A duplicated pair of transcription factors control cell patterning and cell differentiation in maize endosperm development.
Abstract
The aleurone is the outermost layer of cereal endosperm and functions to digest storage products accumulated in starchy endosperm cells as well as to confer important dietary health benefits. Whereas normal maize (Zea mays [Zm]) has a single aleurone layer, naked endosperm (nkd) mutants produce multiple outer cell layers of partially differentiated cells that show sporadic expression of aleurone identity markers such as a viviparous1 promoter-β-glucuronidase transgene. The 15:1 F2 segregation ratio suggested that two recessive genes were involved, and map-based cloning identified two homologous genes in duplicated regions of the genome. The nkd1 and nkd2 genes encode the INDETERMINATE1 domain (IDD) containing transcription factors ZmIDDveg9 and ZmIDD9 on chromosomes 2 and 10, respectively. Independent mutant alleles of nkd1 and nkd2, as well as nkd2-RNA interference lines in which both nkd genes were knocked down, also showed the nkd mutant phenotype, confirming the gene identities. In wild-type kernels, the nkd transcripts were most abundant around 11 to 16 d after pollination. The NKD proteins have putative nuclear localization signals, and green fluorescent protein fusion proteins showed nuclear localization. The mutant phenotype and gene identities suggest that NKD controls a gene regulatory network involved in aleurone cell fate specification and cell differentiation.
Cereal grains are essential for humans as a food source and for value-added industrial materials. The endosperm comprises 70% to 90% of a grain and is the major source of nutrients and feedstock. The outermost layer of the cereal endosperm is a specific cell type called the aleurone. Aleurone cells survive grain desiccation, while starchy endosperm undergoes programmed cell death. At germination, the aleurone layer secretes hydrolases to digest storage molecules (starch and proteins) in the starchy endosperm. The aleurone is relatively protein and lipid rich and also confers most of the dietary benefits attributed to cereal bran (Becraft and Yi, 2011). In particular genotypes of maize (Zea mays [Zm]), anthocyanin pigments specifically accumulate in the aleurone, and this has historically been used as a powerful marker for genetic studies (Cone, 2007). Most cereal grains, including maize, have a single cell layer of aleurone, although barley (Hordeum vulgare) and at least one maize landrace have multilayered aleurone (Wolf et al., 1972; Jestin et al., 2008). Aleurone development involves a decision between aleurone versus starchy endosperm cell fates. Because the aleurone is the outmost layer in the endosperm, it is inferred that a positional signal must be involved in this cell fate decision (Becraft et al., 1996; Becraft and Asuncion-Crabb, 2000; Olsen, 2004a). Cells at the periphery of the endosperm retain the plasticity to become either aleurone or starchy endosperm until the last cell divisions during development (Becraft and Asuncion-Crabb, 2000). Thus, the positional cues that specify aleurone identity are present throughout development and are required to maintain the appropriate cell identities of peripheral endosperm cells.
The molecular mechanisms of aleurone cell differentiation are not well understood. Analysis of aleurone mutant phenotypes and cell lineage studies suggest that positional cues and a hierarchical genetic pathway control aleurone cell fate (Becraft and Asuncion-Crabb, 2000; Olsen, 2004b; Wisniewski and Rogowsky, 2004). Known genes regulating aleurone cell fate include defective kernel1 (dek1), crinkly4 (cr4), and supernumerary aleurone1 (sal1). dek1 is a positive regulator of aleurone cell fate, and its loss-of-function mutant shows an absence of aleurone (Sheridan and Neuffer, 1982; Becraft and Asuncion-Crabb, 2000; Becraft et al., 2002; Lid et al., 2002). DEK1 is a large protein containing 21 predicted transmembrane helices, an extracellular loop region, and a cytoplasmic calpain protease domain (Lid et al., 2002; Wang et al., 2003; Johnson et al., 2008; Liang et al., 2013; Demko et al., 2014). A strong mutant allele such as dek1-1394 eliminates aleurone completely, while the weaker dek1-D shows mosaic aleurone (Becraft et al., 2002). CR4 is a receptor-like kinase, and cr4 mutants show similar, although more sporadic, aleurone phenotypes to dek1 (Becraft et al., 1996). DEK1 and/or CR4 may work as receptors for positional cues that induce and maintain aleurone cell specification. The sal1 mutant has multiple aleurone layers, and the gene encodes a class E vacuolar sorting protein (Shen et al., 2003). SAL1 is hypothesized to act as a negative regulator of CR4 and/or DEK1 (Shen et al., 2003; Tian et al., 2007). Despite having been cloned and studied, the molecular mechanisms by which these proteins specify aleurone identity are not yet understood. Another negative regulator of aleurone cell fate, thick aleurone1 (thk1), was recently reported; the loss-of-function mutant makes four to five layers of aleurone (Yi et al., 2011). The thk1 mutant is epistatic to dek1, and double mutant sectors generated by chromosomal breakage in a dek1 single mutant (aleuroneless) background show multiple layers of aleurone. Although the identity of thk1 is not yet known, the epistasis suggests that it functions in the same pathway as DEK1.
Transcriptional regulation is central to most developmental processes. VIVIPAROUS1 (VP1) is a transcription factor containing a B3 domain that binds the CATGCA DNA element to regulate genes that function in seed maturation (Suzuki et al., 1997). The vp1 gene is required for seed maturation and is the most upstream known transcriptional regulator of the anthocyanin biosynthesis pathway. It is specifically expressed in embryo and aleurone cells and is controlled by abscisic acid (ABA; Cao et al., 2007). ABA and GA3 play major antagonistic roles in controlling seed maturation and dormancy versus germination and vivipary.
INDETERMINATE1 (ID1) is a transcription factor that is important for the flowering response in maize. ID1 is a member of a family of transcriptional regulators containing a conserved C2H2 zinc finger DNA-binding domain called the INDETERMINATE1 domain (IDD). Here, we report a novel aleurone differentiation mutant, naked endosperm (nkd), and show that mutations in duplicated IDD genes, iddveg9 and idd9, cause the nkd phenotype. Gene function was confirmed by the identification of independent mutant alleles and by RNA interference (RNAi)-induced gene knockdown. We propose that NKD proteins function as transcription factors controlling aleurone layer organization and cell differentiation.
RESULTS
The nkd Genes Are Required for Aleurone Cell Fate and Cell Differentiation
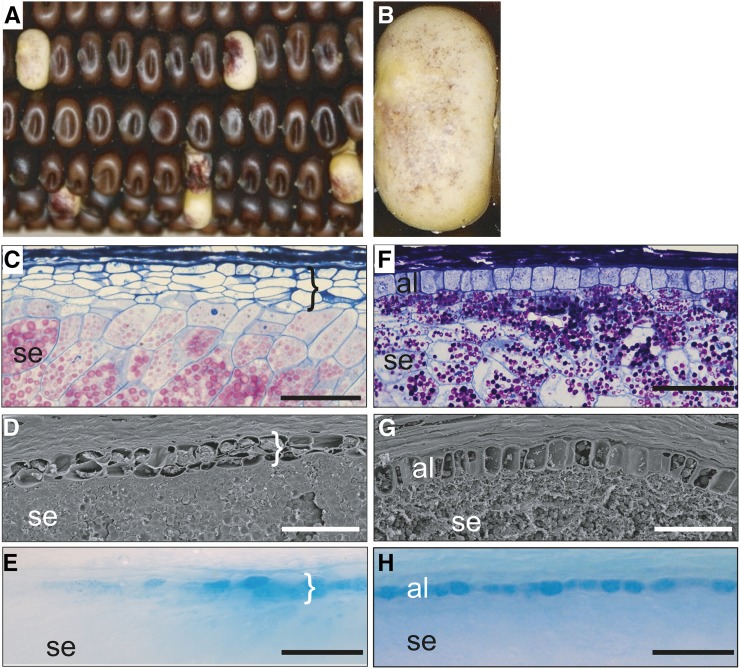
The nkd mutant maize kernels show aleuroneless or mosaic aleurone phenotypes (Becraft and Asuncion-Crabb, 2000). In sections, nkd mutants have multiple (two to five) layers of peripheral endosperm cells that lack starch granules or other features of starchy endosperm (Fig. 1). Yet, most of these cells do not have the typical characteristics of wild-type aleurone cells, such as thick walls, dense cytoplasm, accumulation of anthocyanin pigments, or expression of the viviparous1 promoter (vp1pro)-GUS marker gene. Cells with typical aleurone features do form sporadically within the peripheral layers. Aleurone cells are more likely to form around the silk scar region, following a pattern that has been described for several mosaic aleurone mutants (Becraft et al., 1996, 2002; Becraft and Asuncion-Crabb, 2000; Suzuki et al., 2008; Becraft and Yi, 2011). The nkd mutants show a 15:1 F2 segregation ratio, indicating that two unlinked recessive factors, nkd1 and nkd2, determine this phenotype. Because only the double homozygous mutant shows a mutant phenotype, it is likely that these two genes perform redundant functions.
Figure 1.
The nkd mutant disrupts aleurone differentiation. A and B, nkd mutant kernels show sporadic anthocyanin pigmentation and a 15:1 F2 segregation ratio. C to E, The nkd mutant disrupts aleurone differentiation. The peripheral layer of nkd mutant endosperm is distinct from internal starchy endosperm but has not differentiated the typical attributes of aleurone cells, including the expression of aleurone markers such as vp1. Brackets indicate cells with compromised aleurone characteristics. F to H, Wild-type endosperm contains a single layer of aleurone cells showing densely staining cytoplasm, thick cell walls, cuboidal shape, and the expression of aleurone markers. C and F, Histological sections with starch grains stained pink by periodic acid-Schiff and aleurone darkly stained with Toluidine Blue counterstain. D and G, Scanning electron micrographs. E and H show the expression of a vp1pro-GUS reporter transgene detected by 5-bromo-4-chloro-3-indolyl-β-glucuronic acid staining specifically in aleurone cells. The vp1pro-GUS reporter shows sporadic expression in nkd mutants. al, Aleurone; se, starchy endosperm. Bars = 100 µm.
