Abstract
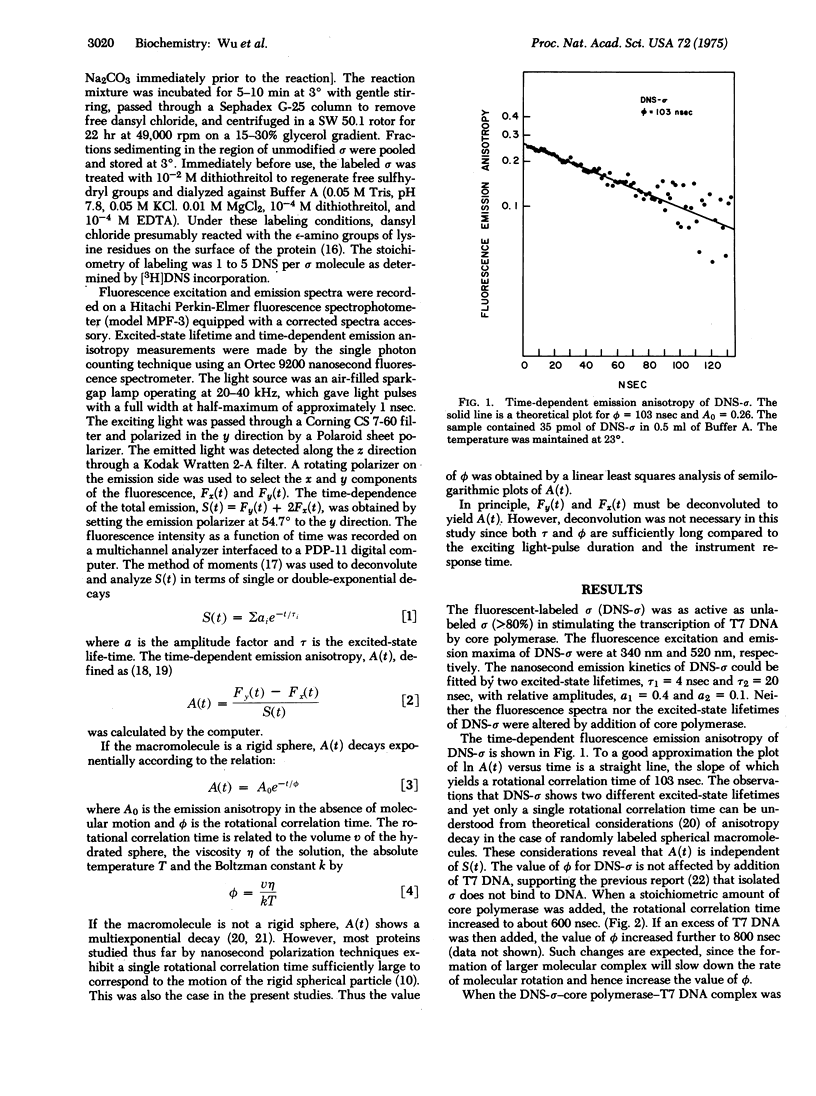
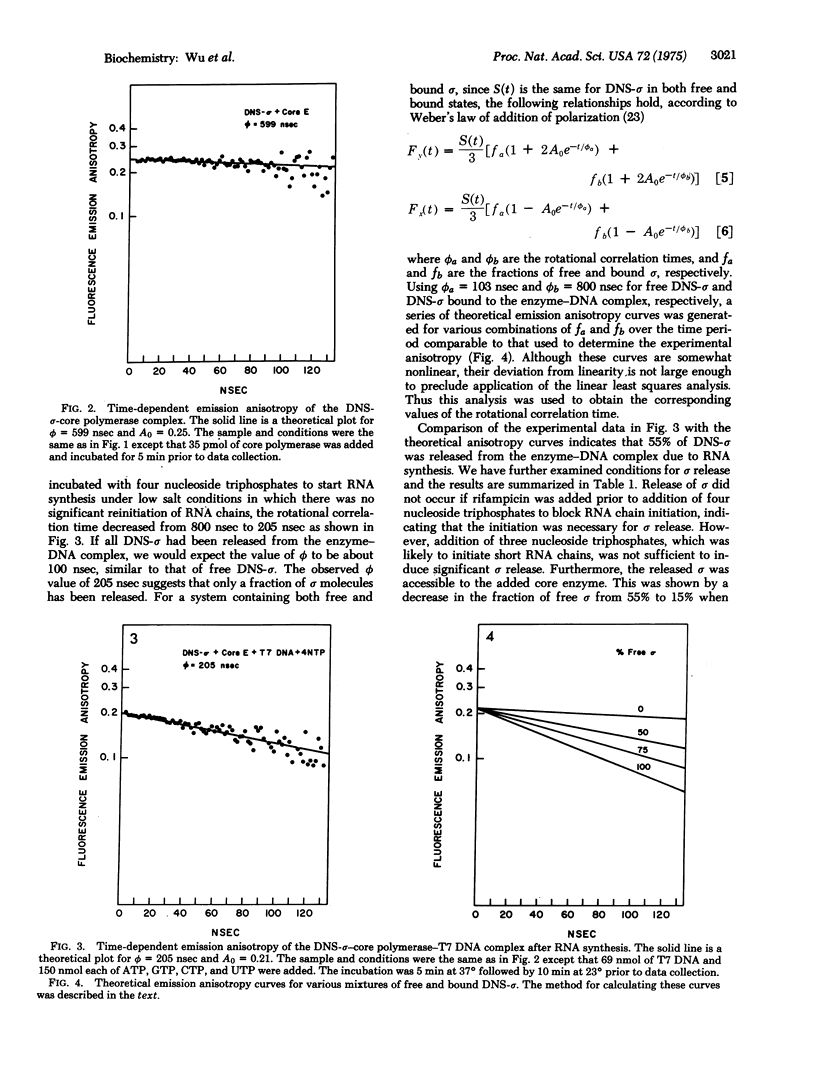
Studies of RNA chain initiation have suggested that the sigma subunit of Escherichia coli RNA polymerase (RNA nucleotidyltransferase; nucleosidetriphosphate: RNA nucleotidyltransferase; EC 2.7.7.6) is released from the enzyme-template complex during transcription and may be reused by another core polymerase. Nanosecond fluorescence depolarization spectroscopy was used to follow the sigma cycle. Isolated sigma subunit labeled with the fluorescent probe dansyl (DNS) chloride bound stoichiometrically to core polymerase and stimulated transcription of phage T7 DNA to the same extent as did unlabeled sigma. DNS-sigma showed an exponential fluorescence anisotropy decay corresponding to a rotational correlation time of about 100 nsec. This value was unaffected by addition of T7 DNA, but increased about 6-fold when core polymerase was added, and increased further when T7 DNA was added. Such increases are expected for the formation of molecular complexes. Using the anisotropy decays for free DNS-sigma and DNS-sigma-core enzyme bound to T7 DNA, we calculated theoretical decay curves for various mixtures of free and bound sigma. Comparison of the observed anisotropy decay with the calculated curves indicated that about 55% of DNA-sigma was released from the enzyme-T7 DNA complex in the presence of four nucleoside triphosphates under low salt conditions. Sigma release did not occur if rifampicin was added prior to addition of four nucleoside triphosphates or if only three nucleoside triphosphates were present. After sigma was released, addition of core polymerase with rifampicin reduced the free sigma to less than 15%, indicating that the released sigma was accessible to the added core enzyme. Thus these studies have provided physical evidence for the sigma cycle during in vitro transcription.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bautz E. K., Bautz F. A., Dunn J. J. E. coli sigma factor: a positive control element in phage T4 development. Nature. 1969 Sep 6;223(5210):1022–1024. doi: 10.1038/2231022a0. [DOI] [PubMed] [Google Scholar]
