Abstract
Background
Short‐term fine particles (PM2.5) exposure is associated with reduced heart rate variability, a strong predictor of cardiac mortality among older people. Identifying modifiable factors that confer susceptibility is essential for intervention. We evaluated whether Toll‐like receptor 2 (TLR2) methylation, a reversible immune‐epigenetic process, and its dietary modulation by flavonoids and methyl nutrients, modify susceptibility to heart rate variability effects following PM2.5 exposure.
Methods and Results
We measured heart rate variability and PM2.5 repeatedly over 11 years (1275 total observations) among 573 elderly men from the Normative Aging Study. Blood TLR2 methylation was analyzed using pyrosequencing. Daily flavonoid and methyl nutrients intakes were assessed through the Food Frequency Questionnaire (FFQ). Every 10 μg/m3 increase in 48‐hour PM2.5 moving average was associated with 7.74% (95% CI: −1.21% to 15.90%; P=0.09), 7.46% (95% CI: 0.99% to 13.50%; P=0.02), 14.18% (95% CI: 1.14% to 25.49%; P=0.03), and 12.94% (95% CI: −2.36% to 25.96%; P=0.09) reductions in root mean square of successive differences, standard deviation of normal‐to‐normal intervals, low‐frequency power, and high‐frequency power, respectively. Higher TLR2 methylation exacerbated the root mean square of successive differences, standard deviation of normal‐to‐normal intervals, low‐frequency, and high‐frequency reductions associated with heightened PM2.5 (Pinteraction=0.006, 0.03, 0.05, 0.04, respectively). Every interquartile‐range increase in flavonoid intake was associated with 5.09% reduction in mean TLR2 methylation (95% CI: 0.12% to 10.06%; P=0.05) and counteracted the effects of PM2.5 on low frequency (Pinteraction=0.05). No significant effect of methyl nutrients on TLR2 methylation was observed.
Conclusions
Higher TLR2 methylation may confer susceptibility to adverse cardiac autonomic effects of PM2.5 exposure in older individuals. Higher flavonoid intake may attenuate these effects, possibly by decreasing TLR2 methylation.
Keywords: epidemiology, epigenetics, heart rate variability, inflammation, nutrition
Introduction
Particulate matter (PM) exposure contributes to an estimated 3.7 million annual deaths worldwide.1 According to the American Heart Association, PM exposure is a modifiable primary contributor to cardiovascular morbidity and mortality, often resulting from acute cardiovascular effects after short‐term exposure peaks.2 The major sources of short‐term fine particles (PM2.5) pollution are combustion activities (motor vehicles, power generation, and industrial processes), biomass burning, and other human activities such as heating and cooking.2 Because prevention of exposure to PM is particularly challenging—more than 80% of people in the United States receive some exposure—it is essential to identify modifiable factors that contribute to individual susceptibility to cardiac effects to aid the development of effective interventions.
Short‐term PM exposure has been associated with reduced heart rate variability (HRV) through epidemiological studies.3–4 Impaired autonomic modulation of the rhythmic activity of the sinus node, as reflected in reduced HRV, represents a pathophysiologic mechanism by which air pollution may lead to cardiac mortality, especially among older individuals.5–6 Notably, HRV shows wide interindividual variability, as well as highly variable responses to PM exposure.7 Therefore, HRV is an ideal early surrogate marker of cardiovascular autonomic dysfunction for identifying underlying factors that may modify susceptibility to cardiovascular effects of PM exposure. Furthermore, growing evidence indicates that systemic inflammation exacerbates HRV disturbances following PM exposure.8 PM with aerodynamic diameter <2.5 μm (PM2.5) is especially deleterious because it penetrates into the alveoli and may act as a stimulus to trigger local cytokine production and systemic inflammation.9 Thus, immunoregulation is critical to limiting immune‐mediated cardiovascular pathology.10
Molecules involved in immunoresponses are attractive as potential modulators of cardiovascular pathology following PM exposure. Toll‐like receptors (TLRs), a group of receptors abundantly expressed on leukocytes, have emerged as crucial first‐responders linking innate and adaptive immunity after environmental challenge.11 Toll‐like receptor 2 (TLR2), in particular, is a unique TLR family member that not only assists the clearance of bacterial components contained in PM via pathogen recognition12 but also modulates the expansion and behavior of regulatory T cells, which are the dominant circulating regulator of immunosuppression.13–14 TLR2‐related immunity is controlled through epigenetic mechanisms: Increased methylation in the TLR2 promoter region is usually associated with TLR2 silencing, while decreased methylation permits TLR2 expression.15 Therefore, maintaining a proper methylation level of the TLR2 gene is critical to ensure protective immunity.