Because the peripheral layers of endosperm in the nkd mutant were distinct from starchy endosperm and sporadically acquired aleurone cell characteristics, the nkd mutant was interpreted as affecting the aleurone cell differentiation process (Becraft and Asuncion-Crabb, 2000). However, the multiple layers of cells distinct from starchy endosperm, in contrast to the normal single layer of aleurone, indicate that nkd function is also required to restrict the number of peripheral cell layers. Therefore, nkd appears required for dual functions in the endosperm periphery, regulating cell fate specification and cell differentiation.
Other Pleiotropic Aspects of nkd Mutants
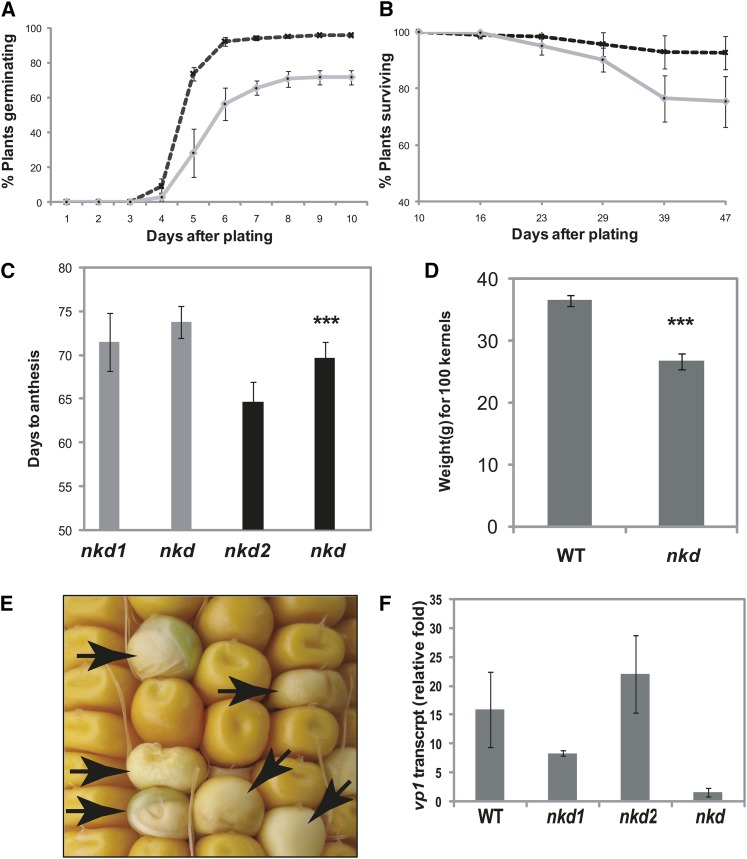
Mutants displayed several additional pleiotropic phenotypes in the kernels and plants (Fig. 2). The endosperms of nkd mutant kernels often display an opaque phenotype instead of the normal translucency. Defects in several endosperm components can result in an opaque phenotype including deficiencies or imbalances in zein storage proteins. HPLC analysis showed no detectable difference in zein profiles between normal and mutant kernels (data not shown); thus, the basis of the endosperm opacity remains unknown.
Figure 2.
Pleiotropic phenotypes of nkd mutants. A and B, Germination rate (A) and survival rate (B) of the wild type (dashed lines) and nkd mutants (solid lines). Segregating F2 kernels were planted in the field. A total of 72% of nkd mutants germinated (96% in the wild type), and 76% of germinated nkd mutants survived (over 93% in the wild type) after 39 d. C, nkd mutants show delayed flowering. nkd double mutants were delayed 5 d to anthesis compared with the nkd2 single mutant (P < 0.001). No significant difference was observed between nkd double mutants and the nkd1 single mutant. Gray and black bars represent two different populations of plants comparing nkd double mutants with nkd1 and nkd2 single mutants, respectively. The number of total leaves did not show any significant difference among the genotypes. D, Hundred-kernel weight was significantly less for nkd mutants than for the wild type (WT; P < 0.001). E, nkd mutant kernels (arrows) are pale yellow and prone to vivipary. F, Quantitative RT-PCR showed decreased amounts of vp1 transcript in nkd mutants.
Mutant kernels are also pale yellow or sometimes nearly white, indicating that they are carotenoid deficient (Fig. 2E). There is also a propensity for vivipary in nkd mutant seeds. The disrupted expression of the vp1pro-GUS marker indicates that nkd functions upstream of vp1 in the endosperm. When mutant embryos were stained for GUS activity, no qualitative difference in staining was detected (data not shown). Expression of vp1, therefore, was assayed by quantitative reverse transcription (RT)-PCR and found to be decreased in mutant kernels compared with the wild type (Fig. 2F).
Seed weight is decreased in the mutant. Hundred-kernel seed weights were measured for nkd mutant and the wild type in five samples from a segregating F2 population. The mutant seed weight (26.8 ± 1.28 g) was only 73% of the wild-type seed weight (36.5 ± 0.95 g). Thus, nkd mutant seed appears to have reduced grain filling for which the basis is not yet known (Fig. 2D).
The nkd mutant seed showed decreased germination rates. Germination rates were assayed by planting triplicates of 230 wild-type and 230 nkd mutant kernels from F2 populations. In the field, mutants showed 72% germination compared with 96% in the wild type, while in the greenhouse, 80% of mutants germinated compared with 97.4% in the wild type (Fig. 2A). Seedling survival rate was lower in the mutant. Only 75% of geminated nkd plants remained viable 45 d after planting, while 93% of wild-type plants survived (Fig. 2B). Approximately one-quarter of the mutant plants were poorly developed and arrested at the two- to three-leaf stage. Mutant plants that survived to maturity were morphologically normal and produced the same number of nodes as the wild type, although they grew slowly and showed a delay of 5 d to anthesis (Fig. 2C).
The nkd Genes Function Independently of thk1 in Aleurone Cell Fate and Downstream of thk1 in Aleurone Cell Differentiation
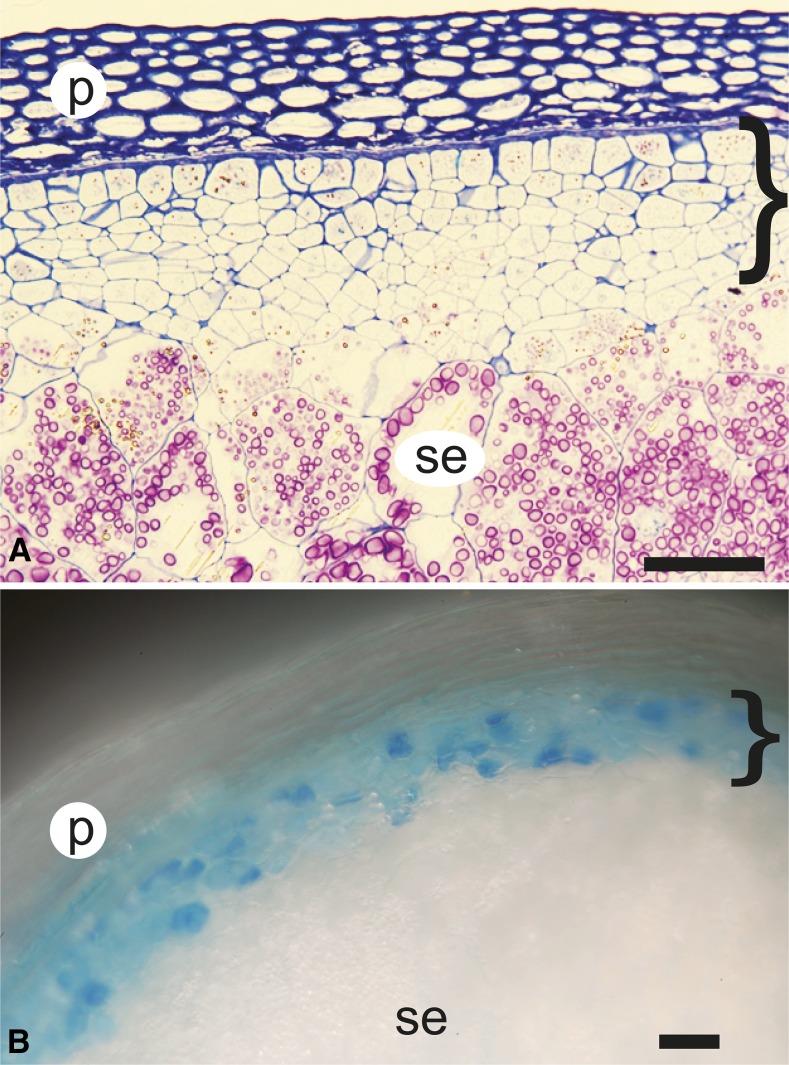
To assess the position of nkd in the genetic regulation of aleurone differentiation, triple mutants were generated with thk1. As described, the thk1 mutant is completely epistatic to dek1, producing a thick aleurone phenotype in double mutants, suggesting that thk1 functions downstream of dek1 to regulate aleurone differentiation (Yi et al., 2011). As shown in Figure 3, nkd1;nkd2;thk1 triple mutants had more peripheral layers than either nkd or thk1, indicating an additive phenotype and suggesting that these genes function independently in regulating the cell fate decisions controlling aleurone layer number. However, the peripheral cells of the triple mutant have compromised aleurone features, including sporadic vp1pro-GUS expression, similar to nkd mutants (Fig. 3B). In contrast, thk1 single mutants show uniformly high levels of vp1pro-GUS expression throughout the multiple aleurone layers (Yi et al., 2011). This suggests that nkd is epistatic to, and therefore downstream of, thk1 with regard to aleurone cell differentiation. Thus, nkd function appears required for at least two different aspects of aleurone differentiation.
