Abstract
Aneuploid genomes, characterized by unbalanced chromosome stoichiometry (karyotype), are associated with cancer malignancy and drug-resistance of pathogenic fungi. The phenotypic diversity resulting from karyotypic diversity endows the cell population superior adaptability. We show here, using a combination of experimental data and a general statistical model, that the degree of phenotypic variation, thus evolvability, escalates with the degree of overall growth suppression. Such scaling likely explains the challenge of treating aneuploidy diseases with a single stress-inducing agent. Instead, we propose the design of an “evolutionary trap” (ET) targeting both karyotypic diversity and fitness. This strategy entails a selective condition “channeling” a karyotypically divergent population into one with a predominant and predictably-drugable karyotypic feature. We provide a proof-of-principle case in budding yeast and demonstrate the potential efficacy of this strategy toward aneuploidy-based azole resistance in Candida albicans. By analyzing existing pharmacogenomics data, we propose the potential design of an ET against glioblastoma.
Keywords: Aneuploidy, budding yeast, pathogenic fungi, cancer, karyotype heterogeneity, evolutionary trap
Introduction
Germline evolution shapes the organismal tree in changing environments during the long course of natural history, while acute environmental fluctuation drives asexual cellular evolution, often associated with dynamic structural changes in the genomes of microbes or cancer cells (Lewontin, 1970; Merlo et al., 2006). Aneuploidy (chromosome copy number imbalance) is a type of genome alteration widely observed during cellular evolution of eukaryotic species, such as laboratory (Chen et al., 2012a; Hughes et al., 2000; Rancati et al., 2008), industrial (Borneman et al., 2011; Chen et al., 2012b; Infante et al., 2003; Kvitek et al., 2008) and pathogenic (Marichal et al., 1997; Ni et al., 2013; Selmecki et al., 2006; Sionov et al., 2010) yeasts, as well as protozoan parasites such as leishmania (Leprohon et al., 2009; Mannaert et al., 2012; Ubeda et al., 2008) and trypanosomes (Llewellyn et al., 2011; Minning et al., 2011). Emerging evidence also points to aneuploidy as an important driver for the evolution of human cancer (Davoli et al., 2013; Holland and Cleveland, 2009; Jones et al., 2010; Ng et al., 2010; Wang et al., 2014). Due to its impact on the expression of many genes, aneuploidy brings about large phenotypic changes that can be either detrimental or beneficial in a karyotype- and environmental condition-dependent manner (Chen et al., 2012b; Pavelka et al., 2010).
Aneuploid populations are often characterized by heterogeneity - the coexistence of many different karyotypes. The genetic diversity provides the raw material for evolutionary selection and endows the aneuploid population high adaptive potential (Burrell et al., 2013; Chen et al., 2012a; Maley et al., 2006). This underscores the exceptional challenge of treating disease-causing cell populations characterized by large karyotype heterogeneity and instability (Gerlinger et al., 2012; Harrison et al., 2014; Lee et al., 2011; Navin et al., 2011; Sotillo et al., 2010). One idea is to find drugs strongly exacerbating a common deficiency of aneuploids irrespective of specific karyotype (Oromendia and Amon, 2014). A study found that among a collection of disomic yeast strains many showed prominent growth defects toward agents that perturb proteome homeostasis such as hygromycin B (a translation inhibitor) or geldanamycin (an Hsp90 inhibitor) (Torres et al., 2007), supporting the notion that a common deficiency of aneuploidy is overloading of the proteome quality control system. However, some of the disomy strains exhibited resistance, as opposed to sensitivity, toward proteotoxic agents, and one of the karyotypic features, increased copy number of chromosome XV (Chr XV), emerged as the adaptive variant when a diploid strain was evolved in the presence of a Hsp90 inhibitor (Chen et al. 2012a). In addition, when a highly heterogeneous aneuploid population was treated with drugs that imposed strong immediate growth inhibition, long-term culturing enabled evolutionary selection for a few variants that eventually rendered the population drug-resistant (Chen et al., 2012a) (also see results of this study). These findings highlight the fallacy of short-term efficacy in drug treatments when dealing with heterogeneous populations already poised for rapid adaptation.
In this study, we aim for an innovative approach that accounts for the evolutionary dynamics and achieves long-term growth suppression or extinction of aneuploid cell populations consisting of a wide spectrum of karyotypes. Our analyses demonstrate on a general level that the evolutionary potential of a heterogeneous aneuploid population escalates under increasing stress but such a population may become highly targetable once the karyotype diversity is drastically confined. These findings led us to design a two-component strategy for treating diseases associated with aneuploid cell populations that targets the population’s adaptability and fitness.
Results
Phenotypic variation scales with growth suppression under diverse stress conditions in budding yeast
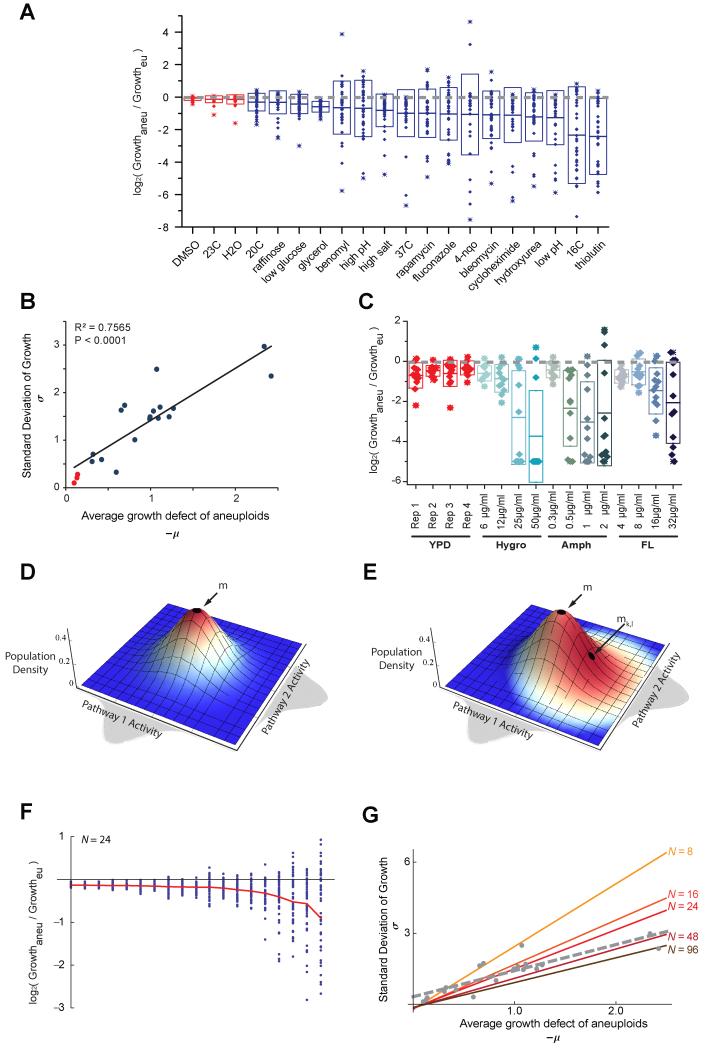
To investigate whether certain stress conditions may be consistently effective toward aneuploids with a wide-spectrum of karyotypes, we performed a re-analysis of the growth data from a previous study subjecting a panel of 38 aneuploid yeast strains (S. cerevisiae) with diverse and random chromosome stoichiometry to phenotypic profiling across a wide range of conditions with varying stress types and levels (Pavelka et al., 2010). (Figure 1A). This analysis revealed a surprising trend: those conditions more toxic to the aneuploidy cohort (lower mean growth ratio) also produced a larger fitness spread among the aneuploids, such that even under highly toxic conditions, while many aneuploids failed to grow, some aneuploids endured or thrived. The standard deviation (spread) and mean of the growth ratio (overall growth suppression) of diverse aneuploids are strongly correlated across diverse stress conditions (Figure 1B). This same trend could also be observed with the set of disomic aneuploid strains isolated in a previous study (Torres et al., 2007) treated with increasing concentrations of different drugs (Figure 1C).
