A single mechanism exists of gibberellin perception for gene expression in rice aleurone cells.
Abstract
Current gibberellin (GA) research indicates that GA must be perceived in plant nuclei by its cognate receptor, GIBBERELLIN INSENSITIVE DWARF1 (GID1). Recognition of GA by GID1 relieves the repression mediated by the DELLA protein, a model known as the GID1-DELLA GA perception system. There have been reports of potential GA-binding proteins in the plasma membrane that perceive GA and induce α-amylase expression in cereal aleurone cells, which is mechanistically different from the GID1-DELLA system. Therefore, we examined the expression of the rice (Oryza sativa) α-amylase genes in rice mutants impaired in the GA receptor (gid1) and the DELLA repressor (slender rice1; slr1) and confirmed their lack of response to GA in gid1 mutants and constitutive expression in slr1 mutants. We also examined the expression of GA-regulated genes by genome-wide microarray and quantitative reverse transcription-polymerase chain reaction analyses and confirmed that all GA-regulated genes are modulated by the GID1-DELLA system. Furthermore, we studied the regulatory network involved in GA signaling by using a set of mutants defective in genes involved in GA perception and gene expression, namely gid1, slr1, gid2 (a GA-related F-box protein mutant), and gamyb (a GA-related trans-acting factor mutant). Almost all GA up-regulated genes were regulated by the four named GA-signaling components. On the other hand, GA down-regulated genes showed different expression patterns with respect to GID2 and GAMYB (e.g. a considerable number of genes are not controlled by GAMYB or GID2 and GAMYB). Based on these observations, we present a comprehensive discussion of the intricate network of GA-regulated genes in rice aleurone cells.
GAs comprise a large family of tetracyclic diterpenoid plant hormones involved in a wide range of plant growth responses, including seed germination, stem elongation, leaf expansion, flowering, and pollen maturation (Richards et al., 2001; Thomas et al., 2005). In the past decade, genetic studies of GA-signaling mutants of rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana) have revealed factors essential for GA perception, including the GA receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1; Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006), the GA signaling repressor protein DELLA (Peng et al., 1997; Itoh et al., 2002), and an F-box protein for DELLA degradation, GID2/SLEEPY1 (SLY1; McGinnis et al., 2003; Sasaki et al., 2003). Functional analyses of these proteins have enabled us to construct a molecular model for GA signaling (Ueguchi-Tanaka et al., 2007; Davière and Achard, 2013; Locascio et al., 2013). In this model, the DELLA protein represses GA action in the absence of GA. When GA is present, the GID1 receptor binds with GA and develops the ability to interact with DELLA. Interaction triggers the degradation of DELLA via the SCFGID2/SLY1 proteasome pathway. The SCF complex is an E3 ligase that consists of S-phase kinase-associated protein1 (Skp1), Cullin, F-box protein, and a RING-H2 motif, which add a polyubiquitin chain to DELLA, thus inducing degradation via the 26S proteasome complex. The ensuing degradation of DELLA triggers GA action.
Prior to the establishment of the GID1-DELLA GA perception system, researchers speculated about the existence of a gibberellin-binding protein (GBP) in the aleurone cells of cereal species. Jelsema et al. (1977) first reported GBP activity in aleurone homogenates derived from wheat (Triticum aestivum) seed. Hooley and colleagues (Hooley et al., 1991; Beale et al., 1992) demonstrated that α-amylase can be induced in aleurone protoplasts by the application of a GA derivative that was impermeable with respect to plasma membranes. When GA was microinjected into the cytoplasm of barley (Hordeum vulgare) aleurone protoplasts, Gilroy and Jones (1994) reported the absence of α-amylase induction. Later, others examined GBPs in the plasma membrane of common wild oat (Avena fatua) aleurone cells using photoaffinity-labeled GA and discovered two GBPs. These were a 60-kD protein localized in the microsomal fraction (Hooley et al., 1993) and a 50-kD protein in the cytosolic fraction (Walker et al., 1994). Two other GBPs of 68 and 18 kD were detected in the plasma membrane fraction of oat (Avena sativa) aleurones using the same photoaffinity-labeling method (Lovegrove et al., 1998). These observations strongly suggest that GA perception occurs in the plasma membrane, which induces α-amylase expression. Thus, there should be a GA receptor in the plasma membrane of cereal aleurone cells. The above-mentioned characteristics of GBPs cannot be explained with respect to GID1; for example, rice GID1 is a 40-kD soluble protein that localizes to the nucleus (not the plasma membrane). Furthermore, the GID1-DELLA interaction is thought to occur primarily in the nucleus because of the nuclear localization of DELLA proteins (Ueguchi-Tanaka et al., 2005). Although the identity of GID1 as the GA receptor has been established for many years, the potential existence of GBP in aleurone cells has persisted. Several reviews and textbooks contain information about an alternative GA perception mechanism that is mediated by GBPs (Hartweck and Olszewski, 2006; Ueguchi-Tanaka et al., 2007; Taiz and Zeiger, 2010). To investigate this alternative mechanism, we examined the expression of GA-regulated genes in half-seeds of rice (Oryza sativa) lacking embryos. This research utilized four GA signaling mutants containing defects in GID1, rice DELLA protein SLENDER RICE1 (SLR1), GA-related F-box protein GID2, and GA-related MYB (GAMYB) proteins (for the presumed functions of these components, see Supplemental Fig. S1). These proteins are encoded by single genes in rice, which facilitated our study of the GA perception system. The comprehensive microarray analysis using these mutants revealed that the genes responsive to GA in the wild-type aleurone cells did not respond to GA in the gid1 and slr1 mutants. This observation clearly demonstrates that GA perception is solely undertaken by the GID1-DELLA system and negates the idea of an alternative system at the level of gene expression. Furthermore, based on transcriptome and quantitative reverse transcription (qRT)-PCR analyses, we present a comprehensive discussion of GA signaling that begins with the perception of GA by GID1 and ends with the expression of downstream genes.
