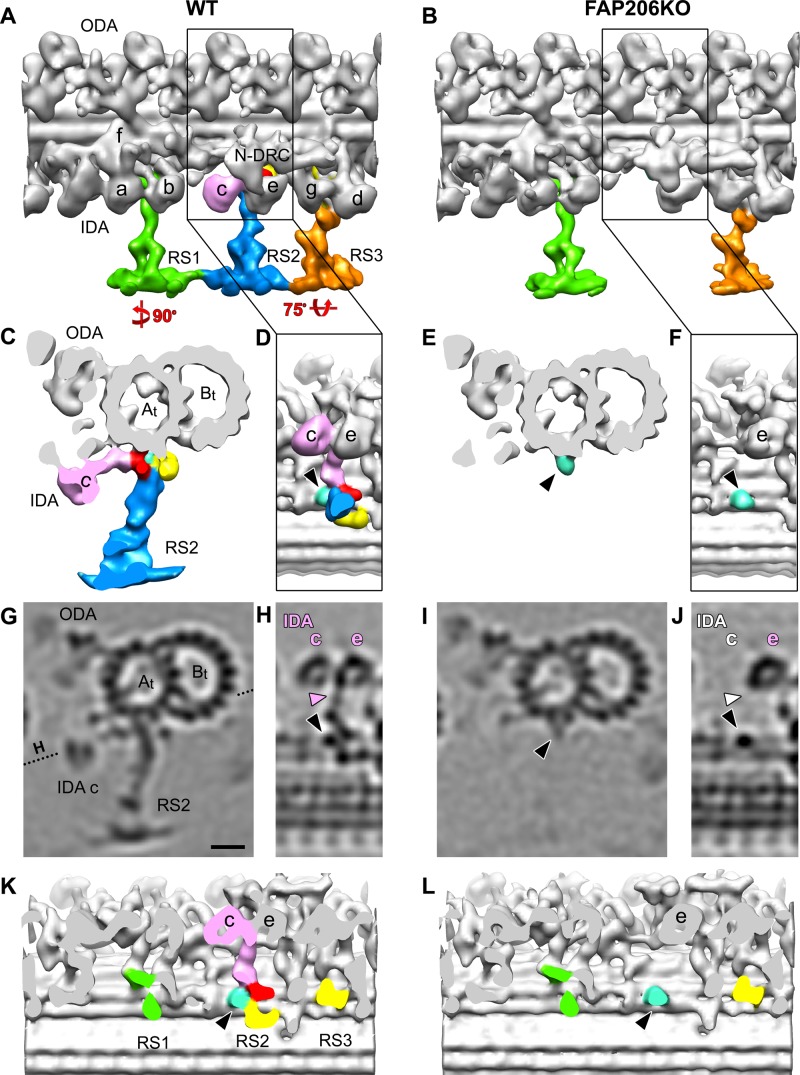
Previous biochemical and genetic studies linked FAP206 to the 96-nm outer doublet repeat (Lin et al., 2011; Gupta et al., 2012). We used cryo–electron tomography and subtomogram averaging to compare the high-resolution three-dimensional (3D) structure of the 96-nm repeat between the wild-type and FAP206-KO axonemes. We reconstructed cryotomograms of 10 wild-type and 7 FAP206-KO axonemes and averaged 1300 and 800 axonemal repeats, respectively. The most striking difference in the subtomogram averages of all repeats is the absence of the middle radial spoke RS2 and its closely associated IDA, dynein c, in the FAP206-KO axonemes (Figure 2 and Supplemental Movies S4 and S5). In the wild-type 96-nm repeat, the RS2 base is attached to the A-tubule by three contacts: the front, back, and side prongs (Figure 2, C, D, H, and K). The front and back prongs (red and yellow in Figure 2, respectively) resemble the corresponding structures in Chlamydomonas (Barber et al., 2012; Lin et al., 2012), whereas the side prong (light blue in Figure 2) is, so far, unique to RS2 of Tetrahymena. In the axonemal repeat of FAP206-KO, the front and back prong, of RS2 are absent, whereas the side prong remains (Figure 2, E, F, J, and L, and Supplemental Movie S6). We conclude that the base of the Tetrahymena RS2 attaches to the microtubule with three prongs, and the assembly of two of these prongs (front and back) depends on FAP206.
FIGURE 2:
Deletion of FAP206 leads to loss of RS2 and associated dynein c in the 96-nm repeat. Isosurface renderings (A–F, K, L) and tomographic slices (G–J) show the averaged 96-nm axonemal repeats of wild type (A, C, D, G, H, K) and FAP206-KO (B, E, F, I, J, L) in longitudinal (A, B, D, F, H, J–L), cross-sectional (C, E, G, I), and bottom views looking from the central pair toward the doublet microtubule (D, F, H, J–L); the dotted line in G indicates the orientation of the tomographic slice shown in H and J. There are three radial spokes: RS1 (green), RS2 (blue), and RS3 (orange) in wild type (A), whereas RS2 is missing in FAP206-KO (B, E, I). The RS2 base is composed of three regions: front (red), back (yellow), and side prongs (light blue and/or black arrowheads). These three regions are connected with each other and form the attachment of RS2 to the doublet A-tubule (At). The back prong of RS2 and the RS3 base (yellow) were previously identified as parts of the CSC (Heuser et al., 2012b). IDA c (pink), which anchors with its tail (pink arrowhead in H) to the A-tubule through the front prong of the RS2 base in WT, is also missing in FAP206-KO (white labels in J, where the IDA c with tail should be). (K, L) Densities representing radial spoke heads and stems were removed to visualize the microtubule-attachment sites of the spoke bases. Note that in the FAP206-KO mutant, the main difference from wild type is the complete absence of the RS2 front prong (red), RS2 back prong (yellow), and dynein c; in contrast, RS1, RS3, and the RS2 side prong (light blue, black arrowhead) appear unaffected in FAP206-KO. All structures shown are subtomogram averages without prior classification analysis. Scale bar, 10 nm.
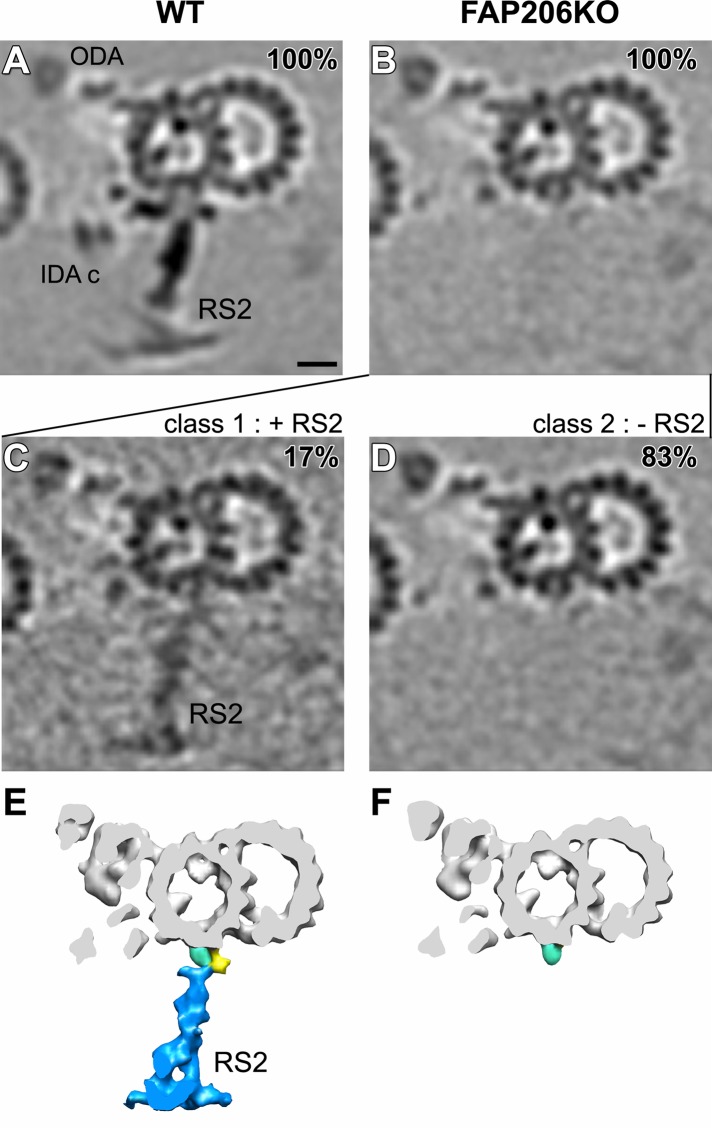
A detailed comparison of the subtomogram averages of all repeats of the FAP206-KO and wild-type axonemes revealed that in FAP206-KO, the electron density of RS1 is unaffected, RS2 is greatly reduced, but a faint signal is still detectable, and RS3 is mildly reduced (Figures 3, A and B, and 4, A and B). A reduced electron density in specific areas of a subtomogram average is an indication that the individual repeats are not identical. Heterogeneity among the individual 96-nm repeats could be either caused by flexibility (i.e., the position of a structure varies between the individual repeats) or because a structure is absent in a subset of repeats. We used automatic image classification (Heumann et al., 2011) to sort the axonemal repeats into homogeneous subgroups before generating class averages. For wild type, we could not identify classes that differed in the structures of radial spokes (Figures 3A and 4A). When we classified for differences in the RS2 structure among the FAP206-KO subtomograms, the majority of axonemal repeats (83%) lacked RS2 entirely (except for the side prong; Figure 3, D and F), whereas the remaining 17% (FAP206-KO [+RS2] class) had a nearly completely assembled RS2 but lacked parts of the microtubule base (Figure 3, C and E). A comparison of the wild-type RS2 structure with the FAP206-KO (+RS2) class revealed a fairly normal stem and head of RS2, whereas the entire front prong and the closely associated dynein c were missing, and the back prong was reduced (compare Figures 2G and 3C, and 2C and 3E; also see Supplemental Movie S7). We estimated the molecular weight of the front prong density that is missing in the FAP206-KO (+RS2) class average as ∼80 kDa (red region in Figure 2, C, D, and K), which is consistent with the predicted (73 kDa) and observed molecular weight (80 kDa; see later discussion of Figure 7D) of FAP206. The simplest explanation of these observations is that FAP206 forms the front prong of RS2.
