Antiphase light and temperature cycles disrupt an auxin-ethylene-induced signaling cascade, leading to reduced hypocotyl elongation.
Abstract
We show that antiphase light-temperature cycles (negative day-night temperature difference [−DIF]) inhibit hypocotyl growth in Arabidopsis (Arabidopsis thaliana). This is caused by reduced cell elongation during the cold photoperiod. Cell elongation in the basal part of the hypocotyl under −DIF was restored by both 1-aminocyclopropane-1-carboxylic acid (ACC; ethylene precursor) and auxin, indicating limited auxin and ethylene signaling under −DIF. Both auxin biosynthesis and auxin signaling were reduced during −DIF. In addition, expression of several ACC Synthase was reduced under −DIF but could be restored by auxin application. In contrast, the reduced hypocotyl elongation of ethylene biosynthesis and signaling mutants could not be complemented by auxin, indicating that auxin functions upstream of ethylene. The PHYTOCHROME INTERACTING FACTORS (PIFs) PIF3, PIF4, and PIF5 were previously shown to be important regulators of hypocotyl elongation. We now show that, in contrast to pif4 and pif5 mutants, the reduced hypocotyl length in pif3 cannot be rescued by either ACC or auxin. In line with this, treatment with ethylene or auxin inhibitors reduced hypocotyl elongation in PIF4 overexpressor (PIF4ox) and PIF5ox but not PIF3ox plants. PIF3 promoter activity was strongly reduced under −DIF but could be restored by auxin application in an ACC Synthase-dependent manner. Combined, these results show that PIF3 regulates hypocotyl length downstream, whereas PIF4 and PIF5 regulate hypocotyl length upstream of an auxin and ethylene cascade. We show that, under −DIF, lower auxin biosynthesis activity limits the signaling in this pathway, resulting in low activity of PIF3 and short hypocotyls.
To ensure optimal growth, plants are able to adapt their physiology and developmental program to accommodate changes in the environment. Light and temperature are two of the strongest environmental signals affecting plant development (for review, see Franklin, 2009). Both signals vary in diurnal cycles and usually oscillate in phase. This natural cycle of warm days and cool nights is referred to as positive day-night temperature difference (+DIF). If the light and temperature cycles are provided in antiphase (cold day and warm night), this is referred to as negative day-night temperature difference (−DIF). The difference between day and night temperatures strongly affects plant growth, and the responses of plants to diurnally fluctuating temperatures are collectively referred to as thermoperiodism (Went, 1944). For many plant species, elongation is stimulated when the positive difference between day and night temperatures increases (Myster and Moe, 1995). In horticulture, excessive elongation growth decreases crop quality, and especially during the seedling stage, excessive elongation of the fragile hypocotyl is unwanted (Grimstad and Frimanslund, 1993; Bakken and Flønes, 1995). Therefore, −DIF is frequently applied in greenhouses to reduce elongation (Myster and Moe, 1995). In Arabidopsis (Arabidopsis thaliana), −DIF inhibits inflorescence stem and leaf elongation, and a 10°C −DIF results in up to a 40% decrease of final leaf length compared with 10°C +DIF (Thingnaes et al., 2003; Bours et al., 2013). Although economically important in horticulture, a full mechanistic understanding of how −DIF specifically affects elongation is still lacking.
The light and temperature signals are translated by plants into different hormonal signals, which regulate specific plant developmental programs but also, frequently show cross talk with each other (Jaillais and Chory, 2010). One of the best studied phytohormones in relation to elongation is auxin (Strader and Nemhauser, 2013). For instance, the rapid elongation of hypocotyls in response to high temperature under continuous light (Gray et al., 1998) depends on the up-regulation of auxin biosynthesis genes (Franklin et al., 2011). Significant cross talk exists between auxin and the gaseous hormone ethylene, because ethylene may affect auxin transport (Zádníková et al., 2010), whereas auxin can induce ethylene biosynthesis (Swarup et al., 2007). The effect of ethylene on plant growth is light dependent; in darkness, ethylene inhibits, whereas in the light, it stimulates hypocotyl elongation (Smalle et al., 1997; Pierik et al., 2006; Zhong et al., 2012).
Light negatively influences elongation by activating phytochrome photoreceptors (Franklin and Whitelam, 2004). After being activated, these proteins stimulate the degradation of growth-stimulating basic helix-loop-helix transcription factors called PHYTOCHROME INTERACTING FACTORs (PIFs; Khanna et al., 2004). PIF proteins have been reported to affect phytohormone biosynthesis and signaling. PIF4 and PIF5, for example, are able to activate auxin biosynthesis genes and/or auxin signaling and therefore, are positive regulators of hypocotyl elongation (Nozue et al., 2007, 2011; Franklin et al., 2011; Hornitschek et al., 2012). In addition to PIF4 and PIF5, PIF3 also promotes elongation in seedlings grown under diurnal light-dark conditions, and previously, it was shown that the stimulatory effect of ethylene on hypocotyl elongation under continuous light is mediated by PIF3 (Zhong et al., 2012). PIF3 directly binds to G boxes in the promoter of other known growth-related transcription factors PIF3-LIKE1 and LONG HYPOCOTYL IN FAR-RED1 and the promoter of the xyloglucan endotransglycosylase-related XYLOGLUCAN ENDOTRANSGLYCOSYLASE7, which supposedly is involved in cell wall growth (Sasidharan et al., 2010; Leivar and Quail, 2011; Soy et al., 2012). However, in the studies mentioned above, the positions of the PIF3, PIF4, and PIF5 transcription factors within the signal transduction pathway that controls growth were not fully established.
In this work, we expand our understanding of hypocotyl elongation by investigating how light-temperature cycles affect the efficiency of the signal transduction pathway for growth. We show that the effect of contrasting diurnal temperature cycles (+DIF and −DIF) on growth relates to cell elongation and not cell division in Arabidopsis hypocotyls and that, under −DIF, both auxin and ethylene become limiting for cell elongation. Results show that reduced signaling under −DIF is linked to reduced activity of auxin biosynthesis genes and that auxin acts upstream of ethylene through transcriptional activation of several 1-aminocyclopropane-1-carboxylic acid (ACC) Synthase (ACS) genes. In addition, our genetic interaction studies show that PIF3, PIF4, and PIF5 act at different positions in this signaling interaction. Auxin activates transcription of PIF3 in an ethylene-dependent fashion, which positions PIF3 downstream in the signaling cascade. In contrast, both PIF4 and PIF5 function upstream in the pathway to elongation by regulating auxin and ethylene signal input. Results show that the relative contribution of the different PIFs varies with conditions; whereas earlier studies indicate an essential role for PIF4 in plant elongation under constant temperature (22°C or 28°C) and +DIF (Nozue et al., 2007; Franklin et al., 2011), our results show that PIF4 is not essential for the growth inhibition response under −DIF.
RESULTS
Reduced Arabidopsis Hypocotyl Cell Elongation under −DIF Can Be Complemented with ACC
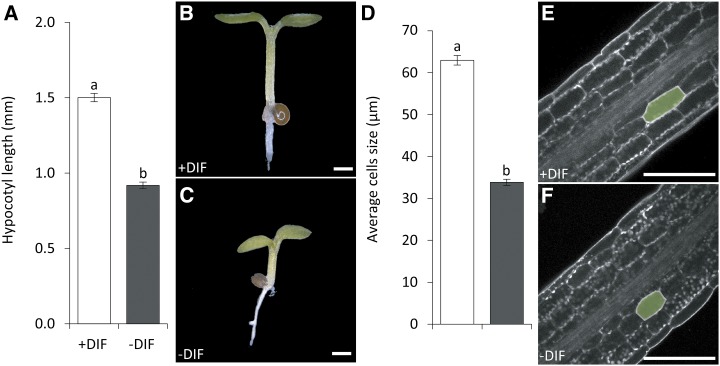
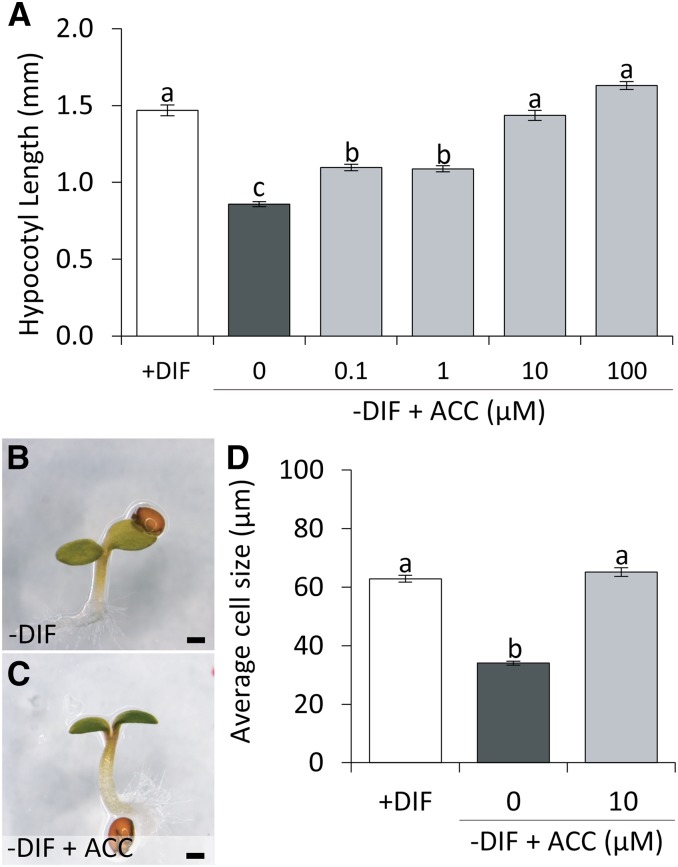
The growth response of Arabidopsis seedlings to −DIF was characterized by comparing seedling growth under +DIF and −DIF diurnal cycles. The −DIF-treated plants showed a 40% reduction in hypocotyl length compared with control-grown (+DIF) seedlings (Fig. 1, A–C). Closer examination of the hypocotyl epidermal cells showed that the reduction in hypocotyl length under −DIF can be attributed to reduced cell elongation rather than reduced cell divisions (Fig. 1, D–F). Previously, we showed that −DIF reduces ethylene sensitivity in Arabidopsis seedlings (Bours et al., 2013). Because of the light-dependent role of ethylene in hypocotyl elongation (Smalle et al., 1997; Pierik et al., 2006; Zhong et al., 2012), we assessed whether ethylene was limiting hypocotyl elongation during the day under −DIF. Indeed, application of the ethylene precursor (ACC) increased hypocotyl length of the Arabidopsis seedlings under −DIF in a dose-dependent manner (Fig. 2, A–C). Analysis of the hypocotyl epidermal cells showed that ACC rescues the hypocotyl-length phenotype under −DIF by enhancing cell elongation (Fig. 2D). Because the action of ethylene is tightly linked to that of auxin (Muday et al., 2012) and auxin has also been linked to the regulation of cell elongation (Chapman et al., 2012; Nakayama et al., 2012), we subsequently investigated the role of auxin and its relation to ethylene in the seedling growth response to −DIF.
