Abstract
The endosperm plays a pivotal role in the integration between component tissues of molecular signals controlling seed development. It has been shown to participate in the regulation of embryo morphogenesis and ultimately seed size determination. However, the molecular mechanisms that modulate seed size are still poorly understood especially in legumes. DASH (DOF Acting in Seed embryogenesis and Hormone accumulation) is a DOF transcription factor (TF) expressed during embryogenesis in the chalazal endosperm of the Medicago truncatula seed. Phenotypic characterization of three independent dash mutant alleles revealed a role for this TF in the prevention of early seed abortion and the determination of final seed size. Strong loss-of-function alleles cause severe defects in endosperm development and lead to embryo growth arrest at the globular stage. Transcriptomic analysis of dash pods versus wild-type (WT) pods revealed major transcriptional changes and highlighted genes that are involved in auxin transport and perception as mainly under-expressed in dash mutant pods. Interestingly, the exogenous application of auxin alleviated the seed-lethal phenotype, whereas hormonal dosage revealed a much higher auxin content in dash pods compared with WT. Together these results suggested that auxin transport/signaling may be affected in the dash mutant and that aberrant auxin distribution may contribute to the defect in embryogenesis resulting in the final seed size phenotype.
Keywords: endosperm, seed size, embryogenesis, Medicago truncatula, auxin
Introduction
During seed development, the tissues that surround the developing embryo play important roles in determining both final seed size and composition, as shown by mutant phenotypes in Arabidopsis thaliana, Medicago truncatula and Pisum sativum (Luo et al., 2005; Adamski et al., 2009; D'Erfurth et al., 2012). In most angiosperms, endosperm development is divided into two phases: (1) a syncytial phase with nuclear divisions without cytokinesis immediately after fertilization; and (2) a cellularization phase, which gives rise to three distinct domains: (i) the micropylar region close to the embryo; (ii) the central region forming the largest part of endosperm; and (iii) the chalazal region involved in the transfer of maternal nutrients to embryo and/or endosperm. In contrast with cereals, the endosperm of most dicotyledonous seeds, including legumes, is a transient structure, undergoing cell breakdown during seed filling to leave only a single layer of endosperm cells at maturity. However, whilst being a non-storage tissue at maturity, the endosperm has an important role to play in nourishing the developing embryo (Ingram, 2010; Hehenberger et al., 2012) and thus indirectly affects final seed content and seed size.
Final seed size has been shown to depend on a complex crosstalk between the endosperm and the integuments during early post-fertilization seed expansion (Garcia et al., 2005). Several genes have been shown to be essential for proper development of both endosperm and embryo. For instance, HAIKU2 (IKU) is a leucine-rich repeat receptor-like kinase (LRR-RLK) expressed specifically in endosperm and iku mutants displayed precocious endosperm cellularization and decrease in embryo cell size, resulting in smaller seeds (Luo et al., 2005). Similar phenotypes regarding endosperm cellularization and embryo cell division have been observed in several mutants such as keule (Assaad et al., 1996), knolle (Lauber et al., 1997), and hinkel (Strompen et al., 2002) implying a link between endosperm cellularization and embryo cytokinesis. A recent study showed that the Arabidopsis peptide ligand CLAVATA3/EMBRYO SURROUNDING REGION 8 (CLE8), which is restricted in its expression to the young embryo and endosperm, regulates both embryo cell divisions and endosperm proliferation (Fiume and Fletcher, 2012). Thus, in addition to its conduction of nutrients, the endosperm appears to coordinate via signal transmission the development of the three seed compartments.
Regarding the role and mode of action of plant hormones during early seed/pod development and specifically in the endosperm, reports are scanty. Auxin is important for several major steps of embryo specification in both the apical and basal domains (Möller and Weijers, 2009). The auxin involved appears to be synthesized in the embryo and distributed throughout the embryo in gradients established by the PIN family of efflux carriers (Cheng et al., 2007; Robert et al., 2013). Cytokinin and auxin has been showed to play a role in endosperm differentiation. High cytokinin concentration due to overproduction of isopentenyltransferase (IPT) caused a mosaic aleurone phenotype (Geisler-Lee and Gallie, 2005). Accumulation of auxin due to auxin transport inhibition by N-1-naphthylphthalamic acid (NPA) caused multiple aleurone layers to develop (Forestan and Varotto, 2011). More recently, the CKX2 gene, encoding a cytokinin oxidase 2 involved in cytokinin degradation, has been shown to be a direct target of the IKU pathway controlling early endosperm growth (Li et al., 2013). Moreover, an auxin (IAA)-deficient maize mutant, de-B18, has 30% less seed mass than the WT due to a defect in early endosperm differentiation (Forestan et al., 2010).
In a previous study we identified several seed-specific TF that were expressed in the endosperm tissue of Medicago truncatula (Verdier et al., 2008). Among them was a DNA-binding with One Finger (DOF) protein, which belongs to the zinc finger TF family, widely distributed within the Viridiplantae but absent from other eukaryotes (Moreno-Risueno et al., 2007). The DOF TFs are characterized by a highly conserved DOF domain of 52 amino acids, essential for DNA binding (Yanagisawa, 2002) and in some cases for protein–protein interactions, although the C-terminal is also involved in this interaction (Diaz et al., 2002). The DOF family is implicated in a wide range of processes essential for seed germination, seed maturation and plant development, including plant defences against pathogens (reviewed in Noguero et al., 2013).
In this study, we have functionally characterized a M. truncatula endosperm-specific DOF TF, called hereafter DASH (DOF Acting in Seed embryogenesis and Hormone accumulation) expressed in the developing seed at the transition phase between embryogenesis and seed filling (Verdier et al., 2008). We have exploited two series of mutants, a TILLING resource created in the M. truncatula reference line A17 (Le Signor et al., 2009), and a Tnt1 transposon insertion library, constructed in the R108 line preferred for genetic transformation of M. truncatula (Cheng et al., 2014). Cytological analyses demonstrated that dash mutation severely affects embryo morphogenesis, and the transcriptomes of loss-of-function mutants highlighted severe changes in molecular processes related to early stages of seed/pod development. Most notably, dash pods accumulated high concentrations of auxin despite their retarded development, suggesting a defect in auxin homeostasis.
Results
DASH encodes an endosperm-specific DOF transcription factor
In a previous study (Verdier et al., 2008), a DOF TF gene (Medtr2g014060, DASH) was identified due to its seed expression restricted to the endosperm. DASH relative expression was evaluated in A17 and R108 M. truncatula seeds between 4 and 14 days after pollination (DAP) by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Figure1a). DASH mRNA was first detected at 6 DAP, corresponding to the embryogenesis phase, in which the embryo is at the globular stage and cell-type differentiation begins. mRNA levels increased by 8 DAP (late globular-heart stage) to reach a maximum abundance at 12 DAP (linear cotyledon stage), when after extensive cell divisions (i.e. embryogenesis), seed storage proteins start to accumulate along with embryo cell expansion (i.e. seed filling) (Gallardo et al., 2007). This expression profile suggested a role for this TF during the terminal phase of embryogenesis. Using the comprehensive dataset available at the M. truncatula Gene Expression Atlas web server (MtGEA, http://mtgea.noble.org) (Benedito et al., 2008), we confirmed that DASH expression (probe set Mtr.21255.1.S1_at) was absent in other plant tissues and that this TF was not expressed during the latest stages of seed development (Figure S1a). qRT-PCR experiments on dissected seed and pod tissues at 12 DAP confirmed that DASH was preferentially expressed in seeds compared with pod wall (Figure S1b).