RESULTS
Expression of Rice α-amylase Genes in Different GA Mutants
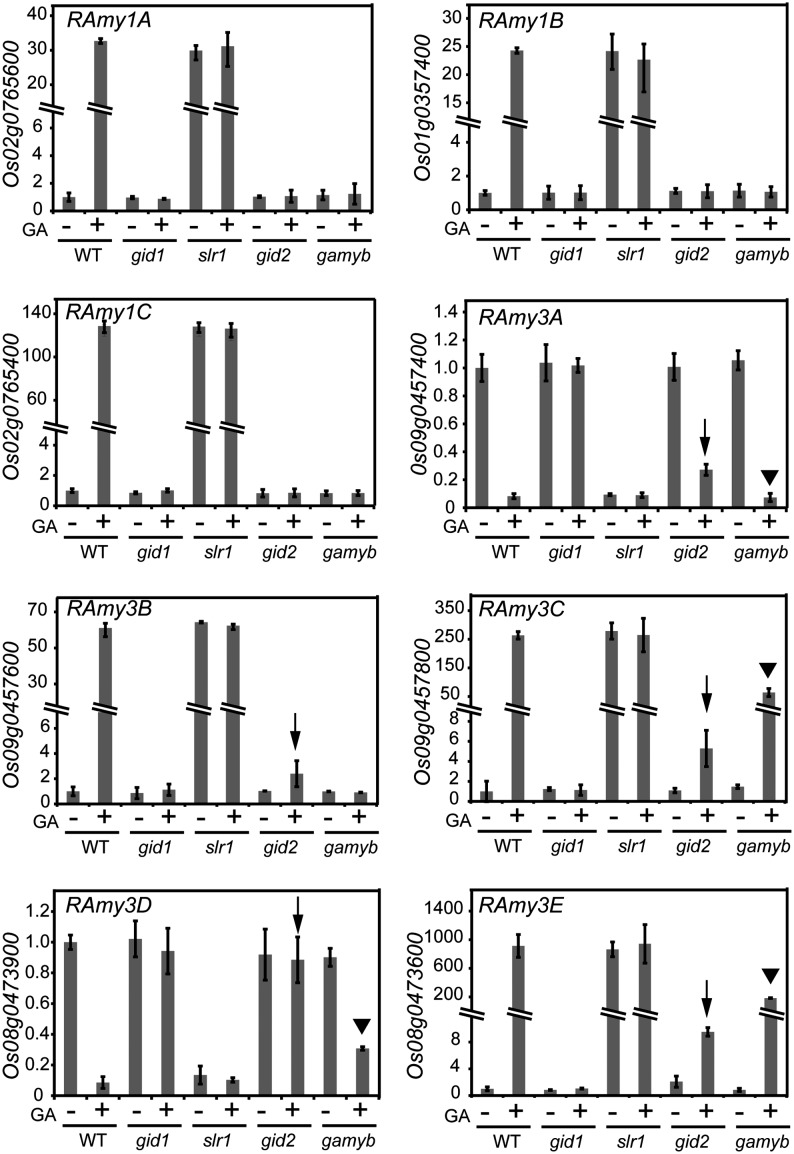
We used halved seeds lacking embryos; these were obtained from rice GA-signaling mutants, namely gid1-4, slr1-1, and gid2-5 (null mutants for GID1, DELLA, and GID2, respectively). We also included gamyb-2, a null mutant for GAMYB, which is an important transcription factor (TF) for GA signaling in cereals (Gubler et al., 1995; Kaneko et al., 2004). We first examined the expression pattern of Rice α-amylase (RAmy) genes in the mutant half-seeds by qRT-PCR, because it has been reported that RAmy genes might be induced in cereal aleurone cells though the GBP activity, as mentioned previously. There are nine genes of RAmy in the rice genome (Rice Annotation Project database; http://rapdb.dna.affrc.go.jp/). Among them, RAmy1A, RAmy1B, RAmy1C, RAmy3B, RAmy3C, and RAmy3E were greatly up-regulated by GA in wild-type seeds (Fig. 1); up-regulation of these RAmy genes did not occur in the gid1 mutant background. In slr1 seeds, regardless of GA treatment, the RAmy genes were expressed at levels similar to the GA-treated wild-type seeds. These results indicate that the induction of RAmy genes completely depends on the GID1-DELLA system. In gid2 seeds, gene expression was generally not induced; exceptions included RAmy3B, RAmy3C, and RAmy3E, which showed slight but significant up-regulation by GA (Fig. 1, arrows). This indicates that the degradation of SLR1 by GID2 is a prerequisite to fully induce the expression of these genes, but some RAmy genes are induced slightly without SLR1 degradation. In the case of gamyb seeds, the expression of RAmy1A, RAmy1B, RAmy1C, and RAmy3B was not induced by GA, whereas RAmy3C and RAmy3E were up-regulated by GA (Fig. 1, arrowheads). These findings indicate that RAmy induction depends on GAMYB; however, there may be a bifurcation for RAmy3C and RAmy3E expression that does not require GAMYB (see “Discussion”). For the RAmy2A gene, it was also up-regulated in the wild type by GA, but only very slightly, and its expression pattern was essentially the same as that of RAmy1 genes (Supplemental Fig. S2).
Figure 1.
Expression of RAmy genes in gid1, slr1, gid2, and gamyb mutant seeds. Embryoless half-seeds of the wild type (WT) and the four GA-related mutants were incubated with (+) or without (−) 10−5 m GA3 for 36 h. RAmy expression was normalized by the Ubiquitin (Ubi) expression in each sample and shown in comparison with that in GA-untreated wild-type seeds. Arrows and arrowheads indicate the induction and suppression of RAmy genes by GA in gid2 and gamyb, respectively. The lack of suppression of RAmy3D in gid2 is also indicated by an arrow. The data represent means ± sd of four replicates.
In contrast to the above-mentioned RAmy genes, the expression of RAmy3A and RAmy3D was down-regulated by GA in the wild-type seeds but not in the gid1 background (Fig. 1). Expression of these two genes did not occur in slr1 seeds regardless of GA treatment. These results indicate that suppression of RAmy3A and RAmy3D by GA is negatively regulated by the GID1-DELLA perception system. As for gid2 seeds, RAmy3A expression was suppressed significantly (but not completely) by GA; RAmy3D expression was not down-regulated in the gid2 background (Fig. 1, arrows). In the gamyb background, the down-regulation of RAmy3A by GA was almost complete, while RAmy3D expression was only partially suppressed (Fig. 1, arrowheads).
In Silico Analysis of GA-Regulated Transcriptomes in Signaling Mutants
Microarray experiments were conducted to examine the change in GA-regulated expression profiles in half-seeds lacking embryos. Based on these results, 615 and 704 probes corresponding to 447 up-regulated and 471 down-regulated genes, respectively, showed significant alterations in gene expression in wild-type seeds (Supplemental Tables S1 and S2, respectively). Using these GA-regulated genes, enrichment analysis of Gene Ontology (GO) terms was performed using agriGO (Du et al., 2010; Supplemental Fig. S3). With regard to the molecular function of GA up-regulated proteins, the GO term “hydrolase activity” was extremely enriched (P = 1.83e−05), indicating the increased predominance of hydrolases, including α-amylase, glucanase, and proteinase, which hydrolyze glycosyl and peptide bonds that reside in various compounds present in germinating seeds. In terms of cellular components, the extracellular region, cell wall, endoplasmic reticulum, and vacuole were significantly enriched. This trend seems logical considering the role of hydrolases and transporters in catabolism, which includes cell wall loosening and endosperm weakening during seed germination.
