Abstract
In nature, cells perform a variety of complex functions such as sensing, catalysis, and energy conversion which hold great potential for biotechnological device construction. However, cellular sensitivity to ex-vivo environments necessitates development of bio-nano interfaces which allow integration of cells into devices and maintain their desired functionality. In order to develop such an interface, the use of a novel Sol Generating Chemical Vapor into Liquid (SG-CViL) deposition process for whole cell encapsulation in silica was explored. In SG-CViL, the high vapor pressure of tetramethyl orthosilicate (TMOS) is utilized to deliver silica into an aqueous medium, creating a silica sol. Cells are then mixed with the resulting silica sol, facilitating encapsulation of cells in silica while minimizing cell contact with the cytotoxic products of silica generating reactions (i.e. methanol), and reduce exposure of cells to compressive stresses induced from silica condensation reactions. Using SG-CVIL, Saccharomyces cerevisiae (S. cerevisiae) engineered with an inducible beta galactosidase system were encapsulated in silica solids and remained both viable and responsive 29 days post encapsulation. By tuning SG-CViL parameters thin layer silica deposition on mammalian HeLa and U87 human cancer cells was also achieved. The ability to encapsulate various cell types in either a multi cell (S. cerevisiae) or a thin layer (HeLa and U87 cells) fashion shows the promise of SG-CViL as an encapsulation strategy for generating cell-silica constructs with diverse functions for incorporation into devices for sensing, bioelectronics, biocatalysis, and biofuel applications.
Introduction
In nature, living cells perform a variety of complex sensing, catalytic, and conversion functions which make them attractive targets for use in a variety of technological applications ranging from sensing,1-3 to biocatalysis,4-6 to atrazine remediation.7 However, environmental conditions (humidity, pH, temperature, nutrient availability) needed by cells to maintain optimal structure and function,8 require strategies for engineering bio-nano interfaces which facilitate cellular integration into devices while maintaining cell function. In order to generate such bio-nano interfaces, researchers have encapsulated cells in inorganic, biocompatible matrices which allow cells to interact with the ex-vivo environment while protecting them from chemical, thermal, and evaporative stresses.9-11 Among the most promising of these approaches are silica matrices prepared through the sol-gel process.9, 12-17
Carturan et. al. pioneered encapsulation of cells in silica by using the sol-gel process to encapsulate genetically engineered Saccharomyces cerevisiae (S. cerevisiae) cells in tetraethyl orthosilicate (TEOS)-based gels.18 In the sol-gel process, an alkoxysilane precursor is hydrolyzed by water, resulting in silanol functional groups which condense to form a silica containing sol. Cells are mixed with this sol which is then aged, leading to formation of a silica gel that encapsulates the cells. Building on the work of Carturan et. al., many groups have demonstrated encapsulation of whole cells using a variety of alkoxysilane precursors.19, 20 While sol-gel derived gels offer great advantages in cell encapsulation, traditional sol-gel routes often require the use of harsh solvents for sol-gel precursors and release harmful reaction byproducts, such as alcohols, which are cytotoxic.21 Researchers have attempted to make encapsulation procedures less cytotoxic by incorporating ameliorants (such as glycerol),22 and using silica precursors with less cytotoxic by- products.23 These methods have shown increased cell viability for encapsulated bacteria and yeast, but more effective encapsulation strategies that can both minimize cell-byproduct contact and reduce compressive stresses associated with condensation reactions are essential to facilitate encapsulation of mammalian cells which have highly desirable biological properties (e.g.: the CANARY cell-based sensing line24) but are inherently more fragile than microorganisms.
In an effort to minimize cell contact with silica precursors and reaction by-products, researchers have employed alcohol-free and vapor deposition approaches. In the alcohol-free aqueous approach, developed by Ferrer et. al.,25 the alcohol released due to TEOS hydrolysis is removed by rotovapor methods. This resulted in an alcohol-free silica sol that was used to encapsulate horseradish peroxidase enzyme while preserving the enzyme's structure. While this approach eliminates alcohol, the tunability of reaction parameters, and therefore silica sol properties, is limited to the initial silica to water ratio, reaction pH, and sol stock dilution. In the vapor deposition approach developed by Carturan et. al.,19 an alkoxysilane gas was flowed over pancreatic islets cells. This approach resulted in silica encapsulation of cells while limiting cell contact with precursors and reaction products due to the transport of these constituents away from cells via the gas flow. While a definite advance in encapsulation methodology, this vapor deposition approach is technically challenging, requires cells to be attached to a scaffold material, and does not ameliorate compressive stresses associated with silica condensation.
Gupta et. al. developed a vapor deposition approach whereby an open chamber containing tetramethyl orthosilicate (TMOS) and a separate open chamber containing a buffered cell suspension are both sealed within a larger third chamber. 26 Within this larger chamber the TMOS vaporizes, forming a concentration gradient that results in deposition of TMOS at the vapor-liquid interface of the cell suspension. Subsequent hydrolysis and condensation of TMOS forms silica particles which deposit onto the suspended cells. The benefits of this process versus the vapor deposition approach of Carturan et. al. are technical simplicity, the ability to coat the entire cell surface in silica, and the minimization of cell contact with silica precursors and toxic byproducts. Using this approach, researchers have achieved whole cell encapsulation of bacteria for development of microbial fuel cells;27 however, to our knowledge this technique has not been used with eukaryotic or mammalian cells demonstrating silica encapsulation with extended viability and retained functionality.
We look to extend the utility of this technique to encapsulate eukaryotic and mammalian (human) cells in silica for generating living hybrid biomaterials capable of performing biological functions. Here we report research using two approaches. In the first approach, termed in-situ Chemical Vapor into Liquid deposition (in-situ CViL), silica generation and encapsulation are achieved using the vapor deposition approach described above, with cells present in the buffer solution during the initial deposition of TMOS vapor, while controlling temperature and agitation. In the second approach, termed sol-generating CViL (SG-CViL), silica is generated using the same vapor deposition approach but is then allowed to age. Cells are mixed with the resulting aged silica solution to further limit cell contact with deleterious reaction constituents, reduce cell exposure to monomeric precursors, and alleviate compressive stresses that result from silica condensation and polymerization. The ability to tune SG-CViL parameters such as reaction temperature, agitation, deposition and aging time potentially offers a greater degree of control of silica sol characteristics, such as silica particle size, than other alcohol free methods. Using these two approaches, S. cerevisiae engineered to express beta-galactosidase in response to a model analyte, as well as human HeLa and U87 cancer cell lines, are encapsulated in silica. The resulting silica coat properties, cell viability and cell responsiveness are determined and demonstrate the suitability of these approaches for silica encapsulation for generating hybrid biomaterials capable of robust biosensing, biocatalytic, bioelectronic and bioremediation functions.
Experimental Methods
Yeast and mammalian cell culture
S. cerevisiae (yeast cells)
Cultures were generated by inoculating 20 mL of YPD rich media (10 g/L yeast extract, 20 g/L peptone, and 20 g/L dextrose) with S. cerevisiae strain YM 2061 (a kind gift from Jim Dover and Mark Johnston, University of Colorado) and incubation in a rotary shaker overnight (18 to 24 hours) at 30°C. The next day the culture (O.D.600 between 0.200 and 0.300) was well mixed and separated into 5 mL aliquots which were pelleted and used for encapsulation. HeLa (human cervical cancer) and U87 (human glioblastoma cancer) cell lines were cultured at 37°C, 5% CO2 atmosphere in T-25 flasks in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (v/v).