Figure 1.

DASH expression during seed development.(a) DASH relative expression levels in Medicago truncatula A17 and R108 through early seed development (in Days After Pollination, DAP). DASH relative transcript abundance was measured by qRT-PCR. Expression levels were obtained by normalization with the MSC27 and PDF2 genes (Verdier et al., 2008). Data are average values ± standard deviation (SD) from three biological replicates carried out in technical replicates.(b) In situ hybridization of 12 DAP seed using a DASH-specific anti-sense probe. The signal (purple colour) is located in the chalazal region of the endosperm (arrowheads). Negative control with the sense probe did not yield to any signal in the endosperm. Dotted squares show background in hilum region, found with both sense and anti-sense probes. Endosperm and embryo are indicated respectively by Eo and Emb and by arrows. Scale bars = 250 μm.
Further exploration of DASH expression was carried out by in situ hybridization on 12 DAP A17 seeds. The endosperm appeared split in two parts: one attached to the seed coat and another surrounding the embryo, as previously observed for early developing M. truncatula seeds (D'Erfurth et al., 2012), the endosperm being delicate and hence difficult to keep intact during steps of tissue preparation for in situ hybridization. DASH expression was restricted to a specific zone of the endosperm corresponding to the chalazal region, close to the hilum, which corresponds to the point of attachment of the seed to the pod (Figure1b).
A distance analysis between DOF proteins from M. truncatula (Noguero et al., 2013) and other species was carried out using the neighbor-joining method. The corresponding tree divided into eight clusters (A–H; Figure S2), which partially match with the classification of Arabidopsis DOF proteins (Yanagisawa, 2002). DASH protein, which belongs to group E, clustered with several soybean and pea DOF TFs. Interestingly none of the members of this group currently has a function ascribed to them.
dash loss-of-function alleles have a seed-lethal or near-lethal phenotype
Two M. truncatula mutant populations were screened to identify mutations in DASH. From the EMS population (le Signor et al., 2009), line EMS109 (G177A) was characterized by the apparition of a stop codon (W59STOP) just after the loop within the DOF domain (Figure S3). This mutation causes a dramatic change in protein sequence by truncation of the DOF domain, resulting in a predicted peptide length of only 59 amino acids instead of 336 for the full-length protein sequence. In addition, two insertion mutant lines were identified from the Tnt-1 insertion mutant population generated in the R108 genotype (Tadege et al., 2008). These insertions were located in the putative promoter region, at −342 bp (mutant NF5285) and at −151 bp (NF6042) upstream of the ATG start codon (Figure S3).
Phenotypes of the different mutants compared with their respective WT lines were studied. While NF6042 and EMS109 exhibited seed-lethal phenotypes, NF5285 displayed a mild seed phenotype in the homozygous state with a reduction of 16% of the mature seed weight (Table1).
Table 1.
Phenotype description of the EMS and Tnt1 dash mutants
| Mutant | Genotype | Seed weight (mg) | Seed weight reduction/ WT (%) | Pod weight (mg) | Number of seeds per pod |
|---|---|---|---|---|---|
| EMS109 | dash/dash a | 1.43 ± 0.042* | 67 | 35.5 ± 1.54* | 6 ± 0.23* |
| dash/DASH | 3.99 ± 0.057* | 9 | 61.1 ± 1.84* | 6 ± 0.28* | |
| DASH/DASH | 4.37 ± 0.072 | 16 | 73.8 ± 2.07 | 8 ± 0.24 | |
| NF5285 | dash/dash | 3.87 ± 0.187* | 10 | 127.2 ± 5.12* | 6 ± 0.3* |
| DASH/DASH | 4.61 ± 0.103 | 169 ± 8.25 | 7 ± 0.3 | ||
| NF6042 | dash/DASH b | 3.53 ± 0.07* | 71.58 ± 5.1 | 5 ± 0.45* | |
| DASH/DASH | 3.94 ± 0.05 | 76.8 ± 3.9 | 8 ± 0.24 |
Seed production in the late stage of the plant life cycle.
Homozygous has a seed-lethal phenotype.
Significant value relative to wild-type (P < 0.05, Student's t-test).
Heterozygous plants with the NF6042 insertion located at −151 bp were analysed by PCR-based genotyping and no homozygous plant could be recovered from the progeny. The observed 2:1 segregation ratio between heterozygous and WT plants among the progeny of self-fertilized heterozygous plants suggested that NF6042 insertion was embryo-lethal at the homozygous state (Figure S4a). To investigate this further, young developing pods of self-fertilized NF6042 heterozygous plants were dissected and small and/or dry seeds were observed within pods in a proportion close to 3:1 (Figure S4b), consistent with Mendelian segregation of a single recessive allele and confirming homozygous seed-lethality. The observed reduction of seed number in mature pods from self-pollinated heterozygotes is also consistent with the segregation of a seed-lethal allele. The same genetic analysis was performed for the EMS109 segregating population and led to similar results (Figure S4a,b). However, one homozygous plant was obtained out of 100 progenies from self-fertilized heterozygous EMS109 plants. Compared with the WT, we observed that the EMS109 mutation in the homozygous state caused pod abortion during the first months of the plant life cycle: flowers appeared normally, but after 10–12 days small developing pods aborted, dried out and detached (Figure2a). These pods were much smaller than the WT pods at the same stage and contained smaller seeds (Figure2b,c). Interestingly, at the end of the plant life cycle (after 2 or 3 months of flowering), EMS109 homozygous plants were able to produce mature pods, which were smaller than WT but contained viable seeds. The weight of these seeds was greatly reduced compared with WT (67%; Table1 and Figure2d) and the number of seeds per pod was also reduced compared with WT plants (Table1). These small seeds gave rise to abnormal cup-shaped cotyledon phenotypes in about 40% of cases (Figure2e), and most of these abnormal seedlings did not give a viable plant.
Figure 2.
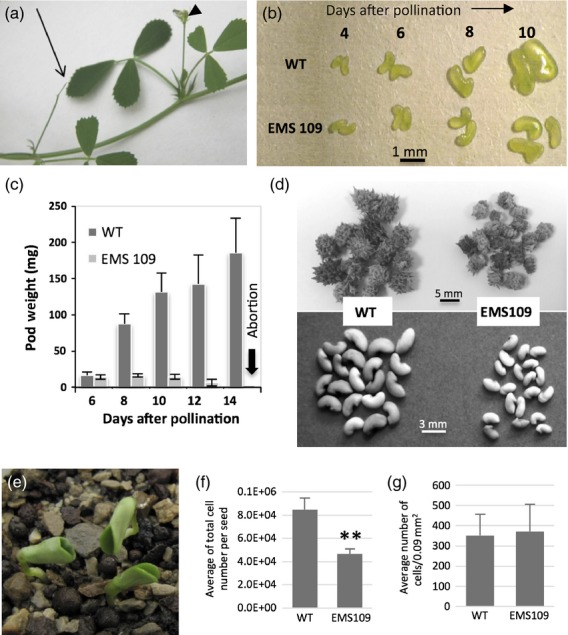
Phenotype of the homozygous EMS109 mutant.(a) Pod abortion during the first months of the plant life cycle.(b) Seed development from 4 DAP to 10 DAP for wild-type (top) and EMS109 mutant (bottom).(c) Kinetics of pod weight from 6 DAP to 12 DAP for wild-type and EMS109.(d) Comparison of mature pod and seed sizes from WT (left) and EMS 109 mutant (right) at the end of the plant life cycle.(e) Abnormal mutant seedlings showing cup-shaped cotyledons.(f) Cell number per seed from WT and EMS 109 mutant at the mature stage.(g) Cell size calculated by the number of cells per 0.9 square mm from WT and EMS 109 mutant at the mature stage.