In the case of GA down-regulated genes, transporter activity was enriched (P = 0.00446), and this indicates the increased molecular function of cation:sugar, solute:hydrogen, and sugar:hydrogen symporters involved in the uptake of sugars into cells. In terms of biological processes, the GO terms “response to endogenous stimulus” and “response to abiotic stimulus” were enriched, which relates to genes stimulated by abscisic acid, such as ABSCISIC ACID-INSENSITIVE1 (ABI1), ABI5, and SNF1-RELATED PROTEIN KINASE6 (Gosti et al., 1999; Finkelstein and Lynch, 2000; Nakashima et al., 2009), supporting cross-talk between GA and ABA during seed germination (Ho et al., 2003).
GA-Regulated Gene Expression in Rice Aleurones Depends on GID1-DELLA
We compared the effect of GA on the expression of GA-regulated genes in wild-type and gid1 seeds to identify those that are not controlled by GID1. In Figure 2A, the y axis shows the log10 ratio of the GA-induced signal intensity change in wild-type seeds. Similarly, the x axis represents the log10 value of the change between GA-treated and untreated gid1 seeds. In this analysis, the clustering of data points (probes) around the y axis (x = 0 line) implies that the GID1-mediated system is the sole mechanism for GA perception. Although most probes were concentrated around the y axis, as indicated by the regression line (Fig. 2A), there were six outliers (Fig. 2, A and B) beyond the 3-fold difference (±0.48 differences in log10 ratio) in cutoff points (Fig. 2A, dashed lines). Thus, we directly examined the change in expression of corresponding genes in wild-type and gid1 seeds by qRT-PCR (Fig. 2C). All of the genes were up- or down-regulated by GA in the wild type at different levels in qRT-PCR, but their expression was not altered significantly in gid1 seeds, where expression was comparable to that in wild-type seeds not treated with GA (Fig. 2C; RAmy3B data are presented in Fig. 1). These results clearly demonstrate that all GA-related changes in expression depend on GID1 activity.
Figure 2.
Contribution of GID1 in GA-dependent gene expression. A, RNAs were independently extracted from wild-type (WT) and gid1 embryoless half-seeds and used for microarray analysis. The y axis represents log10 ratios of the signal intensities of corresponding probes in GA-treated and untreated wild-type seeds, whereas the x axis shows those of gid1 seeds. Probes that were up- or down-regulated by GA more than 3-fold (±0.48 differences in log10 ratios shown by dashed lines) in gid1 are noted as numbered black dots. RAmy3B (probe 1) is indicated. B, List of probes exceeding ±0.48 differences in log10 ratios in gid1. A single gene is often represented by more than one probe. WT ± GA and gid1 ± GA represent fold differences in log10 values between GA-treated and untreated seeds of the wild type and gid1. The annotation for each gene is derived from the Rice Annotation Project database (http://rapdb.dna.affrc.go.jp/). C, qRT-PCR analysis of the genes listed in B shown in the same manner as in Figure 1. The numbers at the top left side of each graph correspond to the same probes presented in A. The qRT-PCR result for RAmy3B is shown in Figure 1. The data presented are means ± sd of four replicates.
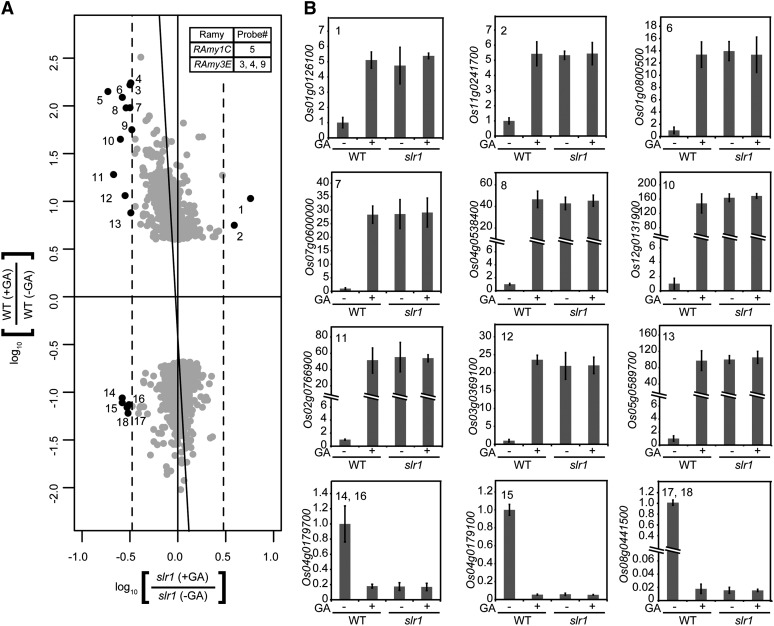
We next examined the involvement of SLR1 in GA perception using the method described above (Fig. 3). Almost all probes were concentrated around the y axis (Fig. 3A), suggesting that SLR1 controls the expression of corresponding genes. However, similar to the results obtained for gid1, 18 outlying probes corresponding to 11 up-regulated genes (including RAmy1C and RAmy3E) and three down-regulated genes were observed (Fig. 3A; Supplemental Table S3). We examined the change in expression for all corresponding genes by qRT-PCR (Fig. 3B). In the wild type, these genes were up- or down-regulated by GA at different levels. In contrast, their expression was not significantly altered in slr1 seeds and was comparable with that of the wild-type seeds treated with GA (Fig. 3B; RAmy1C and RAmy3E data are presented in Fig. 1). These results demonstrate that all GA-related changes in expression depend on SLR1 activity and thus are mediated by the GID1-DELLA-GA perception system.
Figure 3.
Contribution of SLR1 to GA-related gene expression. A, Data are presented essentially as shown in Figure 2A, except that the x axis shows the signal intensity change in slr1 seeds. A table indicating RAmy genes and their corresponding probes is included at top right. B, qRT-PCR analysis of the genes corresponding to the numbered probes in A is shown as the same presentation in Figure 2C. The qRT-PCR results for RAmy1C and RAmy3B are presented in Figure 1. The data represent means ± sd of four replicates. WT, Wild type.