- Belford G. G., Belford R. L., Weber G. Dynamics of fluorescence polarization in macromolecules. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1392–1393. doi: 10.1073/pnas.69.6.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D., Chamberlin M. Physical studies on ribonucleic acid polymerase from Escherichia coli B. Biochemistry. 1970 Dec 22;9(26):5055–5064. doi: 10.1021/bi00828a003. [DOI] [PubMed] [Google Scholar]
- Burgess R. R. Separation and characterization of the subunits of ribonucleic acid polymerase. J Biol Chem. 1969 Nov 25;244(22):6168–6176. [PubMed] [Google Scholar]
- Burgess R. R., Travers A. A., Dunn J. J., Bautz E. K. Factor stimulating transcription by RNA polymerase. Nature. 1969 Jan 4;221(5175):43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J. The selectivity of transcription. Annu Rev Biochem. 1974;43(0):721–775. doi: 10.1146/annurev.bi.43.070174.003445. [DOI] [PubMed] [Google Scholar]
- Gennis L. S., Gennis R. B., Cantor C. R. Singlet energy-transfer studies on associating protein systems. Distance measurements on trypsin, -chymotrypsin, and their protein inhibitors. Biochemistry. 1972 Jun 20;11(13):2517–2524. doi: 10.1021/bi00763a021. [DOI] [PubMed] [Google Scholar]
- Gerard G. F., Johnson J. C., Boezi J. A. Release of the sigma subunit of Pseudomonas putida deoxyribonucleic acid dependent ribonucleic acid polymerase. Biochemistry. 1972 Mar 14;11(6):989–997. doi: 10.1021/bi00756a007. [DOI] [PubMed] [Google Scholar]
- Isenberg I., Dyson R. D. The analysis of fluorescence decay by a method of moments. Biophys J. 1969 Nov;9(11):1337–1350. doi: 10.1016/S0006-3495(69)86456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakow J. S., Fronk E. Azotobacter vinelandii ribonucleic acid polymerase. 8. Pyrophosphate exchange. J Biol Chem. 1969 Nov 10;244(21):5988–5993. [PubMed] [Google Scholar]
- Maitra U., Hurwitz H. The role of DNA in RNA synthesis, IX. Nucleoside triphosphate termini in RNA polymerase products. Proc Natl Acad Sci U S A. 1965 Sep;54(3):815–822. doi: 10.1073/pnas.54.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn D. E., Clarkson K., Kossman C. R., Stonington O. G. Synthesis of ribosomal RNA on a protein-DNA complex isolated from bacteria: a comparison of ribosomal RNA synthesis in vitro and in vivo. J Mol Biol. 1970 Sep 14;52(2):281–300. doi: 10.1016/0022-2836(70)90031-8. [DOI] [PubMed] [Google Scholar]