DASH expression analysis in different mutant lines using qRT-PCR revealed a dramatic decrease of DASH expression in EMS109 mutant seeds (i.e. null mutant, Figure S4d) and a slight decrease in NF5285 mutant seeds at 10 DAP (i.e. knock-down mutant, Figure S4e). No homozygous plant was isolated for the NF6042 line but a decrease of 40% of DASH expression in seeds dissected from heterozygous NF6042 pods was measured by qRT-PCR compared with WT plants at the same stage (Figure S4f)
In order to definitively link the seed phenotype to the dash mutation, complementation of the dash mutant was carried out using the EMS109 mutant line. A construct containing the native promoter sequence fused to the open reading frame of DASH was introduced into the homozygous EMS109 mutant line using Agrobacterium tumefaciens. Among second generation transformant lines containing the pDASH::DASH construct selected in the presence of the selection marker, two lines (i.e. EMS109 Compl A and EMS109 Compl B) displayed complementation of the seed-lethal phenotype and a restoration of the normal seed weight (Figure S5). These complemented lines did not show any defective reproductive phenotypes in the next generations such as those observed in EMS109 mutants.
dash mutants are impaired in embryogenesis
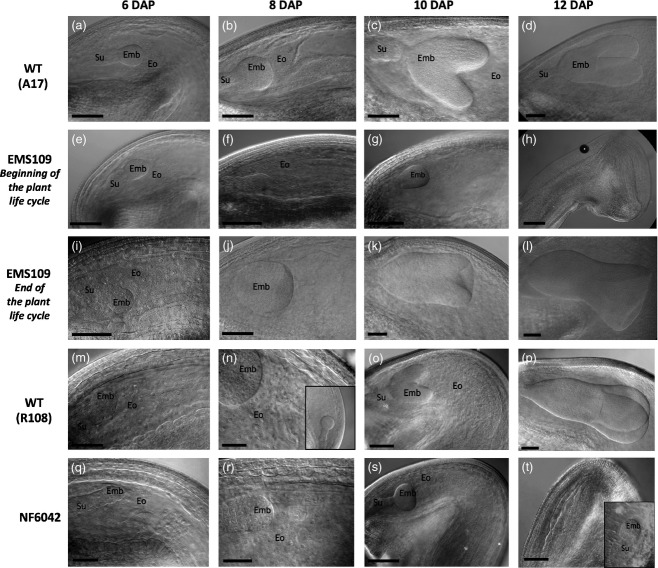
The seed-lethal phenotype prompted us to study in more detail embryo development of the dash mutants. Seeds were dissected from developing pods, cleared and observed using differential interference contrast (DIC) microscopy. A developmental series of WT A17 (A–D) and EMS109 mutant (E–L) cleared seeds is presented in Figure3. Figure3(e–h) shows mutant seeds before abortion harvested from 5-week-old plants (i.e. beginning of the plant cycle) while Figure3(i–l) show seeds harvested on plants at the end of their life cycle that were ultimately able to produce small mature seeds. For seeds harvested from 5-week-old plants, no change was observed in embryo development until 6 DAP (Figure3a,e); whereas at 8 DAP, we observed a delay in embryo development (Figure3b,f), coinciding with the beginning of DASH gene expression. At 10 DAP, WT embryos reached heart stage whereas EMS109 embryos were still at the globular stage, attesting embryo growth arrest (Figure3c,g). Cellularized endosperm appeared between 6 and 10 DAP in WT but gradually degenerated in EMS109 mutants (Figure S6) leading to small seeds, tissue collapse and desiccation (Figure3h). The smallest seeds from developing heterozygous NF6042 pods (Q–T) were also cleared and compared with WT R108 seeds (M–P) in Figure3 and similar results as those seen for EMS109 were observed, with a delay in embryo development from 8 DAP resulting in embryo growth arrest at the globular stage. Regarding the weak allele NF5285, we observed a slight delay in embryo development from 10 DAP onward (Figure S7).
Figure 3.
Embryo growth arrest in EMS109 and NF6042 mutants.Cleared pictures of embryo and endosperm during early seed development. Wild-type A17 seeds (a–d), EMS109 seeds at the beginning of plant life cycle (e–h) and EMS109 seeds escaped from abortion at the end of the plant life cycle (i–l).Wild-type R108 seeds (m–p) and small seeds dissected from heterozygous NF6042 pods (q–t). Emb, embryo; Eo, endosperm; Su, suspensor. Scale bar = 100 μm except for pictures (i, m, n, q, r) (50 μm).
The EMS109 mutant seeds collected at the end of the plant life cycle, that escaped abortion, differed from WT seeds by their smaller size, as previously shown in Figure2d, and their fused cotyledons (Figures2e and 3k,l). By counting the number of embryo cells per square mm in the EMS109 mutant mature seeds that escaped abortion, we did not observe any variation in cell size compared with the WT. However, the total number of cells per seed determined using a haemocytometer after enzymatic digestion was significantly reduced. These results suggest that the number of cell divisions decreased in the mutant embryos, whereas cell elongation was not affected (Figure2f,g).
A transcriptome comparison of WT and EMS109 mutant pods reveals major transcriptional changes in the dash mutant
To get an insight into the processes affected in developing dash seeds, we used the M. truncatula Affymetrix GeneChips for hybridization of RNA extracted from the WT and EMS109 mutant pods at 8 DAP (about 3–4 days before pod abortion). At this stage, both dash and WT embryos are in globular stages with dash embryos being slightly smaller. Following log transformation of the data and t-test correction, a probe set list of 7452 genes differentially expressed between the dash mutant and the WT were selected: 3766 were down-regulated and 3686 up-regulated. The exhaustive list of these genes is presented in Table S1. Schematic pageman representation of under- and over-represented BIN classes [i.e. functional classes (Thimm et al., 2004)] revealed major effects on most metabolic pathways (Table2).