In situ Chemical Vapor into Liquid (in-situ CViL) deposition
Tetramethyl orthosilicate (TMOS) was placed into a 1.2 cm diameter glass dish (TMOS chamber), 2 mL S. cerevisiae in PBS (1X phosphate buffered saline, pH 7.8 to 8.0) was placed in a separate 6.5 cm diameter Pyrex petri dish (sample chamber). Both were placed in a 10 cm diameter glass petri dish (CViL chamber) that was sealed with parafilm. Once sealed, the CViL chamber was placed in a shaking incubator (160 RPM) and TMOS vaporization and subsequent hydrolysis and condensation of TMOS in the sample chamber was allowed to proceed for 1 hour at 30°C or 40°C. Afterwards, the CViL chamber was unsealed, the lid removed, and 1 mL of fresh PBS was added to the cell suspension (3 mL final volume). Cell suspensions were then pelleted and cells were washed 3 times with PBS. Cells were finally resuspended in 1 mL PBS and stored at room temperature.
Sol Generating Chemical Vapor into Liquid (SG-CVIL) deposition
S. cerevisiae (yeast cells) The SG-CViL protocol was identical to in-situ CViL, except that cells were not present in the 2 mL PBS in the sample chamber during TMOS vaporization and deposition. Following the addition of 1 mL of PBS to the sample chamber, the resultant silica sol solution was transferred to a 15 mL conical tube and allowed to age for 15 min at room temperature open to the atmosphere. This last step allows methanol by-products to evaporate, and hydrolysis and condensation reactions to proceed, reducing exposure of cells to monomeric silica and compressive stresses resulting from silica polymerization, both of which are cytotoxic. S. cerevisiae cells were pelleted and then resuspended in the 3 mL CViL solution for 20 min at room temperature and the resulting encapsulated cells were pelleted, washed twice with PBS, and stored in PBS at room temperature. Samples coated in this manner are referred to as SG-CViL-40°C for samples in which the sol solution was generated at 40°C.
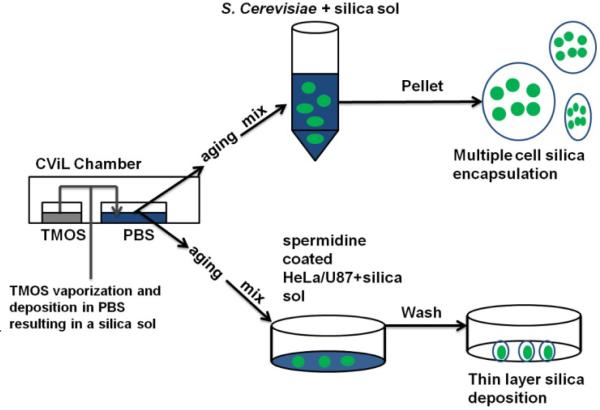
HeLa and U87 cells from approximately 80% confluent cultures were trypsinized and diluted 1:4 (final concentration approximately 300,000 cells/mL) using culture media containing 10% fetal bovine serum. 3 mL of cells were then added to tissue culture dishes (60X15 mm; TRP) and incubated for 18 to 24 hours at culture conditions. The next day cells were stained with the DNA binding dye 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) (10 μM; Sigma) for 30 min, washed twice with PBS and incubated with PBS containing 200 μM spermidine (Sigma) for 5 min at 30°C in a shaking incubator. SG-CViL was performed as described above with the following adaptations: the PBS used in the sample chamber was at the physiologically relevant pH for mammalian cells, pH 7.4. Also, TMOS vaporization and deposition was allowed to proceed for 15 min at 40°C. Post reaction, 1 mL PBS was added to the sample chamber and the sol was aged at 40°C for 15 min in the shaking incubator in open atmosphere. Postaging, spermidine coated HeLa or U87 cells were incubated with 2 mL of SG-CViL solution (+TMOS) or PBS (-TMOS, negative control) for 10 min at 40°C on a shaking incubator in a fume hood. Cells were then washed three times with 1 mL PBS and processed for imaging. A general schematic of the SGCViL process is presented in Fig. 1.
Fig. 1.
Schematic of sol generating chemical vapor into liquid (SG-CViL) silica deposition.
Scanning Electron Microscopy (SEM) and Elemental analysis
10 μL of cell samples (PBS control, SG-CViL-40°C) were pipetted onto 0.1 micron plain nylon membranes (Osmonics magna) and dried overnight in a sterile hood. The next day, membranes were mounted onto SEM pin mounts using carbon tape. Copper tape was fixed to samples to act as a ground, and samples were platinum coated for 1.5 to 2.5 minutes to reduce charging. Samples were imaged at 25 keV with a 50 μA condenser lens setting and 15 mm working distance.
Dynamic light scattering measurements
To examine silica particle growth during aging, dynamic light scattering (DLS) measurements were conducted using a Malvern Zetasizer DLS instrument. SG-CViL was performed using PBS at pH 8 to examine particle growth in pH conditions experienced by S. cerevisiae during encapsulation. TMOS deposition was allowed to proceed for 30 min at 40°C instead of 1 hour. This change was made because the size of silica particles generated at 1 hour TMOS deposition time were too large to fully resolve given the dynamic range available on the Malvern Zetasizer DLS. Post-TMOS deposition, the SG-CViL silica sol was diluted with 1 mL fresh PBS and the average particle size of silica in the diluted SG-CViL silica sol measured. Post measurement, the SG-CViL silica sol was placed in a 15 mL conical tube, aged for 15 min at room temperature, and the average particle size measured again. Results are averages from four independent experiments using student's t-test.
Fluorescence-based silica detection
Silica deposition was also monitored via fluorescence microscopy. S. cerevisiae, cells from overnight culture aliquots (see above) were pelleted and resuspended in a solution of the cell wall staining dye calcofluor white (25 uM calcofluor white, 10 mM HEPES, 2% glucose) for 30 to 45 min. To fluorescently label silica, either 200 μM of 2-(4-pyridyl)-5{[4-(2-dimethylaminoethylaminocarbamoyl)-methoxy]phenyoxazole (PDMPO) or 10 μM Rhodamine B, was placed in the sample chamber before initiation of the SG-CViL process. Fluorescently-labeled silica solution was then mixed with calcofluor-white stained cells. The resulting cell-silica constructs were imaged using a Nikon Eclipse LV100 fluorescence microscope equipped with a Hamamatsu ORCARF camera and Nikon Plan Fluor 10X and 60X objectives using appropriate filters.
Viability analysis
To determine viability of cells, 20 μL of PBS stored cells, SGCViL-40°C samples, as well as live and heat-killed (90°C treatment for 5 min) controls, were pelleted, resuspended in 200 μL of Fungalight viability stain (carboxyfluorescein diacetate (CFDA) and propidium iodide (PI), Invitrogen), and incubated at 30°C for 30 to 45 minutes. 10 to 15 μL of sample was then mounted on a glass microscope slide and imaged.
