Abstract
Despite considerable progress in preventive and therapeutic strategies, myocardial infarction (MI) is one of the leading causes of death throughout the world. A total of 55 susceptibility genes have been identified mostly in European genome-wide association studies (GWAS). Nevertheless, large-scale GWAS from other population could possibly find additional susceptibility loci. To identify as many MI susceptibility loci as possible, we performed a large-scale genomic analysis in Japanese population. To identify MI susceptibility loci in Japanese, we conducted a GWAS using 1666 cases and 3198 controls using the Illumina Human610-Quad BeadChip and HumanHap550v3 Genotyping BeadChip. We performed replication studies using a total of 11 412 cases and 28 397 controls in the Japanese population. Our study identified two novel susceptibility loci for MI: PLCL2 on chromosome 3p24.3 (rs4618210:A>G, P=2.60 × 10−9, odds ratio (OR)=0.91) and AP3D1-DOT1L-SF3A2 on chromosome 19p13.3 (rs3803915:A>C, P=3.84 × 10−9, OR=0.89). Besides, a total of 14 previously reported MI susceptibility loci were replicated in our study. In particular, we validated a strong association on chromosome 12q24 (rs3782886:A>G: P=1.14 × 10−14, OR=1.46). Following pathway analysis using 265 genes related to MI or coronary artery disease, we found that these loci might be involved in the pathogenesis of MI via the promotion of atherosclerosis. In the present large-scale genomic analysis, we identified PLCL2 and AP3D1-DOT1L-SF3A2 as new susceptibility loci for MI in the Japanese population. Our findings will add novel findings for MI susceptibility loci.
Introduction
Myocardial infarction (MI), the severest form of coronary artery disease (CAD), is characterized by the occlusion of the coronary artery, resulting in ischemic damage of the myocardium. The occlusion of the coronary artery is characterized by abrupt plaque rupture with thrombogenesis1 following the atherosclerotic process, which has recently been regarded as a chronic inflammatory condition involving lipid accumulation, abnormal extracellular matrix metabolism, innate immunity with macrophages, and the adaptive immune response with T and B lymphocytes.2 Despite recent dramatic advances in preventive and/or therapeutic strategies, MI is still one of the leading causes of death in Asian populations as well as European populations.
The pathogenesis of MI is considered to be the cumulative effect of multiple genetic and environmental factors. For the genetic aspect, a total of 55 susceptibility loci are known to be associated with CAD. Most of them were identified in European populations.3 Even after large-scale genome-wide association studies (GWAS) in European populations, GWAS in Asian populations have identified novel loci.4, 5, 6, 7 More additional loci are certainly left unidentified, and in such a situation, it remains elusive in total how many susceptibility loci exist. Here, to potently identify as many susceptibility loci as possible, GWAS and replication studies in a total of 44 673 Japanese participants were performed.
Materials and Methods
Subjects
All individuals were Japanese and gave written informed consent to participate in the study, as approved by the ethical committees of the institutional review boards (Table 1).
Table 1. Baseline characteristics of study population.
| Case | Control | |
|---|---|---|
| GWAS (Japanese) | ||
| Source | BioBank Japan | BioBank Japan and MRC |
| Number of samples | 1666 | 3198 |
| Platform | Illumina Human610-Quad | Illumina HumanHap550v3 |
| Male (%) | 86.4 | 54.2 |
| Age (mean±s.d.) | 59.4±10.4 | 52.0±15.1 |
| Hypertension (%) | 84.6 | 24.8 |
| Dyslipidemia (%) | 71.1 | 6.8 |
| Diabetes mellitus (%) | 52.5 | 8.7 |
| Smoking (%) | 78.0 | 52.4 |
| Replication study (Japanese) | ||
| Replication set 1 | ||
| Source | BioBank Japan | BioBank Japan |
| Number of samples | 8422 | 26 659 |
| Platform | Invader assay | Illumina Human610-Quad |
| Male (%) | 81.0 | 49.4 |
| Age (mean±s.d.) | 68.0±9.8 | 62.7±12.2 |
| Hypertension (%) | 74.9 | 34.4 |
| Dyslipidemia (%) | 59.6 | 15.5 |
| Diabetes mellitus (%) | 30.2 | 18.8 |
| Smoking (%) | 70.1 | 49.1 |
| Replication set 2 | ||
| Source | The University of Tokyo Hospital and OACIS | BioBank Japan |
| Number of samples | 2990 | 1738 |
| Platform | Invader assay | HumanOmniExpress and Invader assay |
| Male (%) | 78.7 | 56.7 |
| Age (mean±s.d.) | 64.9±11.4 | 48.1±14.9 |
| Hypertension (%) | 63.8 | 14.0 |
| Dyslipidemia (%) | 48.8 | 5.4 |
| Diabetes mellitus (%) | 35.1 | 6.7 |
| Smoking (%) | 65.2 | 27.4 |
Abbreviation: GWAS, genome-wide association study.
