Abstract
Ubiquitylation is a versatile post-translational modification (PTM). The diversity of ubiquitylation topologies, which encompasses different chain lengths and linkages, underlies its widespread cellular roles. Here, we show that endogenous ubiquitin is acetylated at lysine (K)-6 (AcK6) or K48. Acetylated ubiquitin does not affect substrate monoubiquitylation, but inhibits K11-, K48-, and K63-linked polyubiquitin chain elongation by several E2 enzymes in vitro. In cells, AcK6-mimetic ubiquitin stabilizes the monoubiquitylation of histone H2B—which we identify as an endogenous substrate of acetylated ubiquitin—and of artificial ubiquitin fusion degradation substrates. These results characterize a mechanism whereby ubiquitin, itself a PTM, is subject to another PTM to modulate mono- and polyubiquitylation, thus adding a new regulatory layer to ubiquitin biology.
Keywords: acetylation, mechanism, post-translational modification, ubiquitin
Introduction
Ubiquitylation is an essential post-translational modification (PTM) involved in a variety of biological pathways including protein degradation, signal transduction, DNA repair, and gene regulation 1. Ubiquitin is a 76 amino acid (A.A.) protein that is covalently conjugated to substrates through a cascade involving activating enzymes (E1), conjugating enzymes (E2), and ligases (E3). The widespread biological roles of the ubiquitin system are achieved by the diversity of ubiquitin topologies 2, 3. Ubiquitylation can comprise ubiquitin monomers or polymers. Monoubiquitylation regulates nondegradative events such as gene expression and endocytosis. In addition, ubiquitin can be polymerized through one of its seven lysine (K) residues or its N-terminus within a ubiquitin moiety, resulting in various polyubiquitin chains bearing different linkages. In addition to the best-studied role of K48-chains in the proteasome-mediated protein degradation, the roles of K63-chains in DNA repair and signal transduction, K11-chains in cell cycle regulation, and M1-linked (linear) chains in the NF-κB pathway are being unveiled 2, 4, 5. Thus, diversity in the topologies of ubiquitylation constitutes a “ubiquitin code” that spells out the fate of substrates 3. The ubiquitin code is highly dynamic and restricted to given cellular contexts, as exemplified in the signal-dependent and target-specific assembly of K63-chains or M1-chains.
The integrity of the ubiquitin code is maintained by the cooperation of factors that add, remove, or recognize the code. The addition of specific ubiquitin topologies is mediated by E2 enzymes or by HECT- and RBR-type E3s. CDC34, UBC13-UEV1a, or UBE2S specifically catalyze the elongation of K48-, K63-, or K11-linked chains, respectively 6. UBCH5 mainly initiates the first (mono)ubiquitylation and also synthesizes multiple types of polyubiquitin linkages 7. The trimming of ubiquitin topologies is regulated by certain classes of deubiquitylation enzymes (DUBs), which can remove specific linkages 8. The resulting ubiquitin code is recognized by various decoding proteins that are selective for a specific ubiquitin topology. The hydrophobic patch within the ubiquitin moiety, consisting of Ile44, Leu8, and Val70, is frequently used as the interface with these ubiquitin-binding proteins 9.
Given the importance and complexity of ubiquitylation, we speculated that the ubiquitin code may be regulated by yet other diverse mechanisms. While polyubiquitin chain formation can be viewed as the ubiquitylation of ubiquitin itself, the existence and significance of other ubiquitin-directed PTMs are not well understood. In this study, we searched for possible PTMs on the ubiquitin moiety and found ubiquitin acetylation and phosphorylation. The data revealed a mechanism by which ubiquitin itself is modified by another PTM to regulate the mono- and polyubiquitylation of substrates. Thus, ubiquitin acetylation will add a new layer to the molecular basis of ubiquitin biology.
