Abstract
The wide range of metabolic phenotypes in phenylketonuria is due to a large number of variants causing variable impairment in phenylalanine hydroxylase function. A total of 834 phenylalanine hydroxylase gene variants from the locus-specific database PAHvdb and genotypes of 4181 phenylketonuria patients from the BIOPKU database were characterized using FoldX, SIFT Blink, Polyphen-2 and SNPs3D algorithms. Obtained data was correlated with residual enzyme activity, patients' phenotype and tetrahydrobiopterin responsiveness. A descriptive analysis of both databases was compiled and an interactive viewer in PAHvdb database was implemented for structure visualization of missense variants. We found a quantitative relationship between phenylalanine hydroxylase protein stability and enzyme activity (rs=0.479), between protein stability and allelic phenotype (rs=−0.458), as well as between enzyme activity and allelic phenotype (rs=0.799). Enzyme stability algorithms (FoldX and SNPs3D), allelic phenotype and enzyme activity were most powerful to predict patients' phenotype and tetrahydrobiopterin response. Phenotype prediction was most accurate in deleterious genotypes (≈100%), followed by homozygous (92.9%), hemizygous (94.8%), and compound heterozygous genotypes (77.9%), while tetrahydrobiopterin response was correctly predicted in 71.0% of all cases. To our knowledge this is the largest study using algorithms for the prediction of patients' phenotype and tetrahydrobiopterin responsiveness in phenylketonuria patients, using data from the locus-specific and genotypes database.
Introduction
Phenylketonuria (PKU; OMIM# 261600) is an autosomal recessive metabolic disease caused by a deficiency of hepatic phenylalanine-4-hydroxylase (PAH; EC 1.14.16.1). It is the most common inborn error of amino acid metabolism, affecting about 1 in 10 000 newborns in Europe.1 PAH is a non-heme iron enzyme that catalyzes the rate-limiting step in phenylalanine (Phe) catabolism using molecular oxygen as an additional substrate, iron and tetrahydrobiopterin (BH4) as cofactor. Loss of function variants in the PAH gene therefore lead to an accumulation of Phe in the liver, the blood, and finally in the brain, causing severe mental retardation, behavioral disturbances and psychiatric disorders in untreated patients.1 While a Phe-restricted diet has shown to almost abolish these symptoms, some patients treated from an early age show lower IQ scores than their controls,2 while cognitive outcomes are inversely correlated to blood Phe levels.3 The clear relationship between clinical and metabolic phenotype is the basis of phenotype classification, which is predicated on off-diet blood Phe levels. Most classifications today distinguish between mild hyperphenylalaninemia (MHP; 120–600 μmol/l), mild PKU (600–1200 μmol/l) and classic PKU (>1200 μmol/l),1 while some recognize an additional group of moderate PKU (blood Phe 900–1200 μmol/l). The wide range of metabolic (and clinical) phenotype has been shown mainly to be determined by PAH genotype4, 5 although other factors might play a role as well.6
To date, more than 800 PAH variants (as annotated in the locus-specific database (LSD) PAHvdb; http://www.biopku.org) across the three PAH domains (N-terminal regulatory, central catalytic and C-terminal oligomerization domain) are known. Most patients are compound heterozygotes so that allelic interaction potentially complicates genotype-based prediction of phenotype. The key to comprehend genotype–phenotype relationships and its inconsistencies lies within understanding PKU disease mechanisms: gene variants within the catalytic domain may directly abolish PAH enzyme function,7 while splice-site variants can result in a non-functional truncated protein.8 Missense variants have been reported to primarily result in misfolding of the protein9, 10, 11 which impairs protein stability9, 11 and oligomer assembly.9 Misfolded proteins were observed to aggregate in prokaryote hosts4 and to be degraded in cellular models12 leading to diminished PAH activity. Various algorithms analyzing mutant PAH protein folding, stability and function have been developed to predict metabolic and clinical phenotype. We tested FoldX,13 PolyPhen-2,14 SIFT Blink15 and SNPs3D16 algorithms as well as residual PAH activity (%PAH) for each variant as instruments for phenotype prediction.
Similarly, the issue of BH4 responsiveness in PKU and its prediction have been a topic of intense research.17 Kure et al18 were the first to describe PKU patients responding to oral BH4 administration with lowering of blood Phe levels. Randomized clinical trials19 as well as international multicenter studies20 followed to establish BH4 (FDA-approved drug Kuvan) as a valid treatment option in PKU. Studies have shown that ∼20–30% of all PKU patients and about 60–80% of patients with MHP or mild PKU may benefit from BH4 treatment.21 While multiple mechanisms for BH4 responsiveness have been suggested,22 in many cases BH4 seems to act as a pharmacological chaperone assisting in protein folding, promoting enzyme stability and hindering degradation.23, 24 Response to BH4 has also been shown to depend on initial blood Phe levels21 and seems to require a certain minimal %PAH.25 Additionally, genotype has been closely associated with BH4 responsiveness type although not without inconsistencies.25 In this study we used several prediction algorithms as well as %PAH and allelic phenotype value (APV) to predict disease severity and BH4 responsiveness. The large scale of this study (4181 patients with complete genotype information, 1543 different genotypes, and 267 variants with known APV) allowed for confident identification of predictive factors.
Materials and methods
Databases
PAHvdb is a LSD (http://www.biopku.org/home/pah.asp). A total of 834 PAH variants were tabulated as of December 2013. Allelic phenotype classification was modified from Guldberg's arbitrary assigned values (AV)5 (AV1=classic PKU, AV2=moderate PKU, AV4=mild PKU and AV8=MHP) to a three-category system for APV: APV1=classic PKU, APV3=mild or moderate PKU, and APV8=MHP). Proteins with <3% in vitro %PAH (measured in recombinant mutant proteins expressed in eukaryotic cell systems)26 as well as nonsense and frame-shift variations were defined as null (and assigned APV=1). Splice-site variants affecting invariable ag and gt nucleotides were also considered null while splice-site variants in non-canonical sequence were defined as only putative-null since they can sometimes produce wild-type protein.27 The definition of 156 null-variants (Supplementary Table S1) allowed for the assignment of APVs to their complimentary alleles in hemizygous genotypes. Alleles in homozygous genotypes were assigned APVs whenever there was coinciding phenotype information on at least three patients. Seven variants had conflicting phenotype information and were excluded from APV analysis (see below). All variants were checked with Mutalyzer 2.0.beta-27 (https://mutalyzer.nl/check)28 and PAH genomic sequence.29 The reference accession number for the PAH sequence is ENSG00000171759; RefSeq NM_000277.1. Sequence variants are compliant with the HGVS rules (http://www.hgvs.org/mutnomen/).30
BIOPKU database (http://www.biopku.org/home/biopku.asp) enclosed 4181 patients with full genotype as of January 2014 and phenotype was known for 3374 cases (80.7%). Phenotype classification was based upon highest blood Phe levels before treatment introduction and comprised three categories. BH4 responsiveness was defined as a ≥30% reduction (responder) or 20–30% reduction of blood Phe levels (slow responder) 24–48 h after BH4 administration (usually 20 mg/kg body weight).31 Response classification was based upon blood Phe values at the end of the test (eg T24h or T48h or both) and with a trend for a constant Phe concentrations decrease. Each response category was assigned an arbitrary ordinal value for quantitative analysis: responder=3, slow responder=2 and non-responder=1.