FIGURE 3:
Classification analysis of RS2 reveals that in the absence of FAP206, RS2 can occasionally assemble but lacks the front prong. Cross-sectional tomographic slices (A–D) and isosurface renderings (E, F) of averaged 96-nm axonemal repeats show the presence and absence of RS2 in wild type (WT; A) and FAP206-KO (B–F), respectively. Subtomogram averages of all axonemal repeats (100%) indicate that the density of RS2 is dramatically reduced in FAP206-KO (B) as compared with WT (A). Classification of RS2 resulted in two distinct class averages for FAP206-KO: a large set (83%) of axonemal repeats from FAP206-KO lack RS2 (–RS2), and only the side prong (light blue) remains (D and F). However, in a small subset (17%) of FAP206-KO repeats, RS2 is present (+RS2) yet lacks the front prong density of the RS2 base (C and E); the back prong density also appears slightly reduced. All axonemal repeats from WT showed normal RS2 (A). Scale bar, 10 nm.
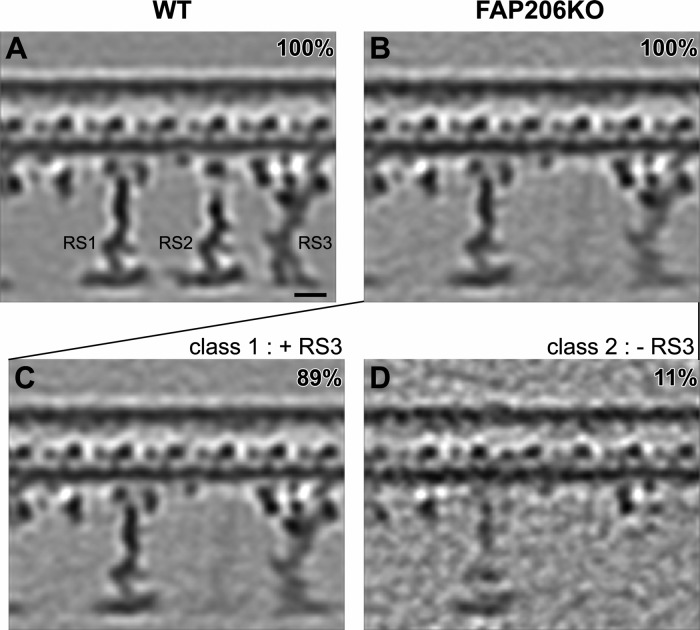
FIGURE 4:
Classification analysis of RS3 reveals that in the absence of FAP206, RS3 is destabilized. Longitudinal tomographic slices (A–D) of averaged 96-nm axonemal repeats show the presence (A–C) and absence (D) of RS3 in WT (A) and FAP206-KO (B–D). The density of RS3 is weaker in the average of all axonemal repeats from FAP206-KO (B) as compared with WT (A). Classification of RS3 resulted in two distinct class averages for FAP206-KO: in the majority (89%) of axonemal repeats from FAP206-KO, RS3 was assembled, whereas a small set (11%) of repeats lacked RS3 (D). All axonemal repeats from WT showed a normal RS3 (A). Scale bar, 10 nm.
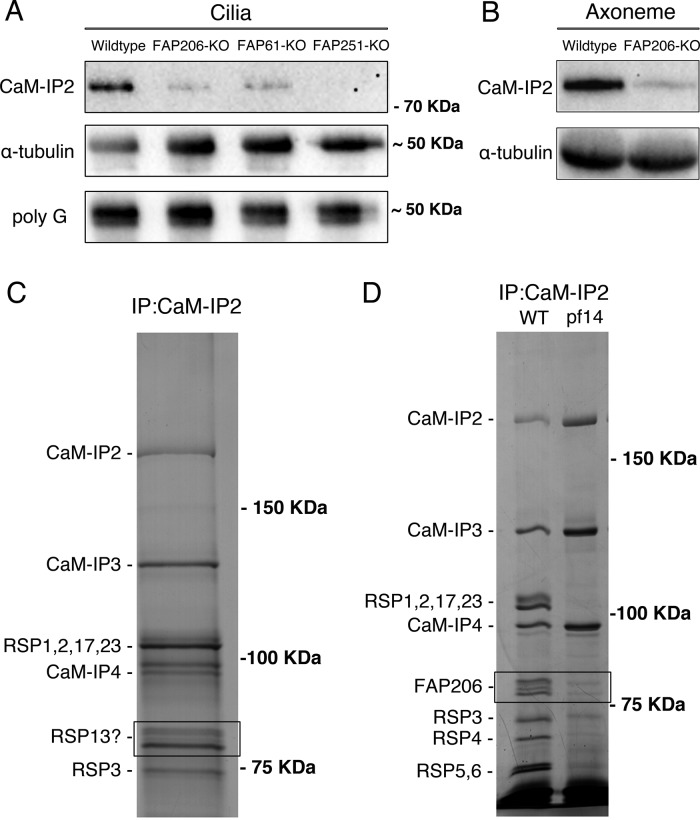
FIGURE 7:
FAP206 interacts with CSC. (A) A Western blot of purified cilia isolated from Tetrahymena strains that are wild type, lack genes encoding the Tetrahymena homologues of CSC proteins (FAP61/CaM-IP3 or FAP251/CaM-IP4), or lack FAP206, probed with antibodies specific to FAP91/CaM-IP2 of C. reinhardtii (top; Dymek et al., 2011), 12G10 monoclonal α-tubulin (middle), or polyglycylated tubulin (bottom). Note that the levels of the anti-FAP91/CaM-IP2 band are strongly reduced in the strains lacking the CSC protein homologues, indicating that the antibodies are specific to the Tetrahymena FAP91 homologue. Furthermore, the levels of the FAP91 homologue are strongly reduced in the strain lacking FAP206, indicating that axonemal assembly of FAP91 and likely other CSC components depends on the presence of FAP206. Images of the entire blots are shown in Supplemental Figure S1. (B) Western blot of axonemes of wild type and FAP206-KO probed with anti-FAP91 and 12G10 anti–α-tubulin antibodies. (C) Silver stained SDS–PAGE gels showing an immunoprecipitate obtained from the radial spoke-enriched supernatants of C. reinhardtii axonemes. The positions of known CSC components and major radial spoke proteins are marked. The two bands (boxed area) that migrated with an apparent molecular weight of 80 kDa were found to contain the Chlamydomonas FAP206 homologue (CHLREDRAFT_171124). (D) Immunoprecipitates obtained with the anti-FAP91/CaM-IP2 antibodies from the radial spoke–enriched supernatant of Chlamydomonas axonemes that were either wild type or spokeless pf14 mutant. Note the absence of the FAP206 bands, indicating that the CSC-FAP206 interaction requires RSP3 as an intermediate.
Clearly, RS1 and RS3 can assemble completely without FAP206, making it unlikely that FAP206 is a part of the RS1 and RS3 structure. However, when classified for differences in the RS3 structure (Figure 4, B–D), in 11% of the FA206-KO repeats, RS3 was missing (Figure 4D); in this subclass, RS2 was also not visible. This indicates a correlation between RS2 and RS3 defects; that is, it appears that RS3 is more likely to be absent from individual 96-nm repeats that also lack RS2, suggesting that in the absence of RS2, RS3 is less stable.