Beta galactosidase activity
To induce beta-galactosidase production, 20 μL of sample (dead cells, PBS control, SG-CViL-30°C, SG-CViL-40°C) was added to either 1 mL PBS or 1 mL induction media (YP +2% galactose). Cells were incubated for 6 hours (the minimum induction time needed for detectable beta-galactosidase activity, Fig. S1) and stored at 4°C until analysis. Activity analysis was conducted by pelleting cells, resuspending in 1 mL Z buffer (60 mM Na2HPO4*7H2O, 40mM NaH2PO4*H2O, 10mM KCl, 1mM MgSO4*7H2O, 50 mM 2-mercaptoethanol), and then adding 25 μL choloroform, 20 μL of 0.1% SDS, and vortexing samples approx. 10 seconds. Samples were then transferred into a 96 well plate, (6 wells/sample; 160 uL/well) and pre-incubated for 5 min at 30°C. Post- incubation, 20 μL of 10 mM substrate, 4-Methylumbelliferyl-β β β β-D-galactopyranoside (MUGal, Sigma), was added to three wells for each sample (3 wells with substrate, three without, per sample) and cells were incubated for 15 min at 30°C. Thereafter, fluorescence was measured using a Wallac Victor 2 fluorescence plate reader. Data are presented as percent normalized arbitrary fluorescent units calculated by dividing the fluorescent signal of analyte exposed cells by the fluorescent signal of non-exposed cells for each condition and normalizing to values for dead (negative control) cells. Results are averages from three independent experiments using student's t-test.
Optical activity measurement
Beta-galactosidase production was induced in S. cerevisiae 24 hours post SG-CViL, with cells being exposed to analyte for 24 hours. Cells were processed for beta-galactosidase activity as described above with 160 μL of cells being added to wells of a 96 well plate. MUGal (20 μL of 10 mM) was then added to one set of wells (+) while a second set received no substrate (−). Cells were then incubated for 15 min at 30°C, exposed to UV light using a transilluminator, and photographed using an EL GEL LOGIC 100 imaging system.
Rhodamine B and APTEOS-FITC silica labeling
To visualize silica deposition on HeLa and U87 cells, Rhodamine B was used in the same manner as in yeast experiments. Additionally, a fluorescently labeled silica alkoxide construct (APTEOS-FITC) was employed.28, 29 Post incubation of cells with SG-CViL silica sol, 5 μL/mL APTEOS-FITC was added to HeLa or U87 cells (final concentration 130 nM) and incubated at culture conditions for 20 to 30 min. Cells were then washed with PBS and imaged.
Results and Discussion
In-situ CViL
In determining the suitability of the CViL process for cellular silica encapsulation, the first approach focused on performing CViL with cells present in the aqueous medium in the sample chamber where TMOS vapor is deposited (in-situ CViL). Under these conditions, hydrolysis of TMOS molecules occurs at the vapor/water interface, followed by condensation of hydrolyzed TMOS to form silica particles and polymeric silica. These particles can then interact with positively charged groups on the cell surface, resulting in silica deposition that encapsulates the cell.
S. cerevisiae cells were subjected to in-situ CViL and stored for 4 days at room temperature in 1.5 mL eppendorf tubes. Cellular activity was then analyzed by interrogating the ability to respond to galactose with de novo production of the enzyme beta-galactosidase. Production of beta-galactosidase is achieved via complex transcription and translation mechanisms which require maintenance of several biochemical pathways on the part of cells. Cells which underwent in-situ CViL showed beta-galacotosidase activity (i.e. responsiveness, active metabolism and ability to synthesize protein) similar to that of dead cells (heat shocked, Fig. S2.), indicating the cells are non-responsive or not viable. The damage to cells upon encapsulation using in-situ CViL may be due to the release of methanol during TMOS hydrolysis. While in-situ CViL significantly reduces cell contact with methanol by allowing evaporation of methanol at the vapor/water interface, methanol generated near cells in the bulk sample solution may damage cells before evaporating. Also, silica particles that are not fully hydrolyzed may deposit on cells and continue to undergo hydrolysis and condensation reactions, leading to localized methanol release at the cell death. su face, destabilizing cell membranes, resulting in cell death. Further, monomeric silica species may pass through the cell surface and deposit within the cells, damaging cellular components and disrupting metabolic processes.17 Finally, as silica species continue to condense in the presence of cells, significant compressive stresses are exerted which can damage cells and result in viability loss. 22 While methanol toxicity, monomeric silica species, and compressive stresses are all possible factors leading to loss of cellular responsiveness, decoupling the impact of each is difficult, necessitating modification of the in-situ technique to reduce the cytotoxic effects of all three factors.
With this finding, the in-situ CViL technique was modified such that silica is first generated using the in-situ CViL approach, but in the absence of cells in the aqueous PBS in the sample chamber. Cells were then mixed with the silica solution following dilution with PBS and a 15 min sol aging step, further reducing cell contact with methanol monomeric and condensing silica species (see Fig. 1). This new approach is termed Sol-Generating Chemical Vapor into Liquid (SG-CViL) deposition.
SG-CViL for multiple cell silica encapsulation
SEM-EDS Si characterization
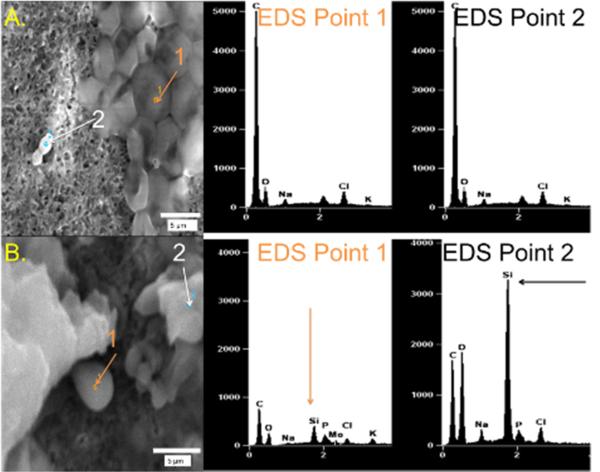
To examine silica generated via SG-CViL, SEM and EDS were used. Fig. 2 contains representative SEM micrographs of S. cerevisiae treated with SG-CViL silica sol solution generated at 40°C (SG-CViL-40oC) or PBS control solution (PBS control). EDS analysis of point 1 (cells) and point 2 (background) in PBS control samples shows no Si peak, whereas analysis of points 1 and 2 in SG-CViL-40°C shows distinct Si peaks. The presence of these Si peaks indicates that vaporized TMOS indeed deposited into PBS and underwent hydrolysis and condensation, generating silica species that associated with the cells. Similar presence of Si peaks was observed with E. coli cells subjected to SG-CViL (Fig. S3). While SEM-EDS shows silica is present, determining cell-silica association from micrographs is difficult.
Fig. 2.
SEM-EDS of SG-CViL treated S. cerevisiae with (A.) PBS only (negative control) and with (B.) TMOS (SG-CViL-40°C). EDS spectra from samples were obtained from two different points: EDS point 1 (orange arrow) in each image corresponds to cells; point 2 (black arrows) corresponds to extracellular material.