Gene variants damage prediction tools
A number of web-based tools for prediction of pathogenic missense variants are currently available.32 We selected four tools frequently used in our laboratories and performed FoldX, PolyPhen-2, SIFT Blink and SNPs3D calculations for all missense variants in PAHvdb. FoldX (http://foldx.crg.es) is a computer algorithm predicting the impact of variants on protein stability.13 FoldX values were calculated using the 1J8U crystal structure (residues 118–424) including the catalytic domain and dimerization motif of human PAH. The 1J8U structure was chosen for its high-resolution (1.5 Å).33 FoldX values were calculated for 422 out of 488 missense variants. The stability of the remaining missense mutations could not be calculated due to missing residues in the PDB file used for the calculations. At present there is no high-resolution 3D structure for the full-length human PAH. The energy parameter VdWDesign was set to 2, which provides a ceiling of 5 kcal/mol for the van der Waals clashes between two atoms. Repair PDB was run in four iterations on 1J8U PDB after each of which ΔΔG was calculated for all possible missense variants. The multiple iterations were performed to get the uncertainty (standard deviation (SD)) in the ΔΔG values, representing the precision of FoldX prediction. Variants leading to a change of ΔΔG>1 kcal/mol were considered destabilizing, those where the change in ΔΔG<−1 kcal/mol counted as stabilizing, while variants in the range in between were considered neutral. Average ΔΔG of the four iterations was used for statistical analysis. PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2) estimates the damaging effect of missense variants based on multiple sequence analysis and by assessing variant effects on 3D structural features of the protein.14 PolyPhen-2 also predicts the likelihood of nucleotide replacements to destroy structural features such as the hydrophobic core or electrostatic interactions of the protein. Continuous output ranges from 0.00 (benign) to 1.00 (probably damaging) with an intermediary category of possibly damaging. We used HumDiv-trained PolyPhen-2 because of its higher accuracy of prediction and sensitivity. The SIFT Blink (http://sift.jcvi.org) algorithm estimates the functional impact of missense variants based solely on multiple sequence analysis.15 It compares the mutant sequence to similar sequences and their functional profile and predicts it to be either deleterious (<0.06) or tolerated (≥0.05). The SNPs3D (http://www.snps3d.org) module assesses the effect of non-synonymous SNPs on protein stability and function based on sequence analysis, structural analysis (using wild-type protein structures as reference) and a Support Vector Machine (SVM).16 A negative SVM Score classifies the substitution as destabilizing to the folded state, while positive scores indicate non-deleterious substitutions.
Predicting allelic phenotype
We established correlations between APV and %PAH, FoldX, SNPs3D, PolyPhen-2 and SIFT Blink values. In a second step, multiple regression analysis was run to predict APV from the aforementioned values.
Linking locus-specific and genotype database
We investigated relationships between patients' genotype and phenotype as well as BH4 responsiveness in (a) deleterious null–null genotypes, (b) functionally hemizygous, (c) compound heterozygous and (d) homozygous genotypes. Since many genotypes were reported with different phenotypes, we first determined an average phenotype (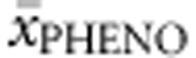 ) for each genotype in BIOPKU. Second, all BIOPKU alleles were assigned their concurrent allelic values from PAHvdb (APV, %PAH, FoldX, SNPs3D, PolyPhen-2 and SIFT Blink values).
) for each genotype in BIOPKU. Second, all BIOPKU alleles were assigned their concurrent allelic values from PAHvdb (APV, %PAH, FoldX, SNPs3D, PolyPhen-2 and SIFT Blink values). 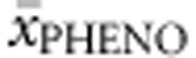 and average BH4 responsiveness (
and average BH4 responsiveness (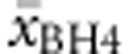 ) were then correlated with (a) genotype in deleterious null–null genotypes, (b) allelic values of the non-null allele in hemizygous genotypes, (c) averaged allelic values (eg
) were then correlated with (a) genotype in deleterious null–null genotypes, (b) allelic values of the non-null allele in hemizygous genotypes, (c) averaged allelic values (eg 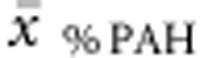 ) in both compound heterozygous and (d) homozygous genotypes. Multiple regression analysis was run to predict
) in both compound heterozygous and (d) homozygous genotypes. Multiple regression analysis was run to predict 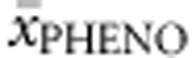 and
and 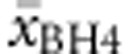 from allelic values. Only genotypes with at least three patients for which phenotype was known were included in this analysis. The same was true for prediction of BH4 response.
from allelic values. Only genotypes with at least three patients for which phenotype was known were included in this analysis. The same was true for prediction of BH4 response.
Statistics
Statistical analysis was performed using IBM SPSS Statistics Version 21 (IBM, Armonk, NY, USA) for Mac. Correlations between ordinal and nominal values were described using χ2-test while ordinal and continuous variables were correlated using Spearman's correlation. Relationships between two continuous variables were evaluated using Pearson's correlation (if normally distributed) and Spearman's correlation (if normality was violated). All correlations are reported as r(N−2) where N=sample size and N−2=degrees of freedom. One-way ANOVA was used to compare means between different groups. Normal distribution was assessed by Shapiro Wilk's test and homogeneity of variance by Levene's Test. Tukey post-hoc analysis was employed to locate significant differences between groups. Whenever homogeneity of variance was violated, ANOVA was replaced by Welch's ANOVA and Tukey test by Games–Howell analysis. Results of regression analysis are reported as F(dfreg/dfres)=obtained F-value, where dfreg=regression degrees of freedom and dfres=residual degrees of freedom. Independence of residuals was assessed by Durbin–Watson statistic.
Results
PAHvdb
Descriptive analysis
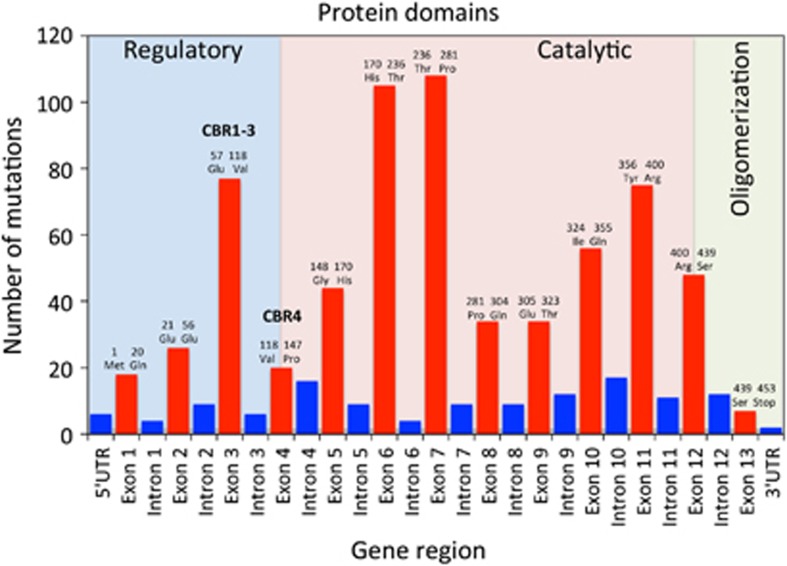
Most (488) of the 834 variants in PAHvdb (58.5%) were missense variants, followed by 133 deletions (15.9%), 114 splice-site variants (13.7%), 50 nonsense variants (6.0%) and 26 insertions (3.1%). Silent variants, ins/del and large genomic rearrangement together accounted for the residual 23 variants (2.8%). Most known variants were located in the catalytic domain (503 variants, 60.3%), followed by intronic and non-coding region locations (124 variants, 14.9%), regulatory domain (148 variants, 167.7%) and oligomerization domain (42 variants, 5.0%). The site with most known variants was exon 7 (catalytic domain, 13.9%) followed by exon 6 (catalytic domain, 13.5%) and exon 3 (regulatory domain, 9.7%) (Figure 1). %PAH was known for 96 variants (11.5%). The values ranged from 0 to 111% with a mean of 35.4% (standard error of the mean (SEM)=3.07; SD 30.09%). APV was known for 267 variants (32.0%), of which 221 were classified as classic PKU (APV1, 82.8%), 25 were associated with mild PKU (APV3, 9.4%) and 21 with MHP (APV8, 7.9%). In seven cases there was conflicting information on APV (Supplementary Table S2).