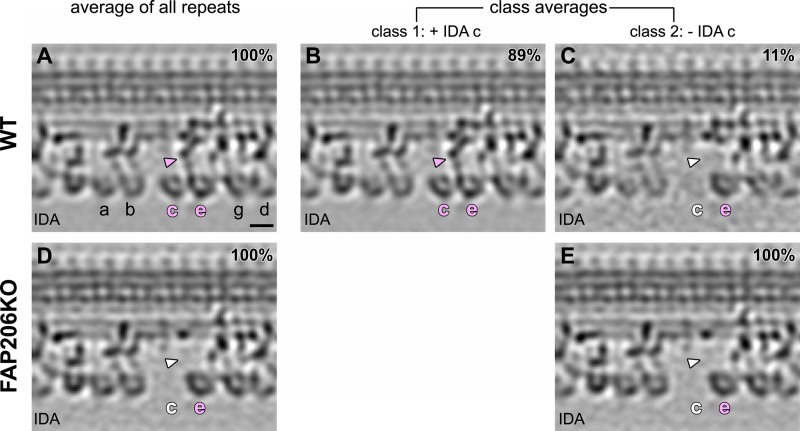
FAP206 is specifically needed for assembly of dynein c
To investigate further the organization of dynein arms, we analyzed the structure of ODAs and IDAs within the 96-nm axonemal repeat. The ODAs are homogeneous in shape and repeat every 24 nm, whereas the row of IDAs is more complex, with multiple subtypes of arms repeating every 96 nm (Bui et al., 2009; Barber et al., 2012; Heuser et al., 2012a). In the subtomogram averages, the structure of the ODAs appeared unaffected by the loss of FAP206 (Figure 2, A and B, and Supplemental Movies S4 and S5). In agreement with previous studies (Nicastro et al., 2006; Pigino et al., 2011; Barber et al., 2012; Bui et al., 2012; Lin et al., 2012) the wild-type repeat shows the ring-shaped AAA motor domains of all IDAs, including the most proximally located, two-headed dynein f (I1), followed by six single-headed dyneins: a (attached to the base of RS1), b (which is doublet specific in Chlamydomonas but seems generally present in Tetrahymena), c (attached to the base of RS2), e (attached to the N-DRC distal of RS2), and g and d (attached near the base of RS3/RS3S; Figure 5A). In the averaged axonemal repeat of FAP206-KO, the IDA array had a single gap that corresponds precisely to the position of dynein c (Figure 5D), whereas assembly of the remaining IDAs was unaffected. Both the head and tail of dynein c were missing in all (100% after classification) 96-nm repeats of FAP206-KO axonemes (Figure 5E). Note that a relatively small fraction of wild-type repeats (11%) also lacked dynein c (Figure 5, B and C). These repeats were randomly distributed along the axoneme length and among the nine doublets (unpublished data). Thus, in the wild type, dynein c is either naturally missing or lost during the axoneme preparation in a small subset of the 96-nm repeats.
FIGURE 5:
Dynein c requires FAP206 for its assembly at the base of RS2. Longitudinal tomographic slices of averaged 96-nm axonemal repeats show the presence (A–B) and absence (C–E) of IDA c in WT (A–C) and FAP206KO (D, E). Classification analysis confirms the complete absence of IDA c (both the head and tail [arrowheads] are missing) from all axonemal repeats from FAP206-KO, whereas the remaining IDAs appear intact (E). The majority of the WT subtomograms have a fully assembled dynein c (B), whereas 11% lack dynein c (C). The subtomograms that lack dynein c are randomly located within all nine outer doublets and were likely extracted during the axoneme isolation procedure. Scale bar, 10 nm.
We took advantage of the 100% penetrant loss of dynein c in the FAP206-KO axoneme to determine the protein composition of dynein c in Tetrahymena. We purified the dynein heavy chain (DHCs) containing the high–molecular weight fraction of axonemes by SDS–PAGE and analyzed it by quantitative label-free mass spectrometry. In the wild-type DHC sample, we found peptides corresponding to 21 of the 25 annotated DHCs of Tetrahymena thermophila (Wilkes et al., 2008). Among the detected 21 DHCs (Table 1), we found both nonaxonemal dyneins, DYH2, the IFT dynein related to the Chlamydomonas Dhc1b (Rajagopalan et al., 2009), and DYH1, earlier implicated in phagocytosis and micronuclear segregation (Lee et al., 1999), which, in light of our finding, could also be involved with cilia; 3 DHCs forming ODAs (DYH3, DYH4, DHY5) that were (as expected) most abundant; DHCs predicted to form the two-headed dynein f (I1;DYH6 and DYH7; Angus et al., 2001; Wood et al., 2007); and 14 DHCs of the 18 DHCs predicted to form single-headed IDAs (Wilkes et al., 2008). Strikingly, among the 14 DHCs of single-headed IDAs detected in the wild-type axonemes, peptides of DYH25, DYH12, and DYH10 were absent in the FAP206-KO axoneme extract (Table 1). Thus, most likely, DYH25, DYH12, and DYH10 collectively constitute the heavy chains of dynein c in Tetrahymena (the assignment of DYH10 as dynein c is done with caution due to its low abundance even in the wild-type extract). In agreement with our conclusion, based on their sequences, DYH25, DYH12, and DYH10 have been assigned to the IAD-3 subgroup of single-headed dyneins, which also contains DHC9, the known dynein c DHC in Chlamydomonas (Yagi et al., 2005; Wickstead and Gull, 2007; Wilkes et al., 2008).
TABLE 1:
Comparative mass spectrometry (MS/MS) analysis of the DHC-containing fractions of wild-type and FAP206-KO axonemes.
| DHC name | Gene ID | Protein accession number | Peptide matched number | |
|---|---|---|---|---|
| CU428 | FAP206-KO | |||
| DYH5 | TTHERM_00486600 | gi|118401102|ref|xp_001032872.1| | 1296 | 1324 |
| DYH4 | TTHERM_00499300 | gi|118378024|ref|xp_001022188.1| | 1216 | 1265 |
| DYH3 | TTHERM_01276420 | gi|118394992|ref|xp_001029853.1| | 1133 | 1115 |
| DYH7 | TTHERM_00912290 | gi|118374012|ref|xp_001020198.1| | 551 | 600 |
| DYH6 | TTHERM_00688470 | gi|118387693|ref|xp_001026949.1| | 581 | 588 |
| DYH15 | TTHERM_00433800 | gi|118356293|ref|xp_001011405.1| | 498 | 492 |
| DYH16 | TTHERM_00558640 | gi|118378501|ref|xp_001022426.1| | 280 | 294 |
| DYH22 | TTHERM_00565600 | gi|118377765|ref|xp_001022060.1| | 315 | 263 |
| DYH11 | TTHERM_00252430 | gi|229595213|ref|xp_001019094.2| | 232 | 216 |
| DYH19 | TTHERM_01027670 | gi|118396733|ref|xp_001030704.1| | 136 | 152 |
| DYH24 | TTHERM_00193520 | gi|118367791|ref|xp_001017105.1| | 141 | 128 |
| DYH25 | TTHERM_00774810 | gi|118376063|ref|xp_001021214.1| | 233 | 0 |
| DYH14 | TTHERM_00492830 | gi|118380021|ref|xp_001023175.1| | 65 | 70 |
| DYH12 | TTHERM_00919540 | gi|118389527|ref|xp_001027847.1| | 53 | 0 |
| DYH20 | TTHERM_00821980 | gi|118398395|ref|xp_001031526.1| | 25 | 15 |
| DYH9 | TTHERM_00947430 | gi|118397291|ref|xp_001030979.1| | 11 | 11 |
| DYH8 | TTHERM_00531870 | gi|118382309|ref|xp_001024312.1| | 7 | 13 |
| DYH23 | TTHERM_00355100 | gi|118354291|ref|xp_001010408.1| | 8 | 8 |
| DYH10 | TTHERM_00420340 | gi|118401939|ref|xp_001033289.1| | 11 | 0 |
| DYH2 | TTHERM_00558310 | gi|118378437|ref|xp_001022394.1| | 6 | 3 |
| DYH1 | TTHERM_00046310 | gi|118363224|ref|xp_001014626.1| | 1 | 0 |
Among the 25 predicted DHCs (based on the genome of T. thermophila (Eisen et al., 2006), we detected peptides that correspond to 21 DHCs in the wild-type axoneme. The DHCs that were predicted but not detected are DYH13, DYH17, DYH18, and DYH21. Of importance, DYH25, DYH12, and DYH10 are detectable in the wild-type axoneme but missing in the FAP206 axoneme. Thus DYH25, DYH12, and possibly DYH10 (the case of DYH10 is less strong due to its low abundance in the wild-type sample) collectively constitute dynein c of Tetrahymena.