Figure 3.
Double mutant analysis between nkd and thk1. A, Histologically stained nkd1;nkd2;thk1 triple mutant kernel. The bracket indicates a region of cells with intermediate characteristics of aleurone and starchy endosperm (se) cells. There are more layers of such cells than in either the nkd (Fig. 1) or thk1 mutant alone (Yi et al., 2011). B, The vp1pro-GUS aleurone marker shows sporadic expression, typical of nkd mutants, in multiple layers of nkd;thk1 mutants. p, Pericarp. Bars = 100 μm.
Map-Based Cloning of the nkd Genes
Crosses with B-A translocation lines revealed that one of the nkd genes, designated nkd1, is located on the short arm of chromosome 2. B-A chromosomes undergo nondisjunction at the second pollen mitosis, generating sperm cells, one hypoploid and one hyperploid for the translocated chromosome arm. Any genes located on the translocated chromosome segment will show distorted segregation ratios (Beckett, 1978). An nkd mutant was crossed to translocation line TB-2Sb, and hypoploid F1 plants were self-pollinated. The F2 showed a 3:1 segregation ratio compared with the expected 15:1 ratio that was observed for translocation lines involving other chromosome arms.
An F2 mapping population was generated by crossing an nkd mutant to the H99 inbred line, and genomic DNA from 62 nkd F2 seedlings was isolated. Insertion deletion polymorphism (IDP) markers (Fu et al., 2006) at 10-centimorgan intervals on the short arm of chromosome 2 were tested for linkage, and nkd1 localized to an approximately 11-centimorgan interval between IDP7746 and IDP1612 (Supplemental Table S1). We increased the number of F2 individuals to 170 and extracted genomic DNA directly from the kernels. To develop markers for fine-mapping, we used B73 genomic DNA sequence (Schnable et al., 2009) and designed primers at the 5′ and 3′ untranslated regions of annotated genes. These primer pairs were screened for polymorphisms evident as size differences on 1% to 2% (w/v) agarose gels between the nkd and H99 parents. Markers polymorphic between parents were applied to the F2 population. About 20% of designed primers were polymorphic. Finally, nkd1 was mapped between markers 266H09-9/10 and 166I20-1/5. These markers had five recombinants out of 328 chromosomes and two recombinants out of 332 chromosomes, respectively. The interval between these two markers was 385 kb in length in B73. According to the maize genome sequence version 53.4a (http://ensembl.gramene.org/Zea_mays/Info/Index), there were 31 annotated genes in this region, 10 of which had no supporting evidence (i.e. no maize EST). Eighteen had weak support such as a single EST, mostly from laser-capture microdissection (LCM)-dissected shoot apical meristem tissue (Emrich et al., 2007; Ohtsu et al., 2007). The remaining three genes with strong support putatively encoded a MutatorDR transposase, a C2H2 zinc finger protein, and a DEAD-like helicase. Only two of these genes have homologs in sorghum (Sorghum bicolor), and only the C2H2 zinc finger protein gene has fully covered EST evidence.
To map the nkd2 locus, we used the Sequenom MassARRAY system in a genome-wide assay of single-nucleotide polymorphism (SNP) markers that are polymorphic between B73 and Mo17 (Fu et al., 2006; Liu et al., 2010). The nkd mutant arose in a mongrel genetic background; therefore, we used two different mapping populations to maximize SNP polymorphisms: BC3F2 for B73 and BC2F2 for Mo17. Bulked segregant analysis was performed by mixing 12 wild-type and 12 nkd kernels each from four different ears (for totals of 48 wild-type and 48 mutant kernels). The results showed linked SNP makers clustered on chromosome 2 and chromosome 10. Linkage to the nkd phenotype was confirmed with an analysis of IDP markers on chromosome 10 (Supplemental Table S2). The nkd2 region on the long arm of chromosome 10 was narrowed to the interval between IDP8526 and IDP8334, which is approximately 1.7 Mb in the B73 genome sequence.
According to the hypothesis that nkd1 and nkd2 genes are duplicate factors, we compared the entire annotated gene sets in the nkd1 and nkd2 regions. Only the C2H2 zinc finger gene from the nkd1 region had a homolog in the nkd2 interval. Therefore, these two genes, ZmIDDveg9 (nkd1; GRMZM2G129261) and ZmIDD9 (nkd2; GRMZM5G884137), were considered as likely candidates.
Sequence Analysis of nkd1 and nkd2 Mutant Alleles
The nkd mutant arose fortuitously in a mongrel genetic background during an unrelated genetic screen. To understand the nature of the mutations, the nkd1 and nkd2 mutant genes were PCR amplified, cloned, and sequenced. For nkd1, a total of 10,876 bp (−3,584 bp 5′ from the start codon to +1,365 bp 3′ of the stop codon) were sequenced, and 65 sequence differences (40 SNPs, 15 deletions, and 10 insertions) were detected relative to the B73 sequence. One mutation caused an amino acid change that is likely to be significant; a C-to-T transition caused an amino acid substitution (CAC→TAC, His→Tyr) at residue 102 (Fig. 4A). This His is part of the C2H2 motif located on the first zinc finger of the IDD domain and is highly conserved among the IDD family (Colasanti et al., 2006). Other changes include a 13-bp deletion in intron 2 and a 26-bp deletion 3′ of the transcribed region. For nkd2, 9,219 bp (−3,233 bp 5′ from the start codon to +876 bp 3′ of the stop codon) were sequenced. Seventy-three sequence differences (44 SNPs, 14 deletions, and 15 insertions) were detected. Of note was a copia-like retrotransposon insertion in the first exon. The insertion site showed a well-conserved 5-bp (CACCG) target site duplication and long terminal repeats.
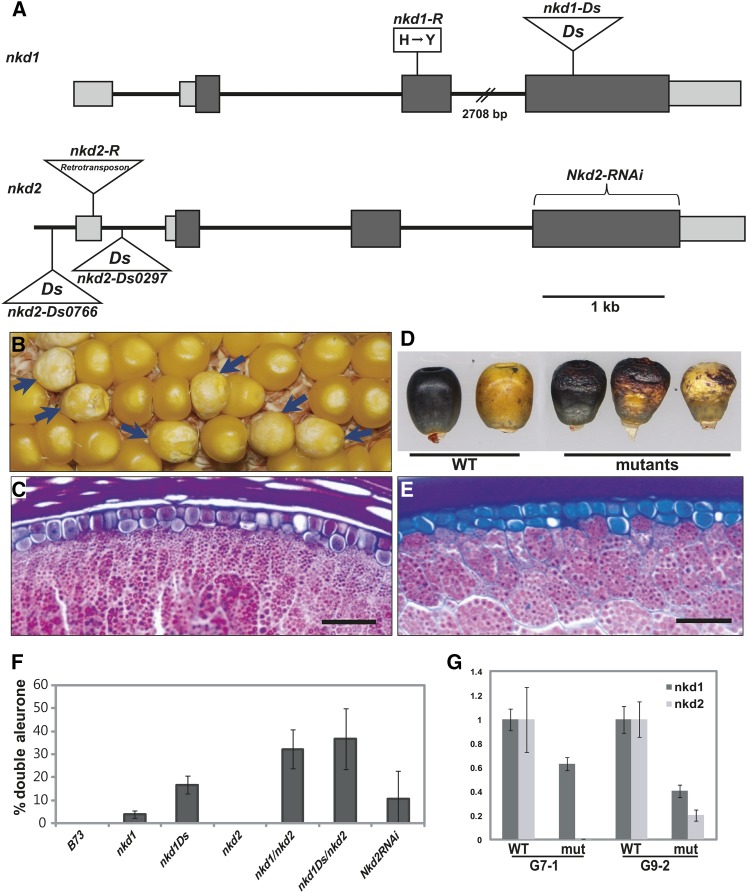
Figure 4.
Genetic confirmation of the nkd1 and nkd2 loci. A, nkd gene structures and alleles. The nkd1-R allele has an amino acid change (H→Y) at residue 102. The nkd1-Ds allele has a Ds insertion in the fourth exon. The nkd2-R allele contains a copia-like retrotransposon insertion in the first exon. The nkd2-Ds alleles have Ds insertions in the putative promoter region and the first intron. RNAi lines were generated using the fourth exon of nkd2. Boxes represent exons, and dark shading indicates a coding region. B, nkd1-Ds/+ self-pollinated ear showed 3:1 segregation for kernels with weak nkd phenotypes and rough-textured opaque endosperm, associated with nkd1-Ds homozygotes. Arrows indicate mutant kernels. C, Section of an nkd1-Ds homozygous kernel shows irregular double-layered aleurone. D, nkd2-RNAi line (G7-1) segregated kernels with wild-type (WT) and nkd phenotypes. Mutant kernels show wrinkled endosperm and sporadic disruption of anthocyanin pigmentation. E, nkd2-RNAi mutant kernel section shows irregular double-layered aleurone. F, Different nkd mutants show different levels of nkd phenotypes in terms of doubled aleurone. The degree of nkd phenotype strength was measured as the percentage of double-layered aleurone. Kernels were scored by counting 150 aleurone cells across the crown of the kernel and calculating the ratio of secondary aleurone cells as a percentage. nkd1-R sometimes showed multiple layers of aleurone. The nkd1-Ds allele has a stronger phenotype than nkd1-R. The phenotype became stronger when nkd2-R was added. G, nkd2-RNAi shows reduced transcript levels for both nkd1 and nkd2 in independent events, G7-1 and G9-2.