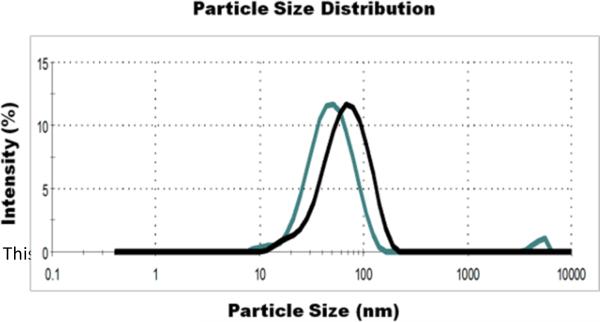
Particle size analysis of silica sol pre- and post-aging
To determine the effect of the aging step on the average particle size of silica particles generated using SG-CViL, dynamic light scattering (DLS) measurements were performed on the SG- CViL sol (30 min TMOS deposition time at 40°C) just after dilution of the silica sol with 1 mL PBS (pre-aging) and after 15 min aging of the diluted silica sol in a 15 mL conical tube at room temperature (post-aging). Fig. 3 is a representative graph of the size distribution of silica particles in the SG-CViL sol pre- and post- sol aging. Pre-aging, the average particle size of the silica sol is approximately 46.5 ± 1.4 nm. When measured post-aging, the average particle size of the silica sol is approximately 57.4 ± 1.1 nm, an increase, on average, of 10.9 nm (p < 0.05, N=4). The increase in particle size indicates that hydrolysis and condensation reactions are occurring rapidly, leading to polymerized silica species. This polymerization step is important due to the fact that, upon hydrolysis of an alkoxysilane, monomeric silica species of less than 2 nm in size are generated.30, 31 When cells are present in PBS during TMOS deposition, as in in-situ CViL, cells encounter these small silica particles, which can then diffuse through the cell membrane, disrupting cellular compartments and metabolic function, thus leading to loss of responsiveness (see Fig. S2). Using SG-CViL, larger silica polymers/particles are generated that cannot diffuse across the cell membrane and damage cells. Additionally, aging the sol allows for more complete condensation of silica and evaporation of methanol released via silica condensation, reducing potential compressive stresses and alcohol toxicity on cells, greatly improving the biocompatibility of the encapsulation process.
Fig. 3.
Representative Dynamic Light Scattering (DLS) measurements of SG-CViL silica sol pre- and post-aging. SG-CViL was performed for 30 min at 40°C. The particle size of the resulting silica sol was measured immediately after dilution with fresh PBS (pre-aging, green line) and after a 15 min aging period (post-aging, black line).
Silica architecture characterization via fluorescent silica labeling
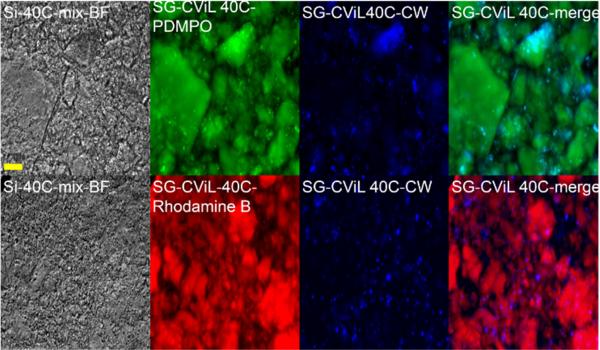
To more fully characterize the interface between cells and the silica deposited from SG-CViL conducted with a 1 hour TMOS deposition time, fluorescence microscopy using silica labeling dyes was employed. Fig. 4 contains representative images of silica encapsulated YM 2061 S. cerevisiae. Silica was labeled with one of two silica labeling dyes;2-(4-pyridyl)-5{[4-(2-dimethylaminoethylaminocarbamoyl)-methoxy]phenyl}oxazole (PDMPO, green emission), which has been previously reported to track silica generation in diatoms, 32 and Rhodamine B (red emission), previously used in labeling silica nanoparticles for biological tracking applications.33, 34 S. cerevisiae cells are labeled with the chitin-specific dye calcofluor white (blue emission). As shown in Fig. 4, green and red fluorescence images show that the size of silica constructs is highly variable, with some being on the order of 100 μM, and others being on the order of a millimeter. The variability of silica-yeast construct size range is attributed to the distribution and settling of in the silica sol during yeast incubation influencing the interaction of growing and polymerizing silica species. However, the exact mechanism for this influence is unclear and is the subject of future work. Images also show that some silica species have defined angular structure, while others are more rounded. The lack of distinguishable structural order of silica species in Fig. 4, along with x-ray diffraction data (data not shown), indicates that overall, SG-CViL conducted at 1 hour TMOS deposition time does not lead to bulk gelation as in other systems,13, 15 but generates silica species with amorphous structure.
Fig. 4.
SG-CViL treated YM 2061 S. cerevisiae cells. Silica stained with the silica specific dyes PDMPO (green) or Rhodamine B (red). Cells were stained with calcofluor white (blue). All images acquired using same magnification. Scale bar = 100 microns.
Merged fluorescence images in Fig. 4 reveal that the punctate blue fluorescence (yeast cells) associates closely with green or red fluorescence, suggesting the yeast cells associate with labeled silica species. Additionally, not all yeast cells are in focus at the same focal depth (data not shown), indicating that yeast are encapsulated throughout the silica matrix. S. cerevisiae cells appear to be unevenly distributed in the matrix, with some silica species containing few cells and others containing large cell clusters (see concentrated blue fluorescence in PDMPO-CW merge image), suggesting that encapsulation of cells is achieved via large scale physical interactions of silica and cells.
Viability analysis of silica encapsulated S. cerevisiae
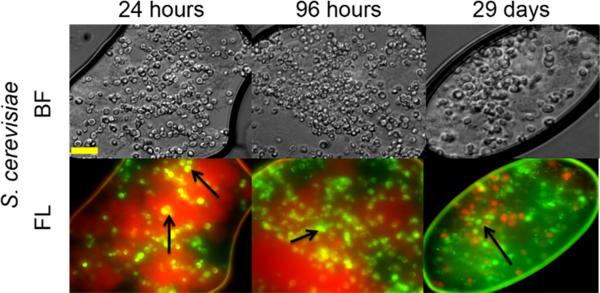
Viability of SG-CViL-40°C samples was determined using fluorescent microscopy and FungaLight live/dead stain (Invitrogen). This assay utilizes two fluorescent dyes, carboxyfluorescein diacetate (CFDA), a membrane permeable esterase substrate which fluoresces green when cleaved by esterases, and propidium iodide, a membrane impermeable DNA intercalating dye which fluoresces red when bound to DNA. Using this assay, green fluorescence indicates cells with esterase activity and membrane integrity; these were counted as viable. Yellow fluorescence indicates cells with esterase activity and compromised cell membranes, and red indicates cells with no esterase activity and compromised membranes; neither of these was counted as viable. Representative fluorescence viability images from silica encapsulated S. cerevisiae cells over time are presented in Fig. 5. Analysis of images indicates 85 ± 2% of encapsulated S. cerevisiae cells are viable 24 hours post encapsulation, 75 ± 10% are viable 96 hours post encapsulation, and 60 ± 15% are viable 29 days post encapsulation. At 29 days post encapsulation, cells routinely appear to be more clustered than cells 24 and 96 hours post encapsulation. This clustering may be due to continued silica condensation, shifting and compacting cells as the system evolves to equilibrium. This appears to lead to a decrease in viability over time as indicated by the 25% reduction in viability for cells analyzed 29 days post encapsulation versus cells analyzed 24 hours post encapsulation. Interestingly, 57 ± 10% of E. coli encapsulated using SG-CViL (Fig. S4), are viable 24 hours post encapsulation and are 47 ± 7% viable 29 days post encapsulation. The greater viability for S. cerevisiae versus E. coli (85% vs. 57%) 24 hours post encapsulation could be due to a small concentration of residual methanol that remains in the silica sol despite the aging step. S. cerevisiae, which are intrinsically resistant to alcohols, may be able to tolerate this small amount of methanol better than E. coli, leading to the increased viability of S. cerevisiae 24 hours post encapsulation. This finding indicates that while SG-CViL can be used to encapsulate various cell types, initial reaction conditions need to be optimized for each cell type in order to achieve the highest biocompatibility. Excitingly, the ability of both encapsulated S. cerevisiae and E. coli to maintain approximately 50% viability 29 days post encapsulation shows that despite initial encapsulation stresses, SG-CViL leads to cell-silica constructs which maintain long term cell viability regardless of the cell type encapsulated.