BioBank Japan samples
For the GWAS, we selected 1681 MI cases from the BioBank Japan project8 using the following criteria: (1) having a coronary angiography and (2) left ventricular asynergy on echocardiography. For replication study 1, we selected 8444 MI cases that satisfied one of the following criteria: (1) one or more total occlusions of coronary arteries on coronary angiography and (2) left ventricular asynergy on echocardiography.
The University of Tokyo Hospital samples
For replication study 2, a total of 201 MI cases were recruited from The University of Tokyo Hospital. The diagnosis of MI required 1 of the following criteria: (1) one or more total occlusions of the coronary arteries on coronary angiography and (2) left ventricular asynergy on echocardiography.
Osaka Acute Coronary Insufficiency study (OACIS) samples
OACIS is a district-based survey that aimed to assess the clinical variables, therapeutic procedures, and consequent clinical events in patients with MI in the Osaka area. For replication study 2, a total of 2789 MI cases were recruited from OACIS. The diagnosis of MI required two of the following three criteria: (1) a clinical history of central chest pressure, pain, or tightness lasting for ≥30 min, (2) ST-segment elevation greater than 0.1 mV in at least 1 standard or 2 precordial leads, and (3) a rise in the serum creatinine kinase concentration to greater than twice the normal laboratory value.
Control subjects
The control subjects were healthy volunteers and GWAS case samples for other diseases in the BioBank Japan project. There was no overlap of genotyped samples between GWAS panel and replication panels in this study. We excluded the samples that were registered for stable angina, unstable angina, or MI throughout the study. We also obtained 906 Japanese control DNAs from volunteers of the Osaka-Midosuji Rotary Club (MRC), Osaka, Japan.
Genotyping and quality control
In the GWAS, we genotyped 1681 case subjects using the Illumina Human610-Quad BeadChip (Illumina, Inc., San Diego, CA, USA) and 3251 control subjects using the Illumina HumanHap550v3 Genotyping BeadChip (Illumina, Inc.) (Figure 1). We excluded samples with a call rate<0.98, single-nucleotide polymorphisms (SNPs) having a call rate<0.99, and SNPs with P-values less than the cutoff value (P<1.0 × 10−6) for the Hardy–Weinberg equilibrium test in the controls. Among the 457 489 common SNPs in both BeadChips, 455 781 SNPs in the autosomal chromosomes passed the quality-control filters. Population stratification for the GWAS data was examined by principal component analysis (PCA).9 The genotype data of the samples along with the European (CEU), African (YRI), and East Asian (JPT and CHB) individuals were obtained from the Phase II HapMap database. We excluded one outlier control sample that was away from the East Asian cluster (Supplementary Figure S1A). In addition, we excluded a total of 15 cases and 52 controls because of gender mismatch or paired closely related samples by allele sharing analysis.
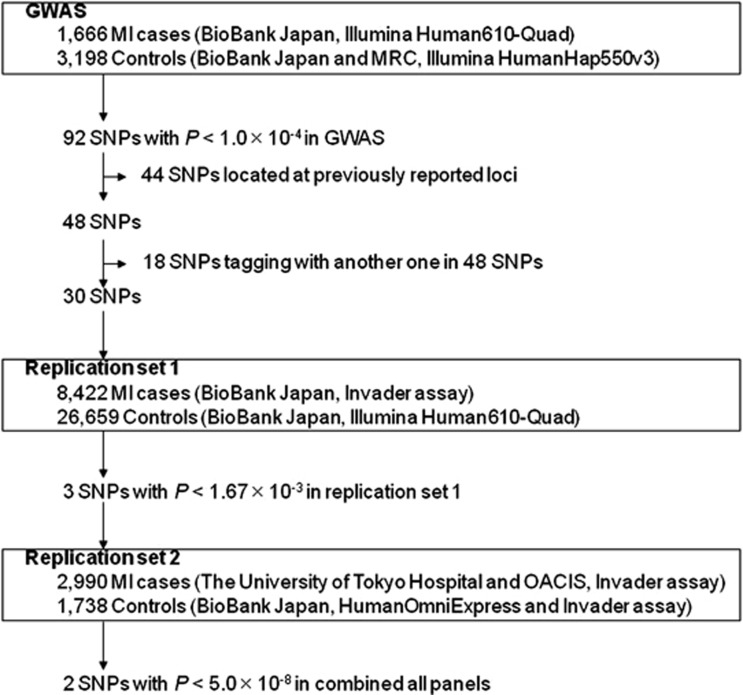
Figure 1.