Genetic Confirmation of the nkd Genes
To test the hypothesis that the candidate genes corresponded to nkd1 and nkd2, public resources were searched, and a Dissociation (Ds) transposon insertion allele (B.S08.0002) for nkd1 was identified from the Activator (Ac)/Ds project (Ahern et al., 2009; Vollbrecht et al., 2010). This allele contained a Ds insertion in the fourth exon (Fig. 4A). This mutant is henceforth referred to as nkd1-Ds, while the original reference allele is designated nkd1-R. The nkd1-Ds heterozygotes (homozygous Nkd2+) were self-pollinated and crossed as pollen donors to plants with the genotype nkd1/+; nkd2/nkd2. Surprisingly one-fourth of the seeds from this cross showed the nkd mutant phenotype. Since nkd2 was uniformly heterozygous in the progeny, we hypothesized that the 3:1 segregation ratio follows the nkd1 genotype and that kernels with nkd1-Ds/nkd1-R would have a mutant phenotype. Twelve wild-type and 12 mutant kernels were genotyped, and all kernels showing an nkd mutant phenotype carried one original nkd1-R allele and one nkd1-Ds allele, while all the wild-type kernels had at least one Nkd1+ allele (Supplemental Fig. S1; Supplemental Methods S1).
The failure of the independent nkd1-Ds allele to complement the original nkd1-R mutant confirmed the identity of nkd1 as IDDveg9. Furthermore, these results also suggested that there may be a dosage requirement for nkd gene function and that the nkd1-R allele may be hypomorphic, while nkd1-Ds is likely to be a stronger loss-of-function allele. Consistent with this, self-pollinating nkd1-Ds/+ showed 3:1 segregation, with mutant kernels showing a weak opaque and rough textured phenotype (Fig. 4B).
Two independent Ds insertion alleles for nkd2 (B.W06.0297 and I.W06.0766) were available from the Ac/Ds project (Ahern et al., 2009; Vollbrecht et al., 2010) and named nkd2-Ds0297 and nkd2-Ds0766, respectively, while the original reference allele is designated nkd2-R. nkd2-Ds0297 has a Ds insertion in the first intron (−499 bp), and nkd2-Ds0766 has a Ds insertion at −1,215 bp in the putative promoter region (Fig. 4A). Neither of these nkd2 mutants showed discernible phenotypes in F2 families from self-pollination. Each of these alleles was crossed to an nkd1-R single mutant, the resulting F1 was self-pollinated, and the F2 showed kernels with mutant phenotypes in about 15:1 segregation ratios (757 wild type:42 nkd for nkd2-Ds0297; 1,062 wild type:84 nkd for nkd2-Ds0766). The lack of a visible kernel phenotype in nkd1-R single mutants and the 15:1 segregation ratio suggest that the nkd mutant phenotype results from nkd1-R;nkd2-Ds double mutant genotypes. Therefore, independent mutations in the IDD9 gene fail to complement the nkd1-R mutant, confirming the identity of nkd2 as IDD9. Finally, double mutants were produced between nkd1-Ds and nkd2-Ds0297, and they showed the same kernel phenotype as the original alleles (Supplemental Fig. S2).
Further confirmation of the identity of the nkd genes came from transgenic RNAi lines developed using nkd2 (IDD9) to down-regulate nkd gene expression. The ID domain encoded by exons 1 to 3 is highly conserved, so to minimize the likelihood of targeting other members of the IDD family, only the fourth exon was used for the RNAi construct. The nkd2 fourth exon shares 87% nucleotide identity with nkd1 but is unique among other IDD genes in maize. This fragment was cloned in forward and reverse orientation under the control of the maize ubiquitin promoter. We obtained T2 seeds from five independent events (G1, G7, G9, G11, and G14), which were crossed to three different inbreds (B73, W22, and H99). Lines from three events showed aleurone phenotypes similar to the original nkd mutant (Fig. 4, D and E). The G7 line showed the strongest phenotype, which segregated 354:281 (wild type:mutant) in the F1 from crosses to the W22 inbred. Each of these three independent lines was analyzed for inheritance of the transgene and cosegregation with the mutant phenotype. All individuals showing the nkd-like aleurone phenotype carried the transgene; however, some kernels carrying the transgene showed normal phenotypes, possibly due to transgene silencing.
The independent lines G7-1 and G9-2 were selected for nkd expression analysis using real-time RT-PCR. In both lines, reductions in both nkd2 and nkd1 transcript abundance were observed in transgenic seedlings with respect to the control, nontransgenic siblings (Fig. 4G). This was expected because of the nucleotide sequence conservation in the fourth exon between the two maize paralogs, leading to concomitant transcript down-regulation. The recapitulation of the nkd mutant seed phenotype by RNAi suppression further supports the identification of the nkd1 and nkd2 genes as IDDveg9 and IDD9, respectively. Because the original report of the nkd mutant (Becraft and Asuncion-Crabb, 2000) predated the first report of the IDD gene family (Kosaki et al., 2004), we use the nkd1 and nkd2 designations.
Characteristics of the IDD Proteins Encoded by the nkd Genes
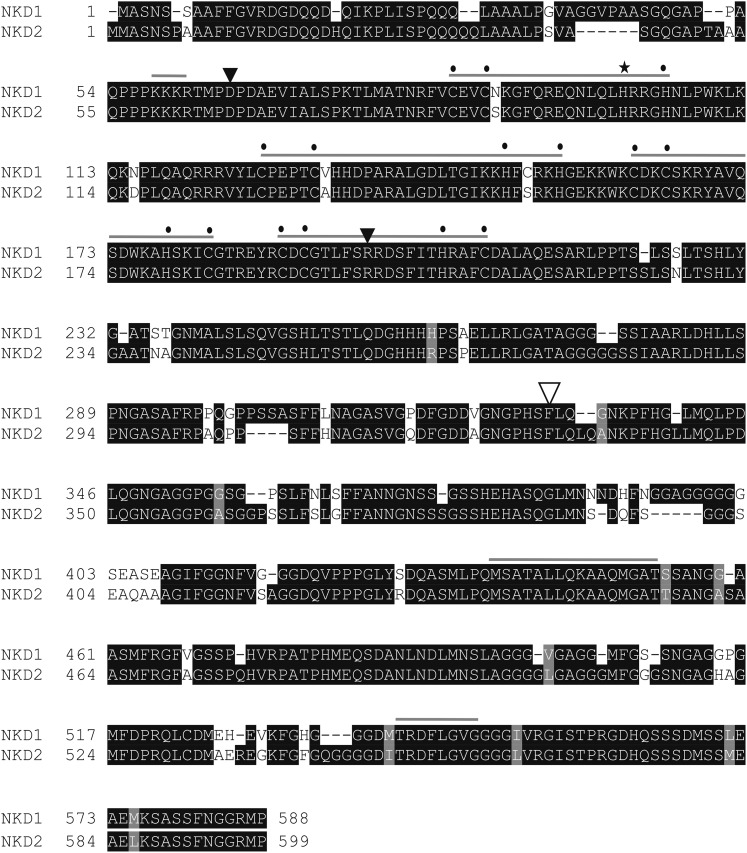
The predicted proteins encoded by the nkd genes contain ID domains, consisting of two C2H2 and two C2HC zinc fingers, that define the IDD family (Colasanti et al., 2006). The founding member of the IDD gene family, ID1, shows a late-flowering phenotype and is the only gene of this family with a known mutant phenotype in maize (Singleton, 1946; Colasanti et al., 1998, 2006). The two nkd genes showed high homology to each other (82% identity/84% positive) in overall amino acid sequence (Fig. 5). The predicted NKD1 protein contains 588 amino acids with a calculated mass of 61 kD. NKD2 contains 599 amino acids with a calculated mass of 62 kD. The ID domains have been shown for several IDD members, including ID1 and NKD1 (IDDveg9), to bind specific DNA sequence motifs, suggesting their likely function as transcription factors (Kozaki et al., 2004). Consistent with this, NKD1 and NKD2 contain predicted N-terminal nuclear localization signals. In addition, the C-terminal domains contain two motifs of unknown function that are each conserved in a subset of IDD family members (Colasanti et al., 2006).
Figure 5.
NKD protein sequences showed 82% identity and 84% positivity. Motifs conserved among IDD family members are designated by light gray bars. Conserved C and H residues of the IDD C2H2 and C2HC zinc finger domains are marked by dots. The starred H was mutated to Y in the nkd1-R allele. Black triangles indicate intron locations, and the white triangle shows the Ds insertion point for the nkd1-Ds allele.