Fig. 5.
Viability of Si matrix encapsulated S. cerevisiae cells (SG-CViL-40°C). 24 hours, 96 hours, and 29 days post encapsulation using CFDA/PI staining. Cells emitting green fluorescence are considered viable; yellow and red emitting cells are considered non-viable. Cells were stained and images taken 24 hours, 96 hours, and 29 days post SG-CViL. Images show that S. cerevisiae cells (black arrows) 60% viability when encapsulated and stored for 29 days. All images taken at same magnification. Scale bar = 20 microns.
Responsiveness of silica encapsulated S. cerevisiae
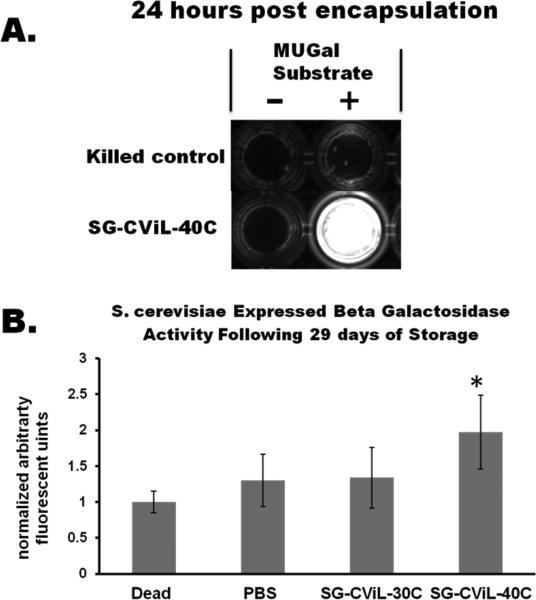
To further characterize the state of silica encapsulated S. cerevisiae cells, beyond membrane integrity and enzymatic activity, we probed the ability of encapsulated cells to respond to the introduction of a small molecule that induces expression of an enzyme. Yeast strain (YM2061), contains a GAL 1 promoter fused to a lacZ gene integrated within the genome. This fusion gene enables yeast to express the enzyme beta-galactosidase (beta-gal) when exposed to the inducer molecule, galactose.35 Using the beta-gal substrate, 4-Methylumbelliferyl-ββββ-D-galactopyranoside (MUGal), beta-galactosidase activity can be measured as MUGal releases a fluorescent molecule when cleaved by beta-gal, giving information on the cells ability to recognize and respond to a model analyteThe ability of silica matrix encapsulated S. cerevisiae to produce active enzyme in response to the presence of a small molecule inducer is presented in Fig. 6. YM 2061 cells (heat killed negative control and SG-Si-40°C encapsulated cells, 24 hours post encapsulation) were exposed to galactose for 24 hours, incubated in MUGal, and imaged over a UV-trans illuminator (Fig. 6A). Encapsulated cells exposed to the inducer produce beta-galactosidase which cleaves the MUGal substrate, producing an intensely fluorescent product. Such a strong response provides evidence that encapsulated cells remain viable and capable of inducible transcription and translation, and can thus be exploited for a multitude of applications including biosensing, biocatalysis, and bioelectronics with an easily visible output signal based upon the presence of multiple components. While the signal obtained here is fluorescence based, this system could be adapted by using cells engineered to produce colorimetric or luminescent signals in response to analyte presence, facilitating development of biosensors with highly tailored functionalities.
Fig. 6.
Optical and quantitative response of silica encapsulated S. cerevisiae to the model analyte galactose. (A) Representative image showing the fluorescent output of cell-silica constructs. Cells were exposed to galactose inducer for 24 hours, incubated with (+) or without (−) MUGal substrate and then exposed to to UV light. (B) Quantification of fluorescence obtained by exposing silica encapsulated S. cerevisiae to galactose following 29 days of storage (23°C) and normalized to the fluorescence measured from heat killed cells. Activity of cells encapsulated using SG-CViL (SG-CViL-40°C) is approximately 100% greater than that of dead cells (p<0.05, *), and approximately 50% greater than that of cells stored in PBS.
The beta-gal activity based-fluorescent signal from cells exposed to various treatments and stored for 29 days at room temperature are shown in Fig. 6B. Values for each treatment were normalized to the fluorescence measured from negative control cells (heat killed). Cells were exposed to analyte for 6 hours (the minimum time needed for detectable response using this construct, Fig. S1). 29 days post encapsulation, at 6 hours of analyte exposure, activity of YM 2061 cells encapsulated using SG-CViL silica sol generated at 40°C (SG-CViL-40°C) is approximately 100% greater than heat killed cells (p<0.05), and approximately 50% greater than PBS control (p=0.08). The statistically significant increase in responsiveness for SG-CViL-40°C versus dead cells 29 days post encapsulation represents a marked improvement over in-situ CViL where cells showed no responsiveness 4 days post encapsulation, indicating SG-CViL is a far more biocompatible process for achieving cellular silica encapsulation
The general trend of higher responsiveness of SG-CViL-40°C encapsulated cells stored in PBS compared to unencapsulated cells stored under identical conditions suggests encapsulation results in a more stable and responsive biological state. This state may result from a higher percentage of encapsulated cells than unencapsulated cells being able to undergo the shift from high proliferation and high metabolic activity to quiescence, a maintenance like resting state where cells can remain viable for extended periods even without nutrients.36 While the exact mechanism for this is not clear, it could be related to how unencapsulated cells and encapsulated cells respond to the environmental factors associated with encapsulation and storage.