Figure 1. Scaling of phenotypic variation with growth suppression in aneuploids under diverse stress conditions.
(A) The growth of 38 aneuploid strains relative to the euploid, as log2ratio of aneuploid growth (OD increase) over the euploid with the nearest ploidy (see Supplemental Information), are binned by growth conditions. Each point in a box plot represents an aneuploidy strain. The half-length of each box represents the standard deviation (SD) of relative growth among aneuploids (σ) and the middle line represents the average (μ). Note that the horizontal dashed line across 0 represents the euploid control. (B) Phenotypic variation among the aneuploids, measured as SD of relative growth (σ), scales with average growth defect of the aneuploid cohort across diverse stress conditions (−μ). (C) The growth of 12 disomy strains relative to the haploid control under increasing concentrations of hygromycin B (Hygro), amphotericin B (Amph) or fluconazole (FL). Box plot representation is as described for (a). (D-E) Schematic representation of the model is shown for the simple case of N = 2 with axes as labeled. Deep blue to deep red code for increasing fitness. (D) Graph represents the stress-free condition, where the euploid is located at the center of the activity field (position m) assumes highest fitness. (E) Graph represents a stress condition, where the optimal fitness point shifts from m to mk,l, reflecting the activity change necessary for adaptation. Consequently, the euploid (located at point m) no longer holds maximal fitness, whereas higher fitness is assumed by certain aneuploids (those occupying redder regions). (F) Example simulation results of the model for 50 random aneuploids under diverse stress conditions (governed by varying type k and magnitude l) for a 24 dimension space (N = 24), with relative growth displayed as the experimental data in A. The red line shows average log2 growth ratio from the simulated aneuploid population. Note the appearance of adaptive aneuploids under high-stress conditions (toward the right of the graph). (G) Simulations of the model with a wide range of N values demonstrate the positive correlation between σ and -μ in various numbers of dimensions. The simulated correlations are shown in colored lines while the experimental data is overlaid in grey.
See also Figure S1.
We used a multi-dimensional model to investigate the theoretical generality of the above experimental observation (Fig. 1D and E, and S1A and B (see Supplemental Information for detailed description of assumptions and simulations). Briefly, we assumed that fitness is governed by N independent pathways, reflecting the modular architecture of cellular systems. We assume that for each individual pathway the distribution of pathway activity across diverse karyotypes assumes a simple normal distribution. Under the stress-free condition, the optimal fitness is reached by the euploid at the peak of the N-dimensional normal distribution (Figure 1D; Supplemental Information), whereas aneuploid populations are located at positions away from the peak, reflecting suboptimal fitness. In the presence of stress, however, the optimal fitness point moves away from the euploid position (Figure 1E). The change in the distance between the position of each karyotype and the optimal fitness point represents the fitness change caused by stress. Simulation of this model with varying values of N revealed two properties of the system remarkably consistent with experimental observations (Figure 1F,G). First, under stress conditions with sufficient magnitude, most aneuploids had lower fitness than the euploid, yet there was always a fraction of aneuploids that assumed higher fitness than the euploid. Second, the absolute value of the mean and standard deviation of the relative fitness satisfied a positive linear correlation across diverse stress conditions and a range of N values (Figure 1F, G and Figure S1C-F). The fact that such a simple model could qualitatively capture the experimental data suggests the observed phenomenon to be a general property of heterogeneous aneuploid populations irrespective of specific stress applied.
Therapeutic compounds elicit phenotypic heterogeneity in human tumor cell lines
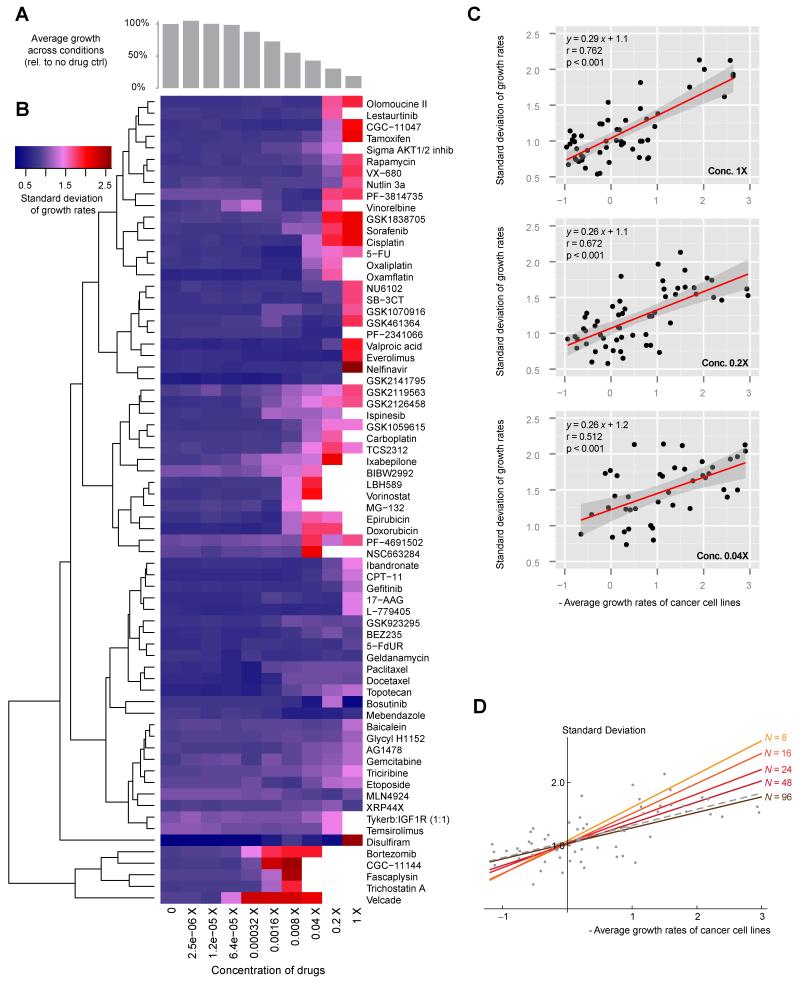
A vast majority of solid tumors are aneuploid: 91.7% of 817 solid tumor cell lines surveyed by Cancer Cell Line Encyclopedia (CCLE) have at least 1 chromosome arm-level copy number variation, whereas 58.9% have more than 10 (See Supplementary Information). Karyotypic heterogeneity has also been observed within a single tumor (Gerlinger et al., 2012; Navin et al., 2011). The relapse fueled by adaptive changes in a tumor remains a major challenge in cancer treatment. A logical extension of our findings is that, in karyotypically heterogeneous cancer cell populations, the treatment suppressing the overall growth may also escalate tumor cells’ phenotypic heterogeneity and potentially adaptability. To examine this possibility, we first investigated a dataset consisting of 54 karyotypically divergent human breast cancer cell lines and 77 different anticancer drugs across 10 concentrations (SU2C breast cancer project (Heiser et al., 2012)). As shown in Figure 2A and B, in drug doses that caused prominent (>50%) overall growth suppression, the spread of growth rates among cell lines treated with most chemicals (~95%) also increased. A few drugs showed more consistent toxicity, but due to the lack of a normal euploid control it is unknown whether this was specific to aneuploid cancer cells. Examination of the correlation between overall growth suppression (−mean) and variation (SD of growth rates) found a consistent positive correlation. Simulation of our multi-dimensional fitness model with varying N values produced quantitatively a similar correlation pattern (Figure.2C,D). Cell lines from other solid tumors (e.g., central nervous system, skin and lung) also demonstrated a similar trend (Figure S2).
Figure 2. Phenotypic heterogeneity of human cancer cell lines positively correlates with increasing growth suppression by therapeutic compounds.