Flowchart summarizing the multistage design of the present study. MRC, Midosuji Rotary Club; OACIS, Osaka Acute Coronary Insufficiency study.
In replication study 1, we selected 92 SNPs that showed GC-corrected P values (PGC)<1 × 10−4 in the GWAS. After excluding 44 SNPs located at previously reported loci, we selected 30 tag-SNPs from the remaining 48 SNPs with pairwise tagging only and an r2 threshold of 0.8 using the Haploview software. We genotyped 8444 case samples using the multiplex PCR-based Invader assay and 26 659 control samples using the Illumina Human610-Quad BeadChip. We excluded 22 case samples because of a low call rate (<0.95).
In replication study 2, 3 SNPs with P<0.05/30=1.67 × 10−3 in replication study 1 (rs4618210:A>G, rs3803915:A>C and rs2467473:A>C) were genotyped in 2990 cases, including 201 from The University of Tokyo Hospital and 2789 from OACIS, using the multiplex PCR-based Invader assay. For the control, we used the genotype data of HumanOmniExpress BeadChip (Illumina, Inc.) for rs3803915:A>C and rs2467473:A>C, whereas we genotyped rs4618210:A>G by the multiplex PCR-based Invader assay, because this SNP was not included in the chip.
Genotype oncordance rate
The genotype concordance rate between the Illumina Human610-Quad BeadChip and the Illumina HumanHap550v3 Genotyping BeadChip was 0.999 among 182 duplicate samples. That between the Illumina Human610-Quad BeadChip and the HumanOmniExpress BeadChip was 0.999 among 780 duplicate samples. Also, the concordance rate between genotype data of the multiplex PCR-based Invader assay and those of the Illumina Human610-Quad BeadChip was 0.994 (8797/8852 genotypings in 6 SNPs).
Genome-wide genotype imputation
Genotype imputation within the GWAS data was performed using MACH.10, 11 Association tests were performed with mach2dat using the fractional dosages output. The 1000 Genomes Project database (phase I JPT, CHB and Han Chinese South (CHS) data, March 2012) was used as a reference panel. SNPs whose minor allele frequency ≥0.01 were used. SNPs with large allele frequency difference between control in GWAS and 1000 Genomes Project were excluded. SNPs with a low quality of imputation (r2<0.9) were excluded. We selected imputed SNPs that showed PGC<1.0 × 10−4 and were located in the newly identified loci. In the region on 3p24.3, we selected 31 imputed SNPs and successfully genotyped 22 out of the 31 SNPs. In the region on 19p13.3, we selected 38 imputed SNPs and successfully genotyped 33 out of the 38 SNPs. These SNPs were genotyped in all GWAS samples using the multiplex PCR-based Invader assay.
Network analysis
Network analysis was performed using the Ingenuity Pathway Analysis software (IPA; Ingenuity Systems, Inc., Redwood City, CA, USA). We selected 4 genes from the newly identified regions in our GWAS, 96 genes from the GWAS catalog of National Human Genome Research Institute (http://www.genome.gov/gwastudies (accessed 1 February 2013)), and 179 genes collected from the Human Gene Mutation Database (HGMD) Professional (2012 December version, BIOBASE). As 13 genes were selected from both the GWAS catalog and the HGMD, we finally selected 266 genes for the network analysis. Genes from the GWAS catalog were collected if the genes were described as having a relationship with ‘coronary heart disease' or ‘myocardial infarction'. Genes from the HGMD were collected if the genes were described as having a relationship with ‘acute coronary syndrome,' ‘coronary artery disease,' ‘coronary heart disease,' ‘myocardial infarction,' or ‘angina'. We mapped 265 of the 266 genes in the IPA database, with the exception of the SMCR3 gene. We considered molecules and/or the relationships available in the Ingenuity Knowledge Base and set the confidence filter to ‘experimentally observed' or ‘highly predicted'. We tried to construct the network directly and/or indirectly as a connection between 4 novel genes and 261 known genes.