Figure 1.
Decreased hypocotyl length under −DIF is caused by reduced cell elongation. A, Average Arabidopsis hypocotyl length after 7 d of growth under +DIF and −DIF (n = 5 × 25). B and C, Bright-field image of representative Arabidopsis seedlings grown for 7 d under +DIF (B) or −DIF (C). Bars = 500 µm. D, Average hypocotyl cell sizes at basal-site hypocotyl scored at 7 d after germination (n = 20 × 25). E and F, Confocal microscopy images of +DIF (E) and −DIF (F) hypocotyl cells. Bars represent means ± se. Bars with different letters differ significantly (P < 0.05). Bars = 100 µm.
Figure 2.
ACC complements hypocotyl elongation under −DIF conditions. A, Average hypocotyl length of 7-d-old Columbia-0 (Col-0) grown under +DIF or −DIF with and without increasing concentrations of ACC (n = 5 × 25). B and C, Bright-field image of representative 7-d-old Arabidopsis seedlings grown under −DIF (B) or −DIF treated with 10 µm ACC (C). Bars = 500 µm. D, Average cell size at the basal part of the hypocotyl for seedlings grown for 7 d under +DIF and −DIF with the addition of 0 or 10 µm ACC (n = 20 × 25). Bars represent means ± se. Bars with different letters differ significantly (P < 0.05).
Endogenous Auxin Levels and Auxin Signaling Are Reduced in −DIF-Grown Seedlings
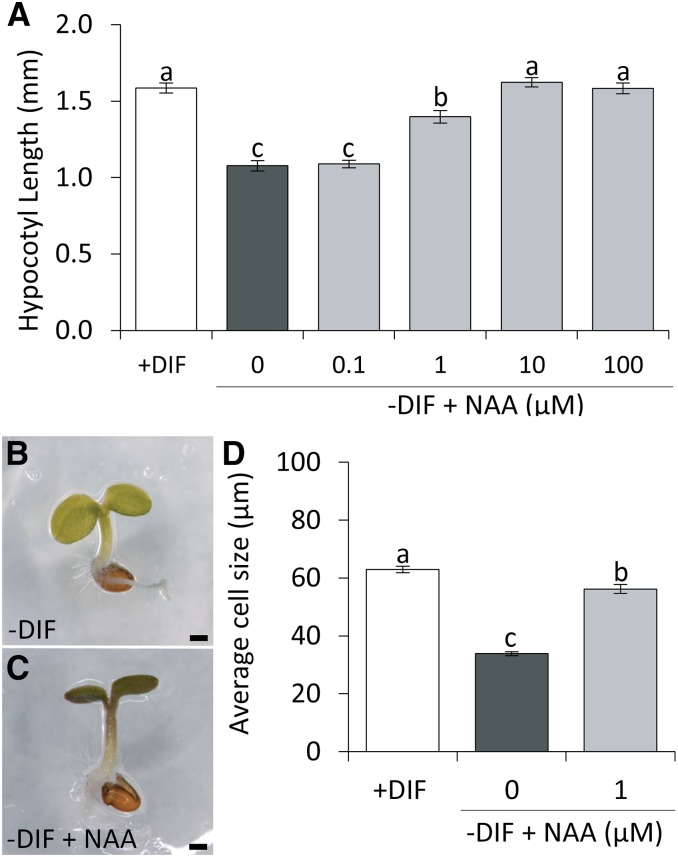
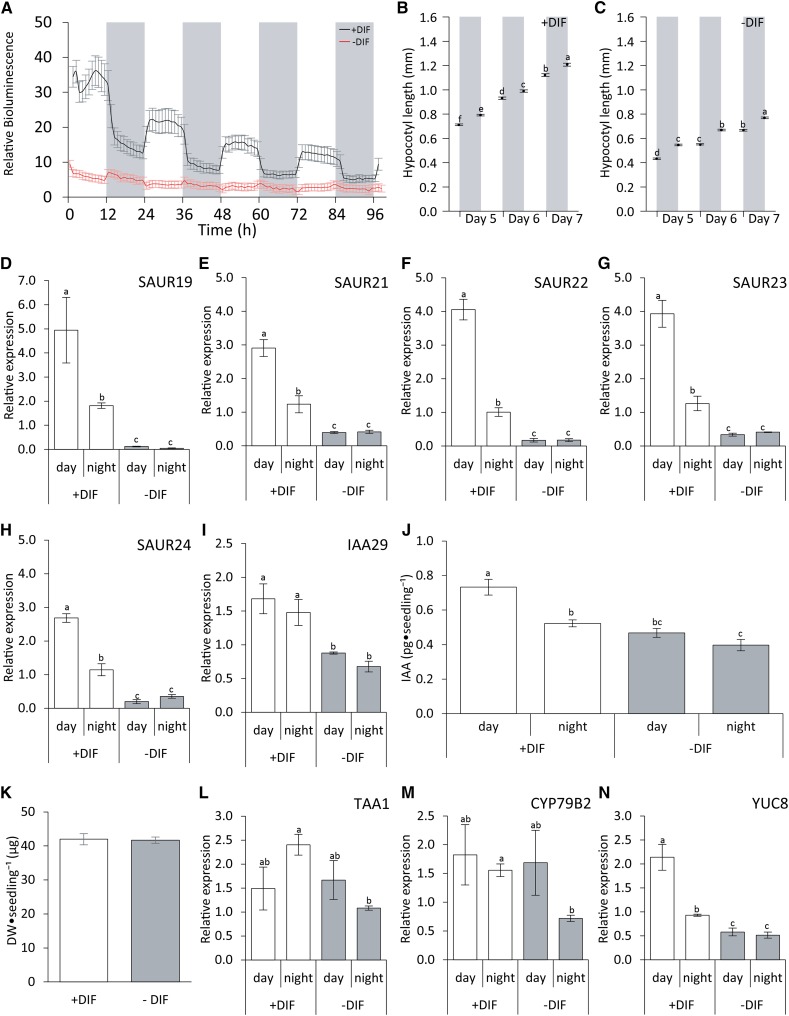
To test the effect of auxin on hypocotyl elongation under −DIF, seedlings were treated with 1-naphthaleneacetic acid (NAA) or indole-3-acetic acid (IAA). Similar to the application of ACC, NAA or IAA also enhanced hypocotyl elongation of seedlings under −DIF in a dose-dependent manner (Fig. 3, A–C; Supplemental Fig. S1). Moreover, examination of seedlings treated with NAA revealed that this effect on hypocotyl length under −DIF is also caused by increased elongation of the hypocotyl cells rather than increased cell division (Fig. 3D). This indicates that, like ethylene, auxin is also limiting for cell elongation under −DIF. To monitor the auxin signaling under +DIF and −DIF, we used enhanced (e) DR5-luciferase (eDR5::LUC) reporter plants (Fig. 4A). The bioluminescence activity of eDR5::LUC is often used as proxy for local auxin signaling in plants (Covington and Harmer, 2007). Results show that, in the control plants (+DIF), eDR5::LUC activity oscillates diurnally, with a phase (high activity) during the day and low activity during the night. Close-up analysis showed that, under +DIF, eDR5::LUC activity was primarily localized in the apical meristem, the cotyledon vasculature, and the cotyledon tips of the seedling (Supplemental Fig. S2). In contrast, −DIF severely reduced eDR5::LUC activity at all of these sites (Supplemental Fig. S2), leading to a complete lack of oscillating eDR5::LUC bioluminescence (Fig. 4A). The results show that the relative eDR5::LUC activity is much more reduced during the day than during the night. To investigate the significance of this observation in relation to elongation, we quantified seedling hypocotyl elongation over the light and dark periods. This analysis shows that, under +DIF, elongation growth occurs during both the photoperiod and the night period (Fig. 4B). In contrast, under −DIF conditions, growth is limited to the night period, and no growth is observed during the cold photoperiod (Fig. 4C). This is indicative of a stronger threshold sensitivity for auxin signaling toward growth during the photoperiod than during the night.
