Abstract
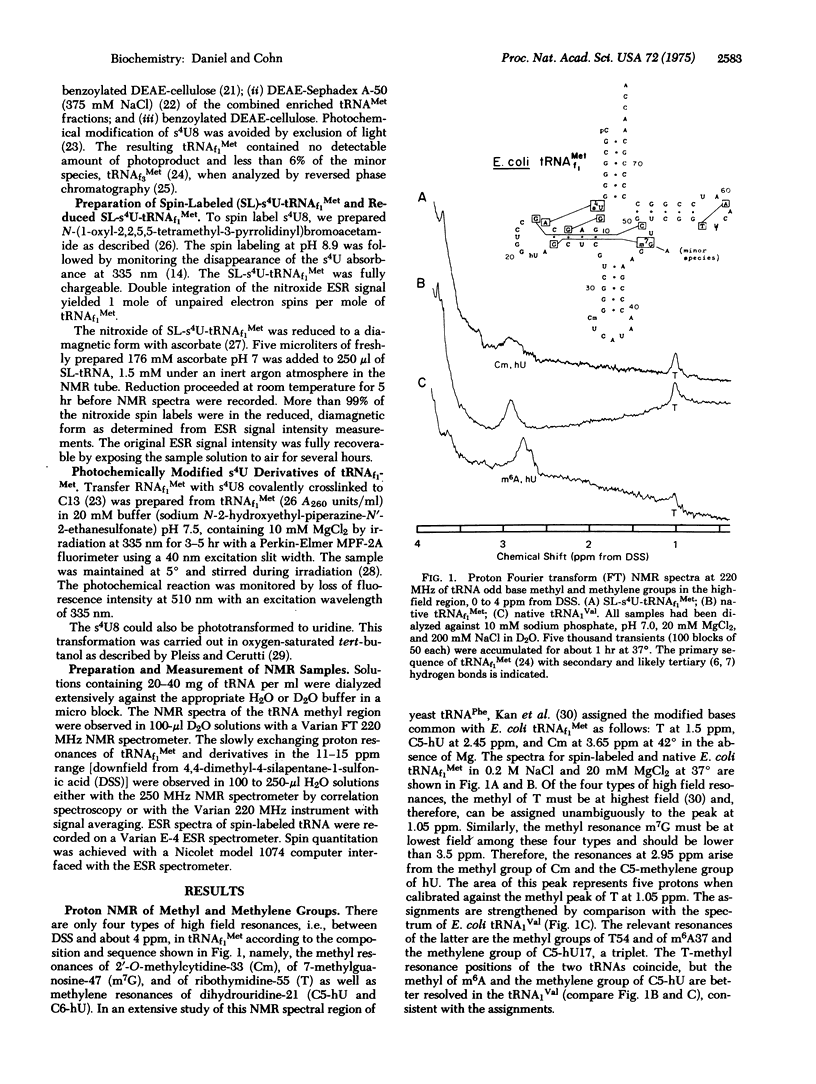
Thiouridine at position 8 (s4U8) of tRNAf1Met was spin-labeled with the nitroxide free radical, N-(1-oxyl-2,2,5,5-Tetramethyl-3-pyrrolidinyl) bromacetamide, for proton nuclear magnetic resonance spectroscopic studies. The well-resolved methyl peak of ribothymidine is unperturbed, but the peak tentatively assigned to the C-5 methylene group of dihydrouridine is considerably broadened in spin-labeled tRNAf1Met. Of the approximately 27 slowly exchanging protons observed in the region between 11 and 15 ppm downfield from 4,4-dimethyl-4-silapentane-1-sulfonic acid, the equivalent of about five protons apparently disappeared in spin-labeled tRNAf1Met. The well-resolved single proton at 14.8 ppm was missing not only in the paramagnetic species, but also in the diamagnetic reduced form of spin-labeled tRNAf1Met, and was unequivocally identified as a hydrogen bond involving s4U8 by comparison of several forms of tRNAf1Met specifically modified at s4U. Evidence that the perturbation of a second single proton resonance at 14.6 ppm (shift and broadening) is coupled to the loss of a tertiary hydrogen bond involving residue 8, arises from the same modified forms. The resolved resonances in the methyl and N-H regions, particularly the resonance at 14.6 ppm as well as the four N-bonded proton resonances at higher field which are broadened solely due to their proximity to the unpaired electron of the spin label, provide specific indicators of the geometry of tRNAf1Met structure in solution. Their observability by nuclear magnetic resonance spectroscopy opens up the possibility of monitoring distance changes among the base residues of spin-labeled tRNAf1Met upon its interaction with aminoacyl-tRNA synthetase and other enzymes.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blanquet S., Iwatsubo M., Waller J. P. The mechanism of action of methionyl-tRNA synthetase from Escherichia coli. 1. Fluorescence studies on tRNAMet binding as a function of ligands, ions and pH. Eur J Biochem. 1973 Jul 2;36(1):213–226. doi: 10.1111/j.1432-1033.1973.tb02903.x. [DOI] [PubMed] [Google Scholar]
- Cramer F. Three-dimensional structure of tRNA. Prog Nucleic Acid Res Mol Biol. 1971;11:391–421. doi: 10.1016/s0079-6603(08)60333-5. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Cole P. E., Hilbers C. W., Shulman R. G. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol. 1974 Jul 25;87(1):63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- Daniel W. E., Jr, Morrisett J. D., Harrison J. H., Dearman H. H., Hiskey R. G. Spin-labeled ribonuclease. A. Selective incorporation of a nitroxide spin label sensitive to active-center geometry. Biochemistry. 1973 Nov 20;12(24):4918–4923. doi: 10.1021/bi00748a017. [DOI] [PubMed] [Google Scholar]