nkd Gene Expression
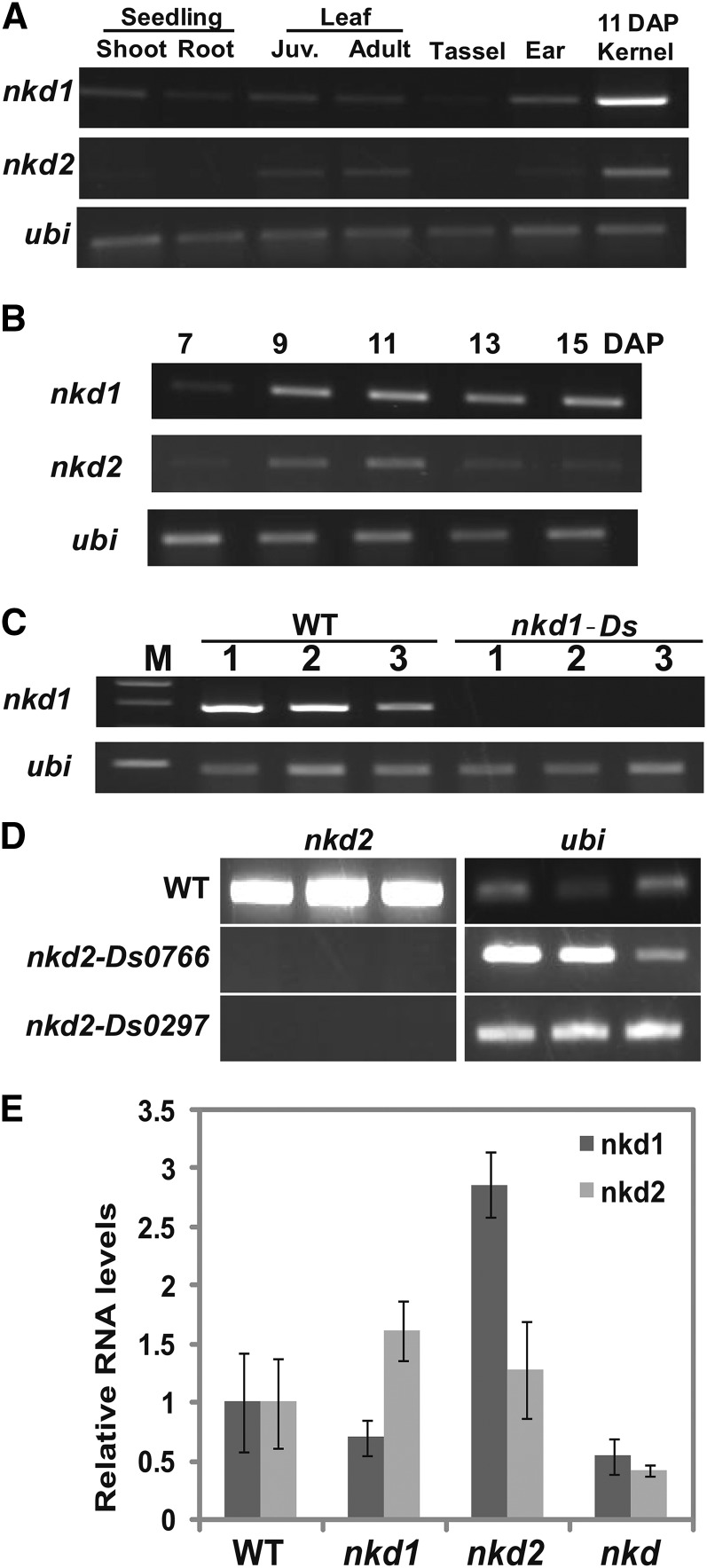
Transcript levels of nkd1 and nkd2 were examined by RT-PCR. A basal level of nkd gene expression was detected in all tissues tested. However, the expression level was very low in root and tassel. The nkd genes were most abundantly expressed in kernels and showed higher transcript levels in endosperm compared with embryo (Fig. 6A). The nkd1 transcript levels gradually increased from 7 d after pollination (DAP) and peaked about 15 DAP, whereas nkd2 peaked at 11 DAP (Fig. 6B).
Figure 6.
Expression analysis of nkd1 and nkd2 transcripts. A to D, RT-PCR analysis. A, The nkd1 transcript was detected in all tissues tested. However, abundance was low in root and tassel. It was most highly expressed in kernels. The nkd2 transcript was detected most highly in kernels and also in leaf and ear tissues. B, In developing kernels, both nkd transcripts were highly expressed at 9 to 11 DAP, after which nkd2 declined. C, The nkd1 transcript was not detected in 11-DAP nkd1-Ds homozygous kernels. D, The nkd2 transcript was not detected in leaves of homozygotes for either nkd2-Ds mutant allele. E, nkd1 and nkd2 transcript levels were quantified by real-time RT-PCR. In nkd-R mutants, both nkd1 and nkd2 transcripts were decreased. Higher expression of nkd1 transcript in the nkd2-R single mutant suggests a potential feedback regulation of the nkd genes. ubi, Ubiquitin; WT, wild type.
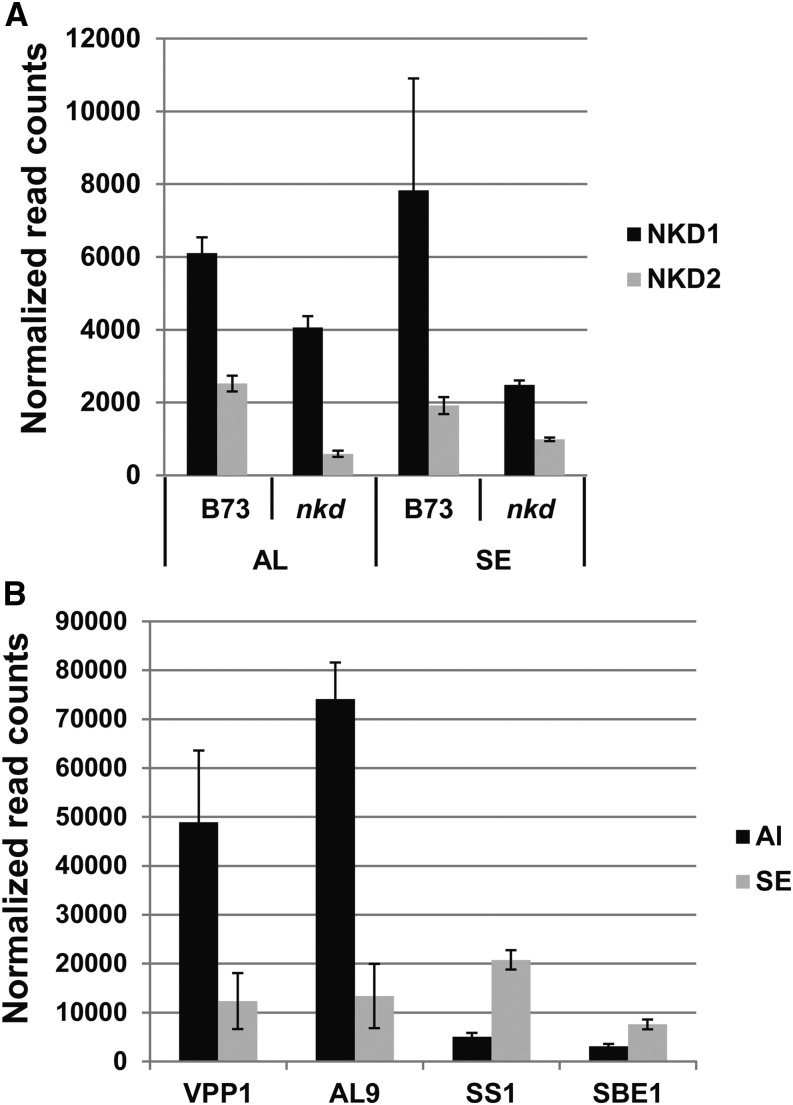
To examine endosperm tissue-specific expression of the nkd genes, aleurone and starchy endosperm cells were isolated by LCM from 15-DAP B73 kernels (Supplemental Fig. S3). As shown in Figure 7, both genes were expressed in aleurone and starchy endosperm. The nkd2 gene was expressed at similar levels in both tissues, while nkd1 was expressed at approximately 3-fold higher levels than nkd2 in aleurone and 4-fold higher levels in starchy endosperm. In the nkd-R mutant (nkd1-R;nkd2-R double homozygote), both genes showed reduced transcript levels in both aleurone and starchy endosperm tissues. To confirm the enrichment for aleurone and starchy endosperm cells in the LCM samples, aleurone- and starchy endosperm-specific markers were tested. Figure 7B shows that the aleurone-specific vacuolar H+-translocating inorganic pyrophosphatase1 (vpp1) and al-9 transcripts are greatly enriched in the aleurone samples, whereas starch synthase1 (ss1) and starch branching enzyme1 (sbe1) transcripts are enriched in the starchy endosperm samples.
Figure 7.
LCM RNA sequencing analysis of nkd transcript levels in endosperm tissues. A, Expression of nkd1 and nkd2 genes in aleurone (AL) and starchy endosperm (SE) cells of wild-type (B73) and nkd1-R;nkd2-R homozygous mutant (nkd) endosperms at 15 DAP. B, LCM tissue purity control showing known tissue-preferential transcripts in B73 endosperm samples. Aleurone markers include vpp1 (Wisniewski and Rogowsky, 2004) and al9, encoding a putative bifunctional inhibitor/lipid transfer protein (Gómez et al., 2009). Starchy endosperm markers include ss1 and sbe1 (Hennen-Bierwagen and Myers, 2013). The data depict normalized mean transcript read counts of genes as determined by RNA sequencing. Error bars represent se from three independent biological replicates.
Transcript levels in nkd mutants were examined by RT-PCR. Figure 6, C and D, shows that transcripts were undetectable in the nkd1-Ds mutant and both nkd2-Ds mutants, suggesting that these are null alleles. Expression was also examined by real-time RT-PCR. As shown in Figure 6E, nkd1 transcript levels were decreased in the nkd1-R single mutant and nkd1-R;nkd2-R double mutant, but increased transcript levels were observed in the nkd2-R single mutant. The nkd2 transcript levels were decreased in the nkd1-R;nkd2-R double mutant, but transcript levels in the nkd2-R single mutant were similar to those in the wild type and increased in the nkd1-R single mutant. That nkd1 and nkd2 transcripts were each more highly expressed in the other’s single mutant background, relative to the wild type, suggests that compensatory mechanisms might regulate transcript levels.
NKD Proteins Localize to Nuclei
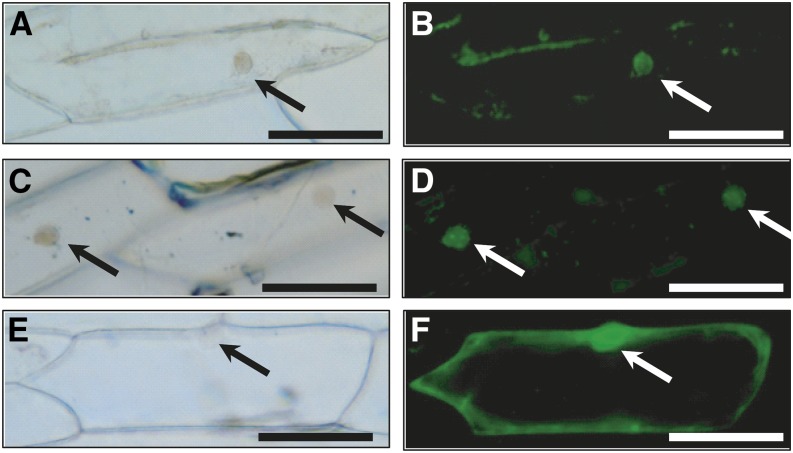
Both predicted NKD proteins contain putative nuclear localization signals (KKKR), as do other IDD proteins. To experimentally verify the nuclear localization of the NKD proteins, NKD-GFP translational fusion constructs were generated and transiently expressed in onion (Allium cepa) bulb epidermal cells. As expected, the fusion proteins showed localization in nuclei, as ascertained by the accumulation of green fluorescence (Fig. 8).