Overexpressed FAP206 decorates nonciliary microtubules in vivo
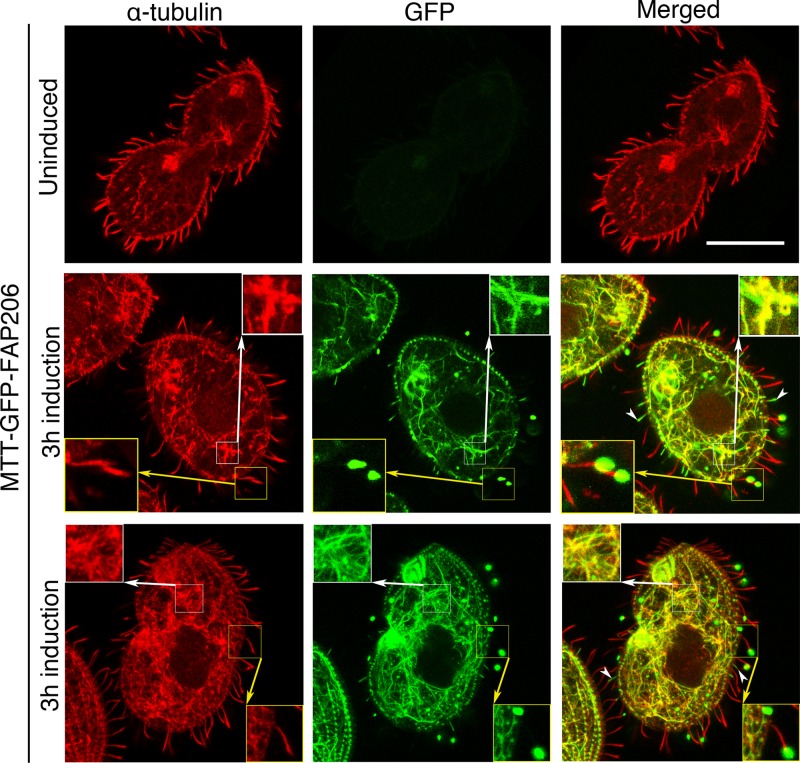
The cryo–electron tomography data place FAP206 in the RS2 front prong, a location close to the surface of the A-tubule of the outer doublet microtubules, opening the possibility that FAP206 might directly interact with the microtubule and thus contribute to RS2 docking. To test whether FAP206 has microtubule-binding activity in vivo, we briefly (3 h) overexpressed GFP-FAP206 fusion protein under the cadmium-inducible promoter in vegetatively growing cells that also expressed the native FAP206. The induced GFP-FAP206 localized to cilia in two distinct patterns. In a subset of cilia that tended to be relatively short (and therefore were likely in the process of assembly during the period of transgene induction), overproduced GFP-FAP206 was distributed along the entire cilium length (Figure 6, white arrowheads), indicating that the tagged protein can be incorporated into the axoneme during cilia assembly in place of the native protein. The second pattern was seen in a subset of cilia that were mostly full length, where GFP-FAP206 was not detectable along most of the cilium length but strongly accumulated at the distal tip (Figure 6, yellow insets). Likely, these full-length cilia were already assembled at the time of induction, and GFP-FAP206 could not be incorporated into the axonemes because its docking sites were occupied by the native protein, which turns over slowly. Strikingly, in the cell body, overexpressed GFP-FAP206 also strongly colocalized with the network of cytoplasmic microtubules (Figure 6, white insets). Thus, when overproduced, GFP-FAP206 colocalizes with nonciliary microtubules, indicating that FAP206 might have a microtubule-binding activity. To this end, we tested whether GFP-FAP206 purified from overproducing Tetrahymena cells can bind to microtubules in vitro, using a sedimentation assay (Goode and Feinstein, 1994). Unfortunately, GFP-FAP206 (and not GFP alone) sedimented on its own, indicating that it undergoes oligomerization or aggregation (unpublished data).
FIGURE 6:
GFP-FAP206 has microtubule-binding activity in vivo. Overexpressed GFP-FAP206 associates with nonciliary microtubules. Cells that overexpress GFP-FAP206 transgene under the MTT1 cadmium-inducible promoter were stained by immunofluorescence using a mix of 12G10 monoclonal anti–α-tubulin and SG polyclonal anti-tubulin antibodies (red) and imaged for GFP (green) using a confocal microscope. Top, a cell before induction; middle and bottom, an interphase cell and a dividing cell, respectively, fixed after 3 h of transgene induction with 2.5 μg/ml CdCl2. The white insets magnify the intracytoplasmic microtubules in the cell body. The yellow insets magnify mature full-length cilia with strong GFP-FAP206 accumulation at the tips. The white arrowheads mark short (likely assembling) cilia with uniform distribution of GFP-FAP206. Scale bar, 20 μm.
FAP206 affects stability of the CSC complex
Recent studies in Chlamydomonas identified CSC as a component of the 96-nm repeat that connects three major axonemal complexes, the two radial spokes RS2 and RS3S, as well as the N-DRC (Dymek and Smith, 2007; Dymek et al., 2011; Heuser et al., 2012b). RNA interference (RNAi) knockdowns of CSC components inhibit the assembly of specific structures near the bases of RS2 and RS3S and destabilize RS2 (Dymek and Smith, 2007; Dymek et al., 2011; Heuser et al., 2012b). These results indicate that the CSC is associated with the base of RS2 and opens a possibility that FAP206 and CSC interact. We used the antibodies generated against the Chlamydomonas CSC component FAP91/CaM-IP2 (Dymek and Smith, 2007) to test whether the absence of FAP206 affects the CSC levels in Tetrahymena. On a Western blot of the wild-type Tetrahymena cilia, the anti-Chlamydomonas FAP91/CaM-IP2 antibodies detected a protein of ∼70 kDa, which is in agreement with the 76 kDa predicted for the Tetrahymena FAP91 homologue (TTHERM_00578560; Figure 7A and Supplemental Figure S1). A corresponding band could not be detected in cilia that were isolated from a Tetrahymena strain lacking a homologue of another component of the CSC complex, FAP251/CaM-IP4 (TTHERM_01262850; Figure 7A). The same band was greatly reduced in cilia from a Tetrahymena strain lacking a homologue of a third CSC protein, FAP61/CaM-IP3 (TTHERM_00641200; Figure 7A). In Chlamydomonas, depletion of FAP61/CaM-IP3 strongly decreases the levels of the axoneme-bound FAP91/CaM-IP2 (Dymek et al., 2011). On the basis of the protein size and dependence on other CSC components for axoneme assembly, we conclude that the anti-Chlamydomonas FAP91/CaM-IP2 antibodies recognize the Tetrahymena homologue FAP91/CaM-IP2. The phenotypes of the Tetrahymena CSC component–knockout strains mentioned are described separately (Urbanska, Song, Joachimiak, Krzemien-Ojak, Koprowski, Hennessey, Jerka-Dziadosz, Fabczak, Gaertig, Nicastro, and Wloga, unpublished data). Of importance, the FAP91 levels were also greatly reduced in the cilia of the Tetrahymena FAP206-KO strain (Figure 7A). A similar strong reduction in the levels of FAP91 was seen in purified FAP206-KO axonemes (Figure 7B). Thus stable assembly of the CSC component FAP91/CaM-IP2 into the axoneme is dependent on FAP206.
To determine whether the CSC and FAP206 are physically linked, we used the C. reinhardtii model, in which CSC and other radial spoke proteins have been well characterized biochemically (Dymek and Smith, 2007; Dymek et al., 2011; Gupta et al., 2012). The anti-FAP91/CaM-IP2 antibodies immunoprecipitated a number of proteins from the wild-type, radial spoke–enriched axonemal extract. As expected, in addition to FAP91/CaM-IP2, the immunoprecipitate contained the partner CSC components—FAP61/CaM-IP3, FAP251/CaM-IP4—and a number of other radial spoke proteins, including RSP1, RSP2, RSP17, RSP23, RSP3, RSP4, RSP5, and RSP6 (Figure 7, C and D). Of importance, using liquid chromatography–tandem mass spectrometry mass spectrometry (LC-MS/MS), we found two of the immunoprecipitated bands located around 80 kDa (previously designated as RSP13; Yang et al., 2006) to contain mainly FAP206 (Figure 7, C and D). The presence of multiple FAP206 bands is not surprising because FAP206 is extensively phosphorylated (Lin et al., 2011). FAP206 is known to be in a complex with RSP3, a major stem component located near the base of RS2 (Wirschell et al., 2008; Gupta et al., 2012). The bands corresponding to FAP206 were greatly reduced in the anti-FAP91/CaM-IP2 immunoprecipitate obtained from an extract of the pf14 Chlamydomonas mutant, which lacks RSP3 and fails to assemble RS1 and RS2 (Figure 7D; Pigino et al., 2011; Lin et al., 2012). The dramatic reduction of FAP206 in the immunoprecipitate of pf14 is in agreement with the lower abundance of FAP206 in the pf14 axonemes that was previously noticed on a two-dimensional gel (Lin et al., 2011). On one hand, this observation could be explained if FAP206 requires RSP3 and presumably the stem of RS2 to assemble onto the axoneme. On the other hand, the same result could be expected if the CSC and FAP206 are associated through an intermediate, RSP3. An earlier study places part of the CSC within or near the back prong of RS2 (Heuser et al., 2012b). Our data would be consistent with FAP206 being associated with CSC indirectly through RSP3 in the radial spoke stem that connects the front and back prongs.