- Dube S. K., Marcker K. A., Clark B. F., Cory S. Nucleotide sequence of N-formyl-methionyl-transfer RNA. Nature. 1968 Apr 20;218(5138):232–233. doi: 10.1038/218232a0. [DOI] [PubMed] [Google Scholar]
- Egan B. Z., Weiss J. F., Kelmers A. D. Separation and comparison of primary structures of three formylmethionine tRNAs from E. coli K-12 MO. Biochem Biophys Res Commun. 1973 Nov 16;55(2):320–327. doi: 10.1016/0006-291x(73)91090-5. [DOI] [PubMed] [Google Scholar]
- Englander J. J., Kallenbach N. R., Englander S. W. Hydrogen exchange study of some polynucleotides and transfer RNA. J Mol Biol. 1972 Jan 14;63(1):153–169. doi: 10.1016/0022-2836(72)90527-x. [DOI] [PubMed] [Google Scholar]
- Favre A., Michelson A. M., Yaniv M. Photochemistry of 4-thiouridine in Escherichia coli transfer RNA1Val. J Mol Biol. 1971 May 28;58(1):367–379. doi: 10.1016/0022-2836(71)90252-x. [DOI] [PubMed] [Google Scholar]
- Fresco J. R., Adams A., Ascione R., Henley D., Lindahl T. Tertiary structure in transfer ribonucleic acids. Cold Spring Harb Symp Quant Biol. 1966;31:527–537. doi: 10.1101/sqb.1966.031.01.068. [DOI] [PubMed] [Google Scholar]
- Hara H., Horiuchi T., Saneyoshi M., Nishimura S. 4-Thiouridine-specific spin-labeling of E. coli transfer RNA. Biochem Biophys Res Commun. 1970 Jan 23;38(2):305–311. doi: 10.1016/0006-291x(70)90713-8. [DOI] [PubMed] [Google Scholar]
- Hilbers C. W., Shulman R. G. Assignment of the hydrogen bonded proton resonances in (Escherichia coli) tRNAGlu by sequential melting. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3239–3242. doi: 10.1073/pnas.71.8.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L. S., Ts'o P. O., von der Haar F., Sprinzl M., Cramer F. NMR study on the methyl and methylene proton resonances of tRNA Phe yeast. Biochem Biophys Res Commun. 1974 Jul 10;59(1):22–29. doi: 10.1016/s0006-291x(74)80168-3. [DOI] [PubMed] [Google Scholar]
- Kearns D. R., Patel D. J., Shulman R. G. High resolution nuclear magnetic resonance studies of hydrogen bonded protons of tRNA in water. Nature. 1971 Jan 29;229(5283):338–339. doi: 10.1038/229338a0. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Sussman J. L., Suddath F. L., Quigley G. J., McPherson A., Wang A. H., Seeman N. C., RICH A. The general structure of transfer RNA molecules. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4970–4974. doi: 10.1073/pnas.71.12.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D., McConnell H. M. Inside-outside transitions of phospholipids in vesicle membranes. Biochemistry. 1971 Mar 30;10(7):1111–1120. doi: 10.1021/bi00783a003. [DOI] [PubMed] [Google Scholar]
- Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969 Nov 22;224(5221):759–763. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- Lightfoot D. R., Wong K. L., Kearns D. R., Reid B. R., Shulman R. G. Assignment of the low field proton nuclear magnetic resonance spectrum of yeast phenylalanine transfer RNA to specific base pairs. J Mol Biol. 1973 Jun 25;78(1):71–89. doi: 10.1016/0022-2836(73)90429-4. [DOI] [PubMed] [Google Scholar]