The relationship between phenotypic variation (SD of growth rates) and average growth suppression was examined using published drug response profiling data from 54 breast cancer cell lines treated with 77 different potential therapeutic compounds over 10 different concentrations (see Supplementary Information). (A) Histograms showing average growth suppression under different drug concentrations. (B) The SD of growth rates caused by each drug under the range of concentrations tested are shown as a heat map. Note that drug concentrations for A and B are aligned, showing the general trend of increasing SD with increasing growth suppression. The clustering is based on Euclidean distances. (C) At 3 concentrations that considerably reduced the overall growth rate (> 50% decrease compared to no drug control in A), the general correlation between μ and σ across different growth conditions is examined. The linear regression line (red) is surrounded by 95 percentile confidence fitting intervals (dark gray area). Note similar fitting parameters across different drug doses. (D) The correlation between SD and average growth suppression is also recapitulated by simulations of the multi-dimensional model with the number of pathways (N) in the range of 48-96. The modeled fitting is shown in colored lines, while the published experimental data are shown in gray.
See also Figure S2.
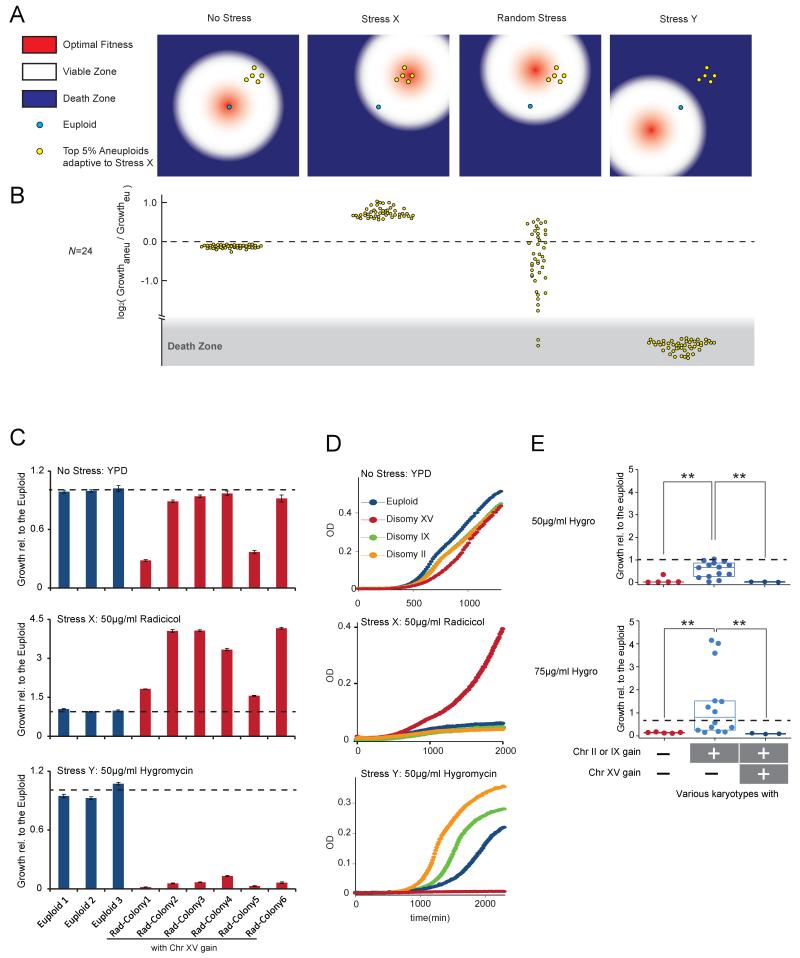
Although model simulations showed that for a given stress (X) a group of aneuploids may be selected for superior fitness, it also predicts that if second stress (Y) is applied to shift the optimal fitness in the direction opposite from stress X, those selected aneuploids would fall into the death zone in the fitness landscape (Figure 3A, B). This implies that if a highly evolvable population with heterogeneous random karyotypes can be “channeled” toward a certain karyotypic characteristic under a designed selection (stress X), thus drastically “shrinking” the population’s evolvability, a second treatment (stress Y) may be added to eradicate the selected singular or limited karyotypes. Together these two treatments would form an “evolutionary trap” (ET) on the elusive aneuploid population. Supporting this idea, for a given specific aneuploidy yeast strain, in most cases one or multiple conditions could be found that caused >80% growth inhibition compared to the euploid control (Figure S1G).
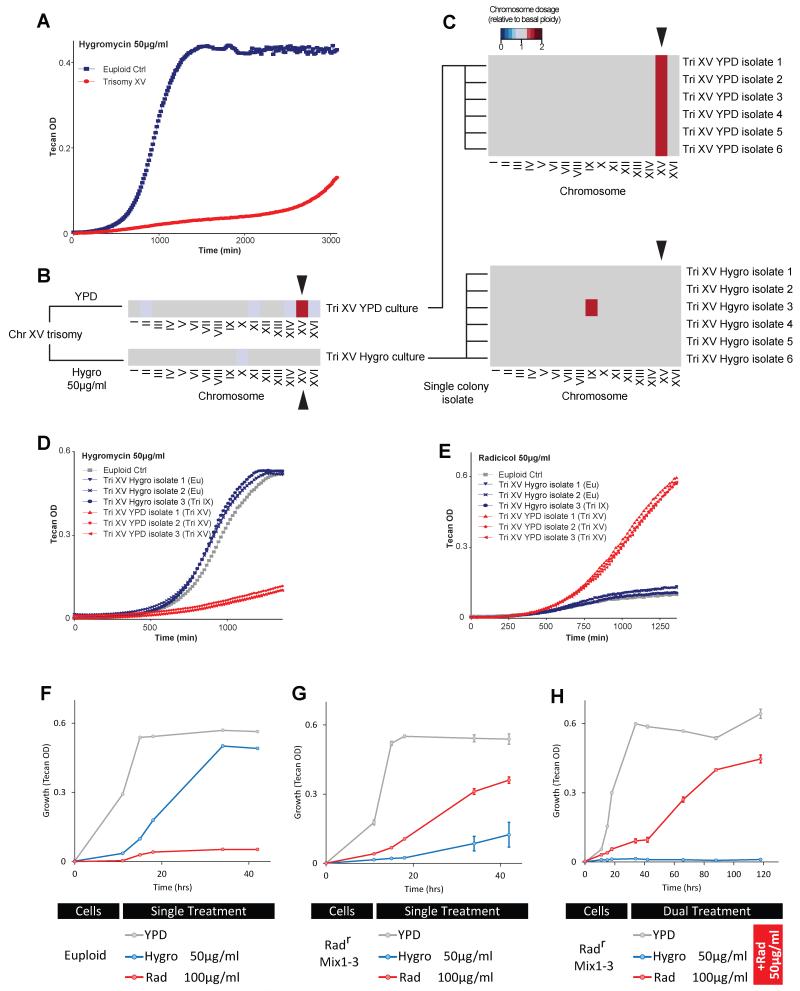
Figure 3. Design and experimental implementation of ET in budding yeast.
(A-B) Model simulations predict cell population adapted to a specific stress (X) through karyotype channeling can be highly targetable by a stress Y that shifts optimal fitness in the direction opposite to X, but not by a second stress in a random direction. (A) Schematic representations of the fitness landscape in a simplified 2D example similar to that in Figure 1D, E but projected to the plane defined by pathway activities. (B) Results of model simulations in a high dimensional fitness space under conditions indicated in A. Note that only the aneuploids selected by Stress X (the top 5% adaptive ones) are shown. A total of 1000 cells were simulated. Each dot represents the relative fitness of an aneuploid cell compared to the euploid. Death zone was defined as having negative growth value. (C) Six independent colonies isolated from radicicol-adapted population aneuploids with gain of Chr XV (as shown in Figure S3F) were grown under indicated conditions until saturation was reached in the fastest growing strain. Histograms show average amount of growth normalized to euploid and standard error of the mean (SEM) derived from 4 replicates. (D) Chr II and Chr IX disomy strains generated previously by genetic manipulation (Torres et al., 2007) exhibit resistance to hygromycin B yet are sensitive to radicicol. (E) Aneuploid strains generated by random triploid meiotic segregation with indicated karyotypic features were culture in different concentrations of hygromycin B. Box plots show growth relative to the euploid control with each dot representing an aneuploid strain. Karyotypes are categorized by their states of Chr II/IX/XV dosage, but other chromosome aneuploidy may also be present in these strains. The amount of growth (OD increase) was normalized to the euploid with the nearest ploidy. The dashed line represents the average of normalized controls (equals to 1). ** indicates p< 0.01 according to Mann-Whitney U test.