Figure 3.
NAA complements hypocotyl elongation under −DIF conditions. A, Average hypocotyl length of 7-d-old Col-0 grown under +DIF or −DIF with and without increasing concentrations of NAA (n = 5 × 25). B and C, Bright-field image of representative 7-d-old Arabidopsis seedlings grown under −DIF (B) or −DIF treated with 1 µm NAA (C). Bars = 500 µm. D, Average cell size at the basal part of the hypocotyl for seedlings grown for 7 d under +DIF and −DIF with the addition of 0 or 1 µm NAA (n = 20 × 25). Bars represent means ± se. Bars with different letters differ significantly (P < 0.05).
Figure 4.
Auxin levels are reduced under antiphase temperature and light cycles (−DIF). A, eDR5::LUC activity in Arabidopsis seedlings. Seedlings were entrained for 3 d, and eDR5::LUC activity was quantified for an additional 4 d under +DIF or −DIF conditions (n = 6 × 30). B and C, Average Arabidopsis hypocotyl length after 5, 6, and 7 d of growth under +DIF and −DIF conditions measured at dusk (0 h; lights on at 0 h) and dawn (12 h; lights off at 12 h; n = 5 × 25) under +DIF (B) and −DIF (C) conditions. D to I, Expression analysis of auxin-responsive genes SAUR19 (D), SAUR21 (E), SAUR22 (F), SAUR23 (G), SAUR24 (H), and IAA29 (I) at midday (6 h; lights on at 0 h) and midnight (18 h; lights off at 12 h; n = 3 × 400). J, Free IAA levels measured in whole seedlings grown for 7 d under +DIF and −DIF at midday (6 h; lights on at 0 h) and midnight (18 h; lights off at 12 h; n = 5 × 400). K, Average dry weight of 7-d-old Arabidopsis seedlings grown under +DIF and −DIF conditions (n = 3 × 108). L to N, Expression analysis of auxin biosynthesis genes TAA1 (L), CYP79B2 (M), and YUC8 (N) at midday (6 h; lights on at 0 h) and midnight (18 h; lights off at 12 h; n = 3 × 400). Data represent means ± se. Bars or data points with different letters differ significantly (P < 0.05).
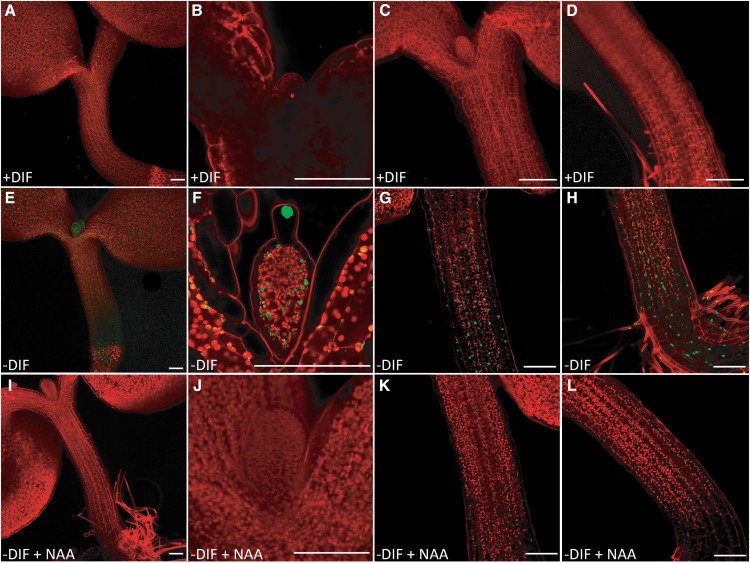
To investigate whether local auxin output signaling is affected, we examined seedlings carrying the Domain II (DII) Venus auxin output reporter. DII Venus produces a constitutive yellow fluorescent protein (YFP) signal in plant cells, which is rapidly degraded in the presence of auxin signaling activity (Brunoud et al., 2012). The DII Venus reporter, therefore, reveals sites of no or reduced auxin signaling. In line with the eDR5 reporter plant results (Supplemental Fig. S2), no YFP signal was observed in the apex or hypocotyl of DII Venus reporter plants grown under +DIF (Fig. 5, A–D), indicative of active auxin signaling in these tissues. In contrast, −DIF-grown DII Venus reporter seedlings showed signal accumulation in the apex (Fig. 5, E and F) and predominantly, the basal part of the hypocotyl (Fig. 5, E, G, and H), indicating no or reduced auxin signaling at these sites compared with seedlings grown under +DIF. After application of 1 μm NAA to the growing medium, the YFP signal in −DIF-grown DII Venus reporter plants disappeared (Fig. 5, I–L), indicating that endogenous auxin levels rather than auxin sensitivity are limiting auxin signaling capacity under −DIF conditions.
Figure 5.
Local auxin signaling is reduced under −DIF because of reduced auxin levels. Confocal image made at midday (6 h) of 7-d-old seedlings carrying the DII::VENUS-YFP reporter. A to D, +DIF conditions: overview (A), leaf primordium (B), apical hypocotyl (C), and basal hypocotyl (D). E to H, −DIF conditions: overview (E), leaf primordium (F), apical hypocotyl (G), and basal hypocotyl (H). I to L, −DIF conditions treated with 1 µm NAA: overview (I), leaf primordium (J), apical hypocotyl (K), and basal hypocotyl (L). Bars in A, E, and I = 200 µm. Bars in B to D, F to H, and J to L = 100 µm.
To substantiate the relationship between eDR5::LUC activity and auxin signaling, the expression levels of several auxin-responsive genes were analyzed at midday (t = 6 h) and midnight (t = 18 h) under both conditions. Small Auxin Up-Regulated (SAUR) genes are known to be regulated by auxin in relation to temperature-induced cell elongation (Franklin et al., 2011; Spartz et al., 2012). All SAURs tested (SAUR19, SAUR21, SAUR22, SAUR23, and SAUR24) showed high relative expression during midday and reduced expression during midnight under control conditions. In contrast, under −DIF, expression of these genes was much lower and not different between day and night (Fig. 4, D–H). In addition, the auxin-responsive gene IAA29 (Sun et al., 2013) was also analyzed, because PIF4 was shown to directly activate the expression of IAA29 (Sun et al., 2013). In our experiments, IAA29 expression did not display any diurnal pattern under +DIF. However, IAA29 transcript levels were reduced by approximately 50% when plants were grown under −DIF (Fig. 4I).
To determine whether the reduced eDR5::LUC activity under −DIF relates to reduced auxin biosynthesis or sensitivity under −DIF, free auxin levels of whole seedlings were analyzed at midday (t = 6 h) and midnight (t = 18 h) for both conditions (Fig. 4J). Because the dry weight of seedlings grown under either condition did not differ (Fig. 4K)—supporting our conclusion that only cell elongation is responsible for the −DIF phenotype—free auxin levels were quantified per seedling. Under +DIF, the level of free auxin during midnight was 30% lower than during midday, confirming that, under control conditions, auxin levels are not constant but show diurnal fluctuations. Under −DIF, auxin levels were much reduced and showed hardly any diurnal fluctuations (Fig. 4J), supporting the eDR5::LUC measurements under −DIF (Fig. 4A). Free auxin during midday was 30% lower than during the same time point under control conditions, whereas the level of free auxin during −DIF midnight was reduced by 25% compared with +DIF midnight.
To investigate whether altered auxin biosynthesis activity is causal for the changes in auxin levels, the expression levels of key genes involved in two different pathways toward auxin production (TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 [TAA1] and CYTOCHROME P450 79B2 [CYP79B2] versus YUCCA8 [YUC8]; Mano and Nemoto, 2012) were analyzed (Fig. 4, L–N). Results show that, under +DIF, TAA1 expression (higher at night) did not correlate well with free auxin levels (lower at night), whereas expression of CYP79B2 did not differ between day and night. Similarly, TAA1 and CYP79B2 expressions were both reduced at night under −DIF, whereas free auxin levels do not differ between day and night under −DIF. In contrast, YUC8 expressions during day and night under both +DIF and −DIF correlated very well with the auxin levels during day and night under these conditions. To examine if the response to −DIF is YUC8 specific, additionally, activity of YUC genes previously examined for their response to high temperature (Sun et al., 2012) was analyzed. Results showed that YUC1 and YUC2 are not detectable in our samples. YUC2 and YUC9 expressions did not differ between +DIF and −DIF, whereas YUC5 showed a significant reduction in response to −DIF during the photoperiod (Supplemental Fig. S3). Therefore, specifically, the YUC8- and YUC5-related pathway seems to be more relevant for auxin production and hypocotyl elongation.