Results
The identification of acetylation and phosphorylation on endogenous ubiquitin
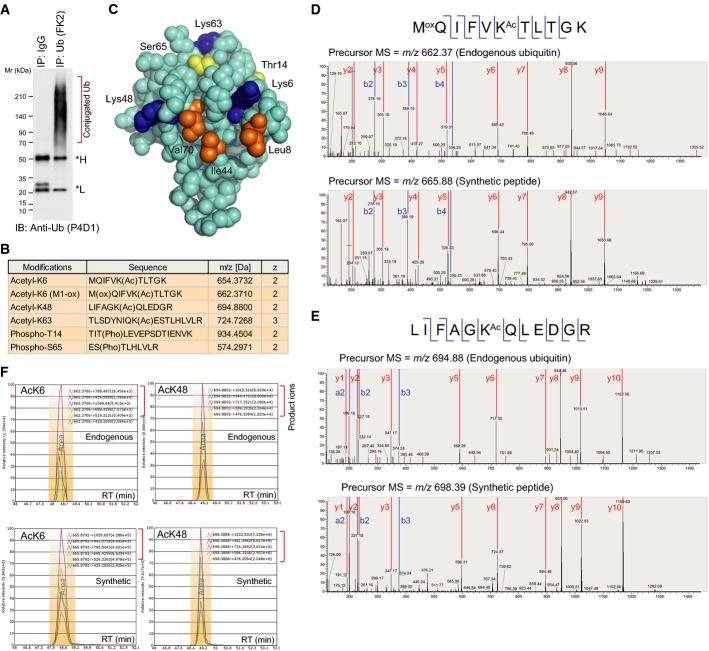
To identify possible PTMs directed to ubiquitin, endogenous ubiquitin conjugated to substrate proteins was immunoprecipitated from cultured human 293F cells (Fig1A). The conjugated ubiquitin (> 70 kD in molecular weight) was subjected to trypsin digestion, and peptides were identified using liquid chromatography–mass spectrometry (LC-MS) 10. A shotgun screening indicated a mass shift corresponding to acetylation at K6 (+42.0106 Da) or phosphorylation at T14 (+79.9663 Da). Next, we performed targeted acquisition of MS/MS spectra for possible acetylation and phosphorylation. As a result, we obtained MS/MS spectra for high-confidence acetylation sites, K6, K48, and K63, and phosphorylation sites, T14 and S65 (Supplementary Figs S1 and S2). These data revealed that ubiquitin was indeed modified by other PTMs (Fig1B).
Figure 1.
- Preparation of conjugated ubiquitin from cell lines. Cell lysates were subjected to immunoprecipitation using an anti-Ub (FK2) antibody. Asterisks indicate the antibody heavy and light chains.
- Summary of the identified PTMs modifying ubiquitin. More details are provided in Supplementary Fig S2.
- Structural view of the ubiquitin modification sites. The images were drawn from PDB 1F9J. Acetylation sites, blue. Phosphorylation sites, yellow. Hydrophobic patch, orange.
- MS/MS spectra identifying acetylation on endogenous ubiquitin at K6 (D) and K48 (E). For each panel, the upper spectra are obtained from cell-derived ubiquitin prepared in (A), and the lower spectra are from synthetic, isotopically labeled AQUA peptides. Identified b and y fragment ions are shown.
- Sample-derived peptides containing acetyllysine at either K6 (left) or K48 (right), and the synthetic, isotopically labeled counterparts co-eluted at the same retention times. The detected fragment ions are listed.
Source data are available online for this figure.
Among the identified ubiquitin PTMs, K6 and K48 residues are located close to the Leu8-Ile44-Val70 hydrophobic patch, while K63, T14, and S65 are at a distance from the patch (Fig1C). We speculated that acetylation of K6 or K48 may modulate interactions involving the hydrophobic patch. Therefore, we explored the molecular function of ubiquitin acetylation at K6 and K48 (hereafter denoted AcK6 and AcK48). To unambiguously confirm K6 and K48 acetylation, standard AQUA (absolute quantification) peptides, harboring acetylation at the target residue and a stable isotope label at a leucine residue (Supplementary Fig S3A) 11, 12, were mixed with endogenous ubiquitin-derived peptides before mass spectrometric analysis. The fragmentation patterns of the AQUA peptides were identical to those of the sample-derived peptides (Fig1D and E). Moreover, extracted ion chromatograms revealed that the corresponding AQUA (heavy) and sample (light) peptides co-eluted at the same retention times (Fig1F). These data successfully confirmed ubiquitin acetylation at K6 and K48.
Ubiquitin acetylation is regulated by multiple classes of histone deacetylases
Next, the levels of AcK6- and AcK48-ubiquitin in cells were quantified using the recently reported parallel reaction monitoring (PRM) approach, an MS/MS-based quantification method using a high-resolution MS instrument 10. We prepared AQUA peptides as internal standards for AcK6, AcK48, and unmodified/GlyGly-bearing peptides, and selected MS/MS fragment ions were used for quantification (Supplementary Fig S3A). Standard curve analyses confirmed that AcK6 and AcK48 could be quantified from 0.1 to 100 fmol (Supplementary Fig S3B and C).
Quantification revealed that endogenous AcK6 and AcK48 represented approximately 0.03 and 0.01%, respectively, of total ubiquitin in 293F cells (Fig2A). These proportions were comparable to those of the M1-linked chains (0.02%), which are known to be dynamically assembled in specific substrates and locations 4. Importantly, ubiquitin acetylation could be quantified from the substrate-conjugated ubiquitin, indicating that “acetyl-ubiquitylation” occurs on substrates. Acetylation was also observed on ubiquitin monomers (Supplementary Fig S3D), suggesting the availability of free acetyl-ubiquitin for ubiquitylation.
Figure 2.