Finding drugs forming an ET against heterogeneous aneuploid budding yeast
To investigate the validity of ET against heterogeneous aneuploids, we first constructed a yeast population with high-degree karyotypic diversity by sporulation of a homozygous pentaploid yeast strain. 28 viable aneuploid meiotic products with ploidy above 1.9N were mixed with isogenic diploid, triploid and tetraploid cells to mimic karyotype heterogeneity observed in pathogenic fungi (Harrison et al., 2014) or human cancers (Mitelman F, 2012). Because most aneuploid karyotypes are unstable (Charles et al., 2010; Pavelka et al., 2010; Zhu et al., 2012), the degree of heterogeneity of this mixed population (heterogeneous mix) is expected to be far greater, as confirmed by a broad DNA content profile that did not show bias toward any specific chromosome gain or loss (Figure S3). To find a selective condition for karyotype channeling, we took advantage of the previous finding that growth in high-concentrations of radicicol, an inhibitor of Hsp90 chaperone, selects for extra chromosome (Chr) XV (Chen et al., 2012a). Treatment of three independent populations of the heterogeneous mix with 50 μg/ml radicicol all predictably led to convergent evolution toward Chr XV gain (Figure S3C, D and E). Six individual colonies derived from a culture that had grown to saturation in radicicol-containing medium were found to vary in karyotypes but all shared Chr XV gain, confirming that radicicol caused a selective sweep in the heterogeneous mix (Figure S3F).
We next performed a screen for chemicals particularly effective against Chr XV trisomy and identified hygromycin B, a translation inhibitor (Singh et al., 1979), as the most potent inhibitor of Chr XV trisomy relative to its effect on the diploid control (Figure S4). We further confirmed potent growth inhibition by hygromycin B for all 6 different karyotypes identified from radicicol-selected cultures sharing Chr XV gain (Figure 3C). We note that even though hygromycin B effectively suppressed the growth of aneuploids that had gained Chr XV, this effect did not extend to all aneuploids: aneuploid strains with increased Chr II or IX dosage but having a normal dosage of Chr XV were associated with superior resistance to hygromycin B compared to the euploid control (Figure 3D,E). However, strains with combined combination of Chr XV, II and IX gains were highly sensitive rather than resistant to hygromycin B (Figure 3E).
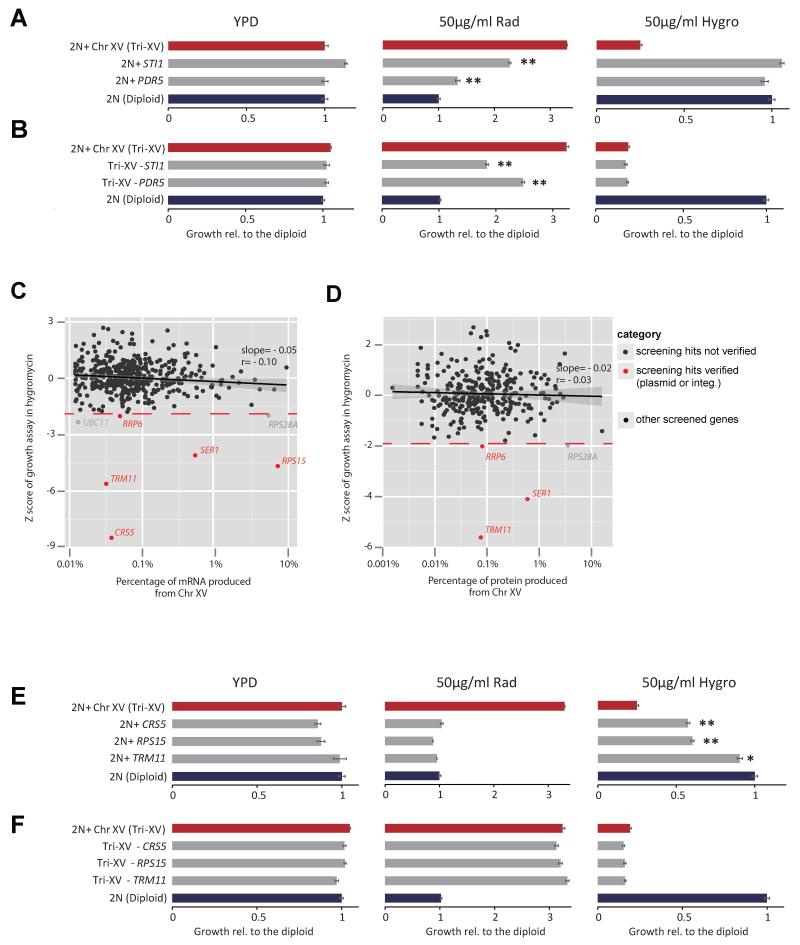
We showed previously that dosage increase of STI1 and PDR5, two genes present on Chr XV, are “driver mutations” underlying Chr XV gain-associated radicicol resistance (Chen et al., 2012a). However, hygromycin B sensitivity was unrelated to dosage alterations of these two genes (Figure 4A, B). To identify genes on Chr XV whose increased copy number could cause hypersensitivity to hygromycin B, we screened each of the 453 genes located on Chr XV with its original promoter and carried on a low-copy (centromeric) plasmid (Ho et al., 2009) and identified 5 genes (CRS5, RPS15, TRM11, RRP6, SER1) (Figure 4C, Figure S5A,B). Integration of one copy of each top three hit (CRS5, RPS15, TRM11) recapitulated up to 50% of the hygromycin B-sensitivity of Chr XV trisomy in a diploid background (Figure 4E). However, the effects of these genes were not additive (data not shown), suggesting that the hygromycin B sensitivity associated with Chr XV gain is an emergent property of gaining many genes carried on this chromosome beyond the three found in the screen. Consistent with this notion, deletion of a copy of each of the three hits (CRS5, RPS15, TRM11) individually did not change hygromycin hypersensitivity of Chr XV trisomy (Figure 4F).
Figure 4. Different sets of genes on Chr XV cause radicicol sensitivity or hygromycin B sensitivity when increased in copy number.
(A-B) Copy number gain (+, by genomic integration) or loss (−, by genomic deletion) assays showing that increased copy numbers of STI1 and PDR5, which are both critical for radicicol resistance, do not contribute to the hygromycin B hypersensitivity. Relative growth compared to the diploid control is reported in bar plots with the SEM derived from 3 replicates. Asterisks denote significant difference from the corresponding control (the diploid or Chr XV trisomy (Tri-XV)) (*, p< 0.05; **, p<0.01; two-tail t-test). (C-D) Each of 453 genes located on Chr XV was transformed into a diploid strain,and Z scores denoting the deviation of growth of each strain from the population average in the presence of 35 μg/ml hygromycin B were plotted against mRNA (C), using RNAseq data, or protein expression abundance (D) (Ghaemmaghami et al., 2003), of each tested gene in the euploid S288c background. Note that protein abundance data were not retrieved for 30% genes (including CRS5 and RPS15). The grey area shows the 95% confidence interval for the linear fitting. 35 μg/ml hygromycin B produced similar growth inhibition to euploid control in SC –ura media compared to euploid control in YPD media. (E) Growth assays showing that copy-number increase (by genomic integration) of 3 genes (CRS5, RPS15, TRM11) on Chr XV were individually sufficient in a diploid euploid context to reproduce enhanced sensitivity to hygromycin B, but not radicicol resistance, contrasting copy number increase for STI1 and PDR5 as shown in A. (F) Growth assays showing that single-copy deletion of none of the 3 genes (CRS5, RPS15, TRM11) alone could rescue Chr XV trisomy from hygromycin B hyper-sensitivity.
See also Figure S5.