Statistical analysis
In the GWAS and replication studies, the statistical significance of the association with each SNP was assessed using a one-degree-of-freedom Cochran–Armitage trend test. Odds ratios (ORs) and confidence intervals (CIs) were calculated using allele 1 as the reference. We combined data from the GWAS and replication studies by the inverse variance method. Heterogeneity across the studies was examined using the Breslow–Day test. We used the Haploview 4.2 Software to analyze the LD values between SNPs. In the imputation analysis, pairwise r2 values were calculated using the R Bioconductor package snpMatrix (version 1.16.2), and the LD map was created using in-house program. Association analysis of imputed data was performed using the single.snp.tests function in the R package snpStats version 1.3.4. Regional association plots were generated using R statistical environment version 2.13.0.
Results
Baseline characteristics of each case–control group are shown in Table 1. Mean age and the ratio of male, hypertension, dyslipidemia, diabetes mellitus, and smoking in the case samples are significantly higher than those of control samples in the GWAS panel as well as in the replication sets.
In the GWAS, when we generated a quantile-quantile plot (Q-Q plot) using all samples, the genomic inflation factor (λGC) was 1.097 (Supplementary Figure S1B). Because we previously reported the existence of a population substructure in the Japanese population,12 we performed PCA using only the JPT and CHB HapMap samples as references (Supplementary Figure S1C). Because we found one main (Hondo) cluster and several sub-clusters in our population, we generated a Q-Q plot and PCA only using the main (Hondo) cluster (Supplementary Figure S1D and E). PCA showed that Hondo cluster was uniform and there might not have population stratification. However, λGC did not show any improvement (1.093). We also calculated the λ1000 adjusted by sample size to be 1.044.13 These results indicate a low possibility of false-positive associations resulting from population stratification. We therefore used λGC-corrected P values (PGC) to adjust for the unknown genetic heterogeneity of the GWAS results.
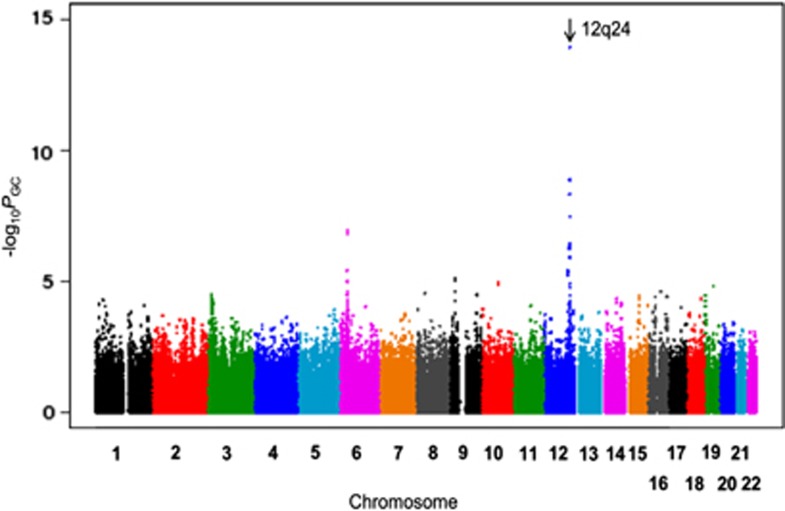
Our GWAS identified a genome-wide significant level of association on chromosome 12q24.12 (rs3782886:A>G, PGC=1.14 × 10−14, odds ratio (OR)=1.46; Figure 2). The regional plot showed that this locus contains many candidate genes, such as BRAP, ALDH2, and SH2B3. The association of any SNPs in this region was not significant after the adjustment of rs3782886:A>G, suggesting this rs3782886 function as a robust marker SNP for this locus (Supplementary Figure S2). We also analyzed the association of previously reported loci for MI or CAD. Among the 55 previously reported loci, we confirmed the association of additional 13 loci3, 4, 5, 6, 7 (P<0.05, Supplementary Table S1).
Figure 2.
Manhattan plot of GWAS result. The genomic-controlled P-values were used. SNPs on chromosome 12q24 showed highly significant association with MI.