DISCUSSION
We show that without FAP206, radial spoke RS2 either fails to assemble or assembles without the front prong, whereas RS1 and RS3 are relatively unaffected. Because FAP206 is stably associated with the axoneme (Figure 1B; Lin et al., 2011; Gupta et al., 2012), it is unlikely that FAP206 is a soluble assembly factor for the front prong of RS2. Most likely, FAP206 is an integral part of the front prong of RS2. This proposed location of FAP206 is consistent with the observed complete loss of dynein c in the FAP206-KO axoneme, an IDA whose tail contacts the RS2 front prong (Barber et al., 2012; Lin et al., 2012).
Overexpressed GFP-FAP206 associated not only with axonemes, but also with cytoplasmic microtubules. This observation, combined with the most likely location of FAP206 at the microtubule-associated base of RS2, suggests that FAP206 has microtubule-binding activity and is one of the adaptors that docks RS2 to the doublet microtubule. The amino acid sequence of FAP206 lacks a recognizable microtubule-binding domain. Further studies are required to identify the FAP206 domains that interface with tubulin, the radial spoke stem (likely via RSP3/LC8; see later discussion), and likely the tail domain of dynein c.
The loss of FAP206 led to slow cell motility and abnormal ciliary waveform. An abnormal waveform is often an indication of a defect in IDAs. Consistently, we show that FAP206 is required for assembly of only one IDA subtype, dynein c. Our proteomic data identify DYH12, DYH25, and, with lower confidence, also DYH10 as DHCs of dynein c. These DHCs could be interchangeable at every dynein c position, or one or more of these DHCs could be specific to an axoneme region (as shown for DHC11 in Chlamydomonas; Yagi et al., 2009) or be restricted to a subtype of Tetrahymena cilia. In Tetrahymena, oral cilia are structurally distinct from locomotory cilia (Williams and Luft, 1968). Furthermore, locomotory cilia have distinct properties, depending on their anteroposterior and dorsoventral positions (Wloga et al., 2006). In Chlamydomonas, a null mutation in DHC9 forming dynein c resulted in slow flagellar motility, as revealed under increased load imposed by a viscous medium (Yagi et al., 2005). Thus the apparently more severe phenotype of the Tetrahymena FAP206-KO is likely a composite of the loss of dynein c and RS2 and partial loss of RS3. Furthermore, the absence of FAP206 could affect N-DRC, whose microtubule attachment is located between RS2 and RS3 (Heuser et al., 2009). A possible effect of the loss of FAP206 on the function of N-DRC is supported by a proteomic study of three different n-drc mutants that showed that FAP206 adopts at least four different phosphorylation states and undergoes major dephosphorylation in the n-drc-mutant axonemes (Lin et al., 2011). Furthermore, the loss of FAP206 and RS2 could affect the function of dynein e, whose tail is associated with the N-DRC base plate (Heuser et al., 2009). Finally, the loss of FAP206 could affect the CSC, based on multiple studies that have linked the CSC to the basal regions of RS2 and RS3.
In Chlamydomonas, RNAi knockdowns of the CSC components lead to losses of specific structural densities, including the back prong of RS2, part of the base of the short spoke RS3S, the N-DRC fork, and dynein e located near RS3 (Dymek et al., 2011; Heuser et al., 2012b). In addition, many 96-nm repeats in the CSC mutants lack RS2, indicating that the CSC is required for stability or assembly of RS2 (Dymek et al., 2011). Our data provide additional evidence in support of a model that the CSC physically links RS2 and RS3. We show that loss of FAP206, an RS2-specific factor, strongly reduces the levels of the CSC component FAP91/CaM-IP2 and results in partial loss of RS3. There is no definitive information about the localization of FAP91/CaM-IP2, but a previous study proposed that it is located close to the RS2 base, contacting or even forming its back prong (Heuser et al., 2012b). In agreement with this prediction, the back prong is absent (or greatly reduced) in most of the FAP206-KO 96-nm repeats.
Our data indicate that RS2 is required for stability of RS3. Unlike Chlamydomonas, Tetrahymena has a full-length RS3 that interacts with RS2 via the spoke heads, which could contribute to lateral stability (Lin et al., 2012). However, the CSC-knockdown phenotypes also indicate that stabilizing interactions occur between RS2 and RS3S, which lacks a spoke head (Dymek et al., 2011; Heuser et al., 2012b). These data, coupled with our observations, strongly indicate that RS2 has a stabilizing effect on RS3 that is mediated by the CSC close to the microtubule surface and spoke bases.
As argued earlier, FAP206 likely forms one of the microtubule-binding interfaces of RS2. So far, the RSP3-LC8 complex has been seen as the most basal part of the radial spoke stem of RS1 and RS2 (Wirschell et al., 2008; Gupta et al., 2012). However, recombinant RSP3 failed to bind to pure microtubules in vitro (Diener et al., 1993). RSP3 binds to pf14 mutant axonemes, and its binding is enhanced by LC8 (Diener et al., 1993; Gupta et al., 2012), a protein that promotes RSP3 dimerization (Gupta et al., 2012). The pf14 axonemes contain most if not all of the basal spoke densities, including at least some FAP206 and CSC (Dymek et al., 2011; Lin et al., 2011; Pigino et al., 2011). It is therefore likely that the basal end of the RS2 stem binds to the axoneme through the docking sites formed by FAP206 and the CSC.
The overproduced GFP-FAP206 accumulated at the tips of mature cilia, indicating that it is efficiently transported by anterograde intraflagellar transport (IFT). Radial spokes are preassembled in the cell body as 12S complexes and assembled into larger 20S complexes within the cilium, possibly at the time of assembly at the axoneme-binding sites (Qin et al., 2004; Diener et al., 2011; Gupta et al., 2012). The 12S complex is shaped like a “7,” suggesting that it contains a longitudinal half of a mature radial spoke stem and head (Diener et al., 2011). In future studies, it will be important to determine whether FAP206 is a part of the 12S radial spoke transport subcomplex. If this is the case, the 12S complexes could fall into two subclasses that are destined to form either RS1 or RS2 (in Tetrahymena and other species with complete radial spoke triplets a similar half-complex could exist for RS3). Alternatively, the 12S subcomplex could lack the basal adapters, and FAP206 could travel in a separate RS2 base complex. Future determination of the composition of the FAP206 cilia-destined transport complex could shed light on the mechanism that prevents premature binding of FAP206-containing complexes at incorrect microtubule sites before its incorporation at the proper RS2 docking site on the axoneme. One possibility is that a cofactor sterically blocks the microtubule-binding domain of FAP206. Another possibility is that the microtubule-binding interface of FAP206 is inhibited by a posttranslational modification that is reversed shortly before its incorporation into the axoneme.
MATERIALS AND METHODS
Strains, culture, gene knockout, overexpression, and phenotypic evaluation of Tetrahymena cells and cilia
Tetrahymena cells were grown in SPPA medium (Gorovsky, 1973). The Tetrahymena thermophila strain CU428 (available from the Tetrahymena Stock Center, Cornell University, Ithaca, NY) was used as a wild type. A knockout strain lacking the FAP206 homologue gene (TTHERM_00820660) was produced by targeting the micronuclear (germline) genomes of mating B2086 and CU428 strains using homologous DNA recombination (Cassidy-Hanley et al., 1997), as described in detail in Dave et al. (2009). Supplemental Table S1 lists primers used to make the knockout-targeting plasmid with portions of FAP206 flanking the neo4 cassette (Mochizuki, 2008). Heterokaryon strains were made, and homozygotes lacking FAP206 in both the macronucleus and micronucleus were obtained as a heterokaryon cross-progeny (Hai and Gorovsky, 1997). The absence of the targeted coding region of FAP206 was confirmed using primers specific to the deleted region listed in Supplemental Table S2.
For GFP tagging of FAP206 expressed in its own genetic locus, a targeting fragment was made for homologous DNA recombination, which inserted a GFP-coding sequence at the 3′ end of the FAP206-coding region using a linked neo3 marker (Shang et al., 2002). The primers used for amplification of the FAP206 genomic fragments that served as homology portions for the native locus targeting are listed in Supplemental Table S3. The resulting native locus targeting plasmid, pFAP206-GFP-NLE, was digested with SacI and SacII to release the targeting fragment and introduced into the macronucleus of starved CU428 cells by biolistic bombardment and selection with 120 μg/ml paromomycin and 2.5 μg/ml CdCl2 (to induce neo3) as described (Gaertig et al., 2013).