- RINDERKNECHT H. A new technique for the fluorescent labelling of proteins. Experientia. 1960 Sep 15;16:430–431. doi: 10.1007/BF02178856. [DOI] [PubMed] [Google Scholar]
- Ruet A., Sentenac A., Fromageot P. On the liberation of sigma and the molecular weight of E. coli RNA polymerase. FEBS Lett. 1970 Dec;11(3):169–171. doi: 10.1016/0014-5793(70)80520-8. [DOI] [PubMed] [Google Scholar]
- Sadowski P. D. Bacteriophage T7 endonuclease. I. Properties of the enzyme purified from T7 phage-infected Escherichia coli B. J Biol Chem. 1971 Jan 10;246(1):209–216. [PubMed] [Google Scholar]
- Smith D. A., Martinez A. M., Ratliff R. L., Williams D. L., Hayes F. N. Template-induced dissociation of ribonucleic acid polymerase. Biochemistry. 1967 Oct;6(10):3057–3064. doi: 10.1021/bi00862a012. [DOI] [PubMed] [Google Scholar]
- Stryer L. Fluorescence spectroscopy of proteins. Science. 1968 Nov 1;162(3853):526–533. doi: 10.1126/science.162.3853.526. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Burgessrr Cyclic re-use of the RNA polymerase sigma factor. Nature. 1969 May 10;222(5193):537–540. doi: 10.1038/222537a0. [DOI] [PubMed] [Google Scholar]
- WEBER G. Polarization of the fluorescence of macromolecules. I. Theory and experimental method. Biochem J. 1952 May;51(2):145–155. doi: 10.1042/bj0510145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F. Y., Wu C. W. Rose Bengal: an inhibitor of ribonucleic acid chain elongation. Biochemistry. 1973 Oct 23;12(22):4343–4348. doi: 10.1021/bi00746a007. [DOI] [PubMed] [Google Scholar]
- Yarbrough L. R., Wu C. W. Role of sulfhydryl residues of Escherichia coli ribonucleic acid polymerase in template recognition and specific initiation. J Biol Chem. 1974 Jul 10;249(13):4079–4085. [PubMed] [Google Scholar]
- Yguerabide J., Epstein H. F., Stryer L. Segmental flexibility in an antibody molecule. J Mol Biol. 1970 Aug;51(3):573–590. doi: 10.1016/0022-2836(70)90009-4. [DOI] [PubMed] [Google Scholar]