Hygromycin B and other proteotoxic agents were previously proposed to enhance the protein quality control deficit due to increased expression of a large number of genes (Oromendia et al., 2012; Torres et al., 2007). However, the genes whose dosage increase gave rise to hygromycin B sensitivity were not all highly expressed genes compared to the other genes located on Chr XV in either euploid or aneuploid with Chr XV gain (p = 0.56 for transcripts, p = 0.43 for proteins, Mann-Whitney U test) (Figure 4C,D and Figure S5C). RPS15, the only highly expressed gene among the hits, encodes one of 30 proteins within the small ribosomal subunit to which hygromycin B binds (Borovinskaya et al., 2008), suggesting that a direct gene-specific cause underlies some of the drug sensitivity. Taken together, different molecular mechanisms are likely to account for radicicol-resistance and hygromycin-hypersensitivity of aneuploid strains with Chr XV gain.
Evolution dynamics of cell populations under single-drug treatment vs ET
Even though hygromycin B alone potently suppressed the growth of Chr XV trisomy strain initially, the treated cell population eventually adapted, as indicated by growth takeoff after ~ 50 hr (Figure 5A). Karyotyping of the adapted Chr XV trisomy culture revealed that the population was a mixture of euploid and aneuploid cells that no longer had Chr XV gain but some now carried an additional copy of Chr IX (Figure 5B,C), consistent with the result shown in Figure 3D,E. This observation reconfirmed the requirement for Chr XV gain in hygromycin B sensitivity and demonstrated the ability of the population to escape single-drug treatment by continued karyotype change. As expected, the condition that selected for Chr XV, i.e. 50 μg/ml radicicol, was highly toxic to all those survivors that had become hygromycin B resistant because they had lost the gained Chr XV (Figure 5D,E), supporting the rationale of combinatorial treatment with both radicicol and hygromycin B.
Figure 5. The combination of radicicol and hygromycin B extincts karyotypically heterogeneous cell population.
(A-E) Chr XV trisomy was able to escape growth inhibition by hygromycin B through loss of the gained Chr XV. (A) The growth (represented by OD reading on a Tecan reader) of both the euploid control and the trisomy XV strain was monitored in media containing 50 μg/ml hygromycin B. (B) The additional copy of Chr XV was lost in hygromycin B culture but not in YPD culture, as shown by the heat map of karyotyping result of the final culture. (C) Karyotypes of 6 single colonies from the trisomy XV culture in YPD or hygromycin are shown, three of which were re-tested for growth in the presence of hygromycin B (D) or radicicol (E). Note radicicol sensitivity was re-established in all three adapted colonies from the trisomy XV culture in hygromycin, whereas isolates from the YPD culture remained radicicol resistant. (F-H) Combination of hygromycin B and radicicol effectively eradicates the radicicol-preselected aneuploid population. (F) Growth curves (as OD600 measured in Tecan) of the diploid control strain under conditions as indicated. Note that 50 μg/ml hygromycin B alone had milder growth suppression compared to 100 μg/ml radicicol. (G) Growth curves of 3 populations preselected independently in the presence of radicicol (Figure S3E) under indicated conditions. (H) Growth curves of the same 3 populations as in (b) under indicated conditions where each drug was combined with 50 μg/ml radicicol. Each data point in B and C shows the mean and SEM from 3 experiments.
See also Figure S6.
Indeed, the combination of both drugs led to the extinction of all 3 independent cultures first subjected to the radicicol selection (Figure 5F-H). The same drug pair was similarly effective against the heterogeneous aneuploid population (heterogeneous mix) when added simultaneously (Figure S6A-E). When the heterogeneous mix was treated with a single chemical, 10 out of 21 tested chemicals (for example, 0.08 μg/ml menadoine and 100μg/ml radicicol) imposed stronger growth suppression than 50 μg/ml hygromycin B (Figure S6C). Hygromycin B also ranked low in suppressing euploid growth (Figure S6A). Yet, when combined with 50 μg/ml radicicol, hygromycin B, but not menadoine or radicicol, led to the extinction of the heterogeneous mix (Figure S6D). Thus, the opposing selective effects of these two drugs on the channeled karyotype (Chr XV gain) imposed an adaptive dilemma for the heterogeneous population, leading to its extinction. A potential pitfall of this ET drug pair is that the diploid with an additional copy of STI1 showed an enhanced level of resistance against 50 μg/ml radicicol yet without causing hypersensitivity to 50 μg/ml hygromycin (Figure 4A). Such effects due to potential single gene gain could be remedied by increasing the concentration (from 50 to 100 μg/ml) of radicicol (Figure S6F, G).
An azole-based ET for human pathogen Candida albicans
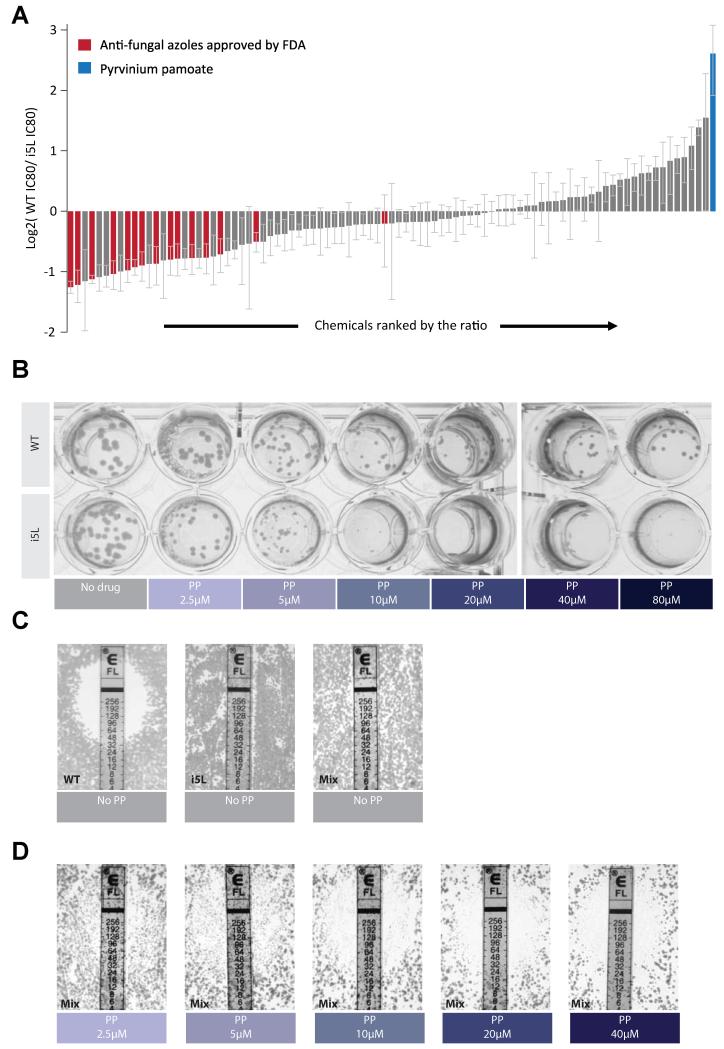
We tested the possibility of using the evolutionary-trap strategy for more effective anti-fungal treatment. It was previously shown that a mechanism for human pathogen Candida albicans to confer resistance to fluconazole, a first-line medicine treating invasive candidiasis in immune-compromised patients, is gain of isochromosome 5L (i(5L), which contains two copies of the left arm of Chr 5) (Selmecki et al., 2006). This aneuploid feature was also recapitulated in a laboratory evolution experiment selecting for fluconazole-resistant C. albicans (Selmecki et al., 2009). Thus, fluconazole may serves as the “selection” drug, which would be used in combination with a second drug specifically targeting i(5L) gain. To this end, we screened a chemical library that contained 1,713 FDA- or other regulatory agency-approved drugs, 580 natural compounds, and 420 other bioactive agents against an i(5L)-containing C. albicans isolate from a 30-year-old male who developed fluconazole-resistant candidemiai(5L) (Marr et al., 1997; Selmecki et al., 2006). We looked for chemicals that showed much elevated potency against the i(5L)-containing strain compared to the euploid control generated by spontaneous i(5L) loss of the original i(5L)-containing strain (Selmecki et al., 2008). Such drugs were likely to be missed in previous screens against euploid C. albicans (Okoli et al., 2009; Spitzer et al., 2011).