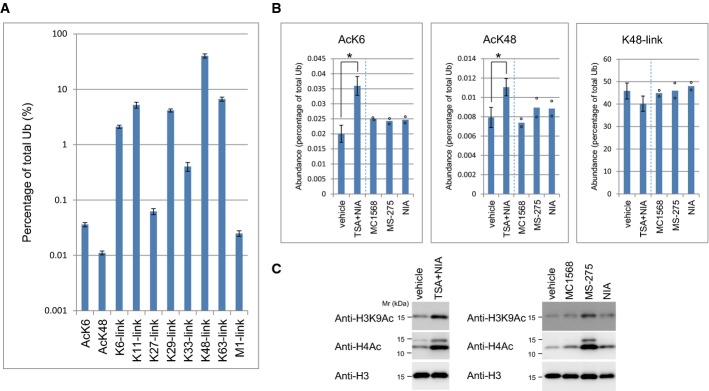
- Cellular abundance of acetylated ubiquitin and polyubiquitin. Ubiquitin conjugates (> 70 kD) immunoprecipitated from 293F cells treated with TSA/NIA and MG132/PR619 were quantified by parallel reaction monitoring (PRM). The K6-locus was used for normalization. Mean ± SD of seven biological replicates is shown.
- Cellular acetylated-ubiquitin levels are upregulated by HDAC inhibitors. Cells treated with the indicated inhibitors were lysed and subjected to anti-Ub immunoprecipitation and subsequent quantification by PRM. Peptide abundance was calculated as a percentage of total ubiquitin. Data represent mean ± SD or individual data points (n = 6, 7, 2, 2, and 2, respectively). *P < 0.01, Student's t-test. TSA/NIA samples are the same as those used in (A).
- Histone acetylation levels. Total lysates were subjected to immunoblotting as indicated.
Source data are available online for this figure.
Combined treatment of 293F cells with trichostatin A (TSA) and nicotinamide (NIA), which inhibit class I/II and class III histone deacetylases (HDACs), respectively, significantly increased levels of AcK6 and, to a lesser extent, AcK48 (Fig2B). By contrast, individual treatment with class-specific HDAC inhibitors, MS-275 (class I), MC1568 (class II), or NIA (class III), only modestly increased ubiquitin acetylation. As expected, histone acetylation was increased markedly by the class I inhibitor, and to a lesser extent by the class II or III inhibitors (Fig2C). These results indicated that ubiquitin acetylation is redundantly regulated by the three classes of HDACs in intact cells.
Functional dissection of ubiquitin acetylation using recombinant acetyl-ubiquitin
As characterized in histones, different PTMs on the same substrate cross talk with each other to constitute the protein PTM code 13. Considering that K6 and K48 residues may modulate ubiquitin-E1/E2/E3 interactions, we asked whether ubiquitin acetylation could modulate its own ubiquitylation.
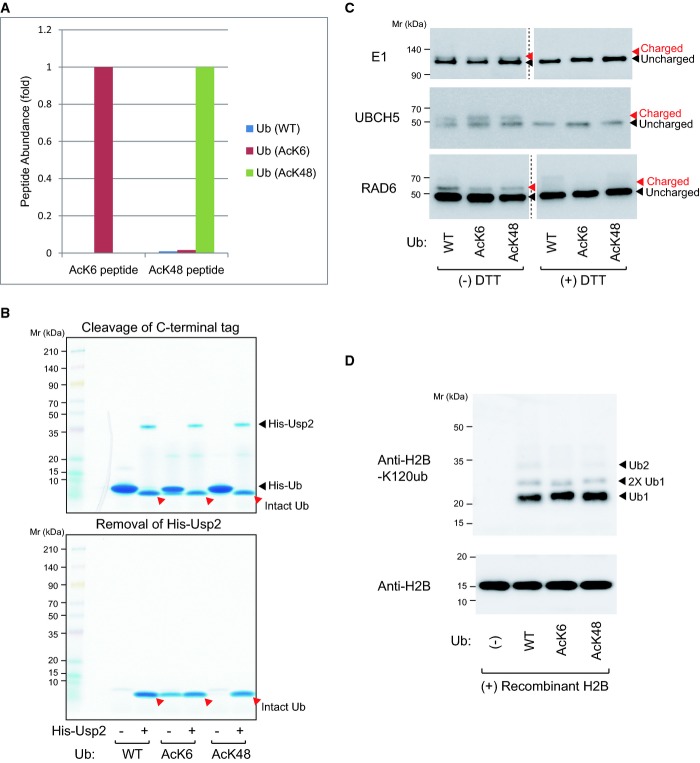
Recombinant acetyl-ubiquitin proteins were synthesized in E. coli with an expanded genetic code. Acetyllysine was incorporated at the positions specified with the UAG codon in the mutant genes 14. MALDI-TOF MS analysis of ubiquitin proteins revealed a mass shift (∽42 Da), confirming that the produced acetylated ubiquitin proteins contained one acetyllysine residue (Supplementary Fig S4A). Subsequent PRM analysis revealed that the recombinant acetyl-ubiquitins were specifically acetylated at the target residues (K6 or K48), while the wild-type ubiquitin was not (Fig3A). The polyhistidine tag at the ubiquitin C-terminus was subsequently removed using USP2 15 to produce intact ubiquitin proteins at near homogeneity (Fig3B).
