Abstract
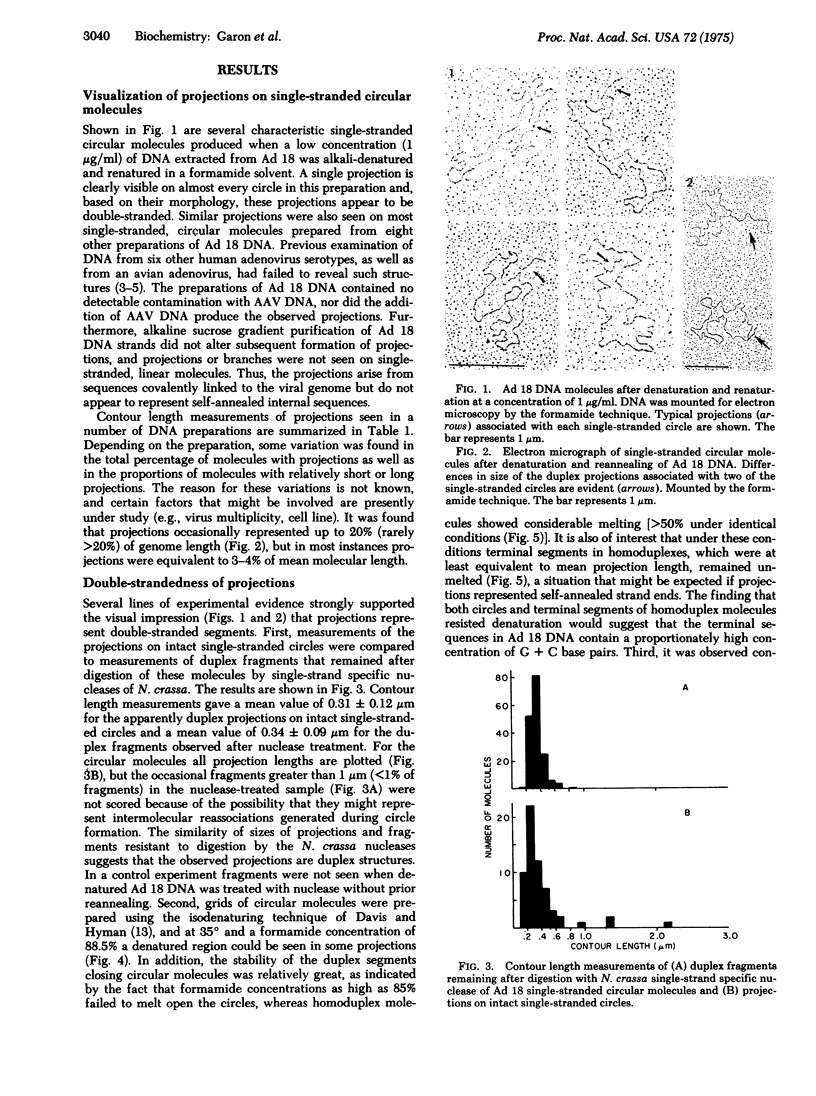
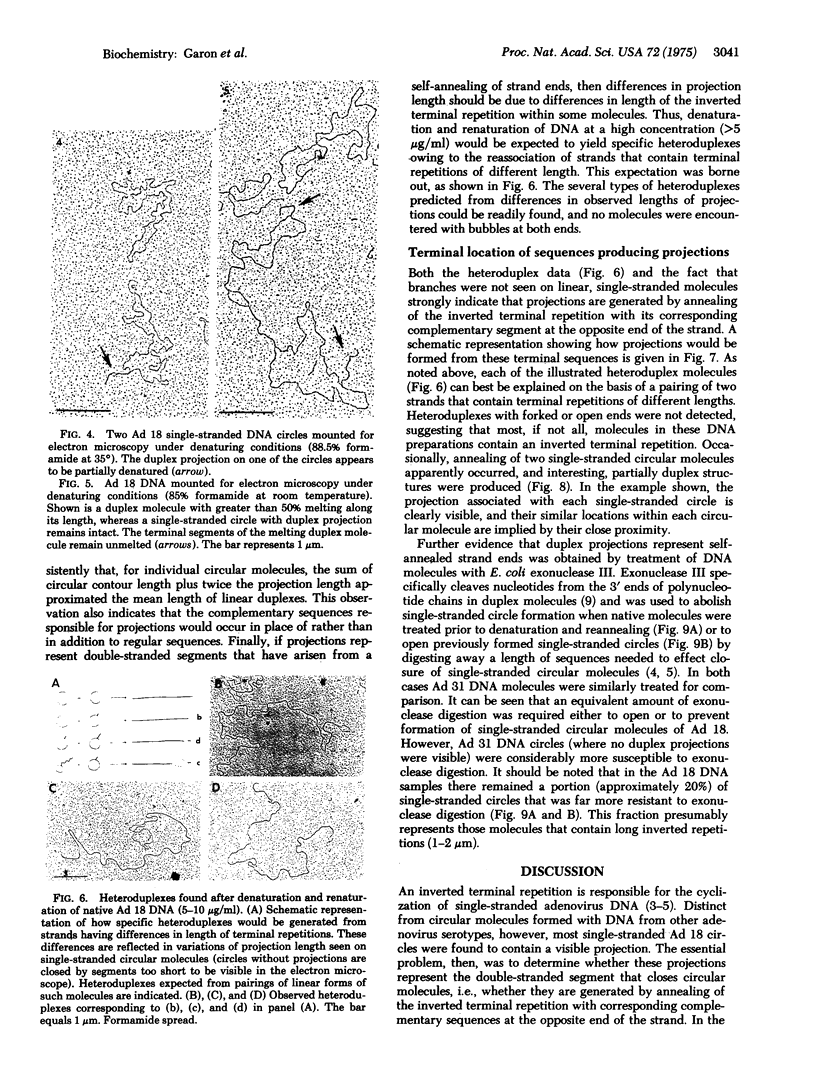
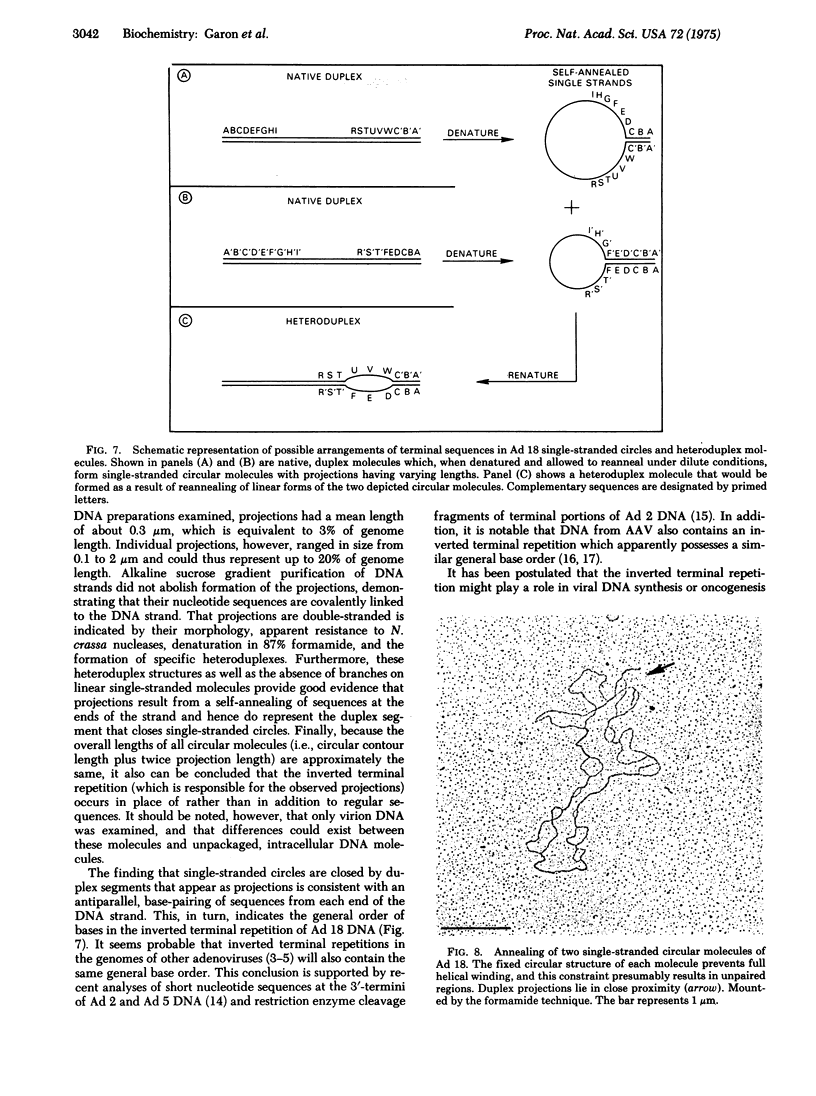
In contrast to the single-stranded circular molecules produced with denatured DNA from other adenoviruses, there was associated with nearly all circular molecules of adenovirus type 18 a visible, duplex projection. These projections had a mean contour length of 0.31 +/- 0.12 mum, equivalent to approximately 3% of genome length. Individual projections ranged in size from 0.1 to 2 mum. Alkaline sucrose gradient purification of single-stranded molecules did not affect formation of these projections, and treatment of a preparation of circular molecules with Neurospora crassa single-strand specific nucleases yielded 0.34 +/- 0.09 mum duplex fragments. Single-stranded circles did not form if a limited number of nucleotides were removed from the 3' ends of native molecules by Escherichia coli exonuclease III digestion prior to denaturation and annealing. In addition, preformed single-stranded circles could be converted to linear molecules by similar treatment. Based on the formation of specific heteroduplex structures when preparations of native DNA were denatured and reannealed and the absence of branches on linear, single-stranded molecules, we conclude that projections are generated by unusually long, inverted terminal repetitions. The repetitious sequences occur in place of rather than in addition to regular sequences. These data provide direct, visual evidence for the arrangement of bases in the inverted terminal repetition of adenovirus DNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berns K. I., Kelly T. J., Jr Letter: Visualization of the inverted terminal repetition in adeno-associated virus DNA. J Mol Biol. 1974 Jan 15;82(2):267–271. doi: 10.1016/0022-2836(74)90344-1. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Garon C. F., Berry K. W., Rose J. A. A unique form of terminal redundancy in adenovirus DNA molecules. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2391–2395. doi: 10.1073/pnas.69.9.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J., Heijneker H. L. Size and location of the transforming region in human adenovirus type 5 DNA. Nature. 1974 Oct 25;251(5477):687–691. doi: 10.1038/251687a0. [DOI] [PubMed] [Google Scholar]
- Green M., Piña M., Kimes R., Wensink P. C., MacHattie L. A., Thomas C. A., Jr Adenovirus DNA. I. Molecular weight and conformation. Proc Natl Acad Sci U S A. 1967 May;57(5):1302–1309. doi: 10.1073/pnas.57.5.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Rose J. A. Simian virus 40 integration site in an adenovirus 7-simian virus 40 hybrid DNA molecule. Proc Natl Acad Sci U S A. 1971 May;68(5):1037–1041. doi: 10.1073/pnas.68.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczot F. J., Carter B. J., Garon C. F., Rose J. A. Self-complementarity of terminal sequences within plus or minus strands of adenovirus-associated virus DNA. Proc Natl Acad Sci U S A. 1973 Jan;70(1):215–219. doi: 10.1073/pnas.70.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills C., Fraser M. J. Different chromatographic forms of Neurospora crassa nucleases specific for single-stranded nucleic acids. Can J Biochem. 1973 Jun;51(6):888–895. doi: 10.1139/o73-110. [DOI] [PubMed] [Google Scholar]