−DIF Disrupts Auxin-Induced Ethylene Biosynthesis
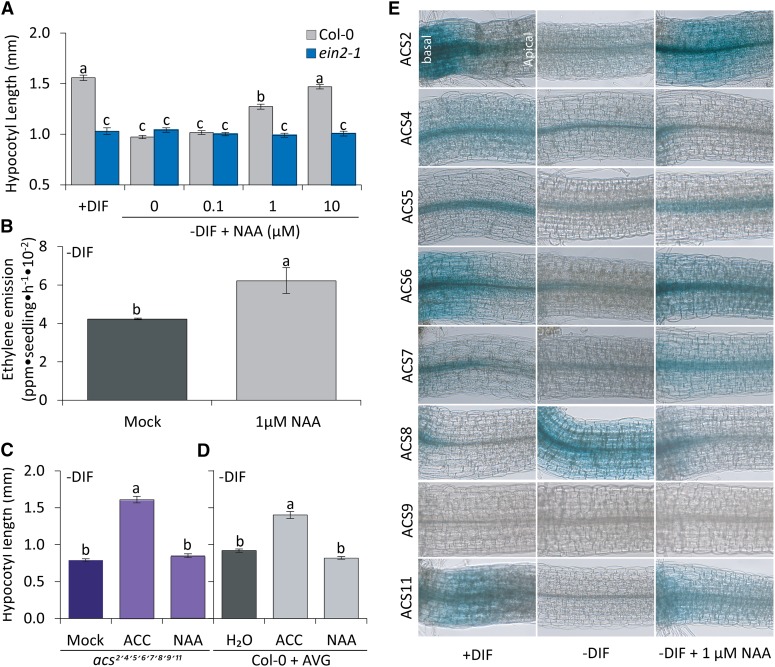
Because both application of auxin and ACC restore hypocotyl growth under −DIF, we used the ethylene signaling mutant ethylene insensitive 2-1 (ein2-1; Guzmán and Ecker, 1990) and the ethylene biosynthesis acs-octuple loss-of-function mutant acs2 acs4 acs5 acs6 acs7 acs9 amiRacs8 (for artificial microRNA ACC synthase8) amiRacs11 (Tsuchisaka et al., 2009) to investigate how auxin and ethylene are linked in this response. The hypocotyl length of the ethylene-insensitive mutant ein2-1 is approximately 40% shorter compared with the wild type when grown under control (+DIF) conditions. When grown under −DIF, its length was not reduced further, which is in contrast to hypocotyl length of wild-type seedlings (Fig. 6A). Moreover, in contrast to wild-type seedlings, NAA application to ein2-1 seedlings did not result in increased hypocotyl length under −DIF (Fig. 6A). This indicates that the auxin regulation of hypocotyl elongation (Fig. 3) requires ethylene signaling. To narrow down the regulatory relationship between auxin and ethylene biosynthesis under −DIF, we quantified how auxin affects ethylene biosynthesis under −DIF conditions. NAA application increased ethylene emission from −DIF-grown seedlings by 40% compared with mock-treated plants (Fig. 6B).
Figure 6.
Auxin acts upstream of ethylene biosynthesis. A, Average hypocotyl length of 7-d-old Arabidopsis seedlings Col-0 (gray) and ein2-1 (blue) with and without NAA. Control represents untreated seedlings developed under +DIF (n = 5 × 25). B, Total ethylene emitted per hour of Arabidopsis seedlings grown under −DIF with and without 1 µm NAA (n = 6 × 30) measured at midday (6 h). C and D, The effect of 10 µm ACC and 1 µm NAA application on hypocotyl elongation in acs2,4,5,6,7,8,9,11 (acs2-1acs4-1acs5-2acs6-1acs7-1acs9-1amiRacs8acs11; purple) seedlings grown for 7 d under −DIF and wild-type seedlings grown in the presence of AVG (gray; n = 5 × 25). Bars represent means ± se. Bars with different letters differ significantly (P < 0.05). E, ACS (2,4,5,6,7,8,9,11)::GUS staining in 7-d-old seedlings in the basal part of the hypocotyl under +DIF, −DIF, and −DIF in the presence of 1 µm NAA performed at midday (6 h).
One of the rate-limiting steps in ethylene biosynthesis is the production of ACC by ACS (Kende, 1993), which is encoded by a multigene family (Tsuchisaka et al., 2009). To determine whether −DIF limits ethylene signaling through auxin-regulated changes in ACS activity, the acs-octuple loss-of-function mutant (Tsuchisaka et al., 2009) was grown under control and −DIF conditions (Fig. 6C; Supplemental Fig. S4). This strong ACC biosynthesis mutant has highly reduced ethylene production (Tsuchisaka et al., 2009), and hypocotyl elongation in this mutant is, therefore, similar to that of the ethylene-insensitive ein2-1 mutant (compare Fig. 6C with Fig. 6A). NAA application to the −DIF-grown acs-octuple mutant did not rescue their hypocotyl growth (Fig. 6, A and C). However, when these mutants were treated with ACC, hypocotyl length was restored to that of the wild type under +DIF conditions (Fig. 6, A and C).
In a complementary assay, we tested the effect of chemical inhibition of ACS activity in wild-type plants by aminoethoxyvinyl-Gly (AVG; Amrhein and Wenker, 1979; Fig. 6D; Supplemental Fig. S4). Application of AVG did not result in a further reduction of wild-type seedling hypocotyl length under −DIF (Fig. 6, A and D). Addition of ACC bypassed the reduced endogenous ACS activity caused by the AVG treatment, and hypocotyl length was restored close to that of the wild type under +DIF (Fig. 6, A and D). NAA application did not increase hypocotyl length in the acs-octuple mutant (Fig. 6C) just like in AVG-treated seedlings under −DIF (Fig. 6D), which is in contrast to wild-type seedlings grown without AVG (Fig. 6A). Combined, these experiments place ethylene signaling downstream of auxin in the elongation response and indicate that, under −DIF, ethylene-induced cell elongation is limited by reduced auxin-regulated ACS activity.
To determine whether a specific member of the ACS family is affected by the day-night temperature difference (DIF) regime, the activities of all functional Arabidopsis ACS genes were analyzed using promoter GUS reporter lines (Tsuchisaka and Theologis, 2004). Under +DIF, the promoters of all ACS genes were active in the basal part of the hypocotyl and the leaf primordia with the exception of ACS9, for which no expression was detected in the seedling (Fig. 6E; Supplemental Fig. S5). −DIF reduced the activity of ACS2, ACS4, ACS5, ACS6, ACS7, and ACS11 in the basal part of the hypocotyl, whereas application of NAA under −DIF restored promoter activity (Fig. 6E). NAA also restored promoter activity of ACS2, ACS4, ACS5, ACS6, and ACS7 but not that of ACS11 in leaf primordia (Supplemental Fig. S5). Only ACS8 showed an opposite behavior, with high activity in the basal hypocotyl under −DIF and low activity under +DIF, whereas NAA under −DIF reduced its activity. ACS8 has, therefore, an opposite response to auxin compared with most other ACS genes. Combined, the results indicate that the reduced auxin levels under −DIF correlate with reduced ACS activity in the zone where reduced cell elongation was observed (Fig. 1D).
PIF3 Acts Downstream of Auxin-Induced Ethylene Biosynthesis
Recently, the transcription factors PIF3, PIF4, and PIF5 were shown to be involved in the regulation of hypocotyl elongation (Nozue et al., 2007; Kunihiro et al., 2011; Soy et al., 2012). Also, it was shown that ACC-induced hypocotyl elongation in the light depends on transcriptional activation of PIF3 (Zhong et al., 2012). To study the relative contribution of individual PIFs in the hypocotyl elongation response, pif single, double, and triple mutants (pif3, pif4, pif5, pif4/pif5, and pif3/pif4/pif5) were grown under +DIF and −DIF. Under +DIF, all pif mutants had shorter hypocotyls than wild-type seedlings (Fig. 7A), whereas under −DIF, the hypocotyl lengths of wild-type and mutant seedlings did not differ (Fig. 7B).
Figure 7.