Figure 3.
- Quantification of the site-specific acetylation of ubiquitin. Acetyllysine is incorporated at the desired residues.
- Purification of intact acetylated ubiquitins. After production of C-terminally His-tagged acetyl-ubiquitin, the tag was cleaved using His-USP2 (upper panel, lanes 2, 4, and 6). Subsequently, His-USP2 and uncleaved His-ubiquitin were absorbed onto Ni+ resin to obtain pure ubiquitin proteins (lower panel, lanes 2, 4, and 6).
- Acetylated ubiquitin retains the ability to be charged to E1/E2. Recombinant ubiquitin is charged to E1 or E2s as indicated. The charging of ubiquitin was analyzed by immunoblotting.
- 3Acetylated ubiquitin can be used to monoubiquitylate histones. Recombinant histone H2B was ubiquitylated by E1, E2 RAD6, and wild-type or acetylated ubiquitin. H2B ubiquitylation was detected by using an anti-H2B-K120ub antibody.
Source data are available online for this figure.
Using the recombinant acetyl-ubiquitins, we first assessed their ability to be charged to E1 or E2 enzymes. In vitro charging assays showed that the charging to E1 (UBA1) and E2 enzymes (UBCH5 or RAD6) was not significantly altered by acetylation (Fig3C). Similarly, we tested the suitability of acetyl-ubiquitins for monoubiquitylation. Since histone H2B is well known as a monoubiquitylation substrate, an H2B ubiquitylation assay was performed using the cognate E2 RAD6 16. Ubiquitylation of H2B occurred almost exclusively as monoubiquitylation, and the levels of monoubiquitylation were not significantly affected by ubiquitin acetylation (Fig3D). These data showed that acetyl-ubiquitins, at least in the contexts we tested, are essentially intact in charging to E1/E2 or acting in monoubiquitylation.
Acetylation of ubiquitin at K6 or K48 represses polyubiquitin chain elongation in vitro
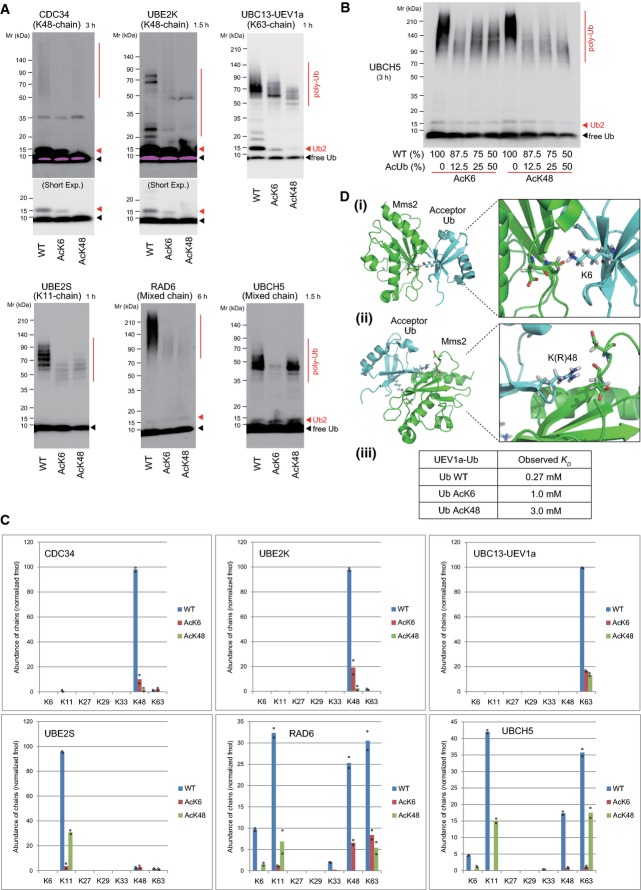
We next asked whether ubiquitin acetylation affected polyubiquitin chain elongation. Chain elongating E2s (CDC34 (K48-link), UBE2K (K48-link), UBC13-Uev1a (K63-link), and UBE2S (K11-link)) and chain-unspecific E2s (UBCH5 and RAD6) 5, 7, 17, 18, 19 were tested in an in vitro ubiquitin chain elongation assay. AcK6 or AcK48 ubiquitins repressed polyubiquitin chain elongation by all of the E2s tested (Fig4A). Assays using less ubiquitin or different antibodies (P4D1) yielded consistent results (Supplementary Fig S4B and C). A titration analysis further demonstrated the inhibitory effect of acetyl-ubiquitin in chain elongation (Fig4B). To mimic nonacetylated lysine or acetyllysine, respectively 20, the ubiquitin K6 residue was replaced with arginine (Ub-K6R) or glutamine (Ub-K6Q). Ub-K6Q similarly inhibited polyubiquitylation (Supplementary Fig S4D).
Figure 4.