- RICHARDSON C. C., LEHMAN I. R., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. II. CHARACTERIZATION OF THE EXONUCLEASE ACTIVITY. J Biol Chem. 1964 Jan;239:251–258. [PubMed] [Google Scholar]
- Rabin E. Z., Preiss B., Fraser M. J. A nuclease from Neurospora crassa conidia specific for single-stranded nucleic acids. Prep Biochem. 1971;1(4):283–307. doi: 10.1080/00327487108081946. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Arrand J. R., Keller W. The length of the terminal repetition in adenovirus-2 DNA. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3829–3833. doi: 10.1073/pnas.71.10.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. J., Bellett A. J. Complementary strands of CELO virus DNA. J Virol. 1975 Mar;15(3):458–465. doi: 10.1128/jvi.15.3.458-465.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. A., Koczot F. Adenovirus-associated virus multiplication. VI. Base compostion of the deoxyribonucleic acid strand species and strand-specific in vivo transcription. J Virol. 1971 Nov;8(5):771–777. doi: 10.1128/jvi.8.5.771-777.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergh P. H., Sussenbach J. S., Roberts R. J., Jansz H. S. The 3'-terminal nucleotide sequences of adenovirus types 2 and 5 DNA. J Virol. 1975 Feb;15(2):268–272. doi: 10.1128/jvi.15.2.268-272.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson J., Dressler D. Adenovirus-2 DNA contains an inverted terminal repetition. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3054–3057. doi: 10.1073/pnas.69.10.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Eb A. J., van Kesteren L. W., van Bruggen E. F. Structural properties of adenovirus DNA's. Biochim Biophys Acta. 1969 Jun 17;182(2):530–541. doi: 10.1016/0005-2787(69)90205-6. [DOI] [PubMed] [Google Scholar]