- Pleiss M. G., Cerutti P. A. Phototransformation of 4-thiouridine in Escherichia coli valine transfer ribonucleic acid to uridine, cytidine, and N 4 -methylcytidine. Biochemistry. 1971 Aug 3;10(16):3093–3099. doi: 10.1021/bi00792a017. [DOI] [PubMed] [Google Scholar]
- Renaud M., Bollack C., Ebel J. P. Interpretation of incomplete aminoacylation of yeast tRNA-Phe: evidence for a phenylalanyl-tRNA synthetase catalyzed transconformation of tRNA-Phe. Biochimie. 1974;56(9):1203–1209. doi: 10.1016/s0300-9084(74)80012-x. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Correlation between three-dimensional structure and chemical reactivity of transfer RNA. Nucleic Acids Res. 1974 Jul;1(7):927–932. doi: 10.1093/nar/1.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Schofield P., Hoffman B. M., Rich A. Spin-labeling studies of aminoacyl transfer ribonucleic acid. Biochemistry. 1970 Jun 9;9(12):2525–2533. doi: 10.1021/bi00814a020. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Goddard J. P. Loss of methionine acceptor activity resulting from a base change in the anticodon of Escherichia coli formylmethionine transfer ribonucleic acid. J Biol Chem. 1973 Feb 25;248(4):1341–1345. [PubMed] [Google Scholar]
- Seno T., Kobayashi M., Nishimura S. Purification of Excherichia coli methionine tRNAF and methionine tRNAM and studies on their biophysical and biochemical properties. Biochim Biophys Acta. 1968 Nov 20;169(1):80–94. doi: 10.1016/0005-2787(68)90010-5. [DOI] [PubMed] [Google Scholar]
- Shulman R. G., Hilbers C. W. Ring-current shifts in the 300 MHz nuclear magnetic resonance spectra of six purified transfer RNA molecules. J Mol Biol. 1973 Jun 25;78(1):57–69. doi: 10.1016/0022-2836(73)90428-2. [DOI] [PubMed] [Google Scholar]
- Sprinzl M. On the structure of phenylalanine tRNA from yeast. Spin-label studies. Eur J Biochem. 1974 Dec 2;49(3):595–605. doi: 10.1111/j.1432-1033.1974.tb03863.x. [DOI] [PubMed] [Google Scholar]
- Suddath F. L., Quigley G. J., McPherson A., Sneden D., Kim J. J., Kim S. H., Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA at 3.0angstroms resolution. Nature. 1974 Mar 1;248(5443):20–24. doi: 10.1038/248020a0. [DOI] [PubMed] [Google Scholar]
- Wong K. L., Kearns D. R. Investigation of the base-pairing structure of the anticodon hairpin from E. coli initiator tRNA by high-resolution nmr. Biopolymers. 1974;13(2):371–380. doi: 10.1002/bip.1974.360130212. [DOI] [PubMed] [Google Scholar]
- Wong K. L., Kearns D. R. NMR evidence for tertiary structure base pair in E. coli tRNA involving S4U8. Nature. 1974 Dec 20;252(5485):738–739. doi: 10.1038/252738a0. [DOI] [PubMed] [Google Scholar]