Involvement of GID2 and GAMYB in GA Signaling
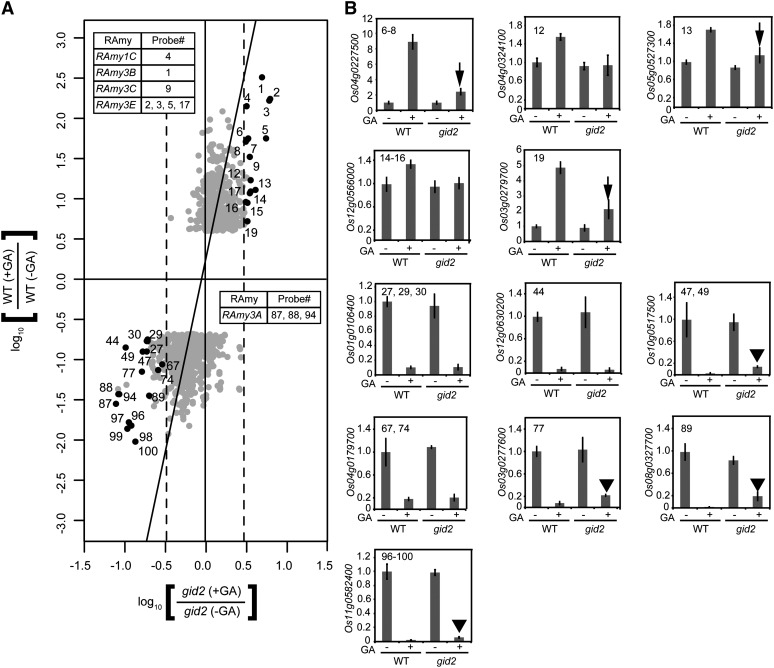
We also examined the involvement of GID2 in the expression of GA-regulated genes using the same approach (Fig. 4A). The probes were concentrated around the y axis, with 100 outliers (Supplemental Table S4). Among these, 22 probes corresponded to 12 GA up-regulated genes. Of these, we examined the expression of nine genes (corresponding to 16 probes; Fig. 4A); four of these were RAmy1C, RAmy3B, RAmy3C, and RAmy3E (Fig. 1). In the qRT-PCR analysis, up-regulation of all the genes by GA was inhibited in gid2 seeds (Fig. 4B), although six genes (RAmy3B, RAmy3C, RAmy3E, Os04g0227500, Os5g0527300, and Os03g0279700) were slightly up-regulated (Figs. 1 and 4B, arrows). These results demonstrate that the GA-dependent up-regulation of most genes is dependent on GID2 function. For the GA down-regulated genes, 49 genes corresponding to 78 probes were identified that deviated from the untreated control by more than 3-fold. Among these, we examined the expression of eight genes corresponding to 18 randomly selected probes that include RAmy3A (Fig. 1). These genes were down-regulated by GA in gid2 seeds to the level observed in GA-treated wild-type seeds (Fig. 4B), whereas the expression of several genes (Os10g0517500, Os03g0277600, Os08g0327700, and Os11g0582400) was significantly but not completely down-regulated (Fig. 4B, arrowheads). These results demonstrate that the expression of several genes is down-regulated by GA independent of GID2, in contrast to gid1 and slr1.
Figure 4.
Contribution of GID2 to GA-related gene expression. A, The data are presented as shown in Figure 2A, except that the x axis shows the signal intensity change in gid2 seeds. Probes examined by qRT-PCR (16 of 22 probes representing up-regulated genes and 18 of 78 probes for down-regulated genes) are demarcated with numbered black dots. B, qRT-PCR expression analysis of genes corresponding to numbered probes in A is shown as the same presentation in Figure 2C. Arrows and arrowheads indicate partial induction and incomplete suppression, respectively, by GA in gid2. The data presented are means ± sd of four replicates. WT, Wild type.
We also examined the involvement of GAMYB (Fig. 5A). The probes were concentrated around the y axis, with a considerable number of outliers in GA down-regulated genes (Supplemental Table S5). For GA up-regulated genes, there were 28 genes corresponding to 31 probes. Among these, the expression of six up-regulated genes, namely RAmy3E (Fig. 1) and Os02g0740400, Os04g0227500, Os05g0527300, Os03g0279700, and Os04g0364800 (Fig. 5B), was examined by qRT-PCR. In general, induction of these genes in GA-treated gamyb was lower than that observed in GA-treated wild-type seeds (Fig. 5B, arrowheads). These results suggest that most of the GA up-regulated genes depend on GAMYB function, although several genes are slightly up-regulated in the absence of GAMYB. As for GA down-regulated genes, there were 64 genes corresponding to 92 probes that deviated from the untreated control by more than 3-fold. In contrast to the up-regulated genes, down-regulated genes were broadly dispersed around the y axis and beyond the y = x (red) line, although the y axis had a greater density of probes (Fig. 5A). qRT-PCR was conducted on seven genes corresponding to 13 probes (dispersed around the y = x line; purple dots), and eight genes corresponding to 10 probes were located between the y = x and x = −0.48 lines (more than 3-fold difference; green dots). All genes clustering around the y = x line (purple dots) were down-regulated by GA in gamyb to the levels observed in GA-treated wild-type seeds, whereas the genes lying between y = x and x = −0.48 (green dots) were significantly but not completely down-regulated in gamyb (Fig. 5B). These results demonstrate that some genes can be down-regulated by GA in the absence of GAMYB.
Figure 5.
Contribution of GAMYB in GA-related gene expression. A, The presentation is essentially the same as in Figure 2A, except that the x axis shows the signal intensity change in gamyb seeds. Probes examined by qRT-PCR are presented as black (up-regulated), purple (down-regulated; dispersed around the y = x line), and green (down-regulated; dispersed between y = x and x = −0.48) numbered dots. B, qRT-PCR expression analysis of genes corresponding to the numbered probes in A is shown as the same presentation in Figure 2C. Arrowheads indicate partial induction in gamyb. The data presented are means ± sd of four replicates. WT, Wild type.
Hierarchical Cluster Analysis of the GA-Signaling Mutants
To elucidate the relationship of the four GA-signaling components, we performed a cluster analysis of GA up- and down-regulated genes by hclust hierarchical clustering (Ben-Hur et al., 2002; Smolkin and Ghosh, 2003). The first group, Up-group1, contained the majority of GA up-regulated genes (435 genes), which includes the six GA up-regulated forms of RAmy (Supplemental Table S6). The Up-group1 genes were generally not induced in the four mutants (Fig. 6A [the expression of two representative genes is denoted with purple lines]; Supplemental Fig. S4, A and B); however, some genes deviated by more than 3-fold (Fig. 6A, dashed lines). Outliers in the gid1 and slr1 mutant backgrounds were directly examined by qRT-PCR, as mentioned previously, which confirmed that their expression was controlled by GID1 and SLR1 (Figs. 2 and 3). In gid2, the change in expression of RAmy genes sometimes deviated by more than a 3-fold increase (Fig. 6A, light blue lines). This observation is consistent with the qRT-PCR results (Fig. 1), namely, that RAmy3B, RAmy3C, and RAmy3E expression was partially induced in gid2. The second group of GA up-regulated genes, Up-group2, was small and contained only 12 genes with expression patterns similar to Up-group1 (Fig. 6B; Supplemental Table S7). The expression patterns of two representative genes from Up-group2 are shown in Supplemental Fig. S4, C and D and Fig. 6B (purple lines). Os04g0364800 showed significant down-regulation in the gamyb mutant in the microarray analysis (Fig. 6B, arrowhead), but qRT-PCR demonstrated no change in expression (Supplemental Fig. S4D, arrowhead). Although Up-group2 has two upward (+) outliers in slr1, qRT-PCR analysis showed that their expression was not significantly different from the wild type (Fig. 3). Taken collectively, cluster analysis confirmed that most GA up-regulated genes fall under the strict control of the four GA-signaling components, GID1, SLR1, GID2, and GAMYB.