To identify additional susceptibility loci for MI, we conducted a replication study (set 1) using an independent set of 8422 MI cases and 26 659 controls from the Japanese population. Among the 92 SNPs that showed PGC<1.0 × 10−4 in our GWAS, we excluded 44 SNPs located at previously reported loci. Then, we calculated the linkage disequilibrium (LD) coefficient (r2) between the remaining SNPs and selected 30 SNPs with the lowest P-value within each region of r2≥0.8. We successfully genotyped all 30 of these SNPs. Three SNPs showed P<0.05/30=1.67 × 10−3 in replication set 1 (Supplementary Table S2): rs4618210:A>G on chromosome 3p24.3 (P=5.18 × 10−7), rs3803915:A>C on chromosome 19p13.3 (P=6.90 × 10−6), and rs2467473:A>C on chromosome 3p24.3 (P=3.80 × 10−4). We further examined the association of these 3 SNPs using an independent set of 2990 MI cases and 1738 controls (replication set 2) from the Japanese population. The SNP rs3803915:A>C showed a nominal association (Prep2=4.74 × 10−3, Table 2), whereas the association of rs2467473:A>C and rs4618210:A>G were not significant (rs2467473, Prep2=0.502; rs4618210, Prep2=0.590). Hence, we combined the results of replication sets 1 and 2 using the inverse variance method. Two SNPs (rs4618210 and rs3803915) showed a significant association after Bonferroni correction (P<0.05/30=1.67 × 10−3, Table 2). When we combined the results of the GWAS and replication studies by the inverse variance method, two SNPs reached a genome-wide significance level of association (rs4618210, combined Pall=2.60 × 10−9, OR=0.91; rs3803915, combined Pall=3.84 × 10−9, OR=0.89). In the control subjects, there was no significant difference in allele frequencies of these two SNPs among healthy volunteers and subjects with other diseases (Supplementary Table S3).
Table 2. Summary of the GWAS and replication studies.
| SNP | Chr | Allele 1/2 | Study | maf_case | maf_ctrl | P | OR (95% CI) | Phet |
|---|---|---|---|---|---|---|---|---|
| rs4618210:g.17099388A>G | 3 | A/G | GWAS | 0.45 | 0.40 | 6.78 × 10−5 | 0.84 (0.77–0.91) | |
| Replication 1 | 0.44 | 0.42 | 5.18 × 10−7 | 0.91 (0.88–0.95) | ||||
| Replication 2 | 0.43 | 0.43 | 0.590 | 0.98 (0.90–1.06) | ||||
| Replication combined | 2.74 × 10−7 | 0.92 (0.89–0.95) | 0.13 | |||||
| ALL | 2.60 × 10−9 | 0.91 (0.88–0.94) | 0.04 | |||||
| rs3803915:g.2111529A>C | 19 | A/C | GWAS | 0.22 | 0.19 | 5.59 × 10−5 | 0.80 (0.72–0.89) | |
| Replication 1 | 0.21 | 0.19 | 6.90 × 10−6 | 0.91 (0.87–0.95) | ||||
| Replication 2 | 0.21 | 0.18 | 4.74 × 10−3 | 0.86 (0.77–0.95) | ||||
| Replication combined | 4.43 × 10−7 | 0.90 (0.87–0.94) | 0.32 | |||||
| ALL | 3.84 × 10−9 | 0.89 (0.86–0.92) | 0.06 |
Abbreviations: CI, confidence interval that was calculated using allele 1 as the reference; Chr, chromosome; GWAS, genome-wide association study; maf, minor allele frequency; OR, odds ratio that was calculated using allele 1 as the reference; SNP, single-nucleotide polymorphism.
The P-values of the GWAS were the genomic-control P-values (PGC). The combined P-values were calculated by the inverse variance method. The P-values of heterogeneities (Phet) across the population were estimated formally using the Breslow–Day test. Chromosome location was based on hg18 (Genome Build 36.3).
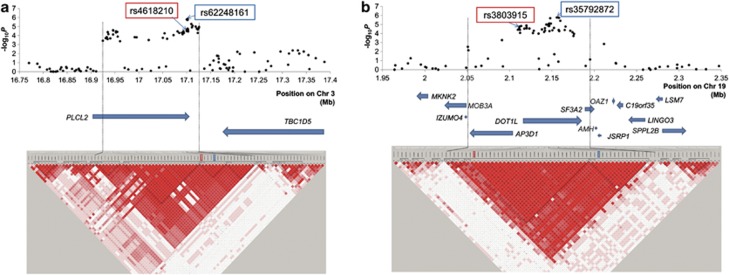
To further investigate the gene(s) that conferred susceptibility to MI, we performed fine mapping analysis of two new loci using GWAS samples. In the region on chromosome 3p24.3, we performed a genome-wide imputation analysis and selected 31 imputed SNPs with PGC<1.0 × 10−4. We genotyped these SNPs in all GWAS samples and showed rs62248161:A>G located at intron 5 of PLCL2 to be the top associated SNP in this region (Figure 3a). The LD block revealed that rs62248161:A>G spans the region from 16.92 to 17.13 Mb, which only contained PLCL2. According to GeneCards (http://www.genecards.org), PLCL2 is expressed in lymphocytes and platelets, which have a pivotal role in the pathogenesis of atherosclerosis.