Table 2.
pageman display of over-represented categories deregulated in 8 DAP dash pods compared with WT. Log2 ratio of mutant data versus WT data from three replicates. Only classes with significant changes according to the normalized Wilcoxon test (= score) are represented. Data from the Medicago Gene Expression Atlas (Benedito et al., 2008) were normalized and added
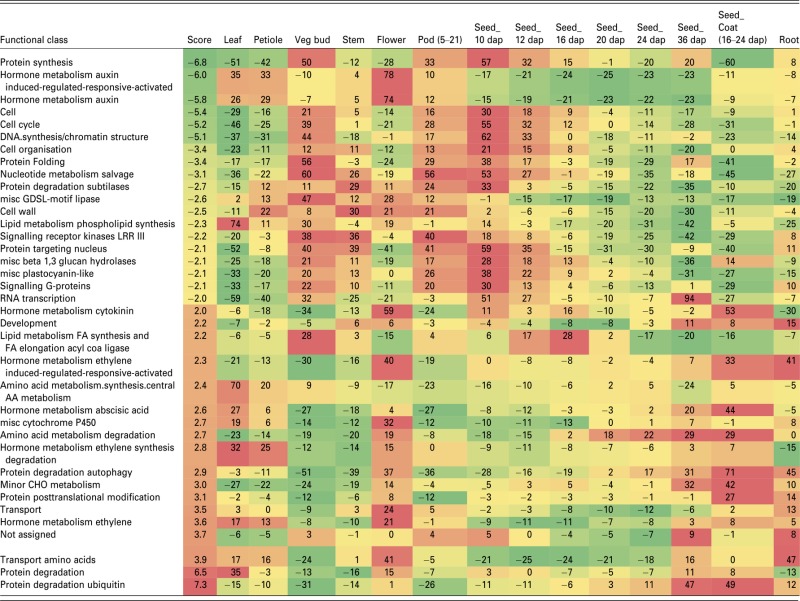 |
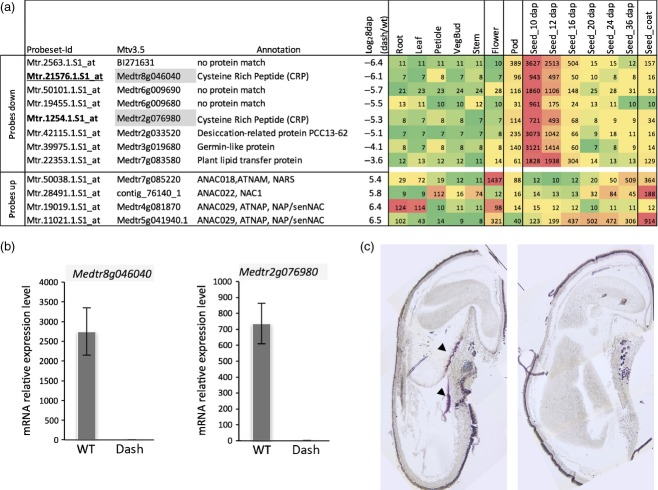
Protein synthesis (mainly ribosomal proteins), auxin pathways and auxin response genes, cell cycle (cyclin-dependent protein kinase regulator) and chromatin structure (nucleosome assembly, DNA binding or DNA replication factor and histone synthesis) were the most down-regulated functional classes in dash (score ≤5, Table2). Besides the down-regulation of specific metabolic pathway/classes, we searched for the most strongly down-regulated genes in dash (log2 expression ratio between dash and WT ≤3.5). A smaller number of genes (12) was identified, of which eight displayed an expression pattern similar to DASH (i.e. seed specific with an expression peaking between 10 and 12 DAP; Figure4a). Two of them (Mtr.1254.1.S1_at, Mtr.21576.1.S1_at) corresponded to short cysteine-rich proteins (CRPs). Down-regulation of these two genes was confirmed in dash seed tissues at 8 DAP by qRT-PCR (Figure4b) and expression of the most deregulated one (Mtr.21576.1.S1_at) was shown by in situ hybridization to be restricted to the chalazal endosperm, a pattern identical to that of DASH (Figures1b and 4c). Although the other three genes (Mtr.19455.1.S1_at, Mtr.2563.1.S1_at and Mtr.50101.1.S1_at) lacked clear annotations, sequences analysis revealed that they encode small peptides possessing signal peptides and several cysteine residues classifying them as CRPs. A further three other genes with expression patterns similar to DASH were down-regulated by 15- to 35-fold in dash pods compared with WT and encode non-specific lipid-transfer protein ns-LPT-like (Mtr.22353.1.S1_at), germin-like (Mtr.39975.1.S1_at) and desiccation-related protein (DRP; Mtr.42115.1.S1_at) (Figure4a).
Figure 4.
A cysteine-rich peptide (CRP) gene expressed in the chalazal endosperm is strongly repressed in the dash mutant seeds.(a) List of the most down-regulated and up-regulated genes in 8 DAP dash pods compared with WT. Tissue expression data from the Medicago truncatula Gene Expression Atlas were added.(b) Down-regulation of the Mtr.21576.1.S1_at and Mtr.1254.1.S1_at probes set (CRP genes Medtr8g046040 and Medtr2g076980 respectively) in dash seeds compared with WT by qRT-PCR. ACTIN PDF2 and GAPDH were used as reference genes for expression levels normalization.(c) In situ hybridization of the CRP gene Medtr8g046040 in the chalazal endosperm (arrowheads) of WT 12 DAP seeds. Hybridization with the control sense probe (right) did not yield to any signal.
The most up-regulated functional classes in dash were protein degradation (mainly E3 ubiquitin ligases) and transport (mainly amino acid transporters orthologous to amino acid permeases, cationic amino acid transporters and proline transporters). Among the 10 most up-regulated genes, four of them corresponded to genes encoding NAC [NAM (no apical meristem), ATAF, CUC (cup-shaped cotyledon)]-domain TFs that were 40–90-fold up-regulated in dash (log2 ratio between 5.4 and 6.5, Figure4a). These TFs are normally expressed in WT flowers and/or during seed filling (16–36 DAP) but are weakly expressed in early stages of seed development (Figure4a), suggesting they could be indirectly repressed by DASH.
Auxin homeostasis and transport are impaired in dash mutants
Genes related to the functional class ‘hormone’, and particularly ‘auxin’, were among the most deregulated in the dash mutant (Table2). Interestingly, most of the auxin-related genes were down-regulated in dash and notably 65% of them encode auxin-induced proteins (Table S2). Homologs of the auxin efflux carriers AtPIN1 (MtPIN4), AtPIN7 (MtPIN1 and MtPIN3) and AtPIN8 (MtPIN8) were also down-regulated in dash. Consistent with these results, the Aux/IAA family was down-regulated in dash. Interestingly, among the few auxin-responsive genes up-regulated in dash was IAA18-LIKE, which alters PIN1 expression in the Arabidopsis embryo (Ploense et al., 2009) Finally, it is noteworthy that the genes related to auxin synthesis were not significantly deregulated in dash suggesting that auxin synthesis is not impaired in dash pods. In order to better understand the link between the dash mutation and the deregulation of auxin-responsive genes observed in the mutant transcriptome, auxin content was measured in EMS109 mutant and WT pods using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). At 8 DAP, the stage used for transcriptomic analysis, IAA content was about 36 times higher in dash pods compared with WT (Figure5a). Higher IAA content was still observed at 10 DAP in mutant pods, just before pod abortion occurring around 12 DAP. Interestingly, we also observed a higher accumulation of the auxin conjugate in 8 and 10 DAP mutant pods compared with WT, which corresponds to an inactive form of auxin potentially involved in IAA catabolism (Figure5b) (Ludwig-Müller, 2011). Similar observations were made for the NF5285 mutant pods, which possessed 11-fold higher auxin content than WT pods (Figure S8).
Figure 5.