Figure 6.
Expression profiles classified by hierarchical cluster analysis. The y axis represents log10 ratios of the signal intensities of corresponding probes of GA-treated and untreated seeds of plants indicated on the x axis, namely the wild type (WT), gid1, slr1, gid2, and gamyb. Threefold differences in intensity (±0.48 differences in log10 ratio) are indicated as dashed lines. Purple lines represent the expression profiles of two representative genes for each group, which were confirmed by qRT-PCR (Supplemental Figs. S4–S7). The expression profiles of RAmy genes are shown as light blue lines in A, those of RAmy1A, RAmy1B, RAmy1C, RAmy3B, RAmy3C, RAmy3E, and RAmy3D are shown in C, and those of RAmy3A are shown in G. Green lines in C and H represent the expression profiles of genes having obvious downward outliers in gid2 and/or gamyb, which were confirmed by qRT-PCR (Supplemental Figs. S5 and S7). The arrowhead in B shows the obvious downward outlier Os04g0364800, which was confirmed by qRT-PCR (Supplemental Fig. S4D). The arrow in D shows Os11g0138300, an obvious downward outlier that was confirmed by qRT-PCR as presented in Figure 5B (probe 120).
In the case of GA down-regulated genes, 355 genes (including RAmy3D) were assigned to Down-group1 (Supplemental Table S8), which is composed of genes showing no change in expression by GA in the four mutants (Fig. 6C). Two representative genes (Fig. 6C, purple lines; Supplemental Fig. S5, A and B) showed no expression change by GA in gid1, gid2, or gamyb and were not expressed in slr1, demonstrating that they are strictly regulated by GID1, SLR1, GID2, and GAMYB. However, there were some downward outliers in the slr1, gid2, and gamyb backgrounds. qRT-PCR demonstrated no actual change in the expression of the outlier, Os04g0179100, in slr1 (Fig. 3B), while the three most down-regulated genes in gid2 (Fig. 6C, green lines) were down-regulated at a level similar to the wild type upon GA treatment (Supplemental Fig. S5, C–E, arrows). On the other hand, the expression of Down-group2, which contains 53 genes (Supplemental Table S9), shows essentially the same pattern as Down-group1 (Fig. 6D). Actually, down-regulation of two representative genes did not occur in gid1, gid2, or gamyb but constitutively occurred in slr1 (Fig. 6D, purple lines; Supplemental Fig. S5, F and G, purple lines). Several genes were downward outliers in the gamyb mutant, such as Os11g0138300 (Fig. 6D, arrow), and this was confirmed by qRT-PCR (Fig. 5B, probe 120).
Down-group3 contains 43 genes (Supplemental Table S10), which did not show significant differences with respect to GA treatment in the gid1, slr1, and gid2 mutants but were down-regulated in gamyb (Fig. 6E). Down-regulation of two representative genes (purple lines) did not occur in gid1 or gid2 but occurred in gamyb, and these genes were constitutively down-regulated in slr1 (Supplemental Fig. S6, A and B, arrows). Down-group4 contained only two genes (Supplemental Table S11), which were down-regulated in gid1 and gamyb in microarray analysis (Fig. 6F). However, qRT-PCR confirmed that down-regulation of these two genes occurred only in gamyb but not in gid1 (Supplemental Fig. S6, C and D, arrows and arrowheads, respectively). Thus, the expression of Down-group4 is essentially the same as that of Down-group3.
Down-group5 is a small group that contains five genes (Supplemental Table S12), including RAmy3A (Fig. 6G, light blue lines). As shown by an analysis of two representative genes (Fig. 6G, purple lines), the expression of Down-group5 was down-regulated in gid2 and gamyb (Supplemental Fig. S7, A and B, arrows). Down-group6 contains 26 genes (Supplemental Table S13), and the expression pattern was similar to that of Down-group5 (Fig. 6H; Supplemental Fig. S7, C and D). Down-group6 contains many genes that were down-regulated by GA by less than 3-fold in the gid2 and gamyb backgrounds, suggesting that these genes are not fully down-regulated in gid2 and gamyb. For example, an expression analysis of two outlier genes (Fig. 6H, green lines; Supplemental Fig. S7, E and F, arrowheads) indicated that many genes in this group are partially down-regulated in the absence of GID2 and GAMYB.
Taken collectively, cluster analysis allowed us to briefly categorize GA down-regulated genes into three categories with respect to their dependence on GID2 and GAMYB. The first category includes genes that are regulated by GID1, SLR1, GID2, and GAMYB, and Down-group1 and Down-group2 include many of these genes. The second category includes genes regulated by GID1, SLR1, and GID2 but not by GAMYB, and these are primarily assigned to Down-group3 and Down-group4. The last category includes genes regulated by GID1 and SLR1 (but not by GID2 or GAMYB); these are generally present in Down-group5 and Down-group6.
Several GA-Suppressed Genes Are Positively Regulated by SLR1
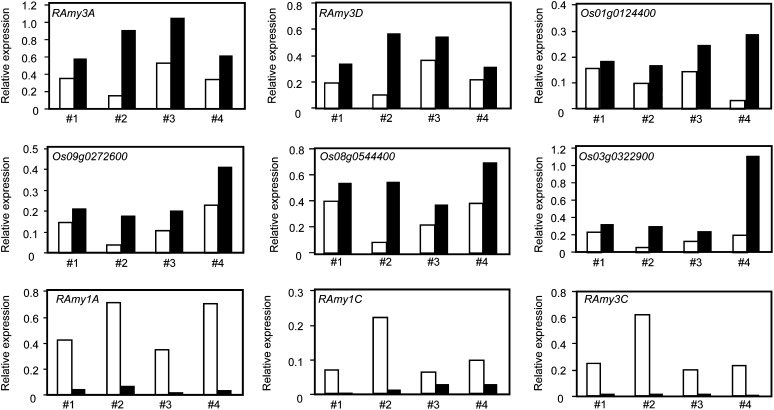
The above results demonstrate that a number of genes (including RAmy3A) within Down-group3, Down-group4, Down-group5, and Down-group6 are down-regulated by GA without the aid of GAMYB. Recently, a novel DELLA function was proposed whereby the DELLA protein interacts with trans-acting factor(s) containing the DNA-binding domain and enhances gene expression by targeting promoter sequences (Zentella et al., 2007; Yoshida et al., 2014). Based on this, we hypothesized that elevated gene expression in the absence of GA may depend on the transactivation activity of SLR1. To examine the effect of SLR1 on the expression of RAmy3A, we attempted a transient gene expression experiment. As an effector, we used SLR1 fused to the transactivation domain of herpes simplex virus protein VP16 (SLR1-VP16; Hirano et al., 2012) to enhance the transcriptional activity of SLR1. After SLR1-VP16 bombardment, the expression of endogenous RAmy3A was examined by qRT-PCR. Bombardment with SLR1-VP16 consistently increased the endogenous expression of RAmy3A relative to the vector control (Fig. 7). We also examined the effect of SLR1-VP16 on RAmy3D expression, largely because RAmy3D was significantly down-regulated by GA in the absence of GAMYB (Fig. 1). As expected, RAmy3D expression was enhanced by SLR1-VP16, and similar results were observed for four other GAMYB-independent, GA down-regulated genes (Fig. 7). This suggested that SLR1 functions as a positive trans-acting factor for the expression of these genes. We also examined the effect of SLR1-VP16 on the expression of GA up-regulated RAmy1A, RAmy1C, and RAmy3C. The expression of RAmy1B, RAmy3B, and RAmy3E was low in GA-untreated seeds and, thus, was not investigated (Supplemental Fig. S8). Although the expression of RAmy1A, RAmy1C, and RAmy3C differed between experiments, bombardment with SLR1-VP16 consistently diminished their expression (Fig. 7).