Figure 1.
Distribution of mutations tabulated in the PAHvdb according to gene region, affected protein domains, amino acid boundaries of the exons and BH4 cofactor binding regions (CBR). Red bars, exons; blue bars, introns.
3D visualization of PAH variants
As a tool to inspect the 3D environment of the amino acid variant sites in PAH, 455 entries in the PAHvdb database were hyperlinked to an interactive viewer built on the iSee concept—a one-stop platform providing interactive visualization of protein structures without the need for extensive user input or prior knowledge of structure file handling.34 This tool enables the user to view the local atomic environment (secondary structure elements, bonding interactions and neighboring residues) for each amino acid in the wild-type protein associated with a missense change. An option is further provided to display, on the wild-type template without further molecular dynamics/energy minimization, the chemical structure of the mutated side-chain. This feature serves to enable the user to discern any difference in the physico-chemical properties between the wild-type and mutated side chains. Example screenshots and first-time user instructions are provided in Supplementary Figure S1.
Residual PAH activity and protein damage
There was a moderate negative correlation between %PAH and the destabilization of the mutant protein (ΔΔG values provided by FoldX; rs (70)=−0.446, P<0.001 (normality was violated)) as well as a moderate positive correlation between %PAH and SNPs3D value (r(89)=0.479, P<0.001). There was no relationship between %PAH and either SIFT Blink or PolyPhen-2 value (Supplementary Figure S2A-D). All alleles with %PAH below 13% (n=15) were associated with classic PKU (APV=1).
Allelic phenotype and variants damage tools
We established correlations between %PAH, FoldX, SNPs3D, PolyPhen-2, SIFT values and APV. There was a strong positive correlation between %PAH and APV (rs (40)=0.799, P<0.01) and mean %PAH differed significantly between APV groups: Welch's F(2,16.110)=50.367, P<0.001. %PAH increased from 3.75 (±4.24%) for APV1 to 40.40 (±15.86%) for APV3 and to 51.69 (±24.95%) for APV8. Post-hoc analysis revealed that the increase from APV1 to APV3 was statistically significant (36.65%, 95% confidence interval (CI): 22.55–50.75, P<0.01), as was the increase from APV1 to APV8 (47.94%, 95% CI (31.60 to 64.28), P<0.01) (Supplementary Figure S3).
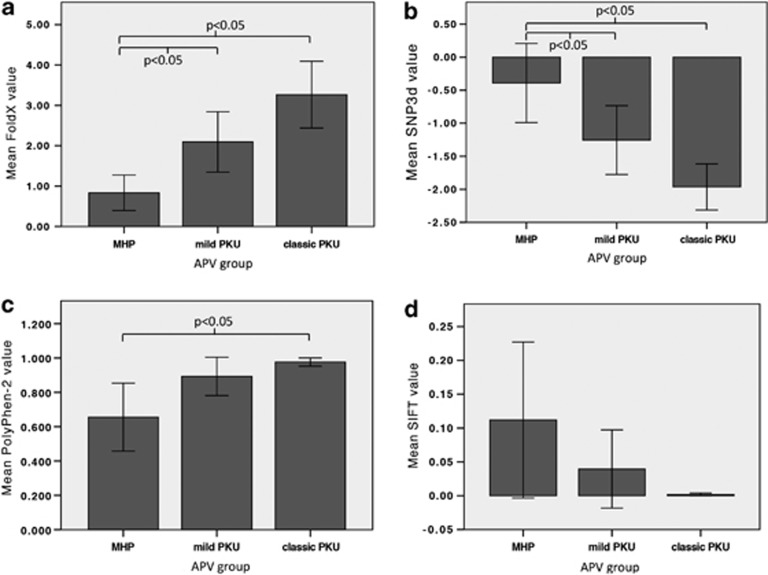
FoldX values correlated moderately with APV (rs(69)=−0.458, P<0.01) and mean FoldX values differed significantly between APV groups. It decreased from 3.27 (±2.52) in APV1 to 2.10 (±1.35) in APV3 to 0.83 (±0.88) in APV8. The decrease from APV1 to APV8 (−2.43, 95% CI: −3.54 to −1.33, P<0.05) was statistically significant as was the decrease from APV3 to APV8 (−1.26, 95% CI: −2.28 to −0.25, P<0.05) (Figure 2a).
Figure 2.
Relationship between different PKU phenotypes and (a) mean FoldX value (rs (70)=−0.446, P<0.001); (b) mean SNPs3D value (rs (89)=0.479, P<0.001); (c) mean PolyPhen-2 value (no relationship); and (d) mean SIFT values (no relationship). Error bars represent the 95% confidence interval.
SNP3D values correlated moderately with APV (rs(83)=0.447, P<0.01) and mean SNPs3D value differed between APV groups (F(2,82)=12.396, P<0.001, ω2=0.211). It increased from −1.96 (±1.11) in APV1 to −1.26 (±1.21) in APV3 to −0.39 (±1.32) in APV8. The increase from APV1 to APV8 (1.57, 95% CI: 0.81–2.33) and the increase from APV3 to APV8 (0.86, 95% CI: 0.01–1.72) were statistically significant (P<0.05) (Figure 2b).
There was a weak to moderate negative correlation between PolyPhen-2 values and APV (rs(83)=−0.411, P<0.01) and mean PolyPhen-2 value differed between APV groups. Only the decrease from APV1 to APV8 (−0.32, 95% CI (−0.56 to −0.08)) was statistically significant (P<0.05), while homogeneity of variances and normality were violated in all APV groups (P<0.001) (Figure 2c). SIFT values (mean 0.04±0.15, SEM=0.02, ranging from 0.00 to 1.00) correlated moderately with APV (rs(83)=0.456, P<0.01), but mean SIFT values did not differ between APV groups (Figure 2d).
Predicting allelic phenotype
Multiple regression analysis was run to predict linearized APV (continuous variable ranging from 1=classic PKU through 3=mild PKU to 8=MHP) from %PAH, FoldX, PolyPhen-2, SNPs3D and SIFT value. We found that %PAH and FoldX together accounted for 64.1%, while %PAH and SNPs3D together accounted for 52.1% of the explained variability in linearized APV, respectively (one outlier c.569T>C was removed because of high leverage). Equations and regression coefficients with standard errors can be found in Supplementary Table S3A.