A primary screen found 100 out of 2713 compounds (Figure S7A) caused at least 80% growth suppression to either the i(5L) strain or the euploid, or both. For this set of compounds,we then determined the concentration causing 80% growth suppression (IC80). i(5L)In the presence of 26 or 11 conditions, the i(5L) strain showed significantly (Z-test, p < 0.05) higher (i.e., more resistant) or lower (more susceptible), respectively, IC80 than the euploid (Figure 6A). As expected, the i(5L) strain exhibited increased resistance toward a panel of 7 other azole derivatives. By contrast, pyrvinium pamoate (PP), a medication for pediatric pinworm infection, strongly suppressed the growth of the i(5L) strain but was ineffective against the euploid even at high concentrations as high as 80 μM after 48 hour culture on agar (Figure 6A, B, S7B). Given that euploid and i(5L)-gained C. albicans could co-exist in patients treated with fluconazole (Selmecki et al., 2008), we tested the combinatorial effect of PP with fluconazole in a 1:1 mixed population of both karyotypes. At concentrations of PP that suppressed the growth of the i(5L) strain, the resistance to fluconazole regressed from over 256 μg/ml back to the same level exhibited by the euploid without PP (Figure 6C,D), supporting the ET rationale for combinatorial anti-fungal treatment.
Figure 6. Pyrvinium pamoate (PP) effectively targets the fluconazole-resistant Candida aneuploid.
(A) Relative IC80 (80% inhibitory concentration) of the diploid vs the i(5L) Candida strain for each of the hits of the primary drug screen (Figure S7A). (B) Images of agar plates showing selective effectiveness of PP toward i(5L) candida. (C) Resistance of the diploid, the i(5L) or the i(5L)+diploid mix population toward fluconazole. (D) PP at concentrations above 10 μM restored the sensitivity of the i(5L)+diploid mix population toward fluconazole in the e-test, in accordance to its singular form’s activity against the i(5L) strain shown in B. Note even though the initial plating density was the same, due to the inhibition of the i(5L) cells, the overall growth was less in D compared to C. Note that our euploid strain also exhibited a reduced susceptibility to fluconazole compared to the clinical E-test standard strain, which may be attributed to other point mutations (such as the hyperactive TAC1) within this strain (Selmecki et al., 2006). All plate images were taken after 48-hour culture.
See also Figure S7.
A potential ET against EGF receptor-driven tumors such as glioblastoma
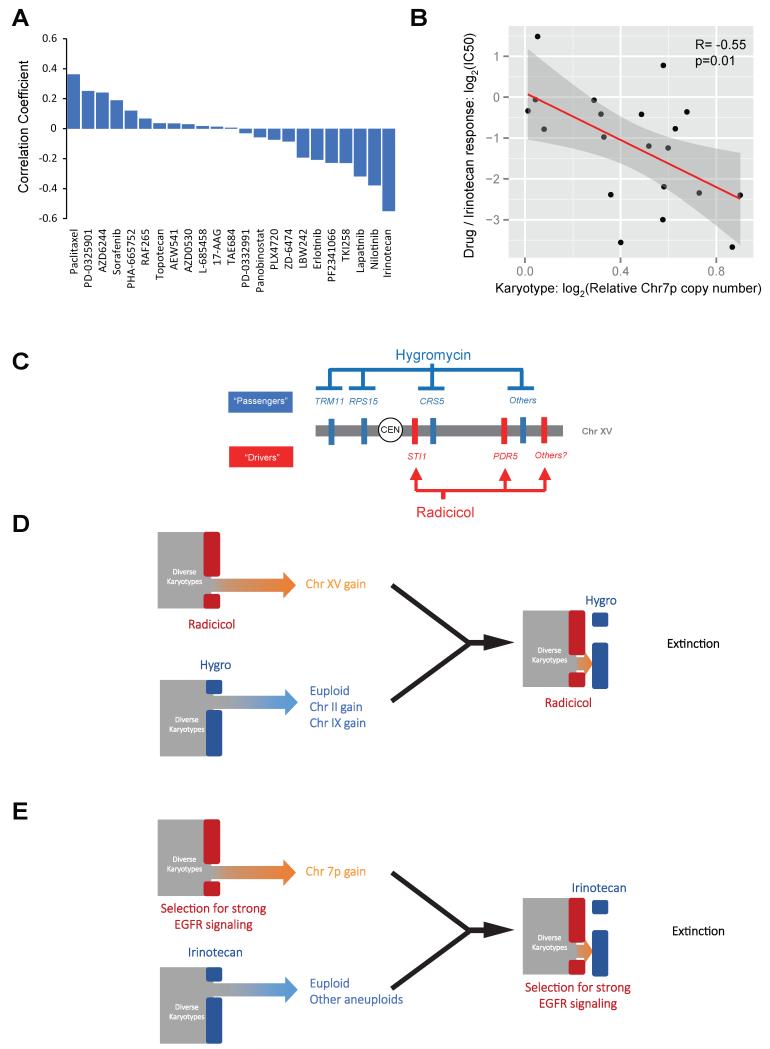
In many human cancers epidermal growth factor receptor (EGFR) signaling is a major contributing factor to aggressive phenotypes (Wheeler et al., 2010). The human EGFR gene is located on the short arm of chromosome 7 (Chr 7p), and Chr 7p gain is a prevalent karyotypic feature across diverse types of human cancers (Beroukhim et al., 2010). In particular, Chr 7 gain was observed in 80% of glioblastoma samples collected from The Cancer Genome Atlas Pilot Project (TCGA) cohort of 219 patients (Bredel et al., 2009; 2008). Within individual tumors, arm-level (mainly Chr 7p) or whole chromosome gain of Chr 7 was detected in nearly all surgical sections examined, suggesting that this event, unlike other heterogeneous genetic alterations, is an early critical event in clonal expansion (Sottoriva et al., 2013). In addition to being a likely product of the selection for EGFR-dependent malignant transformation or adaptation, Chr7p polysomy was also reported in 80% of tumor samples from lung cancer patients that acquired resistance to gefitinib or erlotinib, EGFR kinase inhibitors (Bean et al., 2007). Thus, in designing an ET against EGFR signaling-dependent tumors, gefitinib or erlotinib may be used as the channeling drug that could impose a pharmacological selection for Chr 7p gain beyond the selection during tumorigenesis. To find the pairing drug that may be particularly toxic toward Chr 7p gain, we analyzed the pharmacogenomics data (Barretina et al., 2012) by correlating the dosage of Chr 7p of 29 central nervous system (CNS) tumor cell lines with the response (IC50) to 23 therapeutic compounds (Figure 7A). This analysis identified a significant correlation between increased Chr 7p dosage and increased sensitivity to irinotecan, an FDA-approved drug for treating colon cancer but not yet for glioblastoma (Figure 7B). This result suggests that gefitinib or erlotinib together with irinotecan may form an ET against EGFR-driven tumors, especially glioblastoma where Chr 7p is already highly enriched even without drug treatment (Figure 7E).
Figure 7. A potential drug targeting Chr 7 gain in brain cancer and schematic summary of the mechanism and principle of ET using the yeast example.
(A) Correlation coefficients of drug response (IC50) with Chr 7p dosage in 29CNS tumor cell lines across 23 different therapeutic compounds were plotted as bar graphs. (B) The dot plot illustrates the details of the correlation between dosage of Chr 7p and sensitivity to irinotecan, with each dot showing the drug response and Chr 7p dosage of each cell line. The red line shows linear fitting and the grey area showing the fitting range with 95% confidence interval. Note that a total of 20 cells lines were included here as the IC50 data for 9 cell lines were not available for irinotecan. (C) The molecular makeup of the ET against aneuploidy yeast. (D) Schematic summary of opposing selective effects of radicicol and hygromycin B on Chr XV gain impose an adaptive dilemma for the yeast heterogeneous aneuploidy population. (E) A ET may be established against glioblastoma by opposing selective effects on Chr 7p gain.