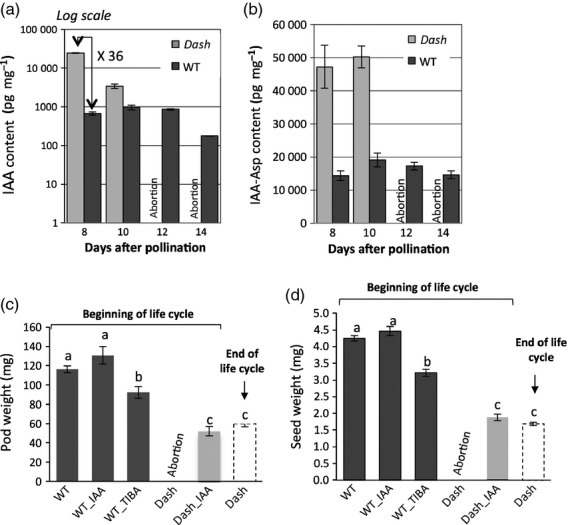
Hormone dosage and treatment effect on pods of EMS109 versus WT.IAA (a) and IAA-Asp (b) dosages in pods of EMS109 and WT. Effect of exogenous IAA and TIBA treatments on pod (c) and seed (d) weights of EMS109 and WT.
In view of the abnormal accumulation of auxin in mutant pods during embryogenesis, we tested the effects of exogenous IAA and TIBA (2,3,5-triiodobenzoic acid) treatments to assess the integrity of IAA polar transport/signaling in mutant lines. Exogenous treatments with IAA on 6 DAP mutant pod led to rescue of the lethal phenotype but only partially complemented the seed/pod size phenotype (Figure5c,d). These treatments mimicked the partial restoration of WT phenotype observed at the end of plant life of EMS109. No significant changes were observed when applying these treatments to WT plants. When applying TIBA, an inhibitor of auxin efflux transport, however, we observed a significant decrease of mature pod and seed weights, suggesting that auxin is required for normal seed development and disturbed auxin transport could be a partial explanation of impaired EMS109 seed development. TIBA treatments of WT pods mimicked the dash phenotype while exogenous IAA restored pod and seed set, consistent with our transcriptomic results that in the mutant pods auxin efflux transport (i.e. the PIN transporters) was impaired.
Discussion
DASH, an integrator between endosperm and embryo development
The M. truncatula gene DASH, which encodes a TF from the DOF family, is specifically expressed in the endosperm during embryogenesis (Figure1). Mutations within the DASH sequence resulted in altered embryo growth (Figure3), either resulting in growth arrest at the globular stage and premature pod abscission (NF6042 and EMS109) or smaller seed size (NF5285). This result is reminiscent of previous data reporting that endosperm cellularization and embryo development are closely related, and probably link via nutrient translocation to determine final seed size (Hehenberger et al., 2012). Contrary to the mini3 mutant of Arabidopsis, which shows precocious endosperm cellularization probably responsible for a reduced seed size (Luo et al., 2005), cellularized endosperm is present in the dash mutant at 6 DAP and 8 DAP (Figure3e,f) but gradually degenerates, leading to tissue collapse, desiccation and finally to abortion (Figures3g,h and S6). EMS109 mutant plants progressively acquire the ability to develop pods and set seeds, albeit small ones, suggesting a partial physiological reversion of the mutant phenotype at the end of plant life (Figure2d). We hypothesize here that a physiological switch occurs to reverse the phenotype at the end of plant cycle, and probably linked to a change in regulation of auxin transport or perception. Interestingly, dramatic switches in biosynthesis, metabolism, signaling and response genes controlling hormone homeostasis have already been demonstrated between leaf development and early leaf senescence (van der Graaff et al., 2006).
The exclusive localization of the DASH mRNA in the chalazal zone endosperm (CZE) is of particular interest (Figure1b). In Arabidopsis, this endosperm region is highly enriched for mRNAs specifically expressed during early seed development (Belmonte et al., 2013). The authors identified transcripts for ubiquitin-dependent protein catabolism genes and enzymes for the biosynthesis of gibberellic acid, abscisic acid, and cytokinin, consistent with other reports showing that hormone metabolism genes are expressed in the CZE (Miyawaki et al., 2004; Hu et al., 2012; Li et al., 2013). Because of the importance of these hormones for seed development, it was proposed that the CZE may serve as a communication hub that integrates developmental processes within the seed (Belmonte et al., 2013). The loss of expression of CZE-specific genes in dash suggests that this endosperm sub-region is severely affected in the mutant, and is consistent with associated developmental effects of the embryo.
DASH, a central regulator of early seed developmental processes
A comparison of the transcriptome of developing pods from WT and EMS109 mutant revealed major changes in expression of genes involved in hormone metabolism, protein synthesis and degradation, cell cycle and DNA/chromatin remodelling (Table2). Many genes expressed early during seed development and involved in cell division were down-regulated in dash (Table2), including cyclin-dependent protein kinase regulators (genes orthologous of CYCD, CYCB, CYCA,), and cytoskeletal proteins (kinesin and phragmoplast-associated kinesin, tubulin, fimbrin). Cyclin and CDK genes have been shown to be over-represented in proliferating endosperm tissue (Day et al., 2008), as syncytial development requires a rapid progression through the cell cycle, suppression of phragmoplast formation and uncoupling of cytokinesis and mitosis. In addition alteration of cell division in the dash mutant was confirmed by the lower number of cells observed in cotyledons of mature seeds (Figure2f). Genes involved in DNA synthesis and cell wall deposition were also mainly down-regulated in dash, which is consistent with the impairment of formation of normal cellular structures observed in dash endosperm (Figure S6). Our transcriptomics data also highlighted a specific class of genes whose expression was greatly reduced in the dash mutant (Figure4). These genes encode small CRPs and interestingly one of them (Medtr8g046040) was found by in situ hybridization to be expressed in the chalazal endosperm, in a pattern identical to DASH suggesting that this gene might possibly be involved in the same pathway. CRPs have been assigned diverse roles and some members of this family are implicated in processes such as fertilization, female gametophyte or seed development (Marshall et al., 2011). Recently, Costa et al. (2014) identified 180 small CRPs expressed in developing seeds of Arabidopsis. They showed a specific family of peptides, called ESF1 (Embryo Surrounding Factor 1), accumulated before fertilization in central cell gametes and thereafter in embryo-surrounding endosperm cells, required for proper early embryonic patterning by promoting suspensor elongation (Costa et al., 2014). In maize, the CRP MEG1 is specifically expressed in the basal endosperm transfer layer, functionally equivalent to the CZE in dicotyledons. MEG1 regulates maternal nutrient translocation into the seed and RNAi lines display smaller seeds (Costa et al., 2012). The Medicago genome contains 682 CRP genes, 52 of which are exclusively expressed in developing seeds (Tesfaye et al., 2013). The possible role of small peptides such as Medtr8g046040, in early endosperm development remains to be studied.
Deregulation of auxin homeostasis in dash
A strict regulation of auxin gradients has been shown to be critical for proper embryo development (Friml et al., 2003). The high concentration of IAA measured in dash mutant pods (Figure5a,b), together with the fact that the class of auxin biosynthesis/catabolism genes was not significantly affected in dash, whereas the class of auxin-induced genes was down-regulated, suggests that auxin transport/distribution may be impaired in the mutants. Down-regulation of auxin response factors in mutant pods accumulating high auxin levels may seem contradictory but is explained by a well documented negative feedback regulatory pathway, which reduces sensitivity of cells towards auxin (Benjamins and Scheres, 2008). Increase of IAA in the dash mutant is accompanied by an increase of IAA-Asp, an inactive form of auxin (Ludwig-Müller, 2011) and is consistent with the down-regulation of IAR33-like gene expression (Medtr2g100560), which has been shown to possess in vitro hydrolase activity against IAA-Asp (Campanella et al., 2008) (Figure5b and Table S2). The content of this auxin conjugate increases in response to elevated IAA contents such as in the sur2 mutant in Arabidopsis (Barlier et al., 2000) consistent with its possible catabolic role to maintain IAA homeostasis (Woodward and Bartel, 2005). A similarly high concentration of auxin in seed and fruit of tomato was observed when auxin transport is inhibited by N-1-naphthylphthalamic acid (NPA) treatment (Pattison and Catala, 2012) at an early stage of development.