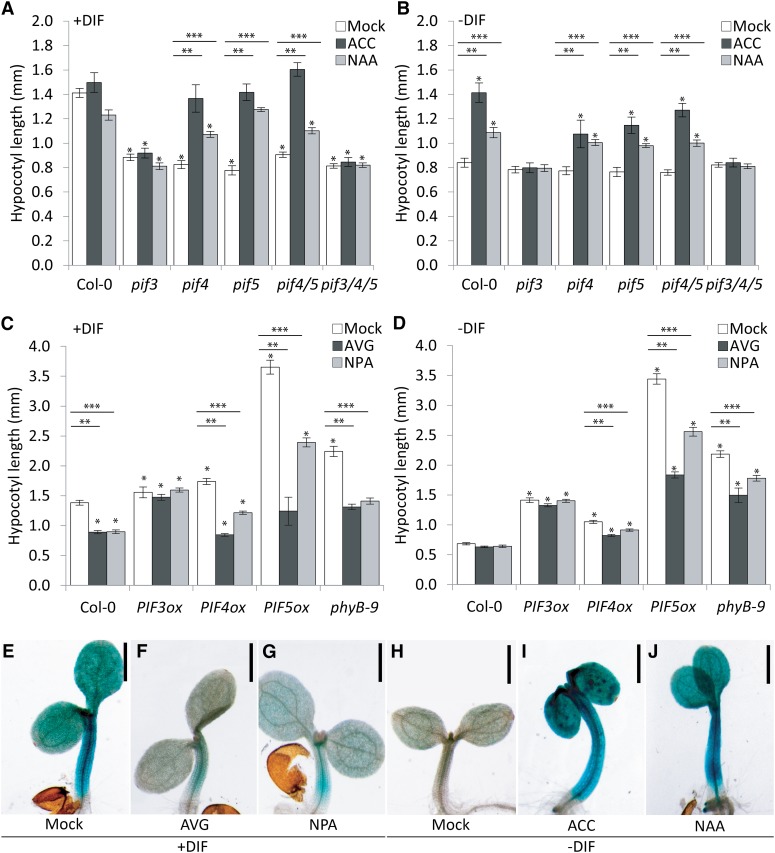
The roles of PIF3, PIF4, and PIF5 in auxin- and ethylene-mediated regulation of hypocotyl elongation. A, The effect of 10 µm ACC and 1 µm NAA application on hypocotyl elongation in Col-0, pif3, pif4, pif5, pif4 pif5, and pif3 pif4 pif5 under +DIF conditions (n = 5 × 25). B, The effect of 10 µm ACC and 1 µm NAA application on hypocotyl elongation in Col-0, pif3, pif4, pif5, pif4 pif5, and pif3 pif4 pif5 under −DIF conditions (n = 5 × 25). C, The effect of 2.5 µm AVG and 1 µm NPA on hypocotyl elongation in Col-0, PIF3ox, PIF4ox, PIF5ox, and phyb-9 under +DIF conditions (n = 5 × 25). D, The effect of 2.5 µm AVG and 1 µm NPA on hypocotyl elongation in Col-0, PIF3ox, PIF4ox, PIF5ox, and phyB-9 under −DIF conditions (n = 5 × 25). E to J, PIF3::GUS promoter activity in 7-d-old seedlings grown under +DIF (E), +DIF in the presence of 2.5 µm AVG (F), +DIF in the presence of 1 µm NPA (G), −DIF (H), −DIF in the presence of 10 µm ACC (I), and −DIF in the presence of 1 µm NAA (J) performed at midday (6 h). *, Significant difference compared with the mock-treated wild type. **, Significant difference between white bars and black bars per genotype. ***, Significant difference between white bars and gray bars per genotype (P < 0.05).
To examine the genetic position of PIF4 and PIF5 in the signal transduction chain relative to auxin and ethylene signaling, the pif mutants were grown under +DIF and −DIF with or without ACC or NAA. Under +DIF, ACC and NAA restored the hypocotyl length of pif4, pif5, and pif4/pif5 to wild-type length. Under −DIF, hypocotyl length of pif4, pif5, and pif4/pif5 did not reach full wild-type length but increased significantly on either application. However, pif3 and pif3/pif4/pif5 hypocotyl lengths were unresponsive to both treatments for both seedlings grown under +DIF (Fig. 7A) and seedlings grown under −DIF (Fig. 7B). Combined, these results indicate that the effect of auxin and ethylene signaling on cell elongation is located downstream of—or parallel to—PIF4 and PIF5 but upstream of PIF3.
To further investigate the role of these PIFs in thermoperiodic hypocotyl elongation, we examined the −DIF responses of PIF3 overexpresser (PIF3ox), PIF4ox, and PIF5ox lines. PIF proteins are degraded by PHYTOCHROME B (PHYB) signaling in the light (Bauer et al., 2004; Park et al., 2004), and as a consequence, the level of endogenous PIF is elevated in phyB mutants (Leivar et al., 2012). Therefore, we also tested how absence of PHYB affects the hypocotyl elongation response under the different DIF treatments. Under +DIF, the hypocotyl lengths of PIF3ox and PIF4ox only marginally increased compared with the wild type. In contrast, both phyB-9 and particularly, PIF5ox displayed a strong increase in hypocotyl length under +DIF (Fig. 7C). The strong response of PIF5ox could be caused by a pleiotropic effect of PIF5ox on PHYB levels (reduced PHYB levels; Khanna et al., 2007), which could make the overexpression of PIF5 affect multiple PIF proteins.
When grown under −DIF, the hypocotyl length of wild-type seedlings strongly decreased as expected (Fig. 7, C and D). However, PIF3ox, PIF5ox, and phyB-9 hypocotyl lengths were not affected by −DIF, indicating that PIF3 and PIF5 are saturating under both DIF conditions. In contrast, the PIF4ox line under −DIF showed a 40% reduction compared with hypocotyl length under +DIF, similar to that for the wild type (Student’s t test P < 0.05). Thus, the PIF4ox line is still sensitive to the effects of −DIF on growth, possibly indicating that the −DIF condition has a stronger effect on PIF4 than on PIF3 and PIF5 (Fig. 7, C and D).
Application of AVG reduced PIF4ox and PIF5ox hypocotyl length, indicating an ethylene component in the elongation response. However, AVG did not affect PIF3ox hypocotyl elongation under either +DIF or −DIF (Fig. 7, C and D). As shown before, phyB-9 showed a strong elongation response under both +DIF and −DIF (Bours et al., 2013), and the treatment with AVG indicated a stronger contribution of ethylene to the elongation response under +DIF than under −DIF (Fig. 7, C and D). Combined, these results indicate that PIF4 and PIF5 function upstream of ethylene, whereas PIF3 acts downstream of ethylene; the results of the phyB-9 mutant with AVG suggest that the contribution of ethylene signaling in the pathway is stronger under +DIF than under −DIF.
We also tested the effect of the polar auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA) in these lines, because previously, it was shown that NPA reduces hypocotyl length of light-grown seedlings in an auxin-dependent manner (Jensen et al., 1998). The application of NPA resulted in reduced hypocotyl length in wild-type seedlings under +DIF but did not further reduce hypocotyl length of seedlings grown under −DIF, indicating that auxin is important for growth under +DIF and that hypocotyl length under −DIF cannot be further reduced (Fig. 7, C and D). In contrast, NPA did not affect hypocotyl length of PIF3ox when grown under either +DIF or −DIF, indicating that, in relation to this phenotype, these lines are insensitive to NPA (Fig. 7, C and D). In contrast, in PIF4ox, PIF5ox, and phyB-9, auxin is required for the elongation response under +DIF, and in these lines, NPA treatment reduced hypocotyl elongation. Under −DIF, the PIF4ox line was only minimally affected by NPA compared with the strong reduction observed in PIF5ox and phyB-9 (Fig. 7, C and D). These results confirm that, under +DIF, PIF4 and PIF5 depend on auxin and that, under −DIF in wild-type plants, auxin becomes limiting for the elongation response. However, for the PIF5ox and phyB-9 mutants, the increased hypocotyl elongation under −DIF is dependent on auxin. The PIF3ox insensitivity to NPA further indicates that PIF3 acts downstream of the auxin signal responsible for elongation.
Because of the important downstream role of PIF3 in the elongation response, we tested how the average PIF3 transcriptional gene activity is affected by DIF treatments. For this purpose, a pPIF3::GUS (Zhong et al., 2012) reporter activity was examined in reporter plants grown under either control or −DIF conditions. Because of the stability of GUS protein, we interpret the GUS staining intensity as indicative for the cumulative PIF3 promoter activity over multiple days. Indeed, less pPIF3::GUS activity was detectable under −DIF compared with +DIF (Fig. 7, E and H). Moreover, this higher PIF3 promoter activity under +DIF was also reduced in response to exogenously applied AVG or NPA (Fig. 7, F and G). In contrast, PIF3 promoter activity under −DIF conditions was up-regulated by ACC and NAA/IAA treatment (Fig. 7, I and J; Supplemental Fig. S6). Combined, the results indicate that PIF3 is an essential downstream target of an auxin-ethylene signal interaction toward elongation growth and that the interaction between diurnal light and temperature cycles regulates the input and signaling capacity of this cascade, possibly through the interaction of other PIFs (e.g. PIF5) and PHYB.
DISCUSSION
In the work presented here, we analyzed how key components of the signaling cascade involved in the growth response of plants (auxin, ethylene, PIFs, and PHYB) function under natural and antiphase light and temperature cycles. We show that antiphase diurnal temperature and light cycles reduce the efficiency in auxin and ethylene signaling for cell elongation in the basal part of the hypocotyl. While under constant light conditions, there is a positive relationship between temperature and growth (Gray et al., 1998; Franklin et al., 2011); under diurnal light-temperature cycles, it is mostly the day (photoperiod) that is most sensitive to temperature. Moreover, we refined the position and the role of these different components within the signal transduction chain. Although we previously identified that, in mature plants, ethylene becomes limiting for leaf elongation growth under −DIF conditions (Bours et al., 2013), we now show that this also holds true for hypocotyl cell elongation and that limited ethylene is a direct consequence of reduced auxin signaling under −DIF. Furthermore, we show that the reduced growth under −DIF can be attributed to limited activity of the growth-promoting PIF transcription factors, for which we have identified distinct positions in the ethylene-auxin signaling toward cell elongation.