Figure 8.
Localization of NKD-GFP fusion protein nuclei of onion cells. Plasmid constructs were biolistically introduced for transient expression. A and B, NKD1-GFP. C and D, NKD2-GFP. E and F, Empty vector GFP control. A, C, and E, Differential interference contrast optics. B, D, and F, GFP fluorescence. Arrows indicate nuclei. Bars = 100 µm.
DISCUSSION
In this study, we identified the nkd1 and nkd2 genes by map-based cloning as duplicated members of the IDD family of zinc finger transcription factors. Because of the mongrel genetic background in which the mutation arose and the high levels of sequence polymorphisms compared with the B73 sequence, it was not possible to unequivocally identify the causal lesions. However, an H→Y substitution in the highly conserved C2H2 motif of the first zinc finger of nkd1 and a retrotransposon insertion in the first exon of nkd2 are likely candidates. An independent nkd1-Ds allele failed to complement the original nkd mutant phenotype, and nkd2-Ds0297 and nkd2-Ds0766 alleles similarly failed to complement the nkd mutant. Furthermore, the mutant phenotype was reproduced by generating nkd2-RNAi transgenic lines, confirming the correct identity of these genes.
The nkd mutant showed two to five layers of undifferentiated aleurone cells as well as floury, opaque endosperm. As reported previously (Becraft and Asuncion-Crabb, 2000), the original mutant segregated 15:1 in the F2 due to the duplicated genes, with only the double homozygous mutant showing a phenotype. A single mutant homozygote for the nkd1-Ds allele showed rough-textured endosperm and undifferentiated aleurone-like cells, and careful examination of the original nkd1-R single mutant showed a subtle effect on aleurone layer number, with sporadic development of double layers. These phenotypes of nkd1 mutants were enhanced as doses of the nkd2 mutant allele were added. The nkd1-Ds allele, which has a Ds transposon insertion within coding sequences of the fourth exon, appears to be a null mutant and showed a stronger phenotype than the nkd1-R reference allele. However, none of the nkd2 single mutants, including the null nkd2-Ds alleles, showed any phenotypic defect. These phenotypic observations combined with the higher expression level of nkd1 transcript (Fig. 7A) suggest that Nkd1+ contributes the majority of the genetic function required for normal endosperm development, while the contribution by Nkd2+ is of secondary importance.
The nkd1 and nkd2 genes are located in a duplicated region of the genome. Interestingly, the syntenic regions of chromosomes 2 and 10 that contain nkd1 and nkd2 also contain the duplicate maize floricaula/leafy homologs (Bomblies et al., 2003; Colasanti et al., 2006). Other cereals, including sorghum and rice (Oryza sativa), have only one copy of the nkd (IDD9) gene, whereas this clade of IDD proteins is absent from Arabidopsis (Arabidopsis thaliana; Colasanti et al., 2006) and other dicot species.
NKD1 and NKD2 have conserved ID domains with two C2H2 and two C2HC zinc finger motifs, consistent with their DNA-binding activity and roles as transcription factors (Kozaki et al., 2004; Colasanti et al., 2006). The nkd1 gene corresponds to ZmIDDveg9, although it is misannotated in the B73 genome assembly version 2. ZmIDDveg9 was cloned by screening a complementary DNA (cDNA) library generated from the vegetative apex using a probe consisting of the region coding the ID domain in id1 (Colasanti et al., 2006). They showed by electrophoretic mobility shift assay that ZmIDDveg9 binds the same DNA sequence elements as ID1, albeit with somewhat different properties. The ZmIDDveg9 transcript was detected in the apical region and immature leaf by RNA blot, but they did not examine kernel samples. In the same study, NKD2 was described as ZmIDD9. Because the nkd mutant (Becraft and Asuncion-Crabb, 2000) was reported prior to the description of the IDD family (Kosaki et al., 2004; Colasanti et al., 2006), we choose to refer to these factors as NKD1 and NKD2.
The expression profiles portrayed at MaizeGDB using public microarray data available at PLEXdb (Winter et al., 2007; Sekhon et al., 2011; Dash et al., 2012; Monaco et al., 2013) were consistent with the results of this study. Transcripts for nkd1 were detected in most tissues examined but most abundantly in kernels. In kernels, nkd1 transcripts accumulated to higher levels in endosperm than in embryo. During kernel development, the expression level gradually increased from 6 to 16 DAP and then plateaued until 24 DAP. Transcript levels for nkd2 showed a similar pattern, although in the endosperm and kernel, the levels tended to be approximately 1 order of magnitude (log2) lower than for nkd1.
The IDD gene family has high conservation in the putative nuclear localization signal (KKKR) as well as the zinc finger motifs of the ID domain. ID1 was nucleus localized in a transfection assay of onion epidermal cells (Wong and Colasanti, 2007). Here, nuclear localization was demonstrated for NKD proteins by detecting NKD-GFP fusion proteins in nuclei of onion epidermal cells in a transfection assay (Fig. 8). Thus, we conclude that NKDs are nucleus-localized proteins and very likely function as transcription factors.
In the nkd1 mutant, nkd2 transcripts were elevated compared with the wild type, whereas in the nkd2 mutant, nkd1 transcripts were elevated (Fig. 6D). This suggests that a feedback mechanism is involved in the compensatory regulation of nkd genes. By aligning promoter regions of nkd1 and nkd2 genomic sequences, it was found that approximately 1.3 kb of nkd1 and 1 kb of nkd2 are conserved with 75% identity, suggesting conserved regulatory functions. We can also find two motifs that resemble the consensus binding sequence defined for ID1 and shown to bind NKD1/IDDveg9 (Colasanti et al., 2006). For nkd1, TTTTGTTGTTTT and TTTTGTTAATCT occur at position −563 with a 58-bp interval, while for nkd2, TTTTGTTGTCTT and TTTTGTTTAATC occur at position −528 with a 60-bp interval. Thus, it is an intriguing possibility that these genes might autoregulate or be controlled by other IDD family members.
The id1 gene is the only other IDD family member with a known mutant in maize. The loss-of-function mutant of id1 causes late flowering or the complete inability to flower in maize (Colasanti et al., 1998). Mutants of IDD8 in Arabidopsis also show delayed flowering time (Seo et al., 2011b). This could be related to altered sugar metabolism, as sugar levels and two Suc synthase gene transcript levels were altered in idd8 mutants; IDD8 directly binds the SUS4 promoter and is itself transcriptionally regulated by sugar levels. The nkd mutant also showed delayed flowering time without any change in total leaf number. It is not yet clear whether late flowering is simply due to slow growth or other flowering time gene regulation.
The maize B73 genome (Schnable et al., 2009) contains 17 IDD gene family members, instead of the 13 originally identified by cDNA cloning (Colasanti et al., 2006). In Arabidopsis, there are 16 IDD genes, and phylogenetic analysis shows that there are monocot- and dicot-specific clades (Colasanti et al., 2006), with the nkd genes belonging to a monocot-specific group (Supplemental Fig. S4). Although IDD proteins share high similarity in their N-terminal ID domains, other parts of the proteins vary substantially. In Arabidopsis, this family performs diverse functions, including carbohydrate metabolism (Seo et al., 2011a), gravitropism (Morita et al., 2006; Tanimoto et al., 2008; Wu et al., 2013), seed germination (Feurtado et al., 2011), and lateral organ morphogenesis (Cui et al., 2013; Reinhart et al., 2013).
Several Arabidopsis IDD genes have been implicated in cellular patterning. JACKDAW and MAGPIE act downstream of SHORTROOT and SCARECROW in establishing radial pattern and cell identities in the root apex (Welch et al., 2007; Hassan et al., 2010; Ogasawara et al., 2011). IDD4 and several other IDD genes are direct targets of regulation by REVOLUTA and KANADI, implicated in establishing the adaxial/abaxial pattern and cell fates in leaves and other lateral organs (Reinhart et al., 2013). It is intriguing to speculate that maize nkd genes might perform an analogous function in controlling cell patterning and differentiation in the endosperm.
The regulation of hormonal processes might provide additional clues to how the nkd genes could potentially regulate endosperm development. Arabidopsis IDD14-1A, IDD15, and IDD16 perform overlapping functions in directing auxin biosynthesis and transport (Cui et al., 2013). In maize endosperm, the auxin transporter ZmPIN-FORMED1 (ZmPIN1) is preferentially expressed in the aleurone, and treatment of developing kernels with the auxin transport inhibitor N-1-naphthylphthalamic acid induced alterations in aleurone cell layer number (Forestan et al., 2010). Furthermore, transgenic expression of isopentenyl transferase, a cytokinin-synthesizing enzyme, showed mosaic aleurone (Geisler-Lee and Gallie, 2005), suggesting that the auxin-cytokinin balance could be important for aleurone differentiation. Recently, Arabidopsis IDD proteins also were found to interact with DELLA proteins and to mediate GA3 activation of gene expression, including SCARECROW-LIKE3 (SCL3; Feurtado et al., 2011; Yoshida et al., 2014; Yoshida and Ueguchi-Tanaka, 2014). Interestingly, the IDD proteins also interacted with SCL3 to mediate its antagonistic function to DELLAs. ABA and GA3 function antagonistically to control seed maturation or germination, and the nkd genes appear to impact this balance. The mutants are sometimes viviparous and show decreased expression of the vp1 gene, required for ABA responses, including aleurone maturation (McCarty et al., 1989, 1991; Cao et al., 2007).