The existence of auxin gradients during fruit growth, with the highest levels of auxin in the seeds, is well documented and, after fruit set, precise spatial and temporal synthesis, transport and action of auxin are required for proper fruit development (Sundberg and Ostergaard, 2009). Auxin distribution is controlled by polar auxin transport mediated by PIN and AUX/LAX proteins, which control cellular auxin efflux and influx respectively (Vanneste and Friml, 2009) and thus the establishment of auxin gradients promoting auxin signaling (Lau and Deng, 2010). Inactivation of auxin efflux carriers is known to result in embryo lethality or severe apical defects depending on the ecotype background (Friml et al., 2003), which is consistent with the down-regulation of four MtPIN transporters in dash mutant (Table S2). Interestingly, among the few auxin-responsive genes up-regulated in dash is IAA18. A gain-of-function mutation of this gene alters PIN1 expression in the apical domain of the embryo in Arabidopsis, indicating that IAA18 disrupts auxin transport and ultimately auxin gradients (Ploense et al., 2009). In the dash mutant, pod growth and seed set were recovered upon external application of IAA directly on immature pods of the dash mutant, and TIBA treatment of WT pods resulted in decreased seed/pod size (Figure5c,d). These results support the hypothesis of a link between the dash mutation and auxin perception and/or transport deficiency: a defect in auxin efflux transport will lead to auxin accumulation in the immature pod, which will severely disturb auxin homeostasis necessary for proper embryo, seed and pod development. Indirect evidence exists for a relationship between seed size determination and IAA. For instance, larger seeds were generated in Arabidopsis by mutation of AUXIN RESPONSE FACTOR2 (ARF2), which mediates gene expression in response to auxin and represses cell division (Schruff et al., 2006). The nature of the relationship between DASH, expressed in endosperm and deregulation of embryo growth, putatively through auxin transport, remains to be clarified. Our data show that dash mutants are affected in IAA accumulation and perception, IAA but do not exclude that other hormonal balances may also be impaired and might impact embryo cell division and ultimately final seed size.
Experimental Procedures
Plant growth conditions and harvest of developing pods
All plants were grown under greenhouse conditions in 1.5 L pots filled with a mix of attapulgite:clay beads (60:40). Temperature was controlled to be 20°C during the day and above 18°C during the night. Artificial lighting was supplied to reach 16 h light per day. The plants were not inoculated with Sinorhizobium sp. bacteria and were automatically supplied with fertilizers (3.5 N/3.1 P/8.6 K).
Screening for dash mutant lines
Medicago truncatula EMS109 mutants was identified from an EMS population containing 4600 M2 lines from cultivar Jemalong A17 by TILLING screening according to Le Signor et al. (2009). Nested PCR was conducted using the following inner primers labelled with IRD-700 and IRD-800 dyes: MtDOF-F2 5′-CCAACCAATAGCAGTAGCAACCG-3′ and MtDOF-R2 5′-GCTGCACCTAGAAAATCCAAAGAT-3′.
The M. truncatula Tnt1 mutant population containing more than 20 000 lines from cultivar R108 was screened as described in Cheng et al. (2014). Tnt1 insertions were identified by nested PCR using gene-specific forward primer sequences 5′-AGGGTCCCATTTCTTTGACTAGT-3′ and 5′-TGACTAGTGCCACCTCATTGTG-3′ and reverse primer sequences 5′-TCACTGAGGAGGATTGAACTCTG-3′ and 5′-TACCATTACCACCAGCACCA-3′.
The DASH gene was sequenced in the mutant lines to confirm and precisely locate the mutation. The EMS and Tnt1 mutant lines were back-crossed with the corresponding WT lines.
In situ hybridization
The method for digoxygenin labelling of RNA probes, tissue preparation and in situ hybridization was described by Coen et al. (1990) with modifications described by Bradley et al. (1993). The DASH (Medtr2g014060) and CRP (Medtr8g046040) probes (1125 and 521 bp respectively) included the whole coding sequence and about 50 bp of the 3′ and 5′ untranslated regions. In vitro transcription templates were amplified from seed cDNA with the forward primer sequence 5′-TGGCACGTCTCTGTTTAGAAAACGC-3′ and the reverse primer sequence
5′-GATTAATTTCTCAGAGCTATGCTTTC-3′ for DASH and with the forward primer sequence
5′-GTCTTTGTAAACCTTTCAAAGTAGCC-3′ and the reverse primer sequence
5′-TTGAGATTGTACAAATATCAACCAC-3′ for the CRP gene. The reverse or the forward primer of each pair contained a tail with the T3 promoter at its 5′ end for the production of the anti-sense or sense probe respectively.
Cell size and cell number measurements
Mature seeds were used for cell size and cell number measurements. For cell size determination, seeds were imbibed 4 h in water then overnight in 20% sucrose dissolved in Tris-buffered saline solution (TBS, pH 7.5) at 4°C. Sections (20 μm) were made by cryo-dissection at −20°C (Leica; http://www.leica-microsystems.com). Sections were stained using toluidine blue and cell density was calculated according to the number of cells per unit area between WT and mutant lines. For cell number determination, imbibed mature seeds were digested for 36 h at 37°C in a enzymatic solution containing 0.45 m sorbitol, 10 mm MgCl2, 1 mm KH2PO4, 20 mm MES (pH 5.6), 0.4% Macerozyme R10 (Phytotechnology; http://www.phytotechlab.com), 1% cellulose Onozuka R10 (Phytotechnology). Cell number of a subset of cell suspension was counted using a haemocytometer and normalized according to the area of the haemocytometer and to the volume of the digestion solution.
Seed clearing
Seeds from plants EMS109, NF6042, NF5285, A17 and R108 were fixed in ethanol/acetic acid (9:1) for 1 h, transferred to 90% ethanol for 1 h then 70% ethanol for 1 h. Treated seeds were transferred onto microscope slides and covered with a chloral hydrate solution (4 g chloral hydrate, 0.5 ml glycerol 100%, 1 ml water) for 4 h and observed with a Zeiss Axiophot photomicroscope equipped with Nomarski DIC optics.
Light microscopy
Developing seeds (8–10 DAP) from dash and WT plants were vacuum-infiltrated overnight at 4°C with a fixative mixture containing 3% (v/v) gluteraldehyde and 2% (w/v) paraformaldehyde in 0.1 m sodium phosphate-buffered medium (pH 7.2). Seeds were then washed and dehydrated in an ethanol series before embedding in Historesin following provider's instructions. Thick sections (1 μm) were stained with 0.1% (w/v) toluidine blue plus 0.5% (w/v) methylene blue prior to examination by bright field microscopy with a microscope (Leica).