Figure 7.
Bombardment with SLR1-VP16 enhances the expression of RAmy3A, RAmy3D, and other GAMYB-independent GA down-regulated genes in aleurone cells. SLR1-VP16 (black bars) and a control plasmid (white bars) were independently bombarded into aleurone cells, and the expression of endogenous genes as revealed by qRT-PCR is shown in comparison with endogenous Ubi expression. The effect of SLR1-VP16 on the expression of RAmy1A, RAmy1C, and RAmy3C, which are up-regulated by GA, was also observed.
DISCUSSION
GID1-DELLA Is the Sole Mechanism for GA Perception in Rice Aleurones
In this work, we investigated the potential existence of an alternative GA receptor as predicted by previous observations (Hooley et al., 1991; Gilroy and Jones, 1994). The alternative GA receptor was expected to have certain biochemical characteristics, such as a plasma membrane location, the ability to perceive GA outside of aleurone cells, and the ability to induce hydrolytic enzymes such as α-amylase (e.g. RAmy genes) after GA perception (Hartweck and Olszewski, 2006; Ueguchi-Tanaka et al., 2007; Taiz and Zeiger, 2010). We examined GA-mediated induction of RAmy genes in rice aleurone cells using mutants for GID1 and the rice DELLA protein SLR1. Six RAmy genes showed enhanced expression in the aleurones of wild-type seeds, whereas none were induced in gid1 seeds (Fig. 1). Furthermore, all genes were expressed in GA-untreated slr1 seeds at levels similar to GA-treated wild-type seeds (Fig. 1). For RAmy3A and RAmy3D, expression was reduced in the presence of GA, and the expression of RAmy3A and RAmy3D in gid1 and slr1 mutants contrasted with that of other RAmy genes. In other words, RAmy3A and RAmy3D were not down-regulated in gid1, and their expression was low in slr1 even in the absence of GA (Fig. 1). These observations clearly demonstrated that GA-mediated expression of all RAmy genes in rice aleurone cells is strictly regulated by the GID1-DELLA system. We also performed a comprehensive analysis of GA-related gene expression by using microarrays and confirmed by qRT-PCR that all GA up- and down-regulated genes are regulated by the GID1-DELLA perception system (Figs. 2 and 3). Thus, GID1-DELLA remains the only perception system known that can mediate GA-dependent gene expression in rice aleurone cells.
Involvement of GID2 and GAMYB in GA-Dependent Expression in Rice Aleurones
We also evaluated the involvement of GID2 and GAMYB in GA-dependent gene expression in rice aleurone cells. In the case of up-regulated genes, expression was not significantly up-regulated by GA treatment in the four mutants (Figs. 2–5), indicating that their expression depends on GID1, SLR1, GID2, and GAMYB (Fig. 8A). In this mechanistic model, the absence of GA allows SLR1 to suppress GAMYB and thus prevents the expression of GA-inducible genes such as RAmy1A, RAmy1B, and RAmy1C. In the presence of GA, SLR1 is degraded through GID2, which results in the activation of GAMYB. However, several genes were partially up-regulated in either gid2 or gamyb mutants (Figs. 4 and 5) or in both mutants (overlapping genes), including RAmy3C, RAmy3E, Os04g0227500, Os05g0527300, Os03g0279700, and Os03g0131200 (Supplemental Tables S4 and S5). The high frequency of overlapping genes suggests that an unknown mechanism may be responsible for the partial dependence on GID2 and GAMYB. This might depend on alternative TFs, which can partially restore the function of GAMYB (Fig. 8B). The partial induction of overlapping genes in gid2 indicates that such TF expression is induced upon GID1-SLR1 interaction and that degradation of SLR1 is not essential. This is consistent with previous reports that the suppressive function of DELLA proteins can be partially deactivated by the GID1-DELLA interaction (Ariizumi et al., 2008; Ueguchi-Tanaka et al., 2008). Thus, it is possible that the GID1-SLR1 interaction may allow the activation of certain TFs, resulting in the induction of some GA up-regulated genes (Fig. 8B, narrow arrow).
Figure 8.
Proposed models for GA-mediated gene expression involving GID1, SLR1, GID2, and GAMYB in rice aleurone cells. A, Up-regulation by GA as mediated by GID1, SLR1, GID2, and GAMYB. B, Up-regulation by GA partially controlled by GID2 and GAMYB. C, Down-regulation by GA under the control of GID1, SLR1, GID2, and GAMYB. D, Down-regulation by GA under the control of GID1, SLR1, and GID2 but not GAMYB (GID2 is essential for down-regulation). E, Down-regulation by GA under the control of GID1 and SLR1 but not GID2 or GAMYB. The typical expression pattern for each group is presented below each model. WT, Wild type.
In the case of down-regulated genes, the situation is more complicated. The expression of down-regulated genes is generally under the control of the four GA signaling factors, which are primarily in Down-group1 and Down-group2. Since GAMYB is considered to be a transactivating factor (Gubler et al., 1995), the down-regulation of genes by GAMYB may be an indirect effect of GAMYB. One possibility is that GAMYB influences carbon metabolites, which are released by catabolic enzymes induced by GA via a GAMYB-mediated mechanism (Fig. 8C). It is well-established that carbon metabolites function to suppress genes in cereal aleurone cells (Chen et al., 2006). An alternative explanation could be that TFs exerting repressive activity on their target genes are positively regulated by GAMYB (Fig. 8C). In this respect, it is important to mention the existence of multiple TFs that are regulated by GAMYB (Supplemental Table S1).