For overexpression of FAP206, the genomic coding region of FAP206 was amplified with addition of MluI and BclI sites (5′-TTAAACGCGTCATGAATTAGCTAGAAGAAATAG-3′ and 5′-TATATGATCATCAATTAGTGTCTTTGTCTCT-3′), cloned into the pMTT1-GFP plasmid, and introduced into the macronuclear BTU1 locus by homologous DNA recombination, followed by complete phenotypic assortment (Gaertig et al., 2013). The resulting strain overexpressed GFP-FAP206 under the MTT1 cadmium-inducible promoter (Shang et al., 2002). To induce overexpression, cells carrying the MTT1-GFP-FAP206 transgene were grown in SPPA to 2 × 105 cells/ml and induced in the same medium with 2.5 μg/ml CdCl2 for 3 h.
The growth rates of Tetrahymena strains were determined by counting cells in 25 ml of SPPA in flask cultures at the initial concentration of 2 × 104 cells/ml and grown at 30°C without shaking. To measure the swimming rates, Tetrahymena cells (at 1 × 104 cells/ml) were recorded for 1 s under a dissecting scope using a Moticam 480 digital camera. To record beating cilia, cells (2 × 105 cells/ml) were video-recorded at 500 frames/s by a 1280 PCI FastCam (Photron, San Diego, CA) on a Nikon Eclipse E600 microscope at 600× total magnification. The microtubule-sliding assays on isolated axonemes in vitro were done as described (Suryavanshi et al., 2010).
Fluorescence microscopy, immunofluorescence, classical electron microscopy, and Western blotting of Tetrahymena
To detect FAP206-GFP (tagged in the native locus) in live cells, we used total internal reflection fluorescence (TIRF) microscopy. A 10-μl amount of cells (2 × 105 cells/ml) in SPPA with 2–3 μM NiCl2 (to slow down the beating of cilia, based on Andrivon, 1974) was placed on a microscopic slide with a 22 × 22 mm #1.5 coverglass and observed using a home-built TIRF system based on a Nikon Eclipse Ti-U inverted microscope equipped with a 60×/numerical aperture 1.49 TIRF objective as described (Engel et al., 2009). To test whether FAP206-GFP is associated with the axoneme, cells expressing FAP206-GFP were extracted with Triton X-100 and fixed with paraformaldehyde (protocol 4.3 in Gaertig et al., 2013). To detect microtubules in cells overproducing GFP-FAP206, Tetrahymena cells were labeled by immunofluorescence (protocol 4.4. in Gaertig et al., 2013) using a mix of primary antibodies: 12G10 monoclonal anti–α-tubulin (1:30; Developmental Hybridoma Bank; Jerka-Dziadosz et al., 2001) and SG polyclonal antibodies against total Tetrahymena tubulin (Guttman and Gorovsky, 1979). For the measurements of the axoneme lengths, cells were labeled by immunofluorescence using a mix of 12G10 anti–α-tubulin and polyG anti–polyglycylated tubulin antibodies (Duan and Gorovsky, 2002) and detected with a mix of secondary antibodies coupled to the same fluorochrome. Fixed cells were imaged in the LSM 710 confocal microscope (Zeiss, Jena, Germany). For reproducibility, we measured the length of all axonemes on 13 cells using confocal optical sections that include the widest diameter of the macronucleus. The length measurements were done using ImageJ 1.46 (National Institutes of Health, Bethesda, MD).
For classical TEM, 2 × 106 cells were suspended in 100 μl of Tris-HCl, pH 7.5, and fixed by addition of 1 ml of 2% glutaraldehyde (in 0.1 M sodium cacodylate buffer, pH 7.2) on ice for 1 h. A 10-μl amount of 1% tannic acid was added, and the cells were incubated on ice for 1 h, washed five times in the cold sodium cacodylate buffer (10 min each on ice), and postfixed with 1 ml of 1% osmium tetroxide for 1 h on ice. The pellet was washed five times in water, followed by dehydration in an ethanol/water concentration series and embedding in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate and analyzed on a JEOL 1200 EX transmission electron microscope (JEOL, Peabody, MA).
For Western blotting, the Tetrahymena cell bodies and cilia were purified from CU428 and FAP206-KO cells after a pH shock deciliation as described in Gaertig et al. (2013). Cilia were loaded in amounts corresponding to 125 × 103 cell equivalents/lane, respectively. Axonemes (purified as described in the next section) were loaded at 5 μg of total protein/lane. The primary antibodies were rabbit polyclonal anti-Chlamydomonas Cam-IP2 antibodies (1:500; Dymek and Smith, 2007), 12G10 (1:1000), and polyG anti–polyglycylated tubulin antibodies (1:10,000). The blots were developed using the Amersham ECL Prime Western blotting detection reagent (GE Healthcare Life Sciences, Pittsburgh, PA). The images of blots were recorded, and bands were quantified using ChemiDoc MP and ImageLab software (Bio-Rad, Hercules, CA).
Axoneme isolation, cryo–electron tomography, and image processing
Axonemes were isolated from Tetrahymena strains CU428 and FAP206-KO as described (Wloga et al., 2008), with minimal modifications. Cells (250 ml at [3–4] × 105 cells/ml) were washed with 10 mM Tris-HCl, pH 7.5, and suspended in 40 ml of 10 mM Tris-HCl, pH 7.5, 50 mM sucrose, 10 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride, and 0.02 mg/ml aprotinin. The cells were deciliated by adding 700 μl of 0.5 M acetic acid and inverting the tube six times during 1 min. Deciliation was stopped by adding 360 μl of 1 M KOH. Subsequent steps were performed at 4°C. The deciliated cells were collected by centrifugation at 1500 × g for 5 min and twice at 1860 × g for 5 min. Cilia were collected by centrifugation at 10,000 × g for 15 min. Cilia were demembranated in 20 ml of HMEEK (30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 25 mM KCl, 5 mM MgSO4, 0.1 mM EDTA, and 1.0 mM ethylene glycol tetraacetic acid, pH 7.4) with 1% IGEPAL CA-630 (Sigma-Aldrich, St. Louis, MO) for 30 min. Demembranated axonemes were collected by centrifugation at 10,000 × g for 10 min and suspended in 200 μl of HMEEK buffer without detergent.
Cryo–electron tomography
Quantifoil holey carbon grids (Quantifoil Micro Tools, Jena, Germany) were glow discharged and then coated with 10-nm colloidal gold (Sigma-Aldrich). After loading the grid in a home-made plunge freezer, 3 μl of axonemes and 1 μl of a 5× concentrated, 10-nm colloidal gold solution were applied to the grid. The grid was blotted for ∼2 s with a filter paper and then immediately plunge frozen in liquid ethane to achieve vitrification. The frozen samples were stored in liquid nitrogen until TEM examination.
Using a cryo-holder (Gatan, Pleasanton, CA) to maintain the sample at a temperature below −140°C, a vitrified sample was transferred into a Tecnai F30 TEM (FEI, Eindhoven, Netherlands) equipped with a field emission gun and a postcolumn energy filter (Gatan). Images were recorded at 300 keV under low-dose conditions and in the zero-loss mode of the energy filter (20-eV slit width). Tilt series of axoneme samples were automatically recorded from −65 to +65º with 1.5–2.5° angular increments using SerialEM software (Mastronarde, 2005). The cumulative electron dose was restricted to ∼100 e/Å2. All data were acquired with a 2k × 2k charge-coupled device camera (Gatan) at −8-μm defocus and at a magnification of 13,500×, resulting in a pixel size of ∼1 nm.
Image processing
The 3D tomograms were reconstructed using the IMOD software package (Kremer et al., 1996) with fiducial alignment and weighted backprojection. Only tomograms of intact and noncompressed or mildly compressed axonemes (6–12 tomograms/strain) were further processed and analyzed. Subtomograms containing 96-nm axonemal repeats were extracted, aligned, and averaged with missing-wedge compensation using the software package Particle Estimation for Electron Tomography (PEET; Nicastro et al., 2006). The University of California, San Francisco, Chimera package (Pettersen et al., 2004) was used for 3D visualization by isosurface rendering and for measuring volume sizes of structural components/defects after normalizing the isosurface rendering threshold to the mass of a doublet microtubule (Heuser et al., 2009). The molecular mass of these volumes was estimated by assuming an average protein density of 1.43 g/cm3 (Quillin and Matthews, 2000). A clustering (unsupervised classification) approach incorporated into PEET (Heumann et al., 2011) was used to analyze the heterogeneity concerning the presence/absence of RS2, RS3, and dynein c in the axonemal repeats of wild-type and FAP206-KO. To focus the classification on the structures of interest, the examined 3D volume was limited to the targeted region using masks.