Figure 3.
Case–control association plots, LD map, and genomic structure of the newly identified regions. LD map and genomic structure of the regions on chromosome 3p24.3 (a) and on 19p13.3 (b). The associated regions are shown between the two black dashed lines. The black dots represent the –log10 P-values obtained from GWAS and genotyping SNPs that showed PGC <1.0 × 10−4 in the imputation. We drew the LD map based on D' values using the genotype data from the cases and controls in the GWAS samples. The arrows indicate the locations of genes. SNPs in red-lined box were identified in GWAS as a marker SNP highly associated with MI, and SNPs in blue-lined box are top associated SNPs among the imputed SNPs.
Similar to our analysis of chromosome 3p24.3, we also performed a fine mapping analysis of the associated region on chromosome 19p13.3. We performed a genome-wide imputation analysis and selected 38 imputed SNPs with PGC<1.0 × 10−4. We genotyped these SNPs in all GWAS samples and showed that rs35792872:A>G in intron 9 of DOT1L had the strongest association in this region (Figure 3b). The LD block revealed that the associated region spanned from 2.05 to 2.20 Mb and contained three genes, DOT1L, AP3D1, and SF3A2. According to GeneCards (http://www.genecards.org), all three genes are expressed in lymphocytes and endothelial cells, which have an important role in the pathogenesis of atherosclerosis.
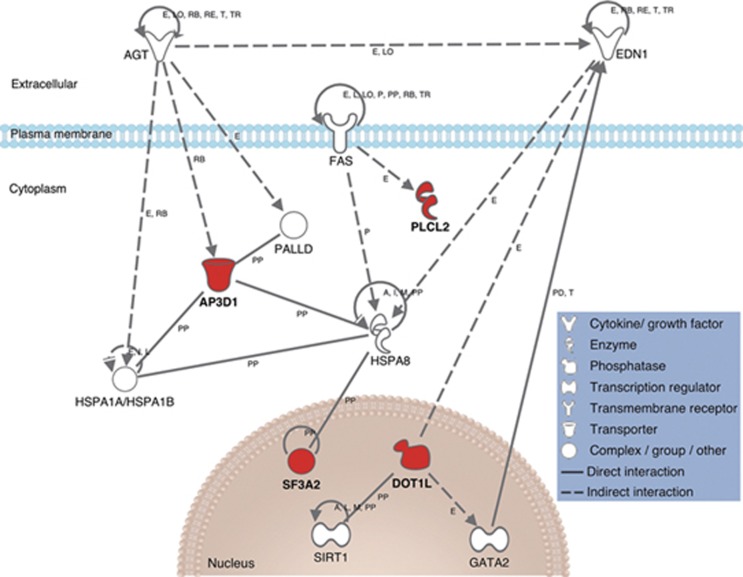
To examine the possible mechanisms responsible for the functional relationship between these four genes and the pathogenesis of MI, we simulated the connection between established MI genes and these four genes identified here (Figure 4, Supplementary Table S4). The expression of PLCL2 was shown to be indirectly affected by FAS. DOT1L was demonstrated to positively regulate GATA2 expression and was shown to be negatively correlated with endothelin-1. Additionally, the binding capacity of AP3D1 was found to be regulated by angiotensinogen.
Figure 4.
Network analysis of MI susceptibility genes. Genes in newly identified regions are drawn in red, and genes taken from the GWAS catalog homepage or HGMD are drawn in white. A, activation; E, expression (includes metabolism/synthesis for chemicals); I, inhibition; L, proteolysis (includes degradation for chemicals); LO, localization; M, biochemical modification; P, phosphorylation/dephosphorylation; PD, protein–DNA binding; PP, protein–protein binding; RB, regulation of binding; RE, reaction; T, transcription; TR, translocation. The expression of PLCL2 was shown to be indirectly affected by FAS. DOT1L was shown to positively regulate GATA2 expression and was shown to be negatively correlated with endothelin-1. The binding capacity of AP3D1 was found to be regulated by angiotensinogen.