Importantly, DNA methylation is reversible, thereby providing unique opportunities for modulation. This feature is critical, particularly for pathologies related to nonpreventable exposures, as it can offer potential interventions to ameliorate exposure‐related disease. Nutrients such as flavonoids and methyl nutrients can modify gene‐specific methylation in opposite directions.16 Flavonoids, the most common group of polyphenolic compounds in the human diet, can lower DNA methylation by reducing DNA methyltransferase activity.17–18 Methyl nutrients such as folic acid, vitamin B12, and methionine, on the other hand, may increase methylation by providing methyl donors.19 Thus, determining how such nutrients alter methylation states can provide insight that may be used to counteract exposure effects.
No study has yet investigated whether DNA methylation and nutrient intake may modify the cardiovascular effects of air pollution exposure. In this study, we hypothesized that programming of TLR2 inactivation by increased methylation contributes to “epigenetic susceptibility” for immune‐mediated PM‐induced cardiovascular responses, as reflected in steeper reductions in HRV following short‐term PM2.5 exposure. We utilized the Normative Aging Study, a cohort of aging men in Eastern Massachusetts, to investigate the relationship between short‐term PM2.5 (48 hours) exposure and HRV, and to examine whether TLR2 methylation in blood leukocytes modifies this relationship. We also examined the effect of flavonoid and methyl nutrients (folic acid, vitamin B12, and methionine) intakes on TLR2 methylation and explored their role as modifiers of the short‐term effects of ambient PM2.5 exposure on HRV. In addition to our primary hypothesis, we examined the effect of black carbon, carbon monoxide, nitrogen dioxide, and ozone on HRV, as well as the relationship between TLR2 methylation and plasma inflammatory markers including interleukin 6 (IL‐6), IL‐8, IL‐1β, tumor necrosis factor α (TNF‐α), TNF‐γ, C‐reactive protein, intercellular adhesion molecule 1, and vascular endothelial growth factor.
Methods
Study Population
The Normative Aging Study is an ongoing prospective cohort of older male participants established in Eastern Massachusetts by the U.S. Veterans Administration.20 For the present study, we excluded participants with incomplete data on PM2.5 exposure and HRV, leaving a total of 573 participants who were community‐dwelling men free of heart arrhythmias at examination (Figure 1). Participants were recalled for visits every 3 to 5 years, and we considered visits conducted since November 2000 (ie, the earliest date for which PM2.5 data were available) through 2011, for a total of 1275 visits (1 visit to 4 visits per participant; average 2.2 visits). Among the 573 participants, 500 participants had methylation data, 513 participants had flavonoid intake data, and 482 participants had methyl nutrients intake data. The study was approved by the institutional review boards of all participating institutions. All subjects provided informed consent.
Figure 1.
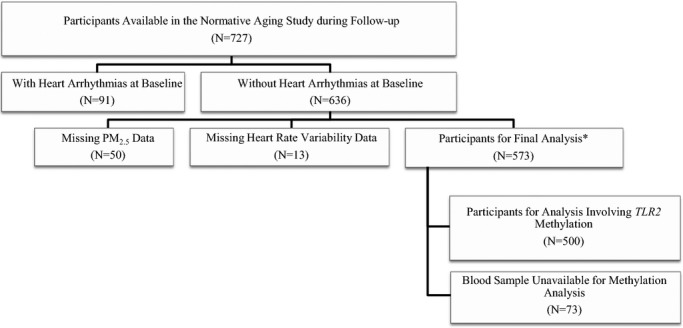
The study participants in the Normative Aging Study, 2000–2011. PM2.5 indicates particulate matter with aerodynamic diameter <2.5 μm; TLR2, Toll‐like receptor 2. *In each analysis, some additional participants may have been excluded due to missing key study variables. We have noted in each table the corresponding sample size.