Mass spectrometry analysis of the dynein-containing axoneme fraction
Axonemes were isolated from Tetrahymena CU428 and FAP206-KO as described and dissolved in the lysis buffer (7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 65 mM dithiothreitol, and 2% IPG buffer, pH 3–10 nonlinear; GE Healthcare) with vigorous stirring for 30 min. To remove insoluble substances, the sample was centrifuged at 45,000 × g for 1 h. Protein concentrations were determined using a 2-D Quant Kit (GE Healthcare). The supernatant was aliquoted and stored at −70°C. A total of 35 μg of axonemal protein/strain was separated on NuPAGE 4-12% Bis-Tris Mini Gels (Novex, Life Technologies, Grand Island, NY). The gels were fixed and stained with Coomassie blue G-250. The portion of the gel corresponding to molecular weights >130 kDa, which was expected to contain all dynein heavy chains, was excised, washed in 50% acetonitrile, and analyzed by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry on a Thermo LTQ-Orbitrap mass spectrometer as described (Chittum et al., 1998; Taniguchi et al., 2002) at the Harvard Microchemistry and Proteomics Analysis Facility, Harvard University.
Immunoprecipitation of CSC in Chlamydomonas
Strain A54-e18 (nit1-1, ac17, sr1, mt+) obtained from P. Lefebvre (University of Minnesota, St. Paul, MN) was used as a wild type. The radial spokeless strain pf14 (cc1032, mt+) was obtained from the Chlamydomonas Genetics Center (Duke University, Durham, NC). Axonemal extracts were prepared, and immunoprecipitations were performed and separated on 7% SDS–PAGE gels according to Dymek and Smith (2007). Two silver-stained bands earlier designated as RSP13 were excised, subjected to in-gel trypsin digestion, and analyzed by LC-MS/MS on an Orbitrap Velos at the University of Massachusetts Medical School (Worcester, MA) mass spectrometry facility.
Supplementary Material
Acknowledgments
We are grateful to Karl Lechtreck (University of Georgia, Athens, GA) for allowing us to use his TIRF system, as well as for many helpful suggestions. We are grateful to Yuyang Jiang for help with the TIRF imaging. The assistance of Mary Ard with the standard TEM (University of Georgia) is acknowledged. We thank William S. Lane (Harvard Microchemistry and Proteomics Analysis Facility, Harvard University, Cambridge, MA) for the mass spectrometry analysis and identification of IDA dyneins. We thank Jianfeng Lin for assistance with sample preparation for mass spectrometry and Chen Xu for training and management of the Brandeis EM facility. This work was supported by funding from the National Institutes of Health (GM089912 to J.G., GM083122 to D.N., GM051173 to W.S.S., GM66919 to E.F.S., and P20RR016475 and P20GM133418 to W.D.). D.W. was supported by Polish Ministry of Science and Higher Education Grant N301 706640, the Marie Curie International Reintegration Grant within the 7th European Community Framework Programme, and an EMBO Installation Grant, Project No. 2331. E.J. was supported by Polish Ministry of Science and Higher Education Grant N303 817840.
Abbreviations used:
- CSC
calmodulin and spoke-associated complex
- 3D
three dimensional
- DHC
dynein heavy chain
- GFP
green fluorescent protein
- IDA
inner dynein arm
- IFT
intraflagellar transport
- N-DRC
nexin-dynein regulatory complex
- ODA
outer dynein arm
- PEET
Particle Estimation for Electron Tomography
- RNAi
RNA interference
- TEM
transmission electron microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1506) on December 24, 2014.
*These authors contributed equally.
REFERENCES
- Andrivon C. Inhibition of ciliary movements by Ni-2+ions in triton-extracted models of Paramecium caudatum. Arch Int Physiol Biochim. 1974;82:843–852. doi: 10.3109/13813457409072332. [DOI] [PubMed] [Google Scholar]
- Angus SP, Edelmann RE, Pennock DG. Targeted gene knockout of inner arm 1 in Tetrahymena thermophila. Eur J Cell Biol. 2001;80:486-497. doi: 10.1078/0171-9335-00178. [DOI] [PubMed] [Google Scholar]
- Barber CF, Heuser T, Carbajal-Gonzalez BI, Botchkarev VV, Jr, Nicastro D. Three-dimensional structure of the radial spokes reveals heterogeneity and interactions with dyneins in Chlamydomonas flagella. Mol Biol Cell. 2012;23:111–120. doi: 10.1091/mbc.E11-08-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ, Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J Cell Biol. 2009;186:437–446. doi: 10.1083/jcb.200903082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol. 2012;198:913–925. doi: 10.1083/jcb.201201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee J, Cole ES, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castleman VH, Romio L, Chodhari R, Hirst RA, de Castro SC, Parker KA, Ybot-Gonzalez P, Emes RD, Wilson SW, Wallis C, et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet. 2009;84:197–209. doi: 10.1016/j.ajhg.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FD, Lee BJ, Hatfield DL. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- Curry AM, Rosenbaum JL. Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil Cytoskeleton. 1993;24:224–232. doi: 10.1002/cm.970240403. [DOI] [PubMed] [Google Scholar]
- Dave D, Wloga D, Gaertig J. Manipulating ciliary protein-encoding genes in Tetrahymena thermophila. Methods Cell Biol. 2009;93:1–20. doi: 10.1016/S0091-679X(08)93001-6. [DOI] [PubMed] [Google Scholar]
- Dentler WL, Cunningham WP. Structure and organization of radial spokes in cilia of Tetrahymena pyriformis. J Morphol. 1977;153:143–151. doi: 10.1002/jmor.1051530110. [DOI] [PubMed] [Google Scholar]
- Diener DR, Ang LH, Rosenbaum JL. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J Cell Biol. 1993;123:183–190. doi: 10.1083/jcb.123.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Curry AM, Johnson KA, Williams BD, Lefebvre PA, Kindle KL, Rosenbaum JL. Rescue of a paralyzed-flagella mutant of Chlamydomonas by transformation. Proc Natl Acad Sci USA. 1990;87:5739–5743. doi: 10.1073/pnas.87.15.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Yang P, Geimer S, Cole DG, Sale WS, Rosenbaum JL. Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken) 2011;68:389–400. doi: 10.1002/cm.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Gorovsky MA. Both carboxy terminal tails of alpha and beta tubulin are essential, but either one will suffice. Curr Biol. 2002;12:313–316. doi: 10.1016/s0960-9822(02)00651-6. [DOI] [PubMed] [Google Scholar]
- Dymek EE, Heuser T, Nicastro D, Smith EF. The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol Biol Cell. 2011;22:2520–2531. doi: 10.1091/mbc.E11-03-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Smith EF. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol. 2007;179:515–526. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Lechtreck KF, Sakai T, Ikebe M, Witman GB, Marshall WF. Total internal reflection fluorescence (TIRF) microscopy of Chlamydomonas flagella. Methods Cell Biol. 2009;93:157–177. doi: 10.1016/S0091-679X(08)93009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J, Wloga D, Vasudevan KK, Guha M, Dentler WL. Methods Enzymol 525, 265–284. 2013. Discovery and functional evaluation of ciliary proteins in Tetrahymena thermophila. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard AR, Diener DR, Rosenbaum JL, Sale WS. Flagellar radial spoke protein 3 is an A-kinase anchoring protein (AKAP) J Cell Biol. 2001;153:443–448. doi: 10.1083/jcb.153.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard AR, Fox LA, Rhea JM, Craige B, Sale WS. Disruption of the A-kinase anchoring domain in flagellar radial spoke protein 3 results in unregulated axonemal cAMP-dependent protein kinase activity and abnormal flagellar motility. Mol Biol Cell. 2006;17:2626–2635. doi: 10.1091/mbc.E06-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Feinstein SC. Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J Cell Biol. 1994;124:769–782. doi: 10.1083/jcb.124.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J Cell Biol. 1985;100:2008–2018. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky MA. Macro- and micronuclei of Tetrahymena pyriformis: a model system for studying the structure and function of eukaryotic nuclei. J Protozool. 1973;20:19–25. doi: 10.1111/j.1550-7408.1973.tb05995.x. [DOI] [PubMed] [Google Scholar]
- Gupta A, Diener DR, Sivadas P, Rosenbaum JL, Yang P. The versatile molecular complex component LC8 promotes several distinct steps of flagellar assembly. J Cell Biol. 2012;198:115–126. doi: 10.1083/jcb.201111041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman SD, Gorovsky MA. Cilia regeneration in starved Tetrahymena: an inducible system for studying gene expression and organelle biogenesis. Cell. 1979;17:307–317. doi: 10.1016/0092-8674(79)90156-9. [DOI] [PubMed] [Google Scholar]