BIOPKU database
Descriptive analysis
A total of 4181 genotypes corresponding to 8362 alleles were analyzed. Of the 1543 different genotypes, 126 were homozygous (8.2%). The most frequent genotype was c.[1066-11G>A][1066-11G>A] found 137 times (3.3%) followed by c.[1222C>T][1222C>T] (3.1%) and c.[782G>A][782G>A] (2.0%) (Table 1). The overall most frequent allele was c.1222C>T (10.3%) followed by c.1066-11G>A (8.7%) and c.782G>A (6.7%) (Table 2).
Table 1. The most frequent genotypes in the BIOPKU database occurring in ≥0.50% of all patients.
| Genotype | Protein variant | Genotype frequency (%) |
|---|---|---|
| c.[1066-11G>A][1066-11G>A] | p.[(?)][(?)] | 3.28 |
| c.[1222C>T]1222C>T] | p.[Arg408Trp][Arg408Trp] | 3.06 |
| c.[782G>A][782G>A] | p.[Arg261Gln][Arg261Gln] | 1.96 |
| c.[782G>A][1222C>T] | p.[Arg261Gln][Arg408Trp] | 1.22 |
| c.[1315+1G>A][1222C>T] | p.[(?)][Arg408Trp] | 1.22 |
| c.[842C>T][842C>T] | p.[Pro281Leu][Pro281Leu] | 1.12 |
| c.[1222C>T][1241A>G] | p.[Arg408Trp][Tyr414Cys] | 1.03 |
| c.[143T>C][143T>C] | p.[Leu48Ser6;[Leu48Ser] | 1.00 |
| c.[1066-11G>A][782G>A] | p.[(?)][Arg261Gln] | 1.00 |
| c.[1315+1G>A][1241A>G] | p.[(?)][Tyr414Cys] | 0.88 |
| c.[1315+1G>A][1315+1G>A] | p.[(?)][(?)] | 0.86 |
| c.[143T>C][1222C>T] | p.[Leu48Ser][Arg408Trp] | 0.84 |
| c.[838G>A][838G>A] | p.[Glu280Lys][Glu280Lys] | 0.74 |
| c.[1066-11G>A][1222C>T] | p.[(?)][Arg408Trp] | 0.74 |
| c.[143T>C][782G>A] | p.[Leu48Ser][Arg261Gln] | 0.62 |
| c.[1066-11G>A][194T>C] | p.[(?)][Ile65Thr] | 0.60 |
| c.[473G>A][1222C>T] | p.[Arg158Gln][Arg408Trp] | 0.57 |
| c.[194T>C][1222C>T] | p.[Ile65Thr][Arg408Trp] | 0.55 |
| c.[1066-11G>A][1241A>G] | p.[(?)][Tyr414Cys] | 0.55 |
| c.[1066-11G>A][143T>C] | p.[(?)][Leu48Ser] | 0.53 |
| c.[1066-11G>A][898G>T] | p.[(?)][Ala300Ser] | 0.50 |
| c.[1055delG][1055delG] | p.[(Gly352Valfs*48)][(Gly352Valfs*48)] | 0.50 |
The reference accession number for the PAH sequence is ENSG00000171759; RefSeq NM_000277.1.
Table 2. The most frequent alleles in the BIOPKU database occurring in ≥0.50% of all patients.
| Nucleotide aberration | Protein variant | Allele frequency (%) |
|---|---|---|
| c.1222C>T | p.Arg408Trp | 10.32 |
| c.1066-11G>A | p.(?) | 8.71 |
| c.782G>A | p.Arg261Gln | 6.67 |
| c.1315+1G>A | p.(?) | 4.64 |
| c.842C>T | p.Pro281Leu | 3.73 |
| c.1241A>G | p.Tyr414Cys | 3.67 |
| c.143T>C | p.Leu48Ser | 3.64 |
| c.194T>C | p.Ile65Thr | 3.17 |
| c.473G>A | p.Arg158Gln | 2.94 |
| c.1208C>T | p.Ala403Val | 2.28 |
| c.1162G>A | p.Val388Met | 1.97 |
| c.728G>A | p.Arg243Gln | 1.91 |
| c.1169A>G | p.Glu390Gly | 1.64 |
| c.898G>T | p.Ala300Ser | 1.55 |
| c.838G>A | p.Glu280Lys | 1.52 |
| c.754C>T | p.Arg252Trp | 1.29 |
| c.1045T>C | p.Ser349Pro | 1.26 |
| c.727C>T | p.(Arg243*) | 1.24 |
| c.165delT | p.(Phe55Leufs*6) | 1.11 |
| c.1042C>G | p.Leu348Val | 1.05 |
| c.441+5G>T | p.(?) | 1.04 |
| c.781C>T | p.(Arg261*) | 1.02 |
| c.1055delG | p.(Gly352Valfs*48) | 0.86 |
| c.814G>T | p.(Gly272*) | 0.85 |
| c.168+5G>C | p.(?) | 0.83 |
| c.331C>T | p.(Arg111*) | 0.78 |
| c.1223G>A | p.Arg408Gln | 0.68 |
| c.721C>T | p.Arg241Cys | 0.66 |
| c.611A>G | p.(Tyr204Cys) | 0.66 |
| c.204A>T | p.Arg68Ser | 0.63 |
| c.1068C>A | p.(Tyr356*) | 0.55 |
| c.734T>C | p.Val245Ala | 0.53 |
| c.722G>A | p.Arg241His | 0.50 |
The reference accession number for the PAH sequence is ENSG00000171759; RefSeq NM_000277.1.
The 8362 variants predominantly affected the catalytic domain (268 residues, 5095 hits, 60.9%), followed by intronic sites (1683 hits, 20.1%), the regulatory domain (142 residues, 1174 variants, 14.0%) and least the oligomerization domain (42 residues, 410 variants, 4.9%). Of the 8362 variants, 6665 (79.7%) hit exonic sites resulting in an average of 14.7 variants per residue if distributed evenly. In reality, however, the catalytic domain was disproportionately affected with 16.4 variants per residue (+1.7 mpr), as was the oligomerization domain (+1.1 mpr), while the regulatory domain was comparatively least affected (−4.7 mpr) (Supplementary Figure S4). Similarly, 6681 exonic variants (including large deletions) being evenly distributed over 1359 nucleotides theoretically result in 4.9 variants per nucleotide (mpn). In reality, however, exon 7 (+ 9.2 mpn) and exon 12 (+8.2 mpn) were disproportionately more affected. Relatively least affected was exon 13 with only three variants total (0.07 mpn) (Supplementary Figure S5).