- Ubiquitin acetylation represses polyubiquitin chain elongation. E1 and the indicated E2 were incubated with wild-type or acetylated ubiquitin. Polyubiquitin chains were analyzed using an anti-Ub antibody (Dako). Short exposure panels allow the visualization of di-ubiquitin (Ub2).
- Inhibitory effect of acetylated ubiquitin in a titration analysis. A total of 2.0 μg recombinant ubiquitin, comprising a mixture of the indicated percentages of wild-type and acetylated ubiquitin, was subjected to an in vitro ubiquitylation assay with an incubation time of 3 h.
- PRM quantification of polyubiquitin linkages. The assembled polyubiquitin chains in (A) were quantified using AQUA peptides. Mean and individual data points of two independent experiments.
- The ubiquitin K6 (i) or K48 (ii) residue is close to the Mms2 interaction surface. Images were drawn from the PDB 1ZGU. (iii) Observed KD of ubiquitin with UEV1a in SPR analysis.
Source data are available online for this figure.
The linkage-specific regulation of polyubiquitylation was monitored using PRM-based quantification (Fig4C). To quantify K11-chains from AcK6 ubiquitin, we used an AQUA peptide encompassing amino acids 1–26, where K6 was acetylated and K11 was GlyGly-modified (Supplementary Fig S5). Intriguingly, ubiquitin acetylation at K6 or K48 significantly decreased specific chain elongation by CDC34, UBE2K, UBE2S, and UBC13-UEV1a. RAD6 and UBCH5 mainly elongated K11, K48, and K63 chains and, to a lesser extent, K6 chains. These chain elongation activities were also repressed in the presence of either AcK6 or AcK48, although the extent of repression varied depending on the combination of acetylation and ubiquitylation sites.
Based on the published structural analyses, the acetylation sites (K6 and K48) appear to be close to the E2 interaction surface. K6 and K48 of the acceptor ubiquitin are close to the interaction surface of UEV1a (Mms2; Fig4D) 17, 21. Similarly, structural (UBCH5 and UBE2K) or modeling (CDC34 and UBE2S) analyses indicate that the K6 residue of noncovalently bound ubiquitin is located at the E2 interaction surface (Supplementary Fig S6A) 5, 7, 22, 23. Our surface plasmon resonance (SPR) analysis revealed that the affinity of noncovalent interactions between UEV1a and ubiquitin is lower with AcK6 or AcK48 (Fig4D and Supplementary Fig S6B). These insights support our findings that acetylation at K6 or K48 represses E2-mediated polyubiquitin chain assembly.
AcK6 controls mono- or polyubiquitylation of UFD substrates and histone H2B
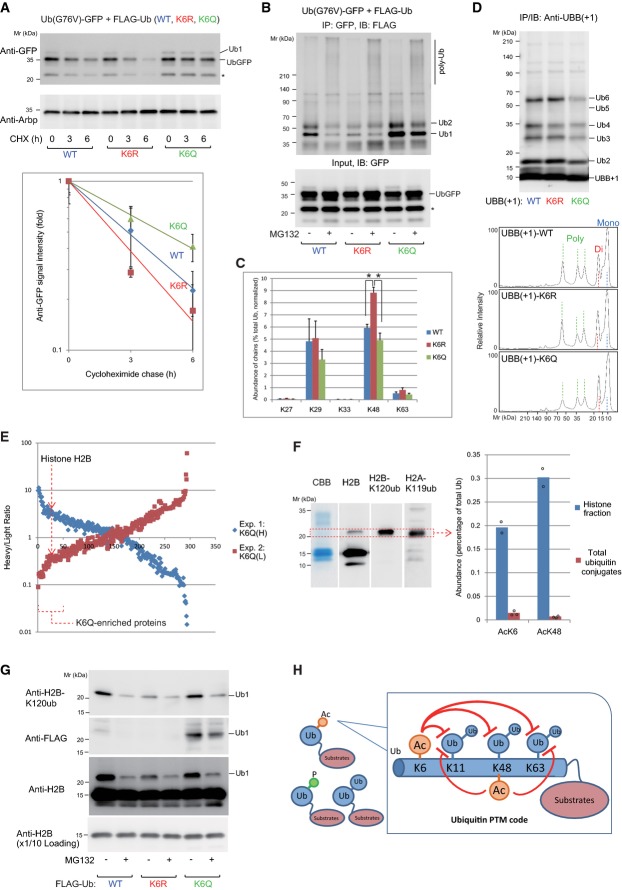
Next, we asked whether ubiquitin acetylation regulates mono- and polyubiquitylation in cells using the ubiquitin fusion degradation (UFD) pathway as a model system 24. Turnover of Ub-G76V-GFP, a model UFD substrate 25, was accelerated by the expression of nonacetylatable Ub-K6R but delayed by that of the acetyl-mimetic Ub-K6Q (Fig5A and Supplementary Fig S7A). Ub-G76V-GFP monoubiquitylated by Ub-K6Q was stabilized, while the levels of total ubiquitin conjugates were not significantly affected by the expression of ubiquitin mutants (Supplementary Fig S7B and C). Consistently, treatment with TSA stabilized Ub-G76V-GFP (Supplementary Fig S7D).