- Hai B, Gorovsky MA. Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc Natl Acad Sci USA. 1997;94:1310–1315. doi: 10.1073/pnas.94.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann JM, Hoenger A, Mastronarde DN. Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J Struct Biol. 2011;175:288–299. doi: 10.1016/j.jsb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Barber CF, Lin J, Krell J, Rebesco M, Porter ME, Nicastro D. Cryoelectron tomography reveals doublet-specific structures and unique interactions in the I1 dynein. Proc Natl Acad Sci USA. 2012a;109:E2067–2076. doi: 10.1073/pnas.1120690109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Dymek EE, Lin J, Smith EF, Nicastro D. The CSC connects three major axonemal complexes involved in dynein regulation. Mol Biol Cell. 2012b;23:3143–3155. doi: 10.1091/mbc.E12-05-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol. 2009;187:921–933. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DR, Habermacher G, Glass DB, Smith EF, Sale WS. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J Cell Biol. 1994;127:1683–1692. doi: 10.1083/jcb.127.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Piperno G, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J Cell Biol. 1981;88:80–88. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Luck DJ. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for Flagellar function. Cell. 1982;28:115–124. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz M, Strzyzewska-Jowko I, Wojsa-Lugowska U, Krawczynska W, Krzywicka A. The dynamics of filamentous structures in the apical band, oral crescent, fission line and the postoral meridional filament in Tetrahymena thermophila revealed by the monoclonal antibody 12G9. Protist. 2001;152:53–67. doi: 10.1078/1434-4610-00043. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 2002;219:115–155. doi: 10.1016/s0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Delmotte P, Robinson ML, Sanderson MJ, Witman GB. Mutations in Hydin impair ciliary motility in mice. J Cell Biol. 2008;180:633–643. doi: 10.1083/jcb.200710162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wisniewski JC, Dentler WL, Asai DJ. Gene knockouts reveal separate functions for two cytoplasmic dyneins in Tetrahymena thermophila. Mol Biol Cell. 1999;10:771–784. doi: 10.1091/mbc.10.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann CB. Structural-functional relationships of the dynein, spokes, and central-pair projections predicted from an analysis of the forces acting within a flagellum. Biophys J. 2003;84:4115–4126. doi: 10.1016/S0006-3495(03)75136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Heuser T, Carbajal-Gonzalez BI, Song K, Nicastro D. The structural heterogeneity of radial spokes in cilia and flagella is conserved. Cytoskeleton (Hoboken) 2012;69:88–100. doi: 10.1002/cm.21000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Tritschler D, Song K, Barber CF, Cobb JS, Porter ME, Nicastro D. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J Biol Chem. 2011;286:29175–29191. doi: 10.1074/jbc.M111.241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck DJ, Huang B, Brokaw CJ. A regulatory mechanism for flagellar function is revealed by suppressor analysis in Chlamydomonas. Prog Clin Biol Res. 1982;80:159–164. doi: 10.1002/cm.970020730. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mochizuki K. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene. 2008;425:79–83. doi: 10.1016/j.gene.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Oda T, Yagi T, Yanagisawa H, Kikkawa M. Identification of the outer-inner dynein linker as a hub controller for axonemal dynein activities. Curr Biol. 2013;23:656–664. doi: 10.1016/j.cub.2013.03.028. [DOI] [PubMed] [Google Scholar]
- Omoto CK, Gibbons IR, Kamiya R, Shingyoji C, Takahashi K, Witman GB. Rotation of the central pair microtubules in eukaryotic flagella. Mol Biol Cell. 1999;10:1–4. doi: 10.1091/mbc.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Computational Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pigino G, Bui KH, Maheshwari A, Lupetti P, Diener D, Ishikawa T. Cryoelectron tomography of radial spokes in cilia and flagella. J Cell Biol. 2011;195:673–687. doi: 10.1083/jcb.201106125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Maheshwari A, Bui KH, Shingyoji C, Kamimura S, Ishikawa T. Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J Struct Biol. 2012;178:199–206. doi: 10.1016/j.jsb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillin ML, Matthews BW. Accurate calculation of the density of proteins. Acta Crystallogr. D Biol Crystallogr. 2000;56:791–794. doi: 10.1107/s090744490000679x. [DOI] [PubMed] [Google Scholar]
- Rajagopalan V, Subramanian A, Wilkes DE, Pennock DG, Asai DJ. Dynein-2 affects the regulation of ciliary length but is not required for ciliogenesis in Tetrahymena thermophila. Mol Biol Cell. 2009;20:708–720. doi: 10.1091/mbc.E08-07-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc Natl Acad Sci USA. 2002;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF, Sale WS. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science. 1992;257:1557–1559. doi: 10.1126/science.1387971. [DOI] [PubMed] [Google Scholar]
- Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgess JM, Chao J, Wong J, Aspin N, Turner JA. Cilia with defective radial spokes: a cause of human respiratory disease. N Engl J Med. 1979;300:53–56. doi: 10.1056/NEJM197901113000201. [DOI] [PubMed] [Google Scholar]
- Suryavanshi S, Edde B, Fox LA, Guerrero S, Hard R, Hennessey T, Kabi A, Malison D, Pennock D, Sale WS, et al. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr Biol. 2010;20:435–440. doi: 10.1016/j.cub.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D'Andrea AD. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- Warner FD, Satir P. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J Cell Biol. 1974;63:35–63. doi: 10.1083/jcb.63.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic. 2007;8:1708–1721. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes DE, Watson HE, Mitchell DR, Asai DJ. Twenty-five dyneins in Tetrahymena: A re-examination of the multidynein hypothesis. Cell Motil Cytoskeleton. 2008;65:342–351. doi: 10.1002/cm.20264. [DOI] [PubMed] [Google Scholar]
- Williams NE, Luft JH. Use of a nitrogen mustard derivative in fixation for electron microscopy and observations on the ultrastructure of Tetrahymena. J Ultrastruct Res. 1968;25:271–292. doi: 10.1016/s0022-5320(68)80074-7. [DOI] [PubMed] [Google Scholar]
- Wirschell M, Yamamoto R, Alford L, Gokhale A, Gaillard A, Sale WS. Regulation of ciliary motility: conserved protein kinases and phosphatases are targeted and anchored in the ciliary axoneme. Arch Biochem Biophys. 2011;510:93–100. doi: 10.1016/j.abb.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Zhao F, Yang C, Yang P, Diener D, Gaillard A, Rosenbaum JL, Sale WS. Building a radial spoke: flagellar radial spoke protein 3 (RSP3) is a dimer. Cell Motil Cytoskeleton. 2008;65:238–248. doi: 10.1002/cm.20257. [DOI] [PubMed] [Google Scholar]
- Witman GB, Plummer J, Sander G. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J Cell Biol. 1978;76:229–746. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Camba A, Rogowski K, Manning G, Jerka-Dziadosz M, Gaertig J. Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Mol Biol Cell. 2006;17:2799–2810. doi: 10.1091/mbc.E05-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Rogowski K, Sharma N, Van Dijk J, Janke C, Eddé B, Bré MH, Levilliers N, Redeker V, Duan J, et al. Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot Cell. 2008;7:1362–1372. doi: 10.1128/EC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CR, Hard R, Hennessey TM. Targeted gene disruption of dynein heavy chain 7 of Tetrahymena thermophila results in altered ciliary waveform and reduced swim speed. J Cell Sci. 2007;120:3075–3085. doi: 10.1242/jcs.007369. [DOI] [PubMed] [Google Scholar]
- Yagi T, Minoura I, Fujiwara A, Saito R, Yasunaga T, Hirono M, Kamiya R. An axonemal dynein particularly important for flagellar movement at high viscosity. Implications from a new Chlamydomonas mutant deficient in the dynein heavy chain gene DHC9. J Biol Chem. 2005;280:41412–41420. doi: 10.1074/jbc.M509072200. [DOI] [PubMed] [Google Scholar]
- Yagi T, Uematsu K, Liu Z, Kamiya R. Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J Cell Sci. 2009;122:1306–1314. doi: 10.1242/jcs.045096. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Song K, Yanagisawa HA, Fox L, Yagi T, Wirschell M, Hirono M, Kamiya R, Nicastro D, Sale WS. The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J Cell Biol. 2013;201:263–278. doi: 10.1083/jcb.201211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, et al. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Owen HA, Yang P. Dimeric heat shock protein 40 binds radial spokes for generating coupled power strokes and recovery strokes of 9 + 2 flagella. J Cell Biol. 2008;180:403–415. doi: 10.1083/jcb.200705069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, O'Toole E, Ghosh S, Mitchell DR. Regulation of flagellar dynein activity by a central pair kinesin. Proc Natl Acad Sci USA. 2004;101:17398–17403. doi: 10.1073/pnas.0406817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkiewicz E, Bukowy-Bieryllo Z, Voelkel K, Klimek B, Dmenska H, Pogorzelski A, Sulikowska-Rowinska A, Rutkiewicz E, Witt M. Mutations in radial spoke head genes and ultrastructural cilia defects in East-European cohort of primary ciliary dyskinesia patients. PLoS One. 2012;7:e33667. doi: 10.1371/journal.pone.0033667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.