In our system, exponential phase yeast go immediately from nutrient rich YPD to nutrient-devoid PBS, giving cells neither the time nor the environmental nutrients required to make the complex metabolic shifts necessary to enter quiescence. Unencapsulated PBS control cells have responsiveness very similar to killed controls as they are unable to make this metabolic shift under these storage conditions and are unable to enter quiescence, resulting in loss of viability for the majority of cells. In contrast, a higher percentage of SG-CViL-40°C-encapsulated cells could be achieving quiescence as a result of activation of pathways associated with recognition of spatial confinement. For example, the pkc1p-MAP kinase pathway responds to cell surface stresses and plays a role in the transition to quiescence.37-40 Cells encapsulated using silica that was generated at 30°C show no difference in responsiveness from unencapsulated controls, and lower responsiveness when compared to SG-CViL-40°C (Fig. 6B). The difference in responsiveness level between SG-CViL-30°C and SG-CViL-40°C is likely due to higher rates of hydrolysis and condensation at higher temperature leading to a higher percentage of fully condensed silica groups (Si-O-Si) versus silanol (Si-OH) groups. A higher silanol group presence in silica generated at 30°C would detrimentally affect cell viability (as silanol groups are cytotoxic19, 22) though Si29 NMR studies are needed to confirm this rational. Also, cells encapsulated using silica generated at 30°C may not be as fully confined as cells encapsulated using silica generated at 40°C. Smaller particles and silica cross-linked to a lesser extent may associate with cells, but not fully confine them. If cellular recognition of spatial confinement is important in the transition of cells to a quiescent-like state, this lack of full confinement may result in a loss of viability.
SG-CViL encapsulation of mammalian cells
Results using SG-CViL to encapsulate S. cerevisiae show that SG-CViL is suitable for encapsulation of multiple cell types in large silica species. While these constructs may prove effective as long term environmental sensors, their utility as rapid detection systems (i.e. for chemical weapons agents) is limited due to potential analyte adsorption to the encapsulation matrix, as well as hindered rates of diffusion of analytes through the matrix to cells. In order to limit adsorption and diffusion complications, deposition of thin layers of silica on cells is preferable. Thin layer silica deposition in S cerevisiae has been achieved using approaches such as evaporation induced self-assembly (EISA)12 and LbL biosilicification.41 While these approaches result in thin layer silica deposition on yeast, the approaches require use of lipid templated silica films (EISA) or reaction conditions (LbL biosilicification) which are not amenable to achieving thin layer silica deposition on adherent mammalian cells. While mammalian cells offer more sophisticated biological functions for use in bio-hybrid systems, their fragility compared to microbes necessitates development of encapsulation techniques which achieve thin layer silica deposition but are milder and more conformal than those used for microbial encapsulation.
Initial experiments to achieve thin layer deposition using SG-CViL were conducted by directly exposing HeLa cells to SG-CViL silica sol generated at 40°C with 15 min TMOS deposition time. TMOS deposition time was reduced from 1 hour to 15 min to generate smaller silica species than those used to encapsulate S. cerevisiae to facilitate deposition of a thinner, more conformal silica coat on the HeLa surface. Similarly, HeLa cells were incubated in SG-CViL solution for 10 min instead of 20 min in order to reduce silica deposition time to achieve thin-layer silica deposition. When HeLa cells were exposed to SG-CViL silica sol, silica deposition and coverage was very low and uneven (Fig. S5). This uneven coating observed in HeLa (Fig. S5) is in contrast to work by Jaroch et. al.,42 who encapsulated murine and human islet cells in silica without a polycation treatment. This is likely due to the higher concentration of silica utilized by Jaroch et. al. (1:16 silica: water mol ratio) compared to that used in SG-CViL (1:1000 silica: water mol ratio). The higher silica concentration results in increased silica condensation and polymerization, leading to film formation. Given these factors, we investigated coating cells in a biocompatible natural polyamine to achieve a more robust thin layer silica deposition.
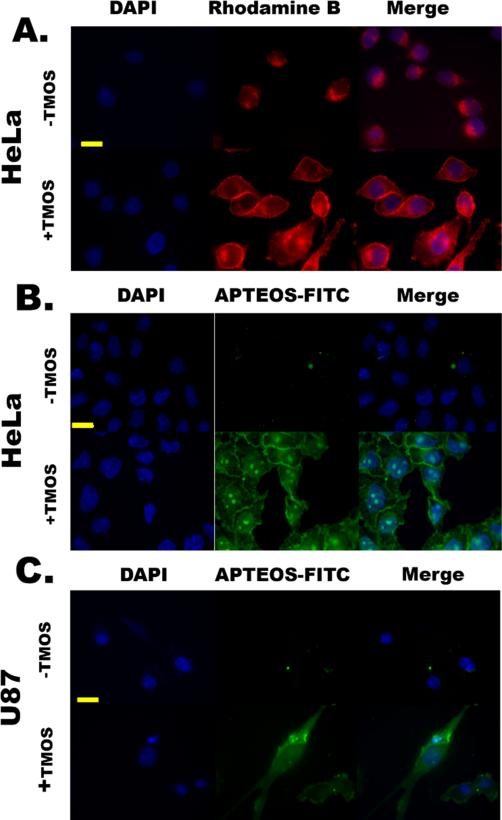
Polyamines play a crucial role in silica formation in diatoms 43 and the presence of polyamines in silica generating systems results in more fully condensed silica.44 Thus, experiments were conducted where cells were first coated with spermidine (a naturally synthesized polyamine) and then exposed to SGCViL. When HeLa cells coated with spermidine were exposed to SG-CViL solution containing Rhodamine B (+TMOS, Fig. 7A), a highly conformal red fluorescence was clearly observable on the exterior of cells. Spermidine likely facilitates this deposition through charge-charge interactions between the positively charged spermidine and negatively charged silica this signal is absent in cells exposed to PBS only containing Rhodamine B (-TMOS. Fig. 7A). To further confirm silica deposition on the cell surface the experiment was repeated utilizing a fluorescent construct, APTEOS-FITC generated by conjugating tetra-ethyl orthosilicate (TEOS) to fluorescein via an amide linker. The TEOS moiety of this construct forms a siloxane bond with silica, resulting in a green fluorescence emission where silica is present in the sample, reducing background fluorescence and improving image resolution and quality.28 When spermidine-coated HeLa underwent SG-CViL and were then exposed to APTEOS-FITC (+TMOS, Fig. 7B) a highly conformal green fluorescence was observed on the cell surface, with no green fluorescence visible on control cells (-TMOS Fig. 7B). These results are in good agreement with the results obtained using Rhodamine B (Fig. 7A). Furthermore, when compared to results in Fig. S5, where +TMOS cells are not coated with spermidine and show non-uniform coating, data in Fig. 7 suggest that spermidine facilitates deposition of silica species from the SG-CViL solution onto the cell surface. Additionally, when silica encapsulated HeLa are returned to culture, they lack visible signs of cell death (i.e. cell detachment) and maintain morphology similar to that of unencapsulated cells 3 days post encapsulation (Fig. S7), suggesting that cells remain viable post encapsulation. Interestingly, it appears that encapsulated HeLa are able to grow, as the cell confluency increases over 72 hours. In order to further demonstrate the benefits of spermidine treatment beyond the promotion of more even silica deposition highlighted in Fig. 7a, we performed an experiment to determine the membrane integrity and enzyme activity (Fig. S8) of cells encapsulated using a higher concentration silica sol with and without spermidine coating. Images show that HeLa encapsulated without a spermidine coating are PI positive, suggesting loss of membrane integrity whereas cells that are coated with spermidine and then encapsulated are PI negative. This indicates the presence of spermidine appears to increase biocompatibility of the encapsulation process by promoting membrane integrity, however the mechanism for this is unclear and bears further investigation. Eleftheriou et. al. recently reported preservation of cell growth and promoter regulation of E. coli in low concentration silica films and developed a novel antibiotic assay platform for utilizing these constructs.45 While growth of HeLa cells encapsulated using SG-CViL silica thin films remains to be quantified, the ability to encapsulate complex mammalian cells while maintaining growth and genetic regulation could lead to development of robust screening platforms for a host of small molecules.