Figure 5.
- Turnover of UbG76V-GFP was delayed by incorporation of the acetylation-mimic K6Q ubiquitin. 293F cells were transfected with UbG76V-GFP together with either wild-type, K6R-, or K6Q-containing FLAG-ubiquitin as indicated, and a cycloheximide chase analysis was performed. Data represent the mean ± SE of quantified UbG76V-GFP band intensities; n = 3. The asterisk indicates a processed form of UbG76V-GFP.
- Incorporation of the acetylation-mimic ubiquitin leads to the accumulation of monoubiquitylated UbG76V-GFP. 293F cells were cotransfected with UbG76V-GFP and the mutant ubiquitins as indicated. Then, the cells were treated with MG132 for 6 h, followed by immunoprecipitation with an anti-GFP antibody. The ubiquitylation of UbG76V-GFP was analyzed using an anti-FLAG antibody. The asterisk indicates a processed form of UbG76V-GFP.
- PRM analysis of UbG76V-GFP prepared as in (B). Data represent mean ± SE; n = 3. *P < 0.05, Student's t-test.
- The acetylation-mimic UBB(+1) represses polyubiquitylation anchored onto UBB(+1). In the lower panel, the band densities were quantified and shown as normalized to the UBB(+1) bands.
- SILAC analysis. FLAG-Ub-K6R or FLAG-Ub-K6Q was expressed in 293F cells cultured in either heavy isotope- or light isotope-containing medium, and cell lysates were immunoprecipitated with an anti-FLAG antibody. The ratio of abundance of each Ub-K6Q/K6R-associated protein is shown from two independent experiments.
- Histones are substrates for acetyl-ubiquitylation. Gel regions corresponding to monoubiquityl-histones were analyzed by PRM. Mean and individual data points; n = 2 for histones, n = 3 for FK2.
- MCF7 cells were transfected with the indicated ubiquitin mutants, and acid-extracted histones were analyzed.
- A proposed model. Acetylation of ubiquitin at K6 and K48 represses polyubiquitylation at K11/K48/K63, constituting the ubiquitin PTM code.
Source data are available online for this figure.
Monoubiquitylated Ub-G76V-GFP was significantly accumulated in cells expressing Ub-K6Q, but decreased in cells expressing Ub-K6R (Fig5B and Supplementary Fig S8A and B). PRM quantification revealed an increase in K48-chains with expression of Ub-K6R (Fig5C). Interaction of the proteasome component C2 (PSMA1) correlated with the levels of polyubiquitylated Ub-G76V-GFP (Supplementary Fig S8C). We also analyzed the ubiquitylation of UBB(+1), a naturally occurring human UFD substrate 22. The ubiquitylation levels of UBB(+1) containing K6Q were decreased as compared to those of UBB(+1) containing K6R or wild-type lysine (Fig5D). These results suggest that AcK6 ubiquitin represses ubiquitin chain elongation in cells.
To identify components of the cellular ubiquitin acetylation-related pathway and its substrates, proteins preferentially interacting with Ub-K6Q were screened using SILAC (stable isotope labeling with amino acids in cell culture; Fig5E). We identified ∽50 proteins that preferentially interacted with K6Q over K6R. Gene ontology analysis of the K6Q-interacting proteins revealed the enrichment of chromosome- or chromatin-related factors, including histone H2B and histone-interacting/-modifying proteins (Supplementary Fig S9). To address whether histones could serve as endogenous substrates for acetyl-ubiquitylation, purified histone fractions were analyzed. We observed a more than tenfold enrichment of AcK6 and AcK48 abundance in the histone fraction versus total ubiquitin conjugates (Fig5F). This indicated that histone H2B, and possibly also H2A, serves as an endogenous substrate of acetyl-ubiquitylation.
Histone H2B is monoubiquitylated at K120 (K120ub), a multi-functional histone PTM regulating histone H3 K4/K79 methylation, transcriptional elongation, and DNA repair response 13, 16. H2B is also polyubiquitylated either at K120 or other residues 26. We found that expression of nonacetylatable Ub-K6R decreased H2B K120ub levels, while that of Ub-K6Q increased them (Fig5G). Because inhibition of the proteasome decreases histone monoubiquitylation to compensate the free ubiquitin pool 27, we could not determine whether the observed stabilization of monoubiquityl-H2B involved the repression of steady-state polyubiquitylation or other mechanisms, such as deubiquitylation. Nonetheless, these results suggested that acetyl-ubiquitin can stabilize the monoubiquitylation state of H2B at certain step(s) in its dynamic regulation.