Discussion
We conducted a genomic analysis in a total of 13 078 MI cases and 31 595 controls and identified a genome-wide significant level of association on 2 novel susceptibility loci for MI: PLCL2 on chromosome 3p24.3 and AP3D1-DOT1L-SF3A2 on chromosome 19p13.3. To our best knowledge, this is the largest study so far in the genomic analysis for MI susceptibility loci in the Asian population.
Importantly, our study validated an association of chromosome 12q24.12. This locus was reported to be associated with CAD,5 intracranial aneurysm,14 ischemic stroke,15 blood pressure,16 kidney function,17 type 1 diabetes,18 serum urate19 and lipid concentration,20 and drinking behavior,21 suggesting that this locus is an important genetic component of cardiovascular diseases and their risk factors. Because top associated SNP (rs3782886:A>G) is closely linked to two functional variants in this region (rs671:A>G in ALDH2 and rs11066001:C>T in BRAP),22 we confirmed that the risk haplotype, which contains several functional variants, will affect the susceptibility of cardiovascular disease and its risk factors.
PLCL2 is a novel phospholipase C-like protein that lacks phospholipase catalytic activity and is expressed in hematopoietic cells.23 Judging from the previous reports that this region was associated with primary biliary cirrhosis24 and multiple sclerosis,25 PLCL2 might have a functional role in the immune system. Indeed, the experimental data on Plcl2-deficient mice indicated that Plcl2 regulates the proliferation of mature B cells in response to B-cell receptor cross-linking.26 PLCL2 is likely to be involved in the pathogenesis of the chronic inflammatory process of MI through the dysregulation of immune system.27
DOT1L, a nucleosomal histone-3 (H3)-specific methyltransferase, mediates the intranucleosomal methylation of H3 at lysine-79, a key pathway in the regulation of cell cycle.28 SF3A2 encodes subunit 2 of the splicing factor 3a protein complex, which is essential for pre-mRNA splicing.29 AP3 is a heterotetrameric adaptor protein involved in the biogenesis of lysosome-related organelles, such as platelet-dense bodies. The ‘mocha' mouse mutant with a null allele of Ap3d1 shows bleeding abnormalities due to the storage pool deficiency in the dense granules of platelets,30 raising the possibility that AP3D1 functions as the modulator of thrombogenesis via platelet function.
We simulated the connection between 261 established MI genes and these 4 genes identified here (Figure 4) to speculate the functional role of newly identified susceptibility loci in the pathogenesis of MI. In our analysis, the expression of PLCL2 was shown to be indirectly affected by FAS.31 Considering that mature B cells undergo FAS-mediated apoptosis,32 it is reasonable to hypothesize that PLCL2 is associated with the regulation of B-cell maturation, leading to the formation of atherosclerotic lesions. DOT1L was shown to positively regulate GATA2 expression,33 which is known to regulate the transcription of endothelial cell-specific genes.34 In particular, DOT1L was shown to be negatively correlated with endothelin-1,35 which has an important role in vasospasm and vasoconstriction. Additionally, the binding capacity of AP3D1 was found to be regulated by angiotensinogen,36 suggesting that AP3D1 might be related to atherosclerosis in concert with a potent atherosclerotic substrate, angiotensin II. Taken together, the candidate genes identified in this study might contribute to the pathogenesis of MI via the promotion of atherosclerosis (and vasospasm) to different extents. Further studies are warranted to clarify the functional roles of these loci and genes in the pathogenesis of MI, providing a valuable starting point for novel biomedical research.37
In conclusion, we performed a large-scale genomic analysis and identified PLCL2 and AP3D1-DOT1L-SF3A2 as new susceptibility loci for MI in the Japanese population. Our findings will add novel findings for MI susceptibility loci.