Hierarchical cluster analysis (Fig. 6) clearly demonstrated that there are certain genes that are down-regulated by GA independent of GID2 or GAMYB, which is clearly different from the GA up-regulated genes. Recent studies suggest that the DELLA protein has two possible roles. One of these functions is an inhibitory effect on the transcription-inducing activity of targets such as PHYTOCHROME-INTERACTING FACTORS (de Lucas et al., 2008; Feng et al., 2008), MYC2 (Hong et al., 2012), and SPLs (Yu et al., 2012). In the other role, DELLA promotes transcriptional activity by collaborating with other TFs carrying DNA-binding domains (Zentella et al., 2007; Yoshida et al., 2014). Moreover, the transactivation activity of SLR1 is important for inducing dwarfism in rice (Hirano et al., 2012). This study showed that SLR1-VP16 enhanced the expression of some genes, including RAmy3A and RAmy3D (Fig. 7). These observations suggest that SLR1 functions as a positive regulator for the expression of GA down-regulated genes. Since DELLA proteins (including SLR1) have been considered to lack DNA-binding domains (Sun et al., 2012; Davière and Achard, 2013), they would need to interact with a TF to modulate DNA binding (Fig. 8, D and E). In accordance with this model, the down-regulation of genes by GA independent of GID2 could be explained by the inhibitory effect of GID1-SLR1 complex formation on the SLR1-TF interaction (Fig. 8E). In contrast, the down-regulation of genes that depend on GID2 suggests that SLR1 degradation is essential to diminish the SLR1-TF interaction (Fig. 8D). Previously, Zentella et al. (2007) comprehensively searched genes down-regulated by GA and up-regulated by DELLA and identified 14 GA-related genes, including GA 20-oxidase (GA biosynthesis), GA 3-oxidase (GA biosynthesis), and GID1s; those authors discussed the functions of DELLA with respect to feedback regulation in GA signaling. Our findings suggest that genes up-regulated by SLR1 are not limited to those involved in GA feedback regulation but also include a diverse array of genes (Supplemental Tables S8–S13). Thus, it is possible that DELLA functions as a transactivator not only in GA feedback regulation but also in other biological processes.
MATERIALS AND METHODS
Plant Material
Seeds of wild-type rice (Oryza sativa ‘Nipponbare’), gid1-4 (Ueguchi-Tanaka et al., 2005), gid2-5 (Sasaki et al., 2003), slr1-1 (Ikeda et al., 2001), and gamyb-2 (Kaneko et al., 2004) were used. Since the mutant plants carrying homozygous alleles are either lethal or sterile, PCR was used to screen for homozygous F2 seeds, and DNA from seed embryos was utilized as a template. Half-seeds lacking embryos were placed in incubation medium (10 mm sodium acetate, pH 5.2, 2 mm CaCl2, with or without 10−5 m GA3) for 36 h at 30°C and then used for RNA extraction.
RNA Isolation and qRT-PCR Analysis
Total RNA was extracted from incubated embryoless half-seeds as described previously (Sambrook et al., 1989). RNA was then treated with Amplification Grade DNase I (Invitrogen) and used for qRT-PCR experiments. More than three biological replicates containing independently isolated RNA samples were analyzed. The first-strand complementary DNA (cDNA) was synthesized from 1 µg of total RNA with the Omniscript reverse transcription kit (Qiagen). The resulting cDNA was diluted 1:20 and used as a template; transcripts were quantified by qRT-PCR with the C1000 Thermal Cycler (Bio-Rad) and the SYBR Green PCR kit (Qiagen). For quantification, a linear standard curve and threshold cycle number versus log (designated transcript level) were constructed from a series of diluted DNA fragments (10−17, 10−19, 10−21, and 10−23 m). Each cDNA sample was subjected to different cycles of PCR amplification (35–40 cycles) to confirm the linear pattern of PCR amplification for each gene. The rice Ubi gene was used as an internal standard for normalizing cDNA concentration variations. The primer sequences used in this study are listed in Supplemental Table S14.
Microarray Hybridization and Data Analysis
An Agilent 44K rice oligoarray (Agilent Technologies), containing 44,000 probes, was used for two-color analysis. Each probe consists of a 60-mer oligonucleotide corresponding to a full-length cDNA of rice. Four biological replicates consisting of independently isolated RNA samples were analyzed. All microarray experiments were performed according to the manufacturer’s manual. The Feature Extraction software (Agilent Technologies) was used to delineate and measure the Cy3 and Cy5 signal intensities of each spot in the array. The resulting data were normalized using the variance-stabilizing normalization algorithm (Huber et al., 2002). To identify differentially expressed genes between GA-untreated and -treated wild-type seeds, the normalized values of log2 signal ratios were analyzed using a simple nonparametric statistical method (rank product method), as described by Breitling et al. (2004). The P value cutoff was set at 0.01, and multiple testing was taken into account using the percentage of false prediction (less than 0.05; Breitling et al., 2004; Hong et al., 2006). Under these conditions, we finally obtained 1,319 probes that corresponded to GA-regulated genes in wild-type seeds (Supplemental Tables S1 and S2). All microarray data from this study were deposited in the Gene Expression Omnibus repository under accession code GSE64290.
Hierarchical Clustering
Array data were analyzed using hierarchical clustering with average linkage and hclust (included in the R software package), which is based on the agglomeration method (Ben-Hur et al., 2002; Smolkin and Ghosh, 2003). The Pearson correlation coefficient was used to measure similarity between gene expression profiles in all microarray data. The clustered dendrogram was divided using the cut-tree function to classify genes according to the expression pattern.
GO Analysis
Probes representing 615 and 704 GA up- and down-regulated genes, respectively, were used for GO analysis with agriGO (Du et al., 2010; http://bioinfo.cau.edu.cn/agriGO/index.php). Fisher’s exact test was used to identify the enriched GO term(s) with a false discovery rate-adjusted P cutoff of less than 0.05. Boxes contained GO terms and descriptions, false discovery rate-adjusted P values, the total number of GO annotated and background genes from GA-regulated genes, and the total number of GO annotated and background genes from the entire array.
DNA Bombardment
DNA bombardment was performed as described by Sutoh and Yamauchi (2003) using wild-type (cv Nipponbare) embryoless grain quadrisections derived from the same seed. For the construction of effector plasmid, we inserted the maize Ubi promoter and nopaline synthase terminator into the HindIII/XbaI and SacI/EcoRI sites of pUC19, respectively, to produce pUbi/pUC19 as a control vector. DNA sequences of SLR1-VP16 (Hirano et al., 2012) were then ligated into the SmaI site of pUbi/pUC19. About 1.5 pmol of plasmid DNA was delivered into aleurone cells and incubated at room temperature for 16 h. For qRT-PCR, RNA was extracted from one embryoless grain quadrisection as mentioned above.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GID1 (Q6L545), SLR1 (AK242577), GID2 (Q7XAK4), and GAMYB (BAF06506).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Simplified model of GA up-regulated gene expression.