Fig. 7.
Encapsulation of spermidine-coated HeLa (A & B) and U87 (C) cells using SG-CViL. All cells were stained with the DNA binding dye DAPI (blue fluorescence). (A.) HeLa cells mixed with SG-CViL solution (+TMOS) containing Rhodamine B (red fluorescence). (B.) HeLa cells exposed to SG-CViL and stained with the silica-binding construct, APTEOS-FITC (green fluorescence). (C.) U87 cells exposed to SG-CViL (+TMOS) and stained with APTEOS-FITC, also showing a highly conformal thin film staining pattern on the cell exterior that is absent in cells exposed to PBS (-TMOS). Images in each image set taken at same magnification. Scale bar = 20 microns.
Similar experiments were conducted using U87, a human brain cancer cell line, to determine if this technique could be extended to other human cell lines. Using APTEOS-FITC, a similar conformal thin film staining pattern was obtained for U87 cells exposed to SG-CViL (+TMOS Figure 8C), including the lack of cell associated green fluorescence in control cells (-TMOS, Fig. 7C). Using SG-CViL and spermidine, Jurkat cells (T-cell lymphoma) were also encapsulated in polymeric silica again templating around cells (Fig. S6). For Jurkat cells, the silica encapsulation was not as conformal as that seen for HeLa and U87 cells, indicating that this technique may require alterations depending on the particular characteristics of the target cell line to ensure fully conformal silica thin film deposition.
Results obtained using HeLa, U87, and Jurkat cells show the suitability of SG-CViL for tailored deposition of silica on various mammalian and human cell lines. While others have encapsulated HeLa cells in silica using a polycationic template,46 encapsulation was achieved on trypsinized cells using a synthetic polyamine. Trypsinization has been shown to alter cellular protein expression such that apoptosis regulating proteins are upregulated.47 Additionally, trypsinization removes cells from the surface, such as near an electrode or on an implant, on which cells may have been previously isolated and maintained. Using SG-CViL, adherent cells (i.e. HeLa and U87) are coated without the need for trypsinization, allowing cells to maintain native morphology, protein expression, and spatial location, all of which are critical components for cells to maintain desired biological functions. The achievement of thin deposition of silica onto the surface of eukaryotic cells opens up the possibility of encapsulating and stabilizing more fragile and relevant cell lines such as CANARY,24 which offer rapid and specific detection capability for biosensor devices. More broadly, the ability to deposit tailored silica on single cells and mechanically restrict their growth could provide a means of studying contact inhibition in mammalian cells. Contact inhibition results from restricted cell growth and its absence plays a major role in carcinogenesis.48 Developing tailorable single cell bio-nano interfaces which mimic the mechanical restriction of contact inhibition could provide a robust tool for researchers to examine contact inhibition- activated cell signaling pathways and their influence on a cells transition from a normal state to a cancerous state.
Conclusions
In conclusion, data presented in this report demonstrate the feasibility of using Sol-Generating Chemical Vapor into Liquid deposition for encapsulation of whole cells in a biocompatible silica matrix to preserve cellular viability, metabolic activity, responsiveness, and inducible gene expression. The SG-CViL process is easily tunable, providing facile control of sol generation, facilitating cellular encapsulation in either a multiple cell or thin layer manner. Overall, the ability to encapsulate varied eukaryotic cell types using SG-CViL offers the possibility to generate cell-silica hybrid materials with a range of functions for biosensing, biocatalysis, biofuels, bioremediation, and platforms for biomedical applications
Supplementary Material
Acknowledgements
The authors wish to thank Jim Dover and Mark Johnston (University of Colorado) for the YM 2061 yeast strain, Gary Chandler for help in SEM sample preparation and analysis, and Dr. Lillya Frolova and Cody Champion for assistance in synthesizing APTEOS-FITC. This project was supported by grants from the Sandia Lab Directed Research and Development (LDRD) Program (Campus Executive Program, 151375), the National Center for Research Resources (5P20RR016480-12), and the National Institute of General Medical Sciences (8 P20 GM103451-12) from the National Institutes of Health (NIH). Sandia National Laboratories is a multi-program laboratory managed and operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the U.S. Department of Energy's National Nuclear Security Administration under contract DE-AC04-94AL85000.
Footnotes
Electronic Supplementary Information (ESI) related to in-situ CViL, S. cerevisiae model system characterization, E. coli and Jurkat encapsulation available. See DOI: 10.1039/b000000x/
References
- 1.Gupta R, Chaudhury NK. Biosens. Bioelectron. 2007;22:2387–2399. doi: 10.1016/j.bios.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 2.Dave BC, Dunn B, Valentine JS, Zink JI. Anal. Chem. 1994;66:A1120–A1127. [Google Scholar]
- 3.Harper JC, Edwards TL, Savage T, Harbaugh S, Kelley Loughnane N, Stone MO, Brinker CJ, Brozik SM. small. 2012;8:2743–2751. doi: 10.1002/smll.201200343. [DOI] [PubMed] [Google Scholar]
- 4.Schrewe M, Julsing MK, Buhler B, Schmid A. Chemical Society Reviews. 2013;42:6346–6377. doi: 10.1039/c3cs60011d. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida A, Hama S, Tamadani N, Fukuda H, Kondo A. Biochemical Engineering Journal. 2012;63:76–80. [Google Scholar]
- 6.Liu C-Q, Deng L, Zhang P, Zhang S-R, Xu T, Wang F, Tan T-W. Journal of Molecular Catalysis B: Enzymatic. 2013;91:1–7. [Google Scholar]
- 7.Chelinho S, Moreira-Santos M, Lima D, Silva C, Viana P, André S, Lopes I, Ribeiro R, Fialho A, Viegas C, Sousa J. J Soils Sediments. 2010;10:568–578. [Google Scholar]
- 8.Murua A, Portero A, Orive G, Hernandez RM, de Castro M, Pedraz JL. J. Control. Release. 2008;132:76–83. doi: 10.1016/j.jconrel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Depagne C, Roux C, Coradin T. Anal. Bioanal. Chem. 2011;400:965–976. doi: 10.1007/s00216-010-4351-y. [DOI] [PubMed] [Google Scholar]