HRV Measurement
HRV was measured in the morning for 7 minutes with a 2‐channel electrocardiography monitor, and time‐domain variability and frequency‐domain variability of the heart rate were obtained as described previously.3 Observations with HRV measurement time <3.5 minutes were excluded. The root mean square of successive differences (rMSSD) and standard deviation of normal‐to‐normal intervals (SDNN) were measured representing the time domain.21 Low‐frequency (LF) (0.04 to 0.15 Hz) and high‐frequency (HF) (0.15 to 0.4 Hz) power were used to represent the frequency domain.21
Blood TLR2 Methylation and Plasma Inflammatory Markers
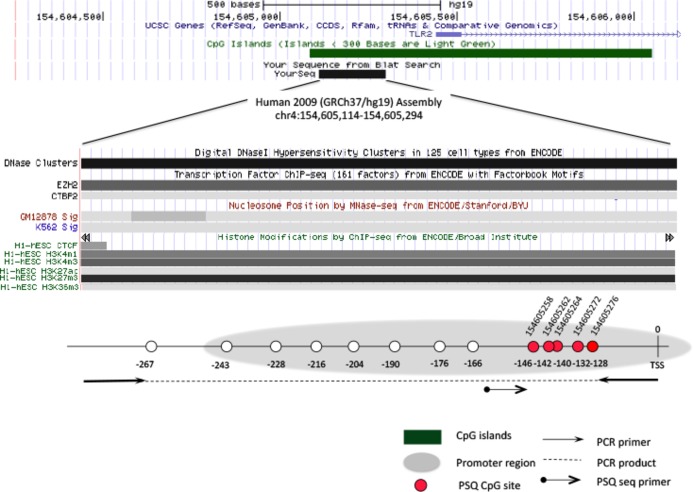
TLR2 methylation was analyzed on blood samples, collected after overnight fasting, at 5 CpG positions within the promoter region (Figure 2) using bisulfite‐treated pyrosequencing, as described previously.22 Exact positions and pyrosequencing primers were reported previously.20 The degree of methylation is reported as the ratio of methylated cytosines over the sum of methylated and unmethylated cytosines (%5mC). Plasma inflammatory markers including IL‐6, IL‐8, IL‐1β, TNF‐α, TNF‐γ, C‐reactive protein, intercellular adhesion molecule 1, and vascular endothelial growth factor were measured using previously described methods.20,22–23
Figure 2.
Schematic view of the genomic structure and CpG dinucleotides selected for analysis in the Toll‐like receptor 2 (TLR2) gene. The chromosomal location of the PCR amplicon for the TLR2 gene pyrosequencing assay is based on the Human Genome Assembly 2009 (GRC37/hg19) (UCSC Genome Browser). The CpG island, gene promoter region, PSQ CpG sites, PCR primer, and PSQ sequence primer location are shown in the figure. TSS indicates the transcription start site; PCR, polymerase chain reaction; PSQ, pyrosequencing. Position 1: CpG154605258; Position 2: CpG154605262; Position 3: CpG154605264; Position 4: CpG154605272; Position 5: CpG154605276.
Air Pollution and Weather Data
PM2.5 exposure levels across the study area were estimated based on a validated hybrid spatiotemporal prediction model,24 which was used in previous epidemiological studies.25–26 This exposure‐assessment method combined real physical measurements from the MODIS (Moderate Resolution Imaging Spectroradiometer) satellite–derived aerosol optical depth and classic land‐use regression methods to predict daily PM2.5 concentration levels across the Boston area from 2000 to 2011, at a 10×10‐km spatial resolution. Kloog and coauthors demonstrated that this method provides more accurate spatial resolution and unbiased predictions compared to prior models27 without compromising temporal resolution.28 For more methodological details of the prediction model please refer to Kloog et al.28–29 The 48‐hour PM2.5 moving averages before each examination were assigned based on the participants' home addresses. The 48‐hour moving averages of outdoor apparent temperature was tabulated based on outdoor air temperature, relative humidity, and wind speed, which were obtained from the Boston airport weather station. Black carbon concentration was measured at the Harvard School of Public Health monitoring site, 1 km from the examination site, using an aethalometer (Magee Scientific, Berkeley, CA).30 Carbon monoxide, nitrogen dioxide, and ozone were obtained from the Massachusetts Department of Environmental Protection local monitoring sites, using a previously described method.30
FFQ and Nutrient Intake
At every visit, detailed information on the typical daily dietary intake of food and beverage items over the previous year was assessed with a self‐administered validated semiquantitative FFQ adapted from the questionnaire used in the Nurses' Health Study. Details on the reproducibility and validity of this FFQ for estimating daily nutrient intakes,31–32 including flavonoid,33 and methyl nutrients (folic acid, vitamin B12, and methionine) intake,21 were published elsewhere. To estimate flavonoid intakes, we constructed a database for assessment of intake of the different flavonoid subclasses using the updated and expanded U.S. Department of Agriculture flavonoid content of foods and the proanthocyanidin databases together with other sources.34–35 Intakes of individual compounds were calculated as the sum of the consumption frequency of each food multiplied by the content of the specific flavonoid for the specified portion size. For foods in the FFQ for which there were no values available in the U.S. Department of Agriculture database, we searched a European database (EuroFIR eBASIS; http://www.eurofir.org) and other sources to ensure all available high‐quality data on flavonoid values could be included in the database. However, although these other sources served as a validation to the U.S. Department of Agriculture database, the addition of the EuroFIR data and the published literature did not contribute to more than 5% to 10% of the overall data used in the calculation of intakes for this analysis. For each participant and visit in this analysis, we derived average daily intakes (mg/day) at each visit of the subclasses commonly consumed in the U.S. diet. The specific subclasses included anthocyanins (cyanidin, delphinidin, malvidin, pelargonidin, petunidin, peonidin), flavanones (eriodictyol, hesperetin, naringenin), flavan‐3‐ols (catechins, gallocatechins, epicatachin, epigallocatechin, epicatachin‐3‐gallate, epigallocatechin‐3‐gallate), flavonols (quercetin, kaempferol, myricetin, isohamnetin), flavones (luteolin, apigenin), and polymers (including proanthocyanidins excluding monomers, theaflavins, and thearubigins). The measure “total flavonoids” represented the sum of these 6 subclasses. Assessment of daily dietary folic acid intake was based on the frequency and dosage information from the FFQ. The average daily total fruit and total vegetable intake (servings/day) was also measured based on FFQ, over the past year. The estimates were made using software developed by the Nurses' Health Study at the Channing Laboratories and processed by Channing Laboratories operators. To avoid undue influence of outliers, we excluded all the questionnaires reporting <500 or >4200 total calories per day. We also truncated flavonoid and folic acid intakes at median±3×(interquartile range). Fewer than 0.5% of observations were excluded.