Complementation of dash mutant plants
A fragment of 1150 bp of the native promoter with the 1011 bp of the coding sequence of MtDASH was amplified by PCR using primers F-TGTAATACTAATGTTTTCTTGACTG and R-CTGAGGAGGATTGAACTCTGACAG. The pDASH::DASH construct was introduced into the pMDC123 vector (Curtis and Grossniklaus, 2003). The leaves of M. truncatula (A17) mutants were infected with A. tumefaciens EHA105 strain harbouring the complementation vector (Cosson et al., 2006). Resistant calluses obtained after 1–1.5 month of selection were transferred onto regeneration medium. The regenerated shoots or plantlets obtained after 3–4 months were transferred to half-strength SH9 medium supplemented with 1 mg/L IAA. All the regenerating cultures were kept at 25°C under fluorescent light (140 μE m−2 s−1) at a photoperiod of 16 h in the growth chamber. After 3–4 weeks, plants were transferred to soil and grown in the greenhouse (16 h light, 390 μE m−2 s−1).
Affymetrix microarray, data extraction, and normalization
Total RNA was extracted using a modified cetyltrimethylammonium bromide method (Verdier et al., 2008). Next, 5 μg of total RNA from each sample was purified (RNeasy MinElute Cleanup kit; Qiagen; http://www.qiagen.com) according to the manufacturer's instructions. The Affymetrix M. truncatula GeneChip Array (Affymetrix; http://www.affymetrix.com) was used for expression analysis during seed development. RNA from three independent biological replicates were analysed for each time point. Probe synthesis/labelling, array hybridization, scanning, and data normalization were performed as described by Benedito et al. (2008). Raw and normalized microarray data were deposited at Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) as GSE58117. To identify probe sets differentially expressed in dash mutant versus WT control, the r package anapuce (J. Aubert, UMR 518 AgroParisTech/INRA) was used. P-values were adjusted by the Benjamini–Hochberg method (Benjamini and Hochberg, 1995). Transcripts were considered as differentially expressed when associated with a P-value ≤0.05. Over-representation analysis were performed using pageman software (Usadel et al., 2006). Data were subjected to a bin-wise Wilcoxon test and resulting P-values adjusted according to Benjamini–Hochberg criteria.
cDNA synthesis and quantitative real-time PCR analysis
Two μg of total RNA were treated with RNase-free DNase RQ1 (Promega; http://www.promega.com) according to the manufacturer's instructions. First-strand cDNA was produced using iScript reverse transcriptase (Bio-Rad; http://www.bio-rad.com) according to the manufacturer's instructions. qRT-PCR was performed on LightCycler 480 (Roche; http://www.usdiagnostics.roche.com) using GoTaq qPCR Master Mix (Promega, Madison, USA). Next, 1 μl cDNA and 200 nm of each gene-specific primer in a final volume of 5 μl, were incubated at: 95°C for 5 min; and 40 cycles of 95°C for 10 sec and 60°C for 1 min. Relative expressions were normalized against ACTIN, GAPDH, MSC27 and PDF2 expressions using the 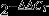 method (Livak and Schmittgen, 2001). Each expression profile was repeated three times (three biological replicates) in three technical repetitions.
method (Livak and Schmittgen, 2001). Each expression profile was repeated three times (three biological replicates) in three technical repetitions.
Phytohormone content measurements and auxin treatments
Pods were ground using a pestle and mortar and 50 mg of tissue powder were used for phytohormone quantification. Phytohormones were extracted using 1 ml of extraction solvent (isopropanol:water:HCl at the ratio 2:1:0.002). Internal standard (50 pg) of d5-IAA was added to the extraction buffer. Extractions were performed by shaking samples at 4°C for 1 h, then 0.5 ml of dichloromethane was added and samples were shaken for another 30 min. After centrifugation at 2900 g (30 min at 4°C), bottom layers were collected and dried under nitrogen. Dry pellets were re-suspended in 0.1 ml methanol and 1 ml of acetic acid 1% then cleaned using conditioned Waters Oasis HLB columns according to manufacturer's instructions (Waters; http://www.waters.com). Hormone samples were eluted from the columns using 1.8 ml of 80% methanol containing 1% acetic acid to glass vials, dried under nitrogen. Dry samples were dissolved in 25 μl methanol and 25 μl 1% acetic acid and 10 μl was injected in an Agilent triple quadrupole LC/MSMS analyser. A reverse phase 1.7 μm UPLC BEH C18 (2.1 × 150 mm) column was used for separation. Data were analysed using masshunter quantitative analysis software (Agilent; http://www.agilent.com).
Individual flowers (50–100) were tagged and developing pods were treated at 6 DAP by immersion in water, IAA (570 μm) or TIBA (200 μm) solution supplemented with a drop of Tween-20. Pods were harvested at maturity and weight measurement was performed on about 20 pods containing seeds.
Distance analysis
The tree was created using full-length DOF coding sequences of proteins from A. thaliana, Glycine max, M. truncatula, P sativum and cereals. These protein sequences were aligned and organized based on sequence similarities of the full-length DOF protein sequences by the clustalw algorithm using CLC Genomics workbench software (CLC bio-CLC sequence viewer 6.2, Aarhus, Denmark; http://www.clcbio.com). Protein sequences are provided in Table S3.
Acknowledgments
We would like to thank Peter Rogowsky and Bertrand Dubreucq for helpful advice, Lloyd Sumner and David Huhman for their help in measuring phytohormone content, and Yuhong Tang and Stacy Allen for their assistance with microarray hybridizations. This project has received funding from the European Union (FP7 grant agreement n° 613551) and the Samuel Roberts Noble Foundation. MN was funded by an INRA BAP - Burgundy Region Ph.D studentship.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Expression of DASH in different plant organs and in seed tissues.
Distance analysis of DOF proteins.
Position of the EMS mutation and of the Tnt1 insertions within the DASH sequence.
Genetic and molecular analysis of the dash mutants.
Complementation of EMS109 mutant.
Microscopic observations of embryo and endosperm at 8 DAP from WT (a, c) and EMS109 (b, d) seeds.
Delay in embryo growth in NF5285 mutant seeds.
IAA dosage on pods of NF5285 versus WT from 8 to 12 DAP.
List of differentially expressed genes at 8 DAP in dash pods compared to WT.
List of differentially expressed genes belonging to the “auxin” functional class.
List of the 96 DOF protein sequences used to compute the distance analysis in Figure S2.