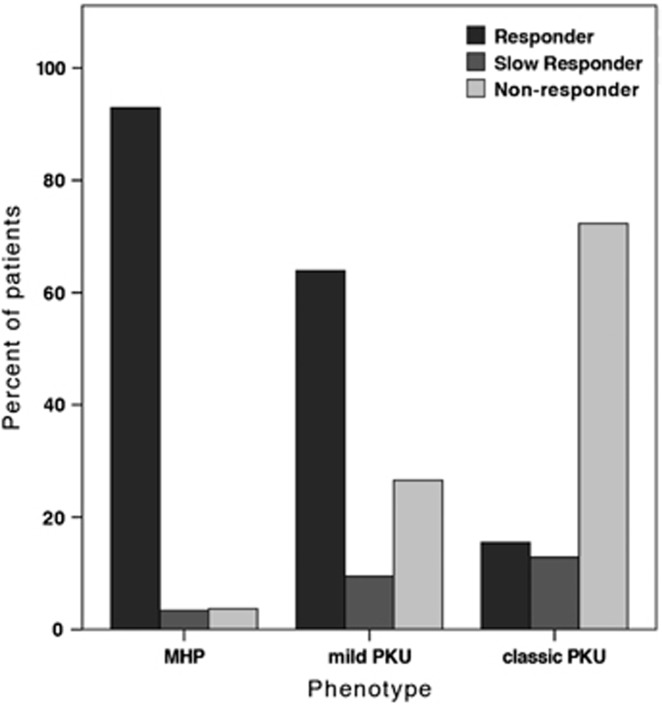
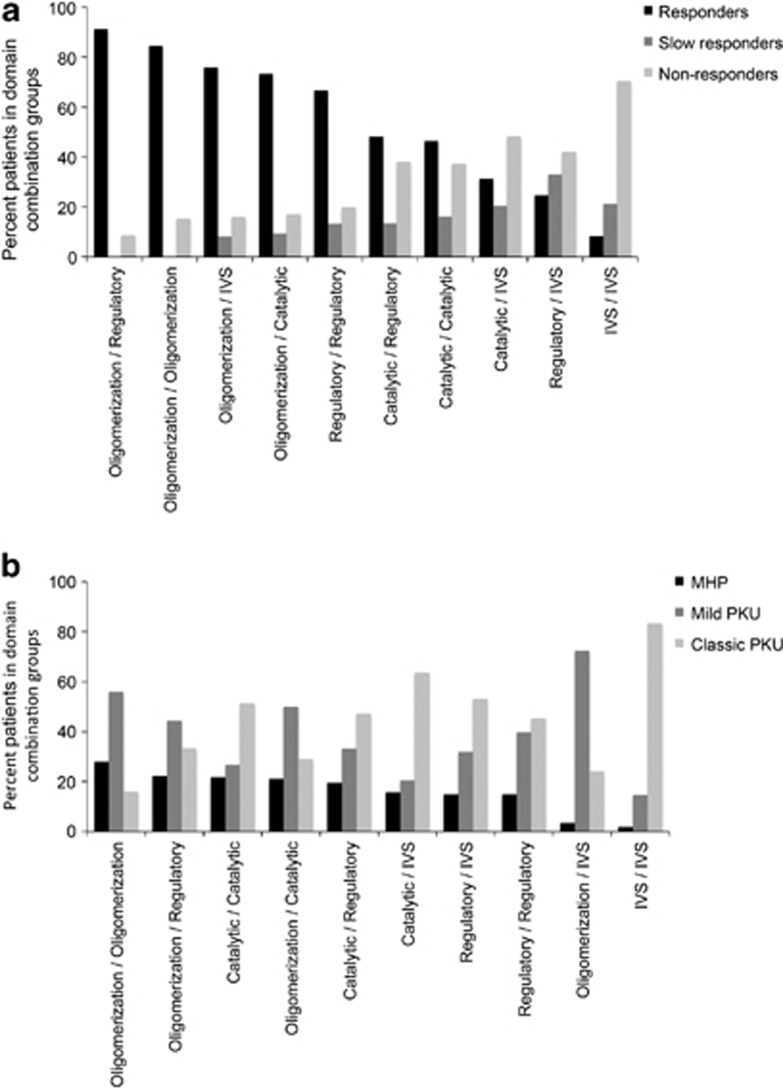
BH4 responsiveness was reported for 2134 out of 4181 patients (51.0%). A total of 936 genotypes were classified as responders (43.8%), 202 as slow responders (9.5%) and 996 as non-responders (46.7%). BH4 responsiveness depended on phenotype: 92.9% of all MHP patients were responders compared with 63.9% in the mild PKU group and 15.5% of classic PKU patients. The latter group included mostly non-responders (71.6%), which applied to only 3.7% of the MHP patients (Figure 3). A Spearman's rank-order correlation revealed a moderate positive correlation between the two variables (rs(1841)=0.621, P<0.01). There was also an association between domain combinations of the genotype and response type. All genotypes with at least one variant in the oligomerization domain showed higher rates of responders than any other domain combination (Figure 4a). To correlate BH4 responsiveness with individual PAH domains, each BIOPKU allele was assigned the genotype's BH4 responsiveness. 77.4% of all variants in the oligomerization domain were associated with a responding phenotype compared with only 51.9% in the regulatory and 47.9% in the catalytic domain. Splice-site variants were part of a responding genotype in 29.9% of all cases. Those being mostly non-homozygous, response was most likely due to the second allele in many cases.
Figure 3.
Percentage of BH4 responders, slow responders and non-responders within PKU phenotype groups.
Figure 4.
(a) Percentage of BH4 responders, slow responders and non-responders in PKU genotypes with different domain combinations; (b) percentage of MHP, mild PKU and classic PKU patients in genotypes with different domain combinations.
Phenotype was known for 3374 out of 4181 genotypes (80.7%). The smallest group was MHP composed of 630 patients (18.7%) followed by mild PKU (30.2%) and classic PKU (51.1%). There was also an association between domain combinations and phenotype: the highest rate of MHP was found in genotypes where both variants affected the oligomerization domain (28.0%) followed by the oligomerization–regulatory (22.2%) and catalytic–catalytic combination (21.8% MHP). Splice site–splice site genotypes showed mostly classic PKU (84.3%) (Figure 4b). Connecting phenotype to individual PAH domains, we found the highest rate of MHP patients in the catalytic domain (20.7%) followed by the oligomerization (18.6%) and regulatory domain (17.5%). Splice-site variants were associated with classic PKU (59.2%).
Predicting disease phenotype
We investigated the possibility of predicting phenotype for genotypes in different constellations: (a) two null-alleles, (b) one null-allele (functionally hemizygous genotypes), (c) two non-null alleles (compound heterozygotes) and (d) two non-null alleles (homozygotes). Potentially, predictive factors were genotype itself and allelic values (APV, %PAH, FoldX, SNPs3D, SIFT Blink and PolyPhen-2).
To determine phenotypic homogeneity of genotypes we considered all cases (n=257) in which phenotype was known for at least three patients (and theoretically all three phenotypes could occur). The majority had a heterogenic phenotype profile (61.1%) with at least two different phenotypes, while in 22 cases (8.6%) even all three phenotypes had been described. Heterogeneity was most abundant in homozygotes (65.0% with two and 15.0% with three different phenotypes described), while deleterious null–null genotypes were most homogenous (57.1% with only one and 40.0% with two described phenotypes). With such heterogeneity at hand it seemed of little use to assign each genotype to one phenotype category. Instead, we determined average phenotype 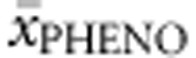 for each genotype. Genotypes with two or less cases were excluded from this analysis (with the exception of deleterious genotypes) to reduce influence of misclassification. Multiple regression analysis was run to predict
for each genotype. Genotypes with two or less cases were excluded from this analysis (with the exception of deleterious genotypes) to reduce influence of misclassification. Multiple regression analysis was run to predict 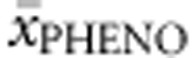 from allelic values. All equations and regression coefficients can be found in Supplementary Table S3A.
from allelic values. All equations and regression coefficients can be found in Supplementary Table S3A.