Discussion
This study revealed that ubiquitin is itself a substrate of yet another PTM. Acetylation at K6/K48/K63 and phosphorylation at T14/S65 were unambiguously identified from endogenous ubiquitin. The identified PTMs are in part consistent with results of other proteomic studies 28. Importantly, these ubiquitin PTMs were identified from ubiquitin fractions conjugated to substrate proteins. This indicates that substrates are modified by acetyl-ubiquitylation or phospho-ubiquitylation. The numerous cellular functions of ubiquitin are achieved by the diversity of ubiquitylation, which includes differences in chain length and linkages 2, 3. Our present findings add PTMs occurring on ubiquitin itself as new factors regulating the diversity of the ubiquitin system.
We found that acetylated ubiquitin was intact in monoubiquitylation but repressed polyubiquitin chain elongation in vitro and stabilized the monoubiquitylation state in intact cells. Our data suggest that acetylation, which neutralizes the positive charges of lysine residues, significantly affects the noncovalent interaction of ubiquitin with E2 enzymes. Moreover, a fraction of ubiquitin may also be acetylated after conjugation to substrates. In such cases, AcK6 and AcK48 may affect ubiquitin interactions with other partners, for example, HECT- and RBR-type E3s in specific chain assembly, various DUBs in chain/substrate-specific processivity 8, and at least 16 classes of ubiquitin-binding domains (UBDs) 9. The protein PTM code, best characterized in histones, dynamically modulates the fate and function of modified proteins 13. Considering that polyubiquitin chain formation represents the ubiquitylation of ubiquitin itself, we propose that ubiquitin is modified by different PTMs that cross talk with each other—acetylation represses ubiquitylation—to constitute a ubiquitin PTM code (Fig5H).
Histone H2B was identified as a substrate for ubiquitin acetylation. Because the global abundance of acetyl-ubiquitin was very low, we speculate that acetyl-ubiquitylation of histones might be locally regulated (e.g. in responsive chromatin regions) to play a role in histone cross talk. Although we could not exclude the possibility of nonenzymatic acetylation of ubiquitin 29, the significant incorporation of acetyl-ubiquitin in monoubiquityl-histone suggests that ubiquitin acetylation is a regulated process.
Independent studies recently reported that ubiquitin S65 phosphorylation activates the E3 activity of Parkin 30, 31, 32. While these reports support our notion that ubiquitin itself is a substrate for other PTMs, our present study revealed that ubiquitin acetylation controls polyubiquitin chain elongation, demonstrating the cross talk of two PTMs (acetylation and ubiquitylation) on ubiquitin. Taken together, the discovery and characterization of ubiquitin PTMs add a new layer to the molecular complexity of the ubiquitin system.
Materials and Methods
Mass spectrometric analyses
Liquid chromatography–mass spectrometry (LC-MS/MS) analyses and quantification of peptides by parallel reaction monitoring (PRM) were performed essentially as previously described 10. The Q Exactive was operated using Xcalibur software (Thermo Fisher Scientific), and fragmentation was performed by HCD (higher energy collisional dissociation). To acquire MS/MS data on peptides of interest, inclusion lists were added in the targeted MS2 mode. Standard peptides: AcK6, MQIFVK[AcK]TL[HeavyL]TGK; AcK48, LIFAGK[AcK]QL[HeavyL]EDGR; and AcK6-ubK11, MQIFVK[AcK]TLTGK[di-GlyGly]TITLEVEPSDTIENV[HeavyV]K, were synthesized by Sigma or Operon. Other unmodified or GlyGly-modified standard peptides were previously described 10. The mass spectrometry datasets are available in PeptideAtlas with identifier PASS00617.
Production of recombinant acetyl-ubiquitin proteins
Ubiquitin variants with acetyllysine were synthesized in E. coli cells expressing a UAG-reading tRNA and an enzyme able to attach the unnatural amino acid to the tRNA 33. The cells were transformed with the variant gene containing UAG in place of a lysine codon at position 6 or 48 and then grown in media supplemented with acetyllysine and nicotinamide at final concentrations of 50 and 5 mM, respectively.
Immunoprecipitation of ubiquitin
Cells were treated with a urea-containing lysis buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1% NP-40, 6 M urea) and sonicated (Handy Sonic, Tomy Seiko). The cell lysates were diluted 10-fold in lysis buffer and then immunoprecipitated with an anti-ubiquitin (FK2) antibody 12.
In vitro ubiquitylation and E1/E2 charging assays
To generate active form of recombinant ubiquitin, 20 μg His-ubiquitin was incubated with 2 μg USP2cc 15 in a deubiquitination buffer (50 mM Tris (pH 7.5), 1 mM DTT) at 37°C overnight. Uncleaved His-ubiquitin and His-USP2 cc were removed from the buffer with Ni-NTA resin (Promega) to yield pure wild-type or acetylated ubiquitin proteins. For the in vitro ubiquitylation assay, 2 μg ubiquitin, 50 ng E1, and the indicated E2 enzymes (400 ng Ubc13, Uev1a, or UBE2S; 800 ng CDC34; 400 ng Rad6; 200 ng UBCH5c; or 400 ng UBE2K) were incubated in a reaction buffer (50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT) at 37°C for the times indicated. For the E1/E2 charging assay, 2 μg ubiquitin, 50 ng E1, and 200 ng of the indicated E2 enzymes were incubated in a reaction buffer at 25°C for 20 or 30 min. Then the samples were divided and boiled in SDS sample buffer either in the presence or absence of 200 mM DTT.