Auxin Acts Upstream of Ethylene in Hypocotyl Elongation through Regulation of ACS Genes
Several results indicate that auxin signaling acts upstream of ethylene biosynthesis and signaling in the thermoperiodic elongation response. NAA application failed to restore hypocotyl elongation in ein2-1 and AVG-treated plants (Fig. 6). In addition, we show that auxin induced expression of most ACS genes, which provides a key step in ethylene biosynthesis (Fig. 6E). The reduced elongation under −DIF correlated with reduced eDR5::LUC activity and increased DII Venus activity, both suggesting that auxin levels and signaling are reduced under −DIF, and this reduced auxin signaling was linked to reduced activity of different members of the ACS gene family at the site of reduced cell elongation. Only the ACS8 promoter activity in seedlings responded opposite to the other ACS genes to −DIF. This up-regulation of ACS8 expression under −DIF has also been observed in rosette leaves (Bours et al., 2013), and indeed, it has been shown that ACS8 expression is suppressed by ethylene signaling (Thain et al., 2004). The elevated ACS8 expression, thus, confirms reduced ethylene signaling under −DIF, but the increased expression of this single ACS gene is apparently not able to compensate for the reduced expression of most other endogenous ACS genes. In contrast, application of ACC to seedlings under −DIF was able to restore the hypocotyl elongation response (Fig. 2).
Auxin measurements showed a diurnal pattern for free auxin levels (high during the day and low at night), and these auxin levels were reduced under −DIF (Fig. 4). Interestingly, the free auxin levels at −DIF day are not significantly different from free auxin levels at +DIF night, whereas there is significant difference in eDR5::LUC activity under these conditions (compare Fig. 6A with Fig. 4A). This suggests that seedlings under +DIF night are more sensitive to auxin or that the reporter activity is differentially influenced by these conditions. However, the latter is less likely, because also, the transcriptional analysis of the auxin-inducible SAUR genes shows differential induction at similar auxin levels under +DIF night and −DIF day (Fig. 4, D–H). Therefore, different thermoperiodic conditions not only affect auxin levels but also, influence auxin signaling efficiency.
Positioning PIFs in the Auxin-Ethylene-Cell Elongation Cascade
The PIFs are highly redundant transcription factors, with overlapping roles in the control of elongation (Zheng et al., 2013). Transcriptional analysis of PIF4 and PIF5 under diurnal light conditions (constant temperature) shows that there are strong oscillations in their expression (Nozue et al., 2007). However, combined with the light-induced degradation of PIF proteins by PHYB, the PIF4 and PIF5 reach peak levels during a limited period at the end of the night, at which time they stimulate maximum elongation (Nozue et al., 2007). Despite the degradation by activated PHYB (Monte et al., 2007; Ni et al., 2014) and the proposed coincidence model between light-induced degradation and circadian expression (Nozue et al., 2007), there is increasing consensus that the PIFs also play a role in regulating growth in the light (Zhong et al., 2012; Yamashino et al., 2013; de Lucas and Prat, 2014). Targets of PIF4 and PIF5 are enriched for genes from the auxin biosynthesis pathway, and indeed, auxin responses are severely affected in the pif4 pif5 double mutant (Nozue et al., 2011).
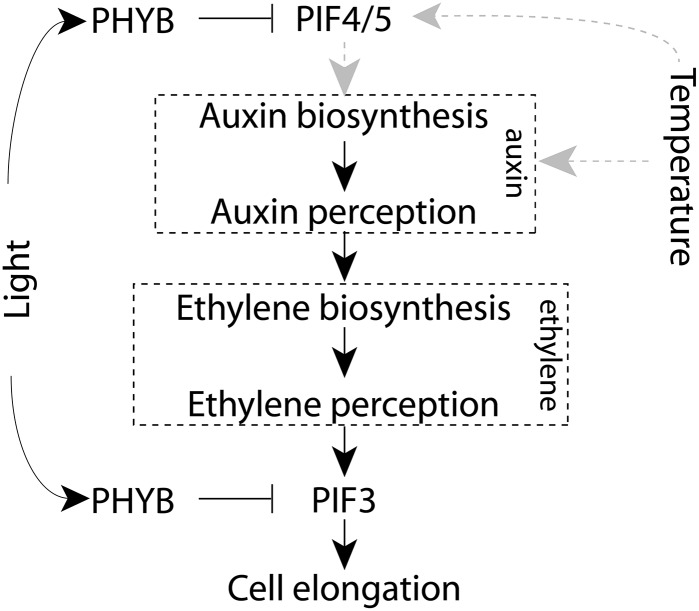
However, the genetic position of these transcription factors relative to the action of auxin and ethylene in elongation growth has not been fully elucidated. We show that the elongation response in pif4, pif5, and pif4/pif5 mutants can be complemented by both auxin and ethylene, indicating that auxin and ethylene act either downstream or independent from PIF4 and PIF5. In contrast, the elongation response of the pif3 and pif3/pif4/pif5 mutants was not complemented by either auxin or ethylene. This indicates that PIF3 functions downstream of auxin and ethylene in the signal transduction chain that regulates thermoperiodic hypocotyl elongation (Fig. 8). Moreover, when ACC application under −DIF conditions was combined with AVG, PIF3 expression could be fully restored (Fig. 7), whereas NAA application combined with AVG only modestly increased PIF3 expression (Fig. 7; Supplemental Fig. S6). This indicates that the effect of auxin on PIF3 expression is indirect and requires ethylene production. The regulation by (indirect) auxin and (direct) ethylene signaling of PIF3 expression during the photoperiod fits well with the finding that the ethylene signaling component EIN3 acts a regulator of PIF3-induced hypocotyl elongation only during the photoperiod. EIN3 was shown to bind to the PIF3 promoter (Zhong et al., 2012), thus inducing growth during the light period and providing a direct link between ethylene signaling and PIF3 activity. Combined, the results indicate that, in wild-type seedlings, the auxin-ethylene-induced PIF3 expression is the main determinant of thermoperiodic hypocotyl elongation.
Figure 8.
Proposed model for the PIF-mediated auxin-ethylene cross talk in relation to hypocotyl elongation under −DIF.
Link between PIF5, PHYB, Auxin, and Ethylene Biosynthesis?
The effect of ectopic expression of PIF genes is complex. Both PIF4 and PIF5 require auxin and ethylene signaling for the elongation response; however, the PIF5ox line shows a very strong growth response and is almost insensitive to −DIF, whereas the PIF4ox line only shows a mild growth increase compared with the wild type, and PIF4ox is still fully sensitive to −DIF (Fig. 7, C and D). This suggests that ectopic expression of PIF5, but not PIF4, may also be able to control growth independent of ethylene. Indeed, differential growth responses independent of ethylene are observed in the mutant ein2-1, which displays hypocotyl elongation in response to high temperature (29°C) under continuous light (Gray et al., 1998). However, this heat-induced growth response was shown to depend specifically on PIF4 (Franklin et al., 2011), whereas ectopic expression of PIF5 seems to be more effective in stimulating growth. Indeed, previously, it has been shown that PIF5ox increases ethylene levels and that PIF5 negatively regulates PHYB levels in the light. The direct effect of PIF5 on ACS genes explains why AVG has a stronger effect on elongation in PIF5ox lines than inhibition of auxin (Fig. 7, C and D). This effect of PIF5ox seems to be independent of the thermoperiodic regime, because PIF5ox elongation was hardly affected by −DIF. The increased ethylene production is a unique feature of the PIF5ox, because this was not observed in PIF3ox and PIF4ox lines (Khanna et al., 2007).
The loss of PHYB function results in increased hypocotyl elongation, which is insensitive to −DIF (Thingnaes et al., 2008; Bours et al., 2013). PIF protein stability is highly dependent on PHYB-mediated degradation (Monte et al., 2007), and a phyB mutant is, therefore, expected to have increased levels of PIF protein. We previously showed that phyB-9 has elevated ethylene emissions and increased ACS expression (Bours et al., 2013). This phenotype is similar to that of PIF5ox, which could suggest that increased PIF5 activity is responsible for the increased ethylene production in phyB-9.
PIF5 has also been shown to positively regulate auxin signaling (Nozue et al., 2011), and the effect of PIF5 on ACS gene expression could, therefore, be through its effect on auxin signaling. However, in the wild type, AVG and NPA have a similar inhibitory effect on hypocotyl elongation (Fig. 7, C and D), whereas in the PIF5ox line, the effect of AVG is larger than that of NPA, indicating that only part of the effect of PIF5ox is through auxin.
Interestingly, PIF5 is also able to directly induce ACS8 expression by binding a G-box domain in the ACS8 promoter in a GA-dependent manner (Gallego-Bartolomé et al., 2011). This indicates that, in addition to an auxin-dependent induction, an additional direct induction of ethylene biosynthesis by PIF5 under +DIF conditions cannot be excluded. However, a negative feedback exists between ACS8 and ethylene signaling (Thain et al., 2004). Thus, the increased expression of ACS8 under −DIF is likely the result of the reduced ethylene sensitivity (Bours et al., 2013) and biosynthesis (Fig. 6E) in seedlings grown under −DIF.