Deleterious genotypes with two null-alleles. A total of 149 genotypes (774 patients) were classified as deleterious. Phenotype was known for 542 patients (70.4%), 501 of which showed classic PKU, while 41 (7.6%) were classified as mild PKU. Of these 41 patients, 18 with mild phenotypes had at least one c.1066-11G>A allele which might, through alternative splicing, lead to a functional protein. The other 23 patients were most likely misclassified. Excluding c.1066-11G>A allele from analysis would therefore raise correct prediction of classic phenotype by double-null-status close to 100%.
Functionally hemizygous genotypes. A total of 154 genotypes were included in the analysis. All non-null alleles were assigned their concurrent FoldX, PolyPhen-2, SIFT Blink, SNPs3D, APV and %PAH values, and multiple regression analysis was run to predict 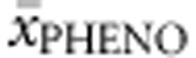 from them. APVnon-null accounted for 76.7% of the variability in
from them. APVnon-null accounted for 76.7% of the variability in 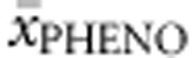 , %PAHnon-null for 32.5% and SNPs3Dnon-null and FoldXnon-null together for 12.4%.
, %PAHnon-null for 32.5% and SNPs3Dnon-null and FoldXnon-null together for 12.4%. 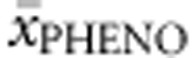 was correctly predicted in up to 94.8%, while 5.2% of all genotypes were classified one category away from
was correctly predicted in up to 94.8%, while 5.2% of all genotypes were classified one category away from 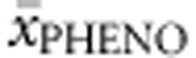 .
.
Compound heterozygotes. Ninety-five genotypes were included in this analysis. All alleles were assigned their concurrent values, which were averaged for the genotype and correlated with 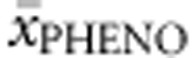 . Multiple regression analysis revealed that
. Multiple regression analysis revealed that 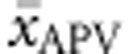 accounted for 66.0% of the explained variability in
accounted for 66.0% of the explained variability in 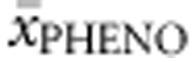 , while
, while 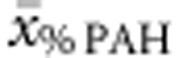 and
and 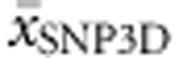 together accounted for 39.4%.
together accounted for 39.4%.
Homozygotes. Twenty-eight homozygous genotypes were included in the analysis. FoldX, PolyPhen-2, SIFT Blink and SNPs3D values as well as APV and %PAH of either allele were correlated with 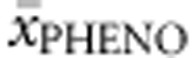 . APV accounted for 81.1% of the explained variability in
. APV accounted for 81.1% of the explained variability in 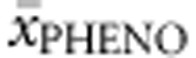 , while %PAH accounted for 27.5% of the explained variability.
, while %PAH accounted for 27.5% of the explained variability.
Predicting BH4 responsiveness
There was also considerable heterogeneity in BH4 responsiveness within genotypes. of all genotypes, 53.8% had a homogenous response profile (27.7% with two types and 18.5% with three response types). This led us to determine average response 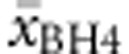 for each genotype. Genotypes with two or less cases were excluded from this analysis (with the exception of deleterious genotypes). Multiple regression analysis was run to predict
for each genotype. Genotypes with two or less cases were excluded from this analysis (with the exception of deleterious genotypes). Multiple regression analysis was run to predict 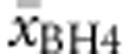 from allelic values. All equations and regression coefficients can be found in Supplementary Table S3B.
from allelic values. All equations and regression coefficients can be found in Supplementary Table S3B.
Deleterious genotypes. All 149 deleterious genotypes (774 patients, 332 for which responsiveness was known) were included in the analysis. Most of the patients for which response was known were non-responders (89.2%), while 10 were classified as responders (3.0%) and 26 as slow responders (7.8%). Five responders and 13 slow responders were part of a genotype including the c.1066-11G>A allele which might, through alternative splicing, lead to a functional protein. All other responding genotypes (n=18) were most likely misclassified as such. Excluding c.1066-11G>A allele from analysis would therefore raise correct prediction of non-responsiveness by double-null-status close to 100%.
Functionally hemizygous genotypes. Seventy-eight hemizygous genotypes were included in the analysis (one outlier c.916A>G/c.1222C>T was removed due to high leverage). Multiple regression analysis revealed that APVnon-null and %PAHnon-null together accounted for 58.4% of the explained variability in BH4 responsiveness.
Compound heterozygotes. A total of 66 compound heterozygous genotypes were included in the analysis. Averaged APV and averaged %PAH together accounted for 39.6% of the explained variability in BH4 responsiveness.
Homozygotes. Sixteen homozygous genotypes were included in the analysis. %PAH accounted for 38.5% and APV for 28.0% of the explained variability in BH4 responsiveness.
Discussion
We have compiled a descriptive analysis of 4181 PKU genotypes in BIOPKU and 834 PAH variants in PAHvdb. Using different algorithms we correlated protein stability, APV and %PAH with patients' phenotype and BH4 responsiveness. The main purpose of our study was to identify factors predictive of allelic and patients' phenotype as well as BH4 responsiveness.
In accordance with previous studies missense variants constituted the largest group of PAH variants. The identification of c.1222C>T as the most frequent allele and c.[1066-11G>A][1066-11G>A] as the most frequent genotype is in agreement with earlier findings.35 The unequal variant density between PAH domains and exons supports the notion of variable residue vulnerability suggesting that nucleotides such as c.1222C, c.143T, c.782G and others are disproportionately more susceptible to alterations.
Missense variants in general have been shown to lead to misfolding of the nascent protein.36 The first large-scale study to investigate and confirm correlations between mutant protein stability (assessed by FoldX algorithm), %PAH and (hemizygous) phenotype was Pey et al.10 They studied 318 PKU-associated missense variants and concluded that decreased protein stability is the main molecular pathogenic mechanism in PKU and the determinant for phenotypic outcome, although additional factors must be considered that may contribute to the patient phenotype. Our study repeats these findings with additional 199 missense variants, supporting the suggested quantitative and causal relationship between enzyme stability, activity and APV. Correlations between FoldX values and APV were similar in the two studies, however, the correlation between our FoldX values and those reported in Pey et al.10 were only moderate (r=0.65). This was most likely due to the different FoldX versions and structures used. The current FoldX program does not include the possibility to test and set an energy penalty (a ceiling for the van der Waals clashes between two atoms) at values higher than 5 kcal/mol. Furthermore, the use of different PDB files (1J8U in this work versus 2PHM, 1PHZ and 2PAH) might have contributed to differing values. We selected the PDB file 1J8U because it covers the region that includes most variants and has the highest resolution for the human enzyme (1.5 Å, vs 3.1 Å for 2PAH). 2PHM and 1PHZ, corresponding to rat PAH, cover a larger section of the sequence including the regulatory ACT domain but have a lower resolution and are further compromised by the need to prepare models of the human enzyme. Using a high-resolution structure is one of the most important criterions for good FoldX prediction of structural impact by variants.13 Compared with the study by Pey et al,10 the use of PDB 1J8U in the present study improves the correlation between ΔΔG values and APV groups significantly (Figure 2a).
APV was best predicted by %PAH, FoldX and SNPs3D algorithms, while PolyPhen-2 and SIFT Blink did not significantly add to the prediction. %PAH showed a strong correlation with APV and distinguished between classic/mild PKU as well as between classic PKU/MHP. Mean %PAH did not differ significantly between mild PKU and MHP indicating other factors might influence phenotype in this range. In accordance with earlier findings, there was a minimum of 13% residual enzyme activity below which APV was usually severe. PolyPhen-2 and SIFT correlations with APV were unstable since data was not normally distributed in either case. The low specificity and lack of correlation with %PAH argues against both tools for phenotype and BH4 responsiveness prediction in PKU.