Surface plasmon resonance (SPR) analysis
Analyses were performed using a BIAcore 3000 instrument (GE Healthcare). His-UEV1a protein was immobilized on the surface of a CM5 sensor chip using an amine coupling kit (GE Healthcare). The analyte (wild-type, AcK6, or AcK48 ubiquitin) was injected at different concentrations (5, 10, 20, 40, or 80 μM) at a flow rate of 5 μl per minute 34.
More detailed methods are provided in the Supplementary Methods.
Acknowledgments
We thank Dr. M. Okada, Dr. S. Yokoyama, Dr. K. Iwai, and Dr. K Aisaki for valuable discussions. We are grateful to Dr. Dantuma for the Ub-GFP vectors and Dr. Rohan Baker for the Usp2 vector. This work was supported in part by JSPS KAKENHI (grant numbers 24112004 and 23657112 to F.O., 24112008 to Y.S., 24112005 to T.O., and 26000014 to K.T.) and Platform for Drug Discovery, Informatics, and Structural Life Science grant from MEXT.
Author contributions
FO designed the project, performed most of the experiments, and wrote the paper. YS contributed to the mass spectrometric analysis and provided reagents and experimental advice. HN and TO performed SPR analysis. HT produced proteins. KS and KO optimized the incorporation of acetyllysine and purified acetyl-ubiquitin. KT and JK provided reagents and suggestions. All authors discussed the results and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9
Supplementary Information
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S4
Source Data for Supplementary Figure S7
Source Data for Supplementary Figure S8
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
References
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. “Protein modifications: beyond the usual suspects” review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- Tsuchiya H, Tanaka K, Saeki Y. The parallel reaction monitoring method contributes to a highly sensitive polyubiquitin chain quantification. Biochem Biophys Res Commun. 2013;436:223–229. doi: 10.1016/j.bbrc.2013.05.080. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Phu L, Izrael-Tomasevic A, Matsumoto ML, Bustos D, Dynek JN, Fedorova AV, Bakalarski CE, Arnott D, Deshayes K, Dixit VM, et al. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol Cell Proteomics. 2011;10:M110.003756. doi: 10.1074/mcp.M110.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Madhani HD. Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 2012;13:619–630. doi: 10.1038/embor.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Hibbert RG, Huang A, Boelens R, Sixma TK. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc Natl Acad Sci USA. 2011;108:5590–5595. doi: 10.1073/pnas.1017516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Saltibus LF, Hau DD, Xiao W, Spyracopoulos L. Structural basis for non-covalent interaction between ubiquitin and the ubiquitin conjugating enzyme variant human MMS2. J Biomol NMR. 2006;34:89–100. doi: 10.1007/s10858-005-5583-6. [DOI] [PubMed] [Google Scholar]
- Ko S, Kang GB, Song SM, Lee JG, Shin DY, Yun JH, Sheng Y, Cheong C, Jeon YH, Jung YK, et al. Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity. J Biol Chem. 2010;285:36070–36080. doi: 10.1074/jbc.M110.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadinata R, Holien JK, Yang G, Parker MW, Papaleo E, Sarcevic B. Molecular and structural insight into lysine selection on substrate and ubiquitin lysine 48 by the ubiquitin-conjugating enzyme Cdc34. Cell Cycle. 2013;12:1732–1744. doi: 10.4161/cc.24818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- Geng F, Tansey WP. Polyubiquitylation of histone H2B. Mol Biol Cell. 2008;19:3616–3624. doi: 10.1091/mbc.E08-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Groothuis TA, Salomons FA, Neefjes J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Wagner GR, Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell. 2014;54:5–16. doi: 10.1016/j.molcel.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- Mukai T, Yanagisawa T, Ohtake K, Wakamori M, Adachi J, Hino N, Sato A, Kobayashi T, Hayashi A, Shirouzu M, et al. Genetic-code evolution for protein synthesis with non-natural amino acids. Biochem Biophys Res Commun. 2011;411:757–761. doi: 10.1016/j.bbrc.2011.07.020. [DOI] [PubMed] [Google Scholar]
- Pastushok L, Spyracopoulos L, Xiao W. Two Mms2 residues cooperatively interact with ubiquitin and are critical for Lys63 polyubiquitination in vitro and in vivo. FEBS Lett. 2007;581:5343–5348. doi: 10.1016/j.febslet.2007.10.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9
Supplementary Information
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S4
Source Data for Supplementary Figure S7
Source Data for Supplementary Figure S8
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5