−DIF Limits Elongation Growth during the Cold Photoperiod
Plant growth is a continuous process. To achieve this continuity, tight regulation of internal hormonal signaling is required. Previous studies showed that, whereas ethylene stimulates growth during the day, it becomes growth limiting during the night (Zhong et al., 2012). Here, we show that, under −DIF, no hypocotyl growth occurs during the cold photoperiod. We show that a disrupted auxin-ethylene cascade is the underlying mechanism that prevents growth-inducing ethylene levels during the day. Because both light and temperature are strong entrainment signals for the clock, the altered growth response under −DIF could be the result of an altered clock function under this condition. PHYB, PIF4, PIF5, ACS, and auxin biosynthesis and signaling genes are all regulated by the circadian clock (Thain et al., 2004; Covington and Harmer, 2007; Nozue et al., 2007). We propose that conflicting input signals result in dysregulation of the PIF-controlled auxin-ethylene signaling cascade. This ultimately leads to a disruption of cellular elongation under −DIF conditions (Fig. 8).
MATERIALS AND METHODS
Plant Material and Growing Conditions
Plant growth under +DIF or −DIF conditions was performed as described previously (Bours et al., 2013). All experiments were performed in automated climate-controlled Weiss (http://www.wkt.com) cabinets (12-h/12-h light-dark cycle). Relative humidity was kept constant at 60% (v/v), and photosynthetic active radiation was 150 µmol m−2 s−1 from white fluorescents tubes (Philips; type T5, color code 840). Ambient temperature cycles for growth under conditions were 22°C (photoperiod) and 12°C (dark period), with a temperature ramp of 0.33°C min−1. For −DIF treatment, the temperature cycles were reversed to −DIF of 12°C (photoperiod) and 22°C (dark period), with a temperature ramp of 0.33°C min−1. All other growth parameters were kept equal to +DIF conditions.
Arabidopsis (Arabidopsis thaliana) seeds were obtained from either the Nottingham Arabidopsis Stock Centre or the authors who first described the line. All transgenic lines and mutants are in Col-0 (N1092) background. Lines used in this research are pACS2::GUS/GFP (N31380), pACS4::GUS (N31381), pACS5::GUS (N31382), pACS6::GUS/GFP (N31383), pACS7::GUS/GFP (N31384), pACS8::GUS/GFP (N31385), pACS9::GUS/GFP (N31386), pACS11::GUS/GFP (N31387), ein2-1 (N3071), eDR5::LUC (Covington and Harmer, 2007), DII::VENUS (Brunoud et al., 2012), acs2 acs4 acs5 acs6 acs7 acs9 amiRacs8 amiRacs11 (Tsuchisaka et al., 2009), phyb-9 (Reed et al., 1993), pif3-7 (N66042), pif4-2 (N66043), pif5-3 (N66044), pif4-2 pif5-3 (Hornitschek et al., 2012), pif3-7 pif4-2 pif5-3 (N66048), PIF3ox (Shin et al., 2007), PIF4ox (Nozue et al., 2007), PIF5ox (Khanna et al., 2007), and pPIF3::GUS (Zhong et al., 2012). All seedlings were surface sterilized and stratified in the dark for 2 or 3 d (4°C) to synchronize germination, and then, they were exposed to treatment conditions for 7 d. All seedlings were grown on petri dishes (6-cm diameter) containing 10 mL of one-half-strength Murashige and Skoog (MS) medium-enriched plant agar (0.22 g L−1 MS [Duchefa] and 8 g L−1 plant agar [Duchefa]) under either +DIF or −DIF conditions as previously described (Bours et al., 2012). Plates were randomized to reduce positional effects. All seedlings were harvested for further use. Corresponding samples were extracted for auxin quantification or quantitative real-time-PCR or freeze-dried (Christ α1-4; LSC freeze dryer) to determine dry weight. Seedling dry weight was determined by measuring the weight of three individual pools of 209 seedlings with an Ohaus Pioneer Precision Balance (0.0001 g of readability). All data sets presented were confirmed in at least two independent trials with similar setups and outcomes.
Hypocotyl Elongation Response to Growth Regulators
Surface-sterilized seeds were grown for 7 d under +DIF or −DIF conditions with different concentrations (0, 0.1, 1, 10, or 100 μm) of NAA (Sigma-Aldrich), IAA (Duchefa), NPA (Sigma-Aldrich), AVG (Sigma-Aldrich), and ACC (Sigma-Aldrich). Used seed batches were of identical age, and Col-0 and ein2-1 were tested as described by Cheng et al. (2009) and did not differ for germination rate or response to NAA under +DIF conditions (Supplemental Fig. S7).
Analysis of Hypocotyl Length
Seedlings were placed in a horizontal position and photographed (with a mounted Nikon D90 camera), and hypocotyl lengths were assessed using ImageJ software (Rasband, 2011).
Histochemical GUS Staining
Histochemical GUS staining of pACS2,4,5,6,7,8,9,11::GUS and pPIF3::GUS reporter hypocotyls grown under +DIF or −DIF was performed as previously described (Kim et al., 2006). Incubation times varied between 10 and 60 min but were always identical within experiments. After staining, fixation, and clearing, individual hypocotyls were imaged.
LUC Imaging and Analysis
LUC imaging and analysis were performed as previously described (Leeuwen et al., 2000) with the following modifications. Seedlings containing the eDR5::LUC reporter construct (Covington and Harmer, 2007) were grown on 0.5 mL of medium in 12-well black plates (Promega). Each well contained 50 seedlings evenly spread out. Seedlings developed in 12-h-white light (approximately 150 μmol m−2 s−1)/12-h-dark cycles for 4 d with +DIF or −DIF temperature cycles; 24 h before the start of imaging, a thin layer of 0.3 mL of d-luciferin (Duchefa) solution (1 mm firefly d-luciferin, sodium salt [Duchefa], and 0.01% [v/v] Tween 80) was added to the media. Seedlings were assayed for bioluminescence by acquiring images with an exposure time of 15 min using a Pixis 1024B (1,024 × 1,024) camera system (Princeston Instruments) equipped with a 35-mm, 1:1.4 Nikkor SLR camera lens (Nikon) fitted with a DT Green filter ring (Image Optics Components Ltd.) to block delayed autofluorescence chlorophyll emissions. Relative bioluminescence was analyzed for each well in each image as mean gray value using Region of Interest manager with ImageJ software. Image background subtraction was performed for each image and each well. Images of LUC activity are depicted with false color scales (blue indicating low activity and red indicating high activity).
Microscopy
For confocal microscopy, hypocotyls of 7-d-old seedlings were incubated for 10 min in 1 mm propidium iodide (SIGMA) and imaged on a Zeiss 700 Axio Imager confocal laser-scanning microscope (Carl Zeiss) using either an EC Plan-Neofluar 40× (numerical aperture 1.30) Oil DIC M27 or an EC Plan-Neofluar 10× (numerical aperture 0.3) Ph1 objective. Samples were excited with 2% of maximum intensity of a 488-nm laser (emission from a 30-mW argon tube) for YFP excitation and 2% of maximum intensity of a 555-nm laser (emission from a 1-mW helium-neon tube) for propidium iodide excitation. Transmission images were simultaneously collected. Single midplane optical sections were selected from z series and compared. Image analysis was done using ZEN 2011 and Adobe Photoshop CS2 (Adobe Systems).
For bright field, 7-d-old seedlings were imaged on a Zeiss Stereo Discovery (A12) with a Plan S 1.0× FWD 81-mm (1–100×) objective. Images were taken with an AxioCam MRc5 (5 MPix camera; Zeiss) and analyzed using AxioVision 4.6 software.
Auxin Analysis
Arabidopsis seedlings grown under either +DIF or −DIF were harvested at midday (6 h; lights on at 0 h) and midnight (18 h; lights off at 12 h), frozen in liquid nitrogen, ground, extracted, and analyzed for auxin content as previously described (Kohlen et al., 2012).
Ethylene Analysis
Thirty seedlings were grown at either +DIF or −DIF for 5 d on 2.5 mL of MS in 10-mL glass vials sealed with micropore tape to allow gas exchange. At the start of the fifth photoperiod, the vials were capped airtight and returned to the experimental conditions. Ethylene was allowed to accumulate in the experimental conditions for 3 d, and ethylene concentrations were quantified in 0.5 mL of headspace using gas chromatography with flame ionization detection.
Gene Expression Analysis
Arabidopsis seedlings grown under either +DIF or −DIF were harvested at midday (6 h; lights on at 0 h) and midnight (18 h; lights off at 12 h). RNA was extracted using the RNeasy Plant Micro Kit (Qiagen), including a subsequent DNaseI treatment (Roche Diagnostics). One microgram of RNA was used for complementary DNA synthesis with the RevertAid H Minus First-Strand cDNA Synthesis Kit (Fermentas). Quantitative PCR was performed using the PowerSYBR-Green PCR Master Mix (Applied Biosystems). Primers are described in Supplemental Table S1. Relative quantification was done using internal standard curves and correcting by the use of the reference gene UBQ5 (Zhong et al., 2012).