Functional orthologies among the IDD genes in dicots and monocots remain to be elucidated, but since the nkd genes belong to a monocot-specific clade, this will not likely be informative of their functions. Studying the molecular mechanism of nkd gene function, including the transcriptional regulation downstream gene networks, will provide new understanding of the control of endosperm cell differentiation and may expand opportunities to improve cereal grain quality.
MATERIALS AND METHODS
Plant Materials
The nkd mutant was crossed to H99 and Mo17 inbred maize (Zea mays) lines and then self-pollinated to produce F2 populations that were used for mapping and germination rate observation. The nkd mutant backcrossed to B73 and Mo17 inbreds three times was used for Sequenom assays. RT-PCR was performed using B73 BC5 introgressed nkd mutant and wild-type B73. The nkd1-Ds allele was identified in stock B.S08.0002, and nkd2-Ds0297 and nkd2-Ds0766 were identified in stocks B.W06.0297 and I.W06.0766 from the Ac/Ds project (Ahern et al., 2009; Vollbrecht et al., 2010).
Genetic Mapping
Homozygous nkd mutants were crossed as females to available B-A translocation stocks involving different chromosome arms (Beckett, 1978). F1 plants were grown, and hypoploids were identified by the stunted plant stature and 50% pollen abortion characteristic of aneuploids. Such plants were self-pollinated, and the resultant ears were examined for segregation ratios of nkd mutant kernels. Successful pollinations were obtained from putative hypoploid plants involving 12 of the 20 chromosome arms (not including 10L). Genomic DNA for mapping was extracted from seedlings or seeds as described (Yi et al., 2011). For Sequenom mapping, DNA samples were submitted to the Iowa State University Genomic Technologies Facility. IDP markers were used for fine-mapping as described (Fu et al., 2006). Additional PCR markers were developed by PCR amplifying the 3′ and 5′ untranslated regions of predicted gene models and identifying size polymorphisms between parents by running amplification products on 2% (w/v) agarose gels. The primers for polymorphic markers are listed in Supplemental Table S3.
Sequencing
For each nkd1 and nkd2 locus, six fragments of around 2 kb were amplified from nkd mutant genomic template DNA. Primers are listed Supplemental Table S3. The PCR amplicons were cloned with the StrataClone PCR Cloning Kit (Agilent) and sequenced at the Iowa State University DNA Facility. At least three different clones were sequenced per fragment and compared with the B73 reference genome sequence. Only when all three clones showed the same difference from the reference sequence was it considered valid.
Subcellular Localization of GFP Fusion Protein
Full-length coding sequences of nkd1 and nkd2 were amplified from cDNA clones (Zm-BFc0139A11 and Zm-BFb0091K24 from the Arizona Genomic Institute) by PCR with primers 2ZNF-Xba1/2ZNF-BamH1 for nkd1 and 10ZNF-Xba1/10ZNF-BamH1 for nkd2 (Supplemental Table S3). XbaI- and BamHI-digested PCR products were then cloned into pJ4GFP-XB (Igarashi et al., 2001) and confirmed by sequencing. Gold particles coated with 1 μg of each plasmid DNA were bombarded into onion (Allium cepa) epidermal cells and incubated 3 h at room temperature. The slides were observed with an Olympus BX-60 microscope equipped with a Chromatek GFP filter and a Jenoptik C-5 camera.
RT-PCR
RNA extraction and cDNA synthesis were performed as described (Myers et al., 2011). The ubiquitin primers (Lee et al., 2009) were used for both RT-PCR and real-time RT-PCR as a control. PCR parameters included 94°C for 4 min, followed by 30 to 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s, and then a final extension at 72°C for 10 min. PCR products were then visualized by 1% (w/v) agarose gel electrophoresis. The existing vp1 primers (Cao et al., 2007) used for RT-PCR and newly designed VP1-RT3 and VP1-RT4 were used for real-time RT-PCR. Quantitative RT-PCR was performed with an Mx4000 (Stratagene) real-time PCR machine and iQ SYBR Green Supermix (Bio-Rad). Primers are listed in Supplemental Table S3.
LCM RNA Sequencing of Developing Maize Kernels
Wild-type (B73) and nkd mutant (fourth-generation backcross to B73) plants were grown in the field in the summer of 2012. Developing kernels were harvested at 15 DAP, fixed in 50% (v/v) ethanol and 5% (v/v) acetic acid, and embedded in paraffin. Endosperm cell types were isolated from sections using an LCM microscope (PixCell II LCM System; Arcturus Engineering). Total RNA was isolated from captured cells using the Arcturus Picopure RNA Isolation Kit (Life Technologies). The total RNA was subjected to a T7 RNA Polymerase-based linear RNA amplification strategy, using the TargetAmp 2-Round aRNA Amplification Kit 2.0 (Epicentre Biotechnologies) to generate a sufficient quantity of RNA as a template for RNA sequencing. The quality of amplified RNA was also checked by bioanalyzer analysis on an Agilent 2100 Bioanalyzer.
Transcript libraries were constructed using the TruSeq RNA sample preparation kit (Illumina). The libraries were indexed using specific adapter sequences and subjected to cluster analysis and 50-bp paired-end sequencing using an Illumina HiSeq 2000 machine at the Iowa State University DNA Facility. The raw sequence data were first quality checked using FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were then aligned to the B73 reference maize genome assembly version 2 using the TopHat short sequence aligner (http://ccb.jhu.edu/software/tophat/index.shtml). Read counts were normalized based on library sizes and analyzed for differentially expressed genes using DESeq software (Anders and Huber, 2010). The maize gene identifiers for the transcripts analyzed (Fig. 7) are as follows: NKD1, GRMZM2G129261; NKD2, GRMZM5G884137; VPP1, GRMZM2G069095; AL9, GRMZM2G091054; SS1, GRMZM2G129451; and SBE1, GRMZM2G088753. The data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (Edgar et al., 2002) and are accessible through Gene Expression Omnibus series accession number GSE61057 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE61057).
RNAi Lines
The nkd2 fourth exon was PCR amplified from B73 template DNA using two sets of primers tailed with different pairs of restriction enzyme sites. Primers 10ZNF3EF-Avr2/10ZNF3ER-Asc1 and 10ZNF3EF-Xma1/10ZNF3ER-Spe1 are listed in Supplemental Table S3. These fragments were cloned into pMCG1005 (McGinnis et al., 2005) in the sense and antisense directions under the control of the maize ubiquitin promoter by using appropriate restriction enzyme digestion, ligation, and sequence confirmation. The vector was transformed into EHA101 by the freeze-thaw method, and construct-containing clones were confirmed by plasmid isolation and PCR. Agrobacterium tumefaciens-mediated transformation was performed at the Iowa State University Plant Transformation Facility. The RNAi plantlets were recovered and crossed to inbred lines for analysis.
Transgenic line genotyping was done by PCR using the forward primer specific to the waxy intron (Waxy GK-1) and the reverse primer complementary to the Nkd2 fourth exon (10ZNF-13). Amplification was performed with GoTaq Green Master Mix (Promega) at an annealing temperature of 52°C, with other parameters as described above. As an internal control, primers specific to the endogenous Nkd1 gene (291F22-5 and 2ZNF-5) were employed with an annealing temperature of 55°C. The amplicon sizes were around 500 and 900 bp for the transgene and the endogenous gene, respectively. For expression analysis, RNA was isolated from 1-week-old seedlings using the RNeasy Plant Mini Kit (Qiagen). All primer sequences are listed in Supplemental Table S3.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XM_008670411 (nkd1) and NM_001165837 (nkd2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. nkd1-Ds complementation test.
Supplemental Figure S2. Endosperm phenotype of nkd1-Ds; nkd2-Ds0297 double mutant.
Supplemental Figure S3. LCM of aleurone and starchy endosperm cells.
Supplemental Figure S4. Maize IDD phylogeny.
Supplemental Table S1. Marker data for nkd1 mapping.
Supplemental Table S2. Marker data for nkd2 mapping.
Supplemental Table S3. PCR primers.
Supplemental Methods S1. Genotyping for nkd1-Ds complementation test.
Supplementary Material
Acknowledgments
We thank members of the Becraft laboratory for helpful discussions of the article and Julie Meyer and Jordon Pace for contributions to the genetic mapping. The following Iowa State University facilities provided technical assistance: the Microscopy and NanoImaging Facility assisted with some of the microscopic preparations, the Genomic Technologies Facility provided the mass array SNP genotyping, the DNA Facility provided sequencing and oligonucleotide synthesis, and the Plant Transformation Facility generated transgenic RNAi lines.
Glossary
- ABA
abscisic acid
- RNAi
RNA interference
- RT
reverse transcription
- LCM
laser-capture microdissection
- SNP
single-nucleotide polymorphism
- DAP
days after pollination
- cDNA
complementary DNA
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–1121738 to P.W.B.).
Articles can be viewed without a subscription.