References
- Adamski NM, Anastasiou E, Eriksson S, O'Neill CM, Lenhard M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl Acad. Sci. USA. 2009;106:20115–20120. doi: 10.1073/pnas.0907024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad FF, Mayer U, Wanner G, Jürgens G. The KEULE gene is involved in cytokinesis in Arabidopsis. Mol. Gen. Genet. 1996;253:267–277. doi: 10.1007/pl00008594. [DOI] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl Acad. Sci. USA. 2000;97:14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, et al. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl Acad. Sci. USA. 2013;110:E435–E444. doi: 10.1073/pnas.1222061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- Benjamins R, Scheres B. Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Campanella J, Smith S, Leibu D, Wexler S, Ludwig-Müller J. The auxin conjugate hydrolase family of Medicago truncatula and their expression during the interaction with two symbionts. J. Plant Growth Regul. 2008;27:26–38. [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wang M, Lee H, Tadege M, Ratet P, Udvardi M, Mysore KS, Wen J. An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol. 2014;201:1065–1076. doi: 10.1111/nph.12575. [DOI] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R. floricaula: A homeotic gene required for flower development in antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Cosson V, Durand P, d'Erfurth I, Kondorosi A, Ratet P. Medicago truncatula transformation using leaf explants. Methods Mol. Biol. (Clifton, N.J.) 2006;343:115–127. doi: 10.1385/1-59745-130-4:115. [DOI] [PubMed] [Google Scholar]
- Costa LM, Yuan J, Rouster J, Paul W, Dickinson H, Gutierrez-Marcos JF. Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr. Biol. 2012;22:160–165. doi: 10.1016/j.cub.2011.11.059. [DOI] [PubMed] [Google Scholar]
- Costa LM, Marshall E, Tesfaye M, et al. Central cell-derived peptides regulate early embryo patterning in flowering plants. Science. 2014;344:168–172. doi: 10.1126/science.1243005. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RC, Herridge RP, Ambrose BA, Macknight RC. Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 2008;148:1964–1984. doi: 10.1104/pp.108.128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Erfurth I, Le Signor C, Aubert G, et al. A role for an endosperm-localized subtilase in the control of seed size in legumes. The New Phytol. 2012;196:738–751. doi: 10.1111/j.1469-8137.2012.04296.x. [DOI] [PubMed] [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martínez M, Isabel-La Moneda I, Carbonero P. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 2002;29:453–464. doi: 10.1046/j.0960-7412.2001.01230.x. [DOI] [PubMed] [Google Scholar]
- Fiume E, Fletcher JC. Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell. 2012;24:1000–1012. doi: 10.1105/tpc.111.094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C, Varotto S. The role of PIN auxin efflux carriers in polar auxin transport and accumulation and their effect on shaping maize development. Mol. Plant. 2011;5:787–798. doi: 10.1093/mp/ssr103. [DOI] [PubMed] [Google Scholar]
- Forestan C, Meda S, Varotto S. ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol. 2010;152:1373–1390. doi: 10.1104/pp.109.150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Gallardo K, Firnhaber C, Zuber H, Hericher D, Belghazi M, Henry C, Kuster H, Thompson R. A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: evidence for metabolic specialization of maternal and filial tissues. Mol. Cell Proteomics. 2007;6:2165–2179. doi: 10.1074/mcp.M700171-MCP200. [DOI] [PubMed] [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell. 2005;17:52–60. doi: 10.1105/tpc.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J, Gallie DR. Aleurone cell identity is suppressed following connation in maize kernels. Plant Physiol. 2005;139:204–212. doi: 10.1104/pp.105.064295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger E, Kradolfer D, Köhler C. Endosperm cellularization defines an important developmental transition for embryo development. Development. 2012;139:2031–2039. doi: 10.1242/dev.077057. [DOI] [PubMed] [Google Scholar]
- Hu YF, Li YP, Zhang JJ, Liu HM, Tian ML, Huang YB. Binding of ABI4 to a CACCG motif mediates the ABA-induced expression of the ZmSSI gene in maize (Zea mays L.) endosperm. J. Exp. Bot. 2012;63:5979–5989. doi: 10.1093/jxb/ers246. [DOI] [PubMed] [Google Scholar]
- Ingram GC. Family life at close quarters: communication and constraint in angiosperm seed development. Protoplasma. 2010;247:195–214. doi: 10.1007/s00709-010-0184-y. [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW. Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant. Biol. 2010;13:571–577. doi: 10.1016/j.pbi.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Signor C, Savois V, Aubert G, et al. Optimizing TILLING populations for reverse genetics in Medicago truncatula. Plant Biotechnol. J. 2009;7:430–441. doi: 10.1111/j.1467-7652.2009.00410.x. [DOI] [PubMed] [Google Scholar]
- Li J, Nie X, Tan JLH, Berger F. Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Natl Acad. Sci. USA. 2013;110:15479–15484. doi: 10.1073/pnas.1305175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the
 method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar] - Ludwig-Müller J. Auxin conjugates: their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011;62:1757–1773. doi: 10.1093/jxb/erq412. [DOI] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl Acad. Sci. USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall E, Costa LM, Gutierrez-Marcos J. Cysteine-rich peptides (CRPs) mediate diverse aspects of cell-cell communication in plant reproduction and development. J. Exp. Bot. 2011;62:1677–1686. doi: 10.1093/jxb/err002. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004;37:128–138. doi: 10.1046/j.1365-313x.2003.01945.x. [DOI] [PubMed] [Google Scholar]
- Möller B, Weijers D. Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 2009;1:a001545. doi: 10.1101/cshperspect.a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Martinez M, Vicente-Carbajosa J, Carbonero P. The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol. Genet. Genomics. 2007;277:379–390. doi: 10.1007/s00438-006-0186-9. [DOI] [PubMed] [Google Scholar]
- Noguero M, Atif RM, Ochatt S, Thompson RD. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013;209:32–45. doi: 10.1016/j.plantsci.2013.03.016. [DOI] [PubMed] [Google Scholar]
- Pattison RJ, Catala C. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 2012;70:585–598. doi: 10.1111/j.1365-313X.2011.04895.x. [DOI] [PubMed] [Google Scholar]
- Ploense SE, Wu MF, Nagpal P, Reed JW. A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development. 2009;136:1509–1517. doi: 10.1242/dev.025932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 2013;23:2506–2512. doi: 10.1016/j.cub.2013.09.039. [DOI] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signaling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- Signor CL, Savois V, Aubert G, et al. Optimizing TILLING populations for reverse genetics in Medicago truncatula. Plant Biotechnol. J. 2009;7:430–441. doi: 10.1111/j.1467-7652.2009.00410.x. [DOI] [PubMed] [Google Scholar]
- Strompen G, Kasmi FEl, Richter S, Lukowitz W, Assaad FF, Jürgens G, Mayer U. The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr. Biol. 2002;12:153–158. doi: 10.1016/s0960-9822(01)00655-8. [DOI] [PubMed] [Google Scholar]
- Sundberg E, Ostergaard L. Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb. Perspect. Biol. 2009;1:a001628. doi: 10.1101/cshperspect.a001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen JQ, He J, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- Tesfaye M, Silverstein KA, Nallu S, et al. Spatio-temporal expression patterns of Arabidopsis thaliana and Medicago truncatula defensin-like genes. PLoS One. 2013;8:e58992. doi: 10.1371/journal.pone.0058992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. MapMan: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, et al. PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge U-I, Kunze R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006;141:776–792. doi: 10.1104/pp.106.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Verdier J, Kakar K, Gallardo K, Le Signor C, Aubert G, Schlereth A, Town CD, Udvardi MK, Thompson RD. Gene expression profiling of M. truncatula transcription factors identifies putative regulators of grain legume seed filling. Plant Mol. Biol. 2008;67:567–580. doi: 10.1007/s11103-008-9320-x. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann. Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7:555–560. doi: 10.1016/s1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of DASH in different plant organs and in seed tissues.
Distance analysis of DOF proteins.
Position of the EMS mutation and of the Tnt1 insertions within the DASH sequence.
Genetic and molecular analysis of the dash mutants.
Complementation of EMS109 mutant.
Microscopic observations of embryo and endosperm at 8 DAP from WT (a, c) and EMS109 (b, d) seeds.
Delay in embryo growth in NF5285 mutant seeds.
IAA dosage on pods of NF5285 versus WT from 8 to 12 DAP.
List of differentially expressed genes at 8 DAP in dash pods compared to WT.
List of differentially expressed genes belonging to the “auxin” functional class.
List of the 96 DOF protein sequences used to compute the distance analysis in Figure S2.