Discussion
The analyses presented above started from an exploration of the general response of karyotypically heterogeneous cell populations to diverse stress conditions and proceeded to the design of a strategy to extinct such cell populations that accounts for their adaptive potential. Growth profiling of yeast aneuploids demonstrated that the phenotypic variation resulting from karyotype diversity scales directly with the degree of average inhibition of population growth under diverse stress conditions. Mathematical simulations based upon a few simple assumptions support the generality of our experimental observation. A more intuitive understanding of this phenomenon may lie in the highly pleiotropic nature of aneuploidy. The finding from yeast aneuploids was also recapitulated in human cancer cell lines derived from highly aneuploid solid tumors: the growth suppressive effects of a majority of therapeutic compounds also significantly correlated with phenotypic heterogeneity in a positive manner. This correlation predicts that a heterogeneous aneuploidy cancer population may harbor large phenotypic variation under stress, fueling rapid adaptive evolution. Frequent association between genetic heterogeneity and poor prognosis has indeed been observed in cancer clinics (Maley et al., 2006; McGranahan et al., 2012). Chromosomal instability (CIN) in mice also promotes tumor development (Sotillo et al., 2007; Weaver et al., 2007), although the extent by which genetic heterogeneity contribute to tumorigenesis in common mouse cancer models remains to be further explored. Beyond aneuploidy, the wide spectrum and large number of genetic modifications in a given cancer genome are likely to augment the pleiotropic effects on cellular pathways caused by aneuploidy.
Our experimental and theoretical findings argue on a general level that the difficulty of suppressing karyotypically heterogeneous cell populations is rooted in the large adaptive potential in the presence of severe stress. However, our findings do not rule out the possible existence of agents with broad inhibitory effects against aneuploids, especially when the karyotypic space is limited. The principle of ET in fact capitalizes on this notion by reducing karyotypic heterogeneity to an exceeding narrow space through an evolutionary process. In our proof-of-principle experiments in budding yeast, this confined karyotypic space is simply gain of Chr XV, which harbors the “driver mutations” for radicicol resistance (selection), while multiple “passenger” genes on Chr XV confer enhanced sensitivity to hygromycin B, the agent for extinction, when increased in copy number (Figure 7C,D). The complex contribution of gene dosage to hygromycin B sensitivity and the requirement for multiple genes to achieve radicicol resistance predicted that an efficient escape from the ET is not easily attainable through single-gene mutations or copy number changes. Although ET is not presumed to be an omnipotent solution for cancer or fungal infections, we argue that, by accounting for the evolutionary trajectory of a cell population, ET may substantially reduce the risk of evolved drug resistance and disease relapse.
The principle of ET is distinct from the idea of targeting a single characteristic trait of aneuploids. We note that both drugs used in the yeast ET, radicicol and hygromycin B, can perturb proteome homeostasis (Oromendia et al., 2012; Singh et al., 1979; Taipale et al., 2010; Torres et al., 2007), but the responses of aneuploids to these drugs are karyotype-specific rather than uniform. The rationale behind the dual drug treatments associated with ET anticipates evolutionary changes and is thus fundamentally different from the idea of synthetic lethality (Kaelin, 2005; Luo et al.) or the use of dual drugs to target different aspects of a common defect of aneuploidy (Tang et al., 2011). ET drugs are not chosen to interfere with redundant processes but rather collaborate to force and intercept a predicted evolutionary trajectory of the genome, which involves changes in the copy number of a chromosome that links pools of genes responding oppositely to the two different drugs (Figure 7C).
Our experiments with Candida provide a possible ET example where PP, which selects against the gain of i(5L), could enhance the efficacy of fluconazole, the prior selection favoring i(5L) gain. PP is extremely well tolerated in pediatric population for gastrointestinal treatment due to the near-zero absorption rate (Smith et al., 1976). Thus, PP may be used to augment fluconazole if the developed azole resistance is determined to be associated with i(5L) gain.
Our analysis of the cancer pharmacogenomics data suggests that irinotecan may form an ET with EGFR inhibitors against human cancers such as glioblastoma. Unlike other tumor types dependent on EGF signaling, glioblastoma responds poorly to EGFR inhibitors with the erlotinib response rate at < 25% (Taylor et al., 2012). We envision that treatment with an EGFR inhibitor may further select for and thus increase the percentage of Chr7-gained cells within the tumor mass. Supporting this idea, it was reported that EGF limitation selects for Chr7 gain in human neural stem cells (Sareen et al., 2009). By analyzing existing pharmacogenomics data in brain tumor cell lines, we identified irinotecan, a FDA-approved chemical that targets topoisomerase 1, as a drug whose potency significantly correlates with Chr 7 gain. Additional experiments will be required to further confirm this correlation and test the efficacy of an ET formed with a combination of gefitinib or erlotinib with irinotecan against glioblastoma.
Experimental Procedures
Cell culture
Budding yeast (strains listed in Table S1 and plasmids/primers used to construct these strains are listed in Table S2/3) was cultured using standard media. C. albicans cells recovered from frozen glycerol stocks were grown on YPD plates. For bulk liquid culture prior to drug sensitivity assays, SC media with additional 80 mg/L uridine was used. RPMI1640 + 0.165M MOPS buffer + 0.2% glucose without bicarbonate (Lonza) was used for 96-well culture according to the Clinical Laboratory and Standards Institute (CLSI) standard M27-A3. For 384 well cultures, additional 2% glucose was added to accelerate the growth. Agar plates were prepared using RPMI1640 with MOPS without bicarbonate powder (US Biological) supplemented with 2% glucose and 1.5% agarose, following the E-test strip manufacturer’s guideline (bioMerieux).
Yeast growth assays
For continuous OD monitoring (e.g. Figure 3D), cultures were set up in 96-well plates sealed with parafilm in a Tecan M200Pro reader with orbital shaking. OD at wavelength 595 nm was taken every 15 min and analyzed by using the Magellan 7 software (Tecan). For cultures whose OD reading was taken intermittently every several hours (e.g. Figure 5F), the culture plates were placed in non-shaking incubator within a humid chamber before each reading after agitation. The OD at wavelength 595 nm was recorded by using a Tecan M200Pro reader and the data files were processed in R. The growth assays lasted until the fastest growing culture reached saturation, at which time the last OD readings of all strains were recorded.
The drug concentrations of hygromycin B and radicicol were adjusted for different media (e.g., YPD vs SC-ura) and/or the strain background, so that the wild-type control show the same growth delay as was observed for the strain RLY2628 in YPD media containing the stated drug concentration. The growth of genetic variants was normalized to the corresponding wild-type controls.
A comprehensive description of all methods used can be found in Supplemental Information.
Supplementary Material
Acknowledgements
We thank A. Amon for providing the set of disomy aneuploid yeast strains and J. Gerton for sharing haploid yeast RNAseq data. This work was supported by NIH grant RO1GM059964 to R.L.
All screening data (on both budding yeast and C. albicans) will be deposited into Open Data Repository (ODR) of the Stowers Institute for Medical Research and become openly available.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
R.L. and G.C. designed the project. G.C. and W.M. performed most budding yeast experiments. G.C. and H.C. performed most Candida experiments with the help from J.B., A.K., B.R., G.C. and R.L. built the model. G.C. and A.K. performed analyses of the human cancer cell line data. J.C., S.M. and L.W. carried out parts of the budding yeast and Candida screens. W.B. performed qPCR karyotyping. J.H. assisted with the flow cytometry. C.S. assisted with the RNAseq data analysis. G.C. and R.L. prepared the manuscript. R.L. supervised the project.