Supplemental Figure S2. qRT-PCR analysis of RAmy2A in embryoless half-seeds of gid1, slr1, gid2, and gamyb.
Supplemental Figure S3. GO analysis of GA-regulated genes.
Supplemental Figure S4. Expression analysis of two representative genes in Up-group1 and Up-group2 by qRT-PCR.
Supplemental Figure S5. Expression analysis of two representative genes in Down-group1 and Down-group2 by qRT-PCR.
Supplemental Figure S6. Expression analysis of two representative genes in Down-group3 and Down-group4 by qRT-PCR.
Supplemental Figure S7. Expression analysis of two representative genes in Down-group5 and Down-group6 by qRT-PCR.
Supplemental Figure S8. Expression analysis of endogenous RAmy1B, RAmy3B, and RAmy3E in comparison with endogenous Ubi expression in embryoless half-seeds in the absence of GA by qRT-PCR.
Supplemental Table S1. List of GA up-regulated probes.
Supplemental Table S2. List of GA down-regulated probes.
Supplemental Table S3. List of probes up- or down-regulated by GA by more than 3-fold in slr1.
Supplemental Table S4. List of probes up- or down-regulated by GA by more than 3-fold in gid2.
Supplemental Table S5. List of probes up- or down-regulated by GA by more than 3-fold in gamyb.
Supplemental Table S6. List of probes categorized under Up-group1.
Supplemental Table S7. List of probes categorized under Up-group2.
Supplemental Table S8. List of probes categorized under Down-group1.
Supplemental Table S9. List of probes categorized under Down-group2.
Supplemental Table S10. List of probes categorized under Down-group3.
Supplemental Table S11. List of probes categorized under Down-group4.
Supplemental Table S12. List of probes categorized under Down-group5.
Supplemental Table S13. List of probes categorized under Down-group6.
Supplemental Table S14. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Y. Nagamura and R. Motoyama (Rice Genome Resource Center of the National Institute of Agrobiological Sciences, Japan) for the use of the rice microarray analysis system and for technical support. We also thank W. Takase and R. Masuda (Bioscience and Biotechnology Center, Nagoya University, Japan) for technical assistance.
Glossary
- GBP
gibberellin-binding protein
- qRT
quantitative reverse transcription
- GO
Gene Ontology
- TF
transcription factor
- cDNA
complementary DNA
Footnotes
This work was supported by the Japan Society for the Promotion of Science (grant no. 23113005 and single-year grant nos. 26・1393) and by the Ministry of Education, Culture, Sports, Science, and Technology (grant nos. 26252001 and 26660278 and Grants-in-Aid from the Network of Centers of Carbon Dioxide Resource Studies in Plants project).
References
- Ariizumi T, Murase K, Sun TP, Steber CM (2008) Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20: 2447–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale MH, Ward JL, Smith SJ, Hooley R (1992) A new approach to gibberellin perception in aleurone: novel hydrophilic, membrane-impermeant, GA-sulphonic acid derivatives induce α-amylase. Physiol Plant 85: A136 [Google Scholar]
- Ben-Hur A, Elisseeff A, Guyon I (2002) A stability based method for discovering structure in clustered data. Pac Symp Biocomput 7: 6–17 [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Chen PW, Chiang CM, Tseng TH, Yu SM (2006) Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of α-amylase genes. Plant Cell 18: 2326–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière JM, Achard P (2013) Gibberellin signaling in plants. Development 140: 1147–1151 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Jones RL (1994) Perception of gibberellin and abscisic acid at the external face of the plasma membrane of barley (Hordeum vulgare L.) aleurone protoplasts. Plant Physiol 104: 1185–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV (1995) Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7: 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck LM, Olszewski NE (2006) Rice GIBBERELLIN INSENSITIVE DWARF1 is a gibberellin receptor that illuminates and raises questions about GA signaling. Plant Cell 18: 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Kouketu E, Katoh H, Aya K, Ueguchi-Tanaka M, Matsuoka M (2012) The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J 71: 443–453 [DOI] [PubMed] [Google Scholar]
- Ho TD, Gomez-Cadenas A, Zentella R, Casaretto J (2003) Crosstalk between gibberellin and abscisic acid in cereal aleurone. J Plant Growth Regul 22: 185–194 [Google Scholar]
- Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J (2006) RankProd: a Bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22: 2825–2827 [DOI] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley R, Beale MH, Smith SJ (1991) Gibberellin perception at the plasma membrane of Avena fatua aleurone protoplasts. Planta 183: 274–280 [DOI] [PubMed] [Google Scholar]
- Hooley R, Smith SJ, Beale MH, Walker RP (1993) In vivo photoaffinity labelling of gibberellin-binding proteins in Avena fatua aleurone. Aust J Plant Physiol 20: 573–584 [Google Scholar]
- Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics (Suppl 1) 18: S96–S104 [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelsema CL, Ruddat M, Morre DJ, Williamson FA (1977) Specific binding of gibberellin A1 to aleurone grain fractions from wheat endosperm. Plant Cell Physiol 18: 1009–1019 [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, et al. (2004) Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Blázquez MA, Alabadí D (2013) Genomic analysis of DELLA protein activity. Plant Cell Physiol 54: 1229–1237 [DOI] [PubMed] [Google Scholar]
- Lovegrove A, Barratt DH, Beale MH, Hooley R (1998) Gibberellin-photoaffinity labelling of two polypeptides in plant plasma membranes. Plant J 15: 311–320 [DOI] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-Ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52: 67–88 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Smolkin M, Ghosh D (2003) Cluster stability scores for microarray data in cancer studies. BMC Bioinformatics 4: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Jones WT, Rikkerink EHA (2012) GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem J 442: 1–12 [DOI] [PubMed] [Google Scholar]
- Sutoh K, Yamauchi D (2003) Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J 34: 635–645 [DOI] [PubMed] [Google Scholar]
- Taiz L, Zeiger E (2010) Plant Physiology, Ed 5 Sinauer Associates, Sunderland, MA [Google Scholar]
- Thomas SG, Rieu I, Steber CM (2005) Gibberellin metabolism and signaling. Vitam Horm 72: 289–338 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Hirano K, Hasegawa Y, Kitano H, Matsuoka M (2008) Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M (2007) Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58: 183–198 [DOI] [PubMed] [Google Scholar]
- Walker RP, Waterworth WM, Beale MH, Hooley R (1994) Gibberellin-photoaffinity labeling of wild oat (Avena fatua L.) aleurone protoplasts. Plant Growth Regul 15: 271–279 [Google Scholar]
- Yoshida H, Hirano K, Sato T, Mitsuda N, Nomoto M, Maeo K, Koketsu E, Mitani R, Kawamura M, Ishiguro S, et al. (2014) DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc Natl Acad Sci USA 111: 7861–7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.