Statistical Methods
Confounding selection and model assumption
For the analysis involving PM2.5 exposure, TLR2 methylation, and HRV, we adjusted for potential confounders, selected based on literature evidence (ie, age, room temperature, outdoor apparent temperature, season [winter/spring–fall/summer], household income, weekday of the visit, and the visit date). To increase efficiency, we also adjusted for the following risk factors for decreased HRV: body mass index (BMI), smoking status (never/former/current), physical activity, fasting glucose, alcohol consumption (<2 drinks/d/≥2 drinks/d), hypertension, and use of calcium channel blockers, β‐blockers, and angiotensin‐converting enzyme inhibitors. For the analysis examining flavonoid and methyl nutrients intake as effect modifier(s) for the association between PM2.5 exposure and HRV, we adjusted for all the covariates listed above, as well as for total fiber, vitamin C, and caloric intakes. To examine the effect of flavonoid and methyl nutrients intakes on TLR2 methylation, we adjusted for age, body mass index, smoking status, total fiber, vitamin C, caloric intake, household income, and physical activity.
HR, rMSSD, SDNN, LF, and HF were log10‐transformed to improve normality and stabilize the variance. All independent variables were fitted as time‐varying covariates. Nonlinear relationships were characterized first using graphical analyses between HRV and all the covariates, as well as between nutrient intakes and TLR2 methylation. When the graphical analyses suggested nonlinear association, higher‐order terms (quadratic, cubic, etc.) were considered in the model, and a likelihood ratio test was used to determine the number of higher‐order terms needed. In the present study, nonlinearity between apparent temperature and HRV was accounted for by the use of linear and quadratic terms. In addition, we utilized the likelihood ratio test to determine whether it is adequate to model the effect modifiers with a linear trend. Additional analysis using spline regression models were also used to confirm linearity.
To account for repeated assessments for many participants, all statistical models were linear mixed‐effect models with a random intercept assigned to each subject, which used compound symmetry covariance structure. A compound symmetry covariance structure was decided upon after comparing the Akaike information criterion (AIC) among models with different covariance structures as well as assessing the fit of the compound symmetry covariance structure to our data by examining the estimated residual covariance matrix. An analysis using a robust/sandwich estimator for the variance gave very similar results and thus analysis without use of robust/sandwich variance estimators were reported in the manuscript. To examine whether any position in the TLR2 promoter region modified the association between PM2.5 exposure and HRV, we fitted a single linear mixed‐effect model with 5 interaction terms included in the model, which are the interactions between each of the 5 TLR2 positions and PM2.5 (in addition to their respective main effects and other covariates) (Model 1). To test the null hypothesis that none of the positions modified the association between PM2.5 exposure and HRV (H0: γ1=γ2=…=γ5=0), the coefficients of these 5 interaction terms were tested all together using a single Wald test, whose null distribution is approximated using an F‐distribution (global test).
As a secondary analysis, we also examined the effect modification by position‐specific TLR2 methylation by including an interaction term between position‐specific TLR2 methylation and PM2.5 in the model (Model 2), only if the global test was significant.
where Yij was the log10 of HRV measures for participant i at visit j, β0 was the overall intercept, bi was the separate random intercept for subject i, and bi~N(0, ϴ),εij~N(0, σ2). TLR2kij was the TLR2 methylation level at the kth position (k=1−5) within the promoter region, for participant i