Our analysis reveals that the majority (61.1%) of all genotypes had a heterogenic phenotype profile. Some of this heterogeneity can likely be attributed to misclassification,37 while it also points to other factors such as individual metabolism influencing blood Phe levels, previously shown in rats.38 Infants are usually screened between days 2 and 7, while samples in the US are usually obtained at 24–48 h when Phe levels might not have peaked yet. While most physicians reported initial blood Phe levels along with attributed severity of PKU in their patients, misclassification could not be entirely excluded where this was not the case. It should be also stressed that PKU is a complex trait disease and that factors other than blood Phe may attribute to the phenotype and thus explain most of outsiders. Thus, the complexity of PKU may be illustrated by (a) different phenotypes that can be accounted for by allelic variation in PAH, (b) the blurring of predicted relationships between genotype and phenotype in PKU, (c) modifier genes and (d) non-genetic factors that contribute to the phenotypes of PKU variants.6 Genotype remains, however, a reliable predictor of patients' phenotype as seen before.39 Our genotype-based approach correctly predicted average phenotype in 89.0% of all genotypes. Not surprisingly, prediction is easiest in deleterious genotypes where double–null-status usually equals classic phenotype.
APV proved to be by far the best overall predictor of patient's phenotype. While average APV was consistently 1 in deleterious genotypes, two severe alleles always excluded MHP phenotype and no MHP allele was ever seen in classic PKU phenotype. Overall, the high predictive value of APV is intuitive as it mirrors blood Phe concentrations most closely. Three factors potentially limit prediction: (1) interallelic complementation in compound heterozygotes (where prediction was least exact), (2) gene dosage in homozygotes and (3) factors other than genotype (eg gene modifiers).
%PAH was consistently<3.0% in deleterious genotypes and an overall moderate predictor of phenotype in other genotype constellations. Accuracy of prediction was higher in compound heterozygotes than in hemizygous and homozygous genotypes.
Overall BH4 responsiveness was 53.3%, which is within the range described in earlier studies.40 The strong association between variants in the oligomerization domain and BH4 responsiveness indicates that BH4 most easily compensates oligomerization defects, which supports a role of BH4 in oligomer assembly as suggested previously.24 Overall, BH4 responsiveness was best estimated by APV and %PAH and correctly predicted in 71.0% of all cases. In accordance with earlier findings,25 BH4 responsiveness required a minimal residual activity of about 20% in hemizygous genotypes, while the threshold was 15% in homozygous and compound heterozygous genotypes. Some splice-site variants (eg, c.1066-3C>T; c.442-5C>G) are clearly associated with mild phenotype and response to BH4.41 It is possible that a BH4-responsive splicing mutation may not be fully penetrant and the gene may produce multiple mRNAs, including some wild-type PAH-mRNA message, and thus result in a small amount of the full-length functional protein. This hypothesis would explain a mild phenotype and BH4 responsiveness in three patients who are homozygous for c.1066-3C>T. The same may be possible for some other splice variants, but was never investigated in detail and is thus only speculative.
To our knowledge this is the largest study using algorithms for the prediction of patients' phenotype and BH4 responsiveness in PKU. Previously frequently used PAH knowledgebase (www.pahdb.mcgill.ca) is no longer up to date (last updated in 2009). Our results using data from the up-to-date LSD PAHvdb and genotype database BIOPKU show that genotype, allelic phenotype, protein stability and residual enzyme activity reliably predict patients' phenotype and BH4 responsiveness. While allelic approaches are limited by interallelic complementation, inconsistencies in genotype–phenotype relationships are most likely due to individual metabolic and genetic properties as well as misclassification of phenotypes. Our findings contribute to a better understanding of relationship between BH4 responsiveness and prediction of disease severity, allowing for a better classification of PKU patients.
Note added to proof
A total of 851 variants were tabulated in the PAHvdb (as of June 2014). Most of them (61.2%) were located in the catalytic domains, followed by the regulatory (16.8%) and oligomerization (5.2%) domains. The rest of the variants were located in non-coding regions of the gene. Out of 851 gene variations, 61.5% were missense variants, 15.0% splice variants, 12.8% deletions, 5.8% nonsense variants, 1.9% insertions, and 3.0% included indels, large deletions and silent variations.
A total of 7453 patients with 1918 different genotypes had been tabulated in BIOPKU as of June 2014. The smallest group was MHP comprising 1161 patients (15.7%), followed by mild PKU (24.1%) and classic PKU (48.2%).
Acknowledgments
Supported by the RD-CONNECT initiative FP7-HEALTH-2012-INNOVATION-1 EU Grant No. 305444 (to NB), the Research Council of Norway, the Western Norway Health Authorities, KG Jebsen Foundation (to AM) and the Fondo de Investigaciones Sanitarias (PI10/00455 to BP). The iSee concept in the PAHvdb is a working collaboration between the Structural Genomics Consortium (registered charity number 1097737) and Molsoft LLC.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Blau N, Van Spronsen FJ, Levy HL. Phenylketonuria. Lancet. 2010;376:1417–1427. doi: 10.1016/S0140-6736(10)60961-0. [DOI] [PubMed] [Google Scholar]
- Koch R, Azen C, Friedman EG, Williamson ML. Paired comparisons between early treated PKU children and their matched sibling controls on intelligence and school achievement test results at eight years of age. J Inherit Metab Dis. 1984;7:86–90. doi: 10.1007/BF01805813. [DOI] [PubMed] [Google Scholar]
- Waisbren SE, Noel K, Fahrbach K, et al. Phenylalanine blood levels and clinical outcomes in phenylketonuria: a systematic literature review and meta-analysis. Mol Genet Metab. 2007;92:63–70. doi: 10.1016/j.ymgme.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Pey AL, Desviat LR, Gamez A, Ugarte M, Perez B. Phenylketonuria: genotype-phenotype correlations based on expression analysis of structural and functional mutations in PAH. Hum Mutat. 2003;21:370–378. doi: 10.1002/humu.10198. [DOI] [PubMed] [Google Scholar]
- Guldberg P, Rey F, Zschocke J, et al. A European multicenter study of phenylalanine hydroxylase deficiency: classification of 105 mutations and a general system for genotype-based prediction of metabolic phenotype. Am J Hum Genet. 1998;63:71–79. doi: 10.1086/301920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriver CR, Waters PJ. Monogenic traits are not simple: lessons from phenylketonuria. Trends Genet. 1999;15:267–272. doi: 10.1016/s0168-9525(99)01761-8. [DOI] [PubMed] [Google Scholar]
- Jennings IG, Cotton RG, Kobe B. Structural interpretation of mutations in phenylalanine hydroxylase protein aids in identifying genotype-phenotype correlations in phenylketonuria. Eur J Hum Genet. 2000;8:683–696. doi: 10.1038/sj.ejhg.5200518. [DOI] [PubMed] [Google Scholar]