References
- Ahern KR, Deewatthanawong P, Schares J, Muszynski M, Weeks R, Vollbrecht E, Duvick J, Brendel VP, Brutnell TP (2009) Regional mutagenesis using Dissociation in maize. Methods 49: 248–254 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett JB. (1978) B-A translocations in maize. I. Use in locating genes by chromosome arms. J Hered 69: 27–36 [Google Scholar]
- Becraft PW, Asuncion-Crabb Y (2000) Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127: 4039–4048 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Li K, Dey N, Asuncion-Crabb Y (2002) The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129: 5217–5225 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR (1996) CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273: 1406–1409 [DOI] [PubMed] [Google Scholar]
- Becraft PW, Yi G (2011) Regulation of aleurone development in cereal grains. J Exp Bot 62: 1669–1675 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Wang RL, Ambrose BA, Schmidt RJ, Meeley RB, Doebley J (2003) Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130: 2385–2395 [DOI] [PubMed] [Google Scholar]
- Cao X, Costa LM, Biderre-Petit C, Kbhaya B, Dey N, Perez P, McCarty DR, Gutierrez-Marcos JF, Becraft PW (2007) Abscisic acid and stress signals induce Viviparous1 expression in seed and vegetative tissues of maize. Plant Physiol 143: 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Tremblay R, Wong AY, Coneva V, Kozaki A, Mable BK (2006) The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V (1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Cone KC. (2007) Anthocyanin synthesis in maize aleurone tissue. InOlsen OA, ed, Endosperm: Development and Molecular Biology. Springer, Berlin, pp 121–140 [Google Scholar]
- Cui D, Zhao J, Jing Y, Fan M, Liu J, Wang Z, Xin W, Hu Y (2013) The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet 9: e1003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA (2012) PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res 40: D1194–D1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demko V, Perroud PF, Johansen W, Delwiche CF, Cooper ED, Remme P, Ako AE, Kugler KG, Mayer KF, Quatrano R, et al. (2014) Genetic analysis of DEFECTIVE KERNEL1 loop function in three-dimensional body patterning in Physcomitrella patens. Plant Physiol 166: 903–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich SJ, Barbazuk WB, Li L, Schnable PS (2007) Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res 17: 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtado JA, Huang D, Wicki-Stordeur L, Hemstock LE, Potentier MS, Tsang EW, Cutler AJ (2011) The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23: 1772–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C, Meda S, Varotto S (2010) ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol 152: 1373–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Wen TJ, Ronin YI, Chen HD, Guo L, Mester DI, Yang Y, Lee M, Korol AB, Ashlock DA, et al. (2006) Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize. Genetics 174: 1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J, Gallie DR (2005) Aleurone cell identity is suppressed following connation in maize kernels. Plant Physiol 139: 204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez E, Royo J, Muñiz LM, Sellam O, Paul W, Gerentes D, Barrero C, López M, Perez P, Hueros G (2009) The maize transcription factor myb-related protein-1 is a key regulator of the differentiation of transfer cells. Plant Cell 21: 2022–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H, Scheres B, Blilou I (2010) JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development 137: 1523–1529 [DOI] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Myers AM (2013) Genomic specification of starch biosynthesis in maize endosperm. InBecraft PW, ed, Seed Genomics. Wiley Blackwell, Ames, IA, pp 123–137 [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13: 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jestin L, Ravel C, Auroy S, Laubin B, Perretant MR, Pont C, Charmet G (2008) Inheritance of the number and thickness of cell layers in barley aleurone tissue (Hordeum vulgare L.): an approach using F2-F3 progeny. Theor Appl Genet 116: 991–1002 [DOI] [PubMed] [Google Scholar]
- Johnson KL, Faulkner C, Jeffree CE, Ingram GC (2008) The phytocalpain defective kernel 1 is a novel Arabidopsis growth regulator whose activity is regulated by proteolytic processing. Plant Cell 20: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A, Hake S, Colasanti J (2004) The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res 32: 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Johnston R, Yang Y, Gallavotti A, Kojima M, Travençolo BAN, Costa LdaF, Sakakibara H, Jackson D (2009) Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol 150: 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Demko V, Wilson RC, Johnson KA, Ahmad R, Perroud P-F, Quatrano R, Zhao S, Shalchian-Tabrizi K, Otegui MS, et al. (2013) The catalytic domain CysPc of the DEK1 calpain is functionally conserved in land plants. Plant J 75: 742–754 [DOI] [PubMed] [Google Scholar]
- Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen OA (2002) The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc Natl Acad Sci USA 99: 5460–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen HD, Makarevitch I, Shirmer R, Emrich SJ, Dietrich CR, Barbazuk WB, Springer NM, Schnable PS (2010) High-throughput genetic mapping of mutants via quantitative single nucleotide polymorphism typing. Genetics 184: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Carson CB, Stinard PS, Robertson DS (1989) Molecular analysis of viviparous-1: an abscisic acid-insensitive mutant of maize. Plant Cell 1: 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- McGinnis K, Chandler V, Cone K, Kaeppler H, Kaeppler S, Kerschen A, Pikaard C, Richards E, Sidorenko L, Smith T, et al. (2005) Transgene-induced RNA interference as a tool for plant functional genomics. InEngelke DR, Rossi JJ, eds, RNA Interference. Elsevier Academic Press, San Diego, pp 1–24 [DOI] [PubMed] [Google Scholar]
- Monaco MK, Sen TZ, Dharmawardhana PD, Ren L, Schaeffer M, Naithani S, Amarasinghe V, Thomason J, Harper L, Gardiner J, et al. (January 23, 2013) Maize metabolic network construction and transcriptome analysis. Plant Genome 6: http://dx.doi.org/10.3835/plantgenome2012.09.0025 [Google Scholar]
- Morita MT, Sakaguchi K, Kiyose S, Taira K, Kato T, Nakamura M, Tasaka M (2006) A C2H2-type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. Plant J 47: 619–628 [DOI] [PubMed] [Google Scholar]
- Myers AM, James MG, Lin Q, Yi G, Stinard PS, Hennen-Bierwagen TA, Becraft PW (2011) Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell 23: 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara H, Kaimi R, Colasanti J, Kozaki A (2011) Activity of transcription factor JACKDAW is essential for SHR/SCR-dependent activation of SCARECROW and MAGPIE and is modulated by reciprocal interactions with MAGPIE, SCARECROW and SHORT ROOT. Plant Mol Biol 77: 489–499 [DOI] [PubMed] [Google Scholar]
- Ohtsu K, Smith MB, Emrich SJ, Borsuk LA, Zhou R, Chen T, Zhang X, Timmermans MCP, Beck J, Buckner B, et al. (2007) Global gene expression analysis of the shoot apical meristem of maize (Zea mays L.). Plant J 52: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA. (2004a) Dynamics of maize aleurone cell formation: the “surface” rule. Maydica 49: 37–40 [Google Scholar]
- Olsen OA. (2004b) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell (Suppl) 16: S214–S227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Liu T, Newell NR, Magnani E, Huang T, Kerstetter R, Michaels S, Barton MK (2013) Establishing a framework for the ad/abaxial regulatory network of Arabidopsis: ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell 25: 3228–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Ryu JY, Jeong EY, Park CM (2011a) Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat Commun 2: 303. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Ryu J, Kang SK, Park CM (2011b) Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J 65: 418–429 [DOI] [PubMed] [Google Scholar]
- Shen B, Li C, Min Z, Meeley RB, Tarczynski MC, Olsen OA (2003) sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proc Natl Acad Sci USA 100: 6552–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan WF, Neuffer MG (1982) Maize developmental mutants: embryos unable to form leaf primordia. J Hered 73: 318–329 [Google Scholar]
- Singleton WR. (1946) Inheritance of indeterminate growth in maize. J Hered 37: 61–64 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9: 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Latshaw S, Sato Y, Settles AM, Koch KE, Hannah LC, Kojima M, Sakakibara H, McCarty DR (2008) The maize Viviparous8 locus, encoding a putative ALTERED MERISTEM PROGRAM1-like peptidase, regulates abscisic acid accumulation and coordinates embryo and endosperm development. Plant Physiol 146: 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto M, Tremblay R, Colasanti J (2008) Altered gravitropic response, amyloplast sedimentation and circumnutation in the Arabidopsis shoot gravitropism 5 mutant are associated with reduced starch levels. Plant Mol Biol 67: 57–69 [DOI] [PubMed] [Google Scholar]
- Tian Q, Olsen L, Sun B, Lid SE, Brown RC, Lemmon BE, Fosnes K, Gruis DF, Opsahl-Sorteberg HG, Otegui MS, et al. (2007) Subcellular localization and functional domain studies of DEFECTIVE KERNEL1 in maize and Arabidopsis suggest a model for aleurone cell fate specification involving CRINKLY4 and SUPERNUMERARY ALEURONE LAYER1. Plant Cell 19: 3127–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Duvick J, Schares JP, Ahern KR, Deewatthanawong P, Xu L, Conrad LJ, Kikuchi K, Kubinec TA, Hall BD, et al. (2010) Genome-wide distribution of transposed Dissociation elements in maize. Plant Cell 22: 1667–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Barry JK, Min Z, Tordsen G, Rao AG, Olsen OA (2003) The calpain domain of the maize DEK1 protein contains the conserved catalytic triad and functions as a cysteine proteinase. J Biol Chem 278: 34467–34474 [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B (2007) Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev 21: 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JP, Rogowsky PM (2004) Vacuolar H+-translocating inorganic pyrophosphatase (Vpp1) marks partial aleurone cell fate in cereal endosperm development. Plant Mol Biol 56: 325–337 [DOI] [PubMed] [Google Scholar]
- Wolf MJ, Cutler HC, Zuber MS, Khoo U (1972) Maize with multilayer aleurone of high protein content. Crop Sci 12: 440–442 [Google Scholar]
- Wong AYM, Colasanti J (2007) Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. J Exp Bot 58: 403–414 [DOI] [PubMed] [Google Scholar]
- Wu X, Tang D, Li M, Wang K, Cheng Z (2013) Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol 161: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G, Lauter AM, Scott MP, Becraft PW (2011) The thick aleurone1 mutant defines a negative regulation of maize aleurone cell fate that functions downstream of defective kernel1. Plant Physiol 156: 1826–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Hirano K, Sato T, Mitsuda N, Nomoto M, Maeo K, Koketsu E, Mitani R, Kawamura M, Ishiguro S, et al. (2014) DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc Natl Acad Sci USA 111: 7861–7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Ueguchi-Tanaka M (2014) DELLA and SCL3 balance gibberellin feedback regulation by utilizing INDETERMINATE DOMAIN proteins as transcriptional scaffolds. Plant Signal Behav 9: e29726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.