References
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean J, Brennan C, Shih J-Y, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ. Whole-Genome Comparison Reveals Novel Genetic Elements That Characterize the Genome of Industrial Strains of <italic>Saccharomyces cerevisiae</italic>. PLoS Genet. 2011;7:e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovinskaya MA, Shoji S, Fredrick K, Cate JHD. Structural basis for hygromycin B inhibition of protein biosynthesis. RNA. 2008;14:1590–1599. doi: 10.1261/rna.1076908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredel M, Scholtens DM, Harsh GR, Bredel C, Chandler JP, Renfrow JJ, Yadav AK, Vogel H, Scheck AC, Tibshirani R, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302:261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- Charles JS, Hamilton ML, Petes TD. Meiotic Chromosome Segregation in Triploid Strains of Saccharomyces cerevisiae. Genetics. 2010;186:537–550. doi: 10.1534/genetics.110.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012a;482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Rubinstein B, Li R. Whole chromosome aneuploidy: Big mutations drive adaptation by phenotypic leap. BioEssays. 2012b;34:893–900. doi: 10.1002/bies.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu Andrew W., Mengwasser Kristen E., Sack Laura M., Yoon John C., Park Peter J., Elledge Stephen J. Cumulative Haploinsufficiency and Triplosensitivity Drive Aneuploidy Patterns and Shape the Cancer Genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. New England Journal of Medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Harrison BD, Hashemi J, Bibi M, Pulver R, Bavli D, Nahmias Y, Wellington M, Sapiro G, Berman J. A Tetraploid Intermediate Precedes Aneuploid Formation in Yeasts Exposed to Fluconazole. PLoS Biol. 2014;12:e1001815. doi: 10.1371/journal.pbio.1001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser LM, Sadanandam A, Kuo W-L, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proceedings of the National Academy of Sciences. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, Natarajan P, Koh JLY, Porter J, Gray CA, Andersen RJ, et al. Nat Biotech. Nature Publishing Group; 2009. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds; pp. 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- Infante JJ, Dombek KM, Rebordinos L, Cantoral JM, Young ET. Genome-Wide Amplifications Caused by Chromosomal Rearrangements Play a Major Role in the Adaptive Evolution of Natural Yeast. Genetics. 2003;165:1745–1759. doi: 10.1093/genetics/165.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Wei G, Sevcikova S, Phan V, Jain S, Shieh A, Wong JCY, Li M, Dubansky J, Maunakea ML, et al. Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia. The Journal of Experimental Medicine. 2010;207:2581–2594. doi: 10.1084/jem.20091071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG. The Concept of Synthetic Lethality in the Context of Anticancer Therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Kvitek DJ, Will JL, Gasch AP. Variations in Stress Sensitivity and Genomic Expression in Diverse <italic>S. cerevisiae</italic> Isolates. PLoS Genet. 2008;4:e1000223. doi: 10.1371/journal.pgen.1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AJX, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, Downward J, Szallasi Z, Tomlinson IPM, Howell M, et al. Chromosomal Instability Confers Intrinsic Multidrug Resistance. Cancer Research. 2011;71:1858–1870. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprohon P, Légaré D, Raymond F, Madore É, Hardiman G, Corbeil J, Ouellette M. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Research. 2009;37:1387–1399. doi: 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. The Units of Selection. Annual Review of Ecology and Systematics. 1970;1:1–18. [Google Scholar]
- Llewellyn MS, Rivett-Carnac JB, Fitzpatrick S, Lewis MD, Yeo M, Gaunt MW, Miles MA. Extraordinary Trypanosoma cruzi diversity within single mammalian reservoir hosts implies a mechanism of diversifying selection. International Journal for Parasitology. 2011;41:609–614. doi: 10.1016/j.ijpara.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong K-K, Elledge SJ. A Genome-wide RNAi Screen Identifies Multiple Synthetic Lethal Interactions with the Ras Oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, Paulson TG, Blount PL, Risques R-A, Rabinovitch PS, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- Mannaert A, Downing T, Imamura H, Dujardin J-C. Adaptive mechanisms in pathogens: universal aneuploidy in Leishmania. Trends in Parasitology. 2012;28:370–376. doi: 10.1016/j.pt.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Marichal P, Vanden Bossche H, Odds FC, Nobels G, Warnock DW, Timmerman V, Van Broeckhoven C, Fay S, Mose-Larsen P. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrobial Agents and Chemotherapy. 1997;41:2229–2237. doi: 10.1128/aac.41.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr KA, White TC, van Burik J-AH, Bowden RA. Development of Fluconazole Resistance in Candida albicans Causing Disseminated Infection in a Patient Undergoing Marrow Transplantation. Clinical Infectious Diseases. 1997;25:908–910. doi: 10.1086/515553. [DOI] [PubMed] [Google Scholar]
- McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–538. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo LMF, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- Minning T, Weatherly DB, Flibotte S, Tarleton R. Widespread, focal copy number variations (CNV) and whole chromosome aneuploidies in Trypanosoma cruzi strains revealed by array comparative genomic hybridization. BMC Genomics. 2011;12:139. doi: 10.1186/1471-2164-12-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, J.B.a.M.F.E. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2012 2012. [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network, T.C.G.A.R. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AP, Hyland CD, Metcalf D, Carmichael CL, Loughran SJ, Di Rago L, Kile BT, Alexander WS. Trisomy of Erg is required for myeloproliferation in a mouse model of Down syndrome. Blood. 2010;115:3966–3969. doi: 10.1182/blood-2009-09-242107. [DOI] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, Heitman J. Unisexual and Heterosexual Meiotic Reproduction Generate Aneuploidy and Phenotypic Diversity <italic>De Novo</italic> in the Yeast <italic>Cryptococcus neoformans</italic>. PLoS Biol. 2013;11:e1001653. doi: 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoli I, Coleman JJ, Tempakakis E, An WF, Holson E, Wagner F, Conery AL, Larkins-Ford J, Wu G, Stern A, et al. Identification of Antifungal Compounds Active against <italic>Candida albicans</italic> Using an Improved High-Throughput <italic>Caenorhabditis elegans</italic> Assay. PLoS ONE. 2009;4:e7025. doi: 10.1371/journal.pone.0007025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia AB, Amon A. Aneuploidy: implications for protein homeostasis and disease. Disease Models & Mechanisms. 2014;7:15–20. doi: 10.1242/dmm.013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes & Development. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. Aneuploidy Underlies Rapid Adaptive Evolution of Yeast Cells Deprived of a Conserved Cytokinesis Motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, McMillan E, Ebert AD, Shelley BC, Johnson JA, Meisner LF, Svendsen CN. Chromosome 7 and 19 Trisomy in Cultured Human Neural Progenitor Cells. PLoS ONE. 2009;4:e7630. doi: 10.1371/journal.pone.0007630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Aneuploidy and Isochromosome Formation in Drug-Resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Molecular Microbiology. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. Acquisition of Aneuploidy Provides Increased Fitness during the Evolution of Antifungal Drug Resistance. PLoS Genet. 2009;5:e1000705. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Ursic D, Davies J. Phenotypic suppression and misreading in Saccharomyces cerevisiae. Nature. 1979;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- Sionov E, Lee H, Chang YC, Kwon-Chung KJ. <italic>Cryptococcus neoformans</italic> Overcomes Stress of Azole Drugs by Formation of Disomy in Specific Multiple Chromosomes. PLoS Pathog. 2010;6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TC, Kinkel AW, Gryczko CM, Goulet JR. Absorption of pyrvinium pamoate. Clin Pharmacol Ther. 1976;19:802–806. doi: 10.1002/cpt1976196802. [DOI] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodríguez E, Teruya-Feldstein J, Cordón-Cardo C, Lowe SW, Benezra R. Mad2 Overexpression Promotes Aneuploidy and Tumorigenesis in Mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman J-M, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences. 2013 doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer M, Griffiths E, Blakely KM, Wildenhain J, Ejim L, Rossi L, De Pascale G, Curak J, Brown E, Tyers M, et al. Cross-species discovery of syncretic drug combinations that potentiate the antifungal fluconazole. Mol Syst Biol. 2011;7 doi: 10.1038/msb.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Tang Y-C, Williams BR, Siegel JJ, Amon A. Identification of Aneuploidy-Selective Antiproliferation Compounds. Cell. 2011 doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TE, Furnari FB, Cavenee WK. Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Current cancer drug targets. 2012;12:197–209. doi: 10.2174/156800912799277557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of Aneuploidy on Cellular Physiology and Cell Division in Haploid Yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Ubeda J-M, Legare D, Raymond F, Ouameur A, Boisvert S, Rigault P, Corbeil J, Tremblay M, Olivier M, Papadopoulou B, et al. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biology. 2008;9:R115. doi: 10.1186/gb-2008-9-7-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014 doi: 10.1038/nature13600. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy Acts Both Oncogenically and as a Tumor Suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nature reviews Clinical oncology. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Pavelka N, Bradford WD, Rancati G, Li R. Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast. PLoS Genet. 2012;8:e1002719. doi: 10.1371/journal.pgen.1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.