Acknowledgments
We thank T Aoi, H Amitani, Y Tsuchiya, and E Kanno for their contributions to the SNP genotyping and their technical assistance. We also thank other staff members of the Laboratory for Genotyping Development, Center for Genomic Medicine, RIKEN. We thank all the subjects for participating in the study and also thank the collaborating physicians for performing the sample collection. We are grateful to the members of BioBank Japan and the Rotary Club of Osaka-Midosuji District 2660 Rotary International in Japan for supporting our study. This work was conducted as a part of the BioBank Japan Project, supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Falk E, Shah PK, Fuster V. Coronary plaque disruption. Circulation. 1995;92:657–671. doi: 10.1161/01.cir.92.3.657. [DOI] [PubMed] [Google Scholar]
- Galkina E, Ley K. Immune and inflammatory mechanisms of atherosclerosis. Annu Rev Immunol. 2009;27:165–197. doi: 10.1146/annurev.immunol.021908.132620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The CARDIoGRAMplusC4D Consortium Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki A, Ozaki K, Sato H, et al. SNPs on chromosome 5p15.3 associated with myocardial infarction in Japanese population. J Hum Genet. 2011;56:47–51. doi: 10.1038/jhg.2010.141. [DOI] [PubMed] [Google Scholar]
- Takeuchi F, Yokota M, Yamamoto K, et al. Genome-wide association study of coronary artery disease in the Japanese. Eur J Hum Genet. 2012;20:333–340. doi: 10.1038/ejhg.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Xu CQ, He Q, et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- Lu X, Wang L, Chen S, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. The BioBank Japan Project. Clin Adv Hematol Oncol. 2007;5:696–697. [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Kabata Y, Nakazono K, Takahashi A, et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet. 2008;83:445–456. doi: 10.1016/j.ajhg.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low SK, Takahashi A, Cha PC, et al. Genome-wide association study for intracranial aneurysm in the Japanese population identifies three candidate susceptible loci and a functional genetic variant at EDNRA. Hum Mol Genet. 2012;21:2102–2110. doi: 10.1093/hmg/dds020. [DOI] [PubMed] [Google Scholar]
- Traylor M, Farrall M, Holliday EG, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, Takeuchi F, Tabara Y, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Sim X, Go MJ, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet. 2012;44:904–909. doi: 10.1038/ng.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V, Howson JM, Smyth DJ, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;7:e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köttgen A, Albrecht E, Teumer A, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45:145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Isono M, Nabika T, et al. Confirmation of ALDH2 as a major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J. 2011;75:911–918. doi: 10.1253/circj.cj-10-0774. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Sato H, Inoue K, et al. SNPs in BRAP associated with risk of myocardial infarction in Asian populations. Nat Genet. 2009;41:329–333. doi: 10.1038/ng.326. [DOI] [PubMed] [Google Scholar]
- Otsuki M, Fukami K, Kohno T, Yokota J, Takenawa T. Identification and characterization of a new phospholipase C-like protein, PLC-L2. Biochem Biophys Res Commun. 1999;266:97–103. doi: 10.1006/bbrc.1999.1784. [DOI] [PubMed] [Google Scholar]
- Mells GF, Floyd JA, Morley KI, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium 2 Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka K, Fukami K, Otsuki M, et al. Role of phospholipase C-L2, a novel phospholipase C-like protein that lacks lipase activity, in B-cell receptor signaling. Mol Cell Biol. 2003;23:7329–7338. doi: 10.1128/MCB.23.20.7329-7338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry HM, Bender TP, McNamara CA. B cell subsets in atherosclerosis. Front Immunol. 2012;3:373. doi: 10.3389/fimmu.2012.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Bennett M, Reed R. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science. 1993;262:105–108. doi: 10.1126/science.8211113. [DOI] [PubMed] [Google Scholar]
- Kantheti P, Qiao X, Diaz ME, et al. Mutation in AP-3 δ in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- Kobayashi SD, Voyich JM, Braughton KR, DeLeo FR. Down-regulation of proinflammatory capacity during apoptosis in human polymorphonuclear leukocytes. J Immunol. 2003;170:3357–3368. doi: 10.4049/jimmunol.170.6.3357. [DOI] [PubMed] [Google Scholar]
- Fukuyama H, Adachi M, Suematsu S, et al. Requirement of Fas expression in B cells for tolerance induction. Eur J Immunol. 2002;32:223–230. doi: 10.1002/1521-4141(200201)32:1<223::AID-IMMU223>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Feng Y, Yang Y, Ortega MM, et al. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood. 2010;116:4483–4491. doi: 10.1182/blood-2010-03-276501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JJ, Wang T, Cox JE, et al. GATA2 is associated with familial early-onset coronary artery disease. PLoS Genet. 2006;2:e139. doi: 10.1371/journal.pgen.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Liu K, Wu H, et al. Spironolactone rescues Dot1a-Af9-mediated repression of endothelin-1 and improves kidney injury in streptozotocin-induced diabetic rats. PLoS One. 2012;7:e47360. doi: 10.1371/journal.pone.0047360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, McClatchy DB, Shukla AK, et al. Functional specialization of β-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci USA. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita H. Human genomics in cardiovascular medicine: implications and perspectives. Circ J. 2013;77:876–885. doi: 10.1253/circj.cj-13-0126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.