- Marvit J, DiLella AG, Brayton K, Ledley FD, Robson KJ, Woo SL. GT to AT transition at a splice donor site causes skipping of the preceding exon in phenylketonuria. Nucleic Acids Res. 1987;15:5613–5628. doi: 10.1093/nar/15.14.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamez A, Perez B, Ugarte M, Desviat LR. Expression analysis of phenylketonuria mutations. Effect on folding and stability of the phenylalanine hydroxylase protein. J Biol Chem. 2000;275:29737–29742. doi: 10.1074/jbc.M003231200. [DOI] [PubMed] [Google Scholar]
- Pey AL, Stricher F, Serrano L, Martinez A. Predicted effects of missense mutations on native-state stability account for phenotypic outcome in phenylketonuria, a paradigm of misfolding diseases. Am J Hum Genet. 2007;81:1006–1024. doi: 10.1086/521879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersting SW, Kemter KF, Staudigl M, et al. Loss of function in phenylketonuria is caused by impaired molecular motions and conformational instability. Am J Hum Genet. 2008;83:5–17. doi: 10.1016/j.ajhg.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doskeland AP, Flatmark T. Recombinant human phenylalanine hydroxylase is a substrate for the ubiquitin-conjugating enzyme system. Biochem J. 1996;319:941–945. doi: 10.1042/bj3190941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Peng H, Peck D, White DA, Christ SE. Tract-based evaluation of white matter damage in individuals with early-treated phenylketonuria. J Inherit Metab Dis. 2013;37:237–243. doi: 10.1007/s10545-013-9650-y. [DOI] [PubMed] [Google Scholar]
- Heintz C, Cotton RG, Blau N. Tetrahydrobiopterin, its mode of action on phenylalanine hydroxylase and importance of genotypes for pharmacological therapy of phenylketonuria. Hum Mutat. 2013;34:927–936. doi: 10.1002/humu.22320. [DOI] [PubMed] [Google Scholar]
- Kure S, Hou DC, Ohura T, et al. Tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency. J Pediatr. 1999;135:375–378. doi: 10.1016/s0022-3476(99)70138-1. [DOI] [PubMed] [Google Scholar]
- Levy H, Milanowski A, Chakrapani A, et al. Efficacy of sapropterin dihydrochloride (tetrahydrobiopterin, 6R-BH4) for reduction of phenylalanine concentration in patients with phenylketonuria: a phase III randomized placebo-controlled study. Lancet. 2007;370:504–510. doi: 10.1016/S0140-6736(07)61234-3. [DOI] [PubMed] [Google Scholar]
- Keil S, Anjema K, van Spronsen FJ, et al. Long-term follow-up and outcome of phenylketonuria patients on sapropterin: A retrospective study. Pediatrics. 2013;131:e1881–e1888. doi: 10.1542/peds.2012-3291. [DOI] [PubMed] [Google Scholar]
- Fiege B, Blau N. Assessment of tetrahydrobiopterin (BH4)-responsiveness in phenylketonuria. J Pediatr. 2007;150:627–630. doi: 10.1016/j.jpeds.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Blau N, Erlandsen H. The metabolic and molecular bases of tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency. Mol Genet Metab. 2004;82:101–111. doi: 10.1016/j.ymgme.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Erlandsen H, Pey AL, Gamez A, et al. Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations. Proc Natl Acad Sci USA. 2004;101:16903–16908. doi: 10.1073/pnas.0407256101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pey AL, Perez B, Desviat LR, et al. Mechanisms underlying responsiveness to tetrahydrobiopterin in mild phenylketonuria mutations. Hum Mutat. 2004;24:388–399. doi: 10.1002/humu.20097. [DOI] [PubMed] [Google Scholar]
- Zurflüh MR, Zschocke J, Lindner M, et al. Molecular genetics of tetrahydrobiopterin responsive phenylalanine hydroxylase deficiency. Hum Mutat. 2008;29:167–175. doi: 10.1002/humu.20637. [DOI] [PubMed] [Google Scholar]
- Waters PJ, Parniak MA, Nowacki P, Scriver CR. In vitro expression analysis of mutations in phenylalanine hydroxylase: linking genotype to phenotype and structure to function. Hum Mutat. 1998;11:4–17. doi: 10.1002/(SICI)1098-1004(1998)11:1<4::AID-HUMU2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Heintz C, Dobrowolski SF, Andersen HS, Demirkol M, Blau N, Andresen BS. Splicing of phenylalanine hydroxylase (PAH) exon 11 is vulnerable: molecular pathology of mutations in PAH exon 11. Mol Genet Metab. 2012;106:403–411. doi: 10.1016/j.ymgme.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- Kwok SC, Ledley FD, DiLella AG, Robson KJ, Woo SL. Nucleotide sequence of a full-length complementary DNA clone and amino acid sequence of human phenylalanine hydroxylase. Biochemistry. 1985;24:556–561. doi: 10.1021/bi00324a002. [DOI] [PubMed] [Google Scholar]
- Taschner PE, den Dunnen JT. Describing structural changes by extending HGVS sequence variation nomenclature. Hum Mutat. 2011;32:507–511. doi: 10.1002/humu.21427. [DOI] [PubMed] [Google Scholar]
- Blau N, Belanger-Quintana A, Demirkol M, et al. Optimizing the use of sapropterin (BH4) in the management of phenylketonuria. Mol Genet Metab. 2009;96:158–163. doi: 10.1016/j.ymgme.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Castellana S, Mazza T. Congruency in the prediction of pathogenic missense mutations: state-of-the-art web-based tools. Brief Bioinform. 2013;14:448–459. doi: 10.1093/bib/bbt013. [DOI] [PubMed] [Google Scholar]
- Andersen OA, Flatmark T, Hough E. High Resolution crystal structures of the catalytic domain of human phenylalanine hydroxylase in its catalytically active Fe(II) form and binary complex with tetrahydrobiopterin. J Mol Biol. 2001;314:266–278. doi: 10.1006/jmbi.2001.5061. [DOI] [PubMed] [Google Scholar]
- Lee WH, Atienza-Herrero J, Abagyan R, Marsden BD. SGC—structural biology and human health: a new approach to publishing structural biology results. PLoS One. 2009;4:e7675. doi: 10.1371/journal.pone.0007675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisensmith RC, Goltsov AA, O'Neill C, et al. Recurrence of the R408W mutation in the phenylalanine hydroxylase locus in Europeans. Am J Hum Genet. 1995;56:278–286. [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- Blau N, Belanger-Quintana A, Demirkol M, et al. Management of phenylketonuria in Europe: survey results from 19 countries. Mol Genet Metab. 2010;99:109–115. doi: 10.1016/j.ymgme.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Salter M, Knowles RG, Pogson CI. Quantification of the importance of individual steps in the control of aromatic amino acid metabolism. Biochem J. 1986;234:635–647. doi: 10.1042/bj2340635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayaalp E, Treacy E, Waters PJ, Byck S, Nowacki P, Scriver CR. Human phenylalanine hydroxylase mutations and hyperphenylalaninemia phenotypes: a metanalysis of genotype-phenotype correlations. Am J Hum Genet. 1997;61:1309–1317. doi: 10.1086/301638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernegger C, Blau N. High frequency of tetrahydrobiopterin-responsiveness among hyperphenylalaninemias: a study of 1919 patients observed from 1988 to 2002. Mol Genet Metab. 2002;77:304–313. doi: 10.1016/s1096-7192(02)00171-3. [DOI] [PubMed] [Google Scholar]
- Desviat LR, Pérez B, Bèlanger-Quintana A, et al. Tetrahydrobiopterin responsiveness: results of the BH4 loading test in 31 Spanish PKU patients and correlation with their genotype. Mol Genet Metab. 2004;82:157–162. doi: 10.1016/j.ymgme.2004.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.