- 10.Depagne C, Masse S, Link T, Coradin T. J. Mater. Chem. 2012;22:12457–12460. [Google Scholar]
- 11.Amoura M, Nassif N, Roux C, Livage J, Coradin T. Chem Commun (Camb) 2007 doi: 10.1039/b711380c. DOI: 10.1039/b711380c, 4015-4017. [DOI] [PubMed] [Google Scholar]
- 12.Harper JC, Khirpin CY, Carnes EC, Ashley CE, Lopez DM, Savage T, Jones HDT, Davis RW, Nunez DE, Brinker LM, Kaehr B, Brozik SM, Brinker CJ. ACS Nano. 2010;4:5539–5550. doi: 10.1021/nn101793u. [DOI] [PubMed] [Google Scholar]
- 13.Harper JC, Lopez DM, Larkin EC, Economides MK, McIntyre SK, Alam TM, Tartis MS, Werner-Washburne M, Brinker CJ, Brozik SM, Wheeler DR. Chemistry of Materials. 2011;23:2555–2564. [Google Scholar]
- 14.Perullini M, Jobbagy M, Moretti MB, Garcia SC, Bilmes SA. Chemistry of Materials. 2008;20:3015–3021. [Google Scholar]
- 15.Perullini M, Jobbágy M, Soler-Illia GJAA, Bilmes SA. Chemistry of Materials. 2005;17:3806–3808. [Google Scholar]
- 16.Perullini M, Rivero MM, Jobbágy M, Mentaberry A, Bilmes SA. Journal of Biotechnology. 2007;127:542–548. doi: 10.1016/j.jbiotec.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Kaehr B, Townson JL, Kalinich RM, Awad YH, Swartzentruber BS, Dunphy DR, Brinker CJ. Proceedings of the National Academy of Sciences. 2012;109:17336–17341. doi: 10.1073/pnas.1205816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carturan G, Campostrini R, Dire S, Scardi V, Dealteriis E. Journal of Molecular Catalysis. 1989;57:L13–L16. [Google Scholar]
- 19.Carturan G, Dal Toso R, Boninsegna S, Dal Monte R. J. Mater. Chem. 2004;14:2087–2098. [Google Scholar]
- 20.Jaroch D, McLamore E, Zhang W, Shi J, Garland J, Banks MK, Porterfield DM, Rickus JL. Biotechnol. Bioeng. 2011;108:2249–2260. doi: 10.1002/bit.23195. [DOI] [PubMed] [Google Scholar]
- 21.Avnir D, Coradin T, Lev O, Livage J. J. Mater. Chem. 2006;16:1013–1030. [Google Scholar]
- 22.Nassif N, Roux C, Coradin T, Rager MN, Bouvet OMM, Livage J. J. Mater. Chem. 2003;13:203–208. [Google Scholar]
- 23.Gill I, Ballesteros A. Trends in Biotechnology. 2000;18:282–296. doi: 10.1016/s0167-7799(00)01457-8. [DOI] [PubMed] [Google Scholar]
- 24.Rider TH, Petrovick MS, Nargi FE, Harper JD, Schwoebel ED, Mathews RH, Blanchard DJ, Bortolin LT, Young AM, Chen JZ, Hollis MA. Science. 2003;301:213–215. doi: 10.1126/science.1084920. [DOI] [PubMed] [Google Scholar]
- 25.Ferrer ML, del Monte F, Levy D. Chemistry of Materials. 2002;14:3619–3621. [Google Scholar]
- 26.Gupta G, Rathod SB, Staggs KW, Ista LK, Abbou Oucherif K, Atanassov PB, Tartis MS, Montano GA, Lopez GP. Langmuir. 2009;25:13322–13327. doi: 10.1021/la903475d. [DOI] [PubMed] [Google Scholar]
- 27.Luckarift HR, Sizemore SR, Roy J, Lau C, Gupta G, Atanassov P, Johnson GR. Chem Commun (Camb) 2010;46:6048–6050. doi: 10.1039/c0cc01255f. [DOI] [PubMed] [Google Scholar]
- 28.Doussineau T, Smaïhi M, Mohr GJ. Advanced Functional Materials. 2009;19:117–122. [Google Scholar]
- 29.Van Blaaderen A, Vrij A. Langmuir. 1992;8:2921–2931. [Google Scholar]
- 30.Iler RK. The chemistry of silica: solubility, polymerization, colloid and surface properties, and biochemistry. 1979.
- 31.Carcouët CCMC, van de Put MWP, Mezari B, Magusin PCMM, Laven J, Bomans PHH, Friedrich H, Esteves ACC, Sommerdijk NAJM, van Benthem RATM, de With G. Nano Letters. 2014;14:1433–1438. doi: 10.1021/nl404550d. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu K, Del Amo Y, Brzezinski MA, Stucky GD, Morse DE. Chemistry & Biology. 2001;8:1051–1060. doi: 10.1016/s1074-5521(01)00072-2. [DOI] [PubMed] [Google Scholar]
- 33.Liong M, France B, Bradley KA, Zink JI. Advanced Materials. 2009;21:1684–1689. [Google Scholar]
- 34.Park K-S, Tae J, Choi B, Kim Y-S, Moon C, Kim S-H, Lee H-S, Kim J, Kim J, Park J, Lee J-H, Lee JE, Joh J-W, Kim S. Nanomedicine: Nanotechnology, Biology and Medicine. 2010;6:263–276. doi: 10.1016/j.nano.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Hovland P, Flick J, Johnston M, Sclafani RA. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- 36.Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. Microbiology and Molecular Biology Reviews. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray JV, Ogas JP, Kamada Y, Stone M, Levin DE, Herskowitz I. The EMBO Journal. 1997;16:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamada Y, Jung US, Piotrowski J, Levin DE. Genes & development. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 39.Lew DJ. Current opinion in genetics & development. 2000;10:47–53. doi: 10.1016/s0959-437x(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 40.Krause SA, Gray JV. Current biology. 2002;12:588–593. doi: 10.1016/s0960-9822(02)00760-1. [DOI] [PubMed] [Google Scholar]
- 41.Yang SH, Lee KB, Kong B, Kim JH, Kim HS, Choi IS. Angewandte Chemie-International Edition. 2009;48:9160–9163. doi: 10.1002/anie.200903010. [DOI] [PubMed] [Google Scholar]
- 42.Jaroch DB, Lu J, Madangopal R, Stull ND, Stensberg M, Shi J, Kahn JL, Herrera-Perez R, Zeitchek M, Sturgis J, Robinson JP, Yoder MC, Porterfield DM, Mirmira RG, Rickus JL. Mouse and human islets survive and function after coating by biosilicification. 2013 doi: 10.1152/ajpendo.00081.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sumper M, Kroger N. J. Mater. Chem. 2004;14:2059–2065. [Google Scholar]
- 44.Belton DJ, Patwardhan SV, Perry CC. J. Mater. Chem. 2005;15:4629–4638. [Google Scholar]
- 45.Eleftheriou NM, Ge X, Kolesnik J, Falconer SB, Harris RJ, Khursigara C, Brown ED, Brennan JD. Chemistry of Materials. 2013;25:4798–4805. [Google Scholar]
- 46.Lee J, Choi J, Park JH, Kim M-H, Hong D, Cho H, Yang SH, Choi IS. Angewandte Chemie International Edition. 2014;53:8056–8059. doi: 10.1002/anie.201402280. [DOI] [PubMed] [Google Scholar]
- 47.Huang H-L, Hsing H-W, Lai T-C, Chen Y-W, Lee T-R, Chan H-T, Lyu P-C, Wu C-L, Lu Y-C, Lin S-T. Journal of biomedical science. 2010;17:36. doi: 10.1186/1423-0127-17-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puliafito A, Hufnagel L, Neveu P, Streichan S, Sigal A, Fygenson DK, Shraiman BI. Proceedings of the National Academy of Sciences. 2012;109:739–744. doi: 10.1073/pnas.1007809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.