Statistical Analysis
When appropriate, data were subjected to the Student's t test (Microsoft Excel). All other data were subjected to one-way ANOVA. Individual differences were then identified using a post hoc Tukey test (P < 0.05). All analyses were performed using SAS_9.20 (http://www.sas.com/).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. IAA complements hypocotyl elongation under −DIF conditions.
Supplemental Figure S2. Auxin response as visualized by eDR5::LUC bioluminescence.
Supplemental Figure S3. Expression analysis of YUC auxin biosynthesis genes.
Supplemental Figure S4. Auxin and ethylene application in combination with ethylene insensitivity.
Supplemental Figure S5. ACS::GUS promoter activity.
Supplemental Figure S6. PIF3::Columbia-0 (Col-0; promoter activity under −DIF.
Supplemental Figure S7. Germination rate of Col-0 and ein2-1 seeds.
Supplemental Table S1. Primer sequences used for gene expression analysis.
Supplementary Material
Acknowledgments
We thank Johanna Rienks (Bonn University), Francel Verstappen (Wageningen University and Research Centre), Tatsiana Charnikhova (Wageningen UR, The Netherlands), and Alexandra Kalde (Max Planck Institute for Plant Breeding Research) for technical assistance and Amaury de Montaigu (MPIPZ, Germany) and Maarten Koornneef (MPIPZ, Germany) for critical reading of this article.
Glossary
- AVG
aminoethoxyvinyl-Gly
- Col-0
Columbia-0
- DIF
day-night temperature difference
- −DIF
negative day-night temperature difference
- +DIF
positive day-night temperature difference
- MS
Murashige and Skoog medium
- NAA
1-naphthaleneacetic acid
- NPA
1-N-naphthylphthalamic acid
Footnotes
This work was supported by the Alexander von Humboldt Foundation (research fellowship no. 1146256 to W.K.), the Royal Botanical Society of The Netherlands and the Hugo de Vries Foundation (Hugo de Vries Award to W.K.), and the Top Technological Institute Green Genetics (grant no. 2CFL009RP to A.v.d.K.).
References
- Amrhein N, Wenker D (1979) Novel inhibitors of ethylene production in higher plants. Plant Cell Physiol 20: 1635–1642 [Google Scholar]
- Bakken AK, Flønes M (1995) Morphology and field performance of Brassica transplants propagated under different day and night temperature regimes. Sci Hortic (Amsterdam) 61: 167–176 [Google Scholar]
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, et al. (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bours R, Muthuraman M, Bouwmeester H, van der Krol A (2012) OSCILLATOR: a system for analysis of diurnal leaf growth using infrared photography combined with wavelet transformation. Plant Methods 8: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bours R, van Zanten M, Pierik R, Bouwmeester H, van der Krol A (2013) Antiphase light and temperature cycles affect PHYTOCHROME B-controlled ethylene sensitivity and biosynthesis, limiting leaf movement and growth of Arabidopsis. Plant Physiol 163: 882–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M (2012) Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 7: e36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Chiang MH, Hwang SG, Lin PC (2009) Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol Biol 71: 61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Prat S (2014) PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol 202: 1126–1141 [DOI] [PubMed] [Google Scholar]
- Franklin KA. (2009) Light and temperature signal crosstalk in plant development. Curr Opin Plant Biol 12: 63–68 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC (2004) Light signals, phytochromes and cross-talk with other environmental cues. J Exp Bot 55: 271–276 [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Arana MV, Vandenbussche F, Zádníková P, Minguet EG, Guardiola V, Van Der Straeten D, Benkova E, Alabadí D, Blázquez MA (2011) Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J 67: 622–634 [DOI] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimstad SO, Frimanslund E (1993) Effect of different day and night temperature regimes on greenhouse cucumber young plant production, flower bud formation and early yield. Sci Hortic (Amsterdam) 53: 191–204 [Google Scholar]
- Guzmán P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Chory J (2010) Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Biol 17: 642–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M (1998) Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol 116: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. (1993) Ethylene biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 44: 283–307 [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH (2004) A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E, Quail PH (2007) The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19: 3915–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Franceschi VR, Davin LB, Lewis NG, Salinas J, Sanchez-Serrano JJ (2006) Beta-glucuronidase as reporter gene: advantages and limitations. InSalinas J, Sanchez-Serrano JJ, eds, Arabidopsis Protocols, Vol 323 Humana Press, Totowa, NJ, pp 263–273 [DOI] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Lammers M, Pollina T, Tóth P, Haider I, Pozo MJ, de Maagd RA, Ruyter-Spira C, Bouwmeester HJ, et al. (2012) The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol 196: 535–547 [DOI] [PubMed] [Google Scholar]
- Kunihiro A, Yamashino T, Nakamichi N, Niwa Y, Nakanishi H, Mizuno T (2011) Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol 52: 1315–1329 [DOI] [PubMed] [Google Scholar]
- Leeuwen W, Hagendoorn MM, Ruttink T, Poecke R, Plas LW, Krol A (2000) The use of the luciferase reporter system forin planta gene expression studies. Plant Mol Biol Re 18: 143–144 [Google Scholar]
- Leivar P, Monte E, Cohn MM, Quail PH (2012) Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually negative phyB-PIF feedback loop. Mol Plant 5: 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano Y, Nemoto K (2012) The pathway of auxin biosynthesis in plants. J Exp Bot 63: 2853–2872 [DOI] [PubMed] [Google Scholar]
- Monte E, Al-Sady B, Leivar P, Quail PH (2007) Out of the dark: how the PIFs are unmasking a dual temporal mechanism of phytochrome signalling. J Exp Bot 58: 3125–3133 [DOI] [PubMed] [Google Scholar]
- Muday GK, Rahman A, Binder BM (2012) Auxin and ethylene: collaborators or competitors? Trends Plant Sci 17: 181–195 [DOI] [PubMed] [Google Scholar]
- Myster J, Moe R (1995) Effect of diurnal temperature alternations on plant morphology in some greenhouse crops; a mini review. Sci Hortic (Amsterdam) 62: 205–215 [Google Scholar]
- Nakayama N, Smith RS, Mandel T, Robinson S, Kimura S, Boudaoud A, Kuhlemeier C (2012) Mechanical regulation of auxin-mediated growth. Curr Biol 22: 1468–1476 [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH (2014) A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Nozue K, Harmer SL, Maloof JN (2011) Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol 156: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Kim J, Lee Y, Shin J, Oh E, Chung WI, Liu JR, Choi G (2004) Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol 45: 968–975 [DOI] [PubMed] [Google Scholar]
- Pierik R, Tholen D, Poorter H, Visser EJW, Voesenek LACJ (2006) The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci 11: 176–183 [DOI] [PubMed] [Google Scholar]
- Rasband WS (2011) ImageJ. U.S. National Institutes of Health, Bethesda, MD,. http://imagej.nih.gov/ij/ (January 1, 2011)
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R, Chinnappa CC, Staal M, Elzenga JTM, Yokoyama R, Nishitani K, Voesenek LA, Pierik R (2010) Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol 154: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Smalle J, Haegman M, Kurepa J, Van Montagu M, Straeten DV (1997) Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci USA 94: 2756–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, González-Schain N, Sentandreu M, Prat S, Quail PH, Monte E (2012) Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J 71: 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Nemhauser JL (2013) Auxin 2012: a rich mea ho’oulu. Development 140: 1153–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Zhai Q, Li C (2013) PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25: 2102–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GTS, Sandberg G, Bhalerao R, Ljung K, Bennett MJ (2007) Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain SC, Vandenbussche F, Laarhoven LJJ, Dowson-Day MJ, Wang ZY, Tobin EM, Harren FJM, Millar AJ, Van Der Straeten D (2004) Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol 136: 3751–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingnaes E, Torre S, Ernstsen A, Moe R (2003) Day and night temperature responses in Arabidopsis: effects on gibberellin and auxin content, cell size, morphology and flowering time. Ann Bot (Lond) 92: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingnaes E, Torre S, Moe R (2008) The role of phytochrome B, D and E in thermoperiodic responses of Arabidopsis thaliana. Plant Growth Regul 56: 53–59 [Google Scholar]
- Tsuchisaka A, Theologis A (2004) Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol 136: 2982–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A (2009) A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183: 979–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Went FW. (1944) Plant growth under controlled conditions. II. Thermoperiodicity in growth and fruiting of the tomato. Am J Bot 31: 135–150 [Google Scholar]
- Yamashino T, Nomoto Y, Lorrain S, Miyachi M, Ito S, Nakamichi N, Fankhauser C, Mizuno T (2013) Verification at the protein level of the PIF4-mediated external coincidence model for the temperature-adaptive photoperiodic control of plant growth in Arabidopsis thaliana. Plant Signal Behav 8: e23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zádníková P, Petrásek J, Marhavý P, Raz V, Vandenbussche F, Ding Z, Schwarzerová K, Morita MT, Tasaka M, Hejátko J, et al. (2010) Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novák O, Dai X, Zhao Y, Ljung K, Noel JP, Chory J (2013) Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat Chem Biol 9: 244–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wang L, Xi Y, Li J, Quail Peter H, Deng Xing W, Guo H (2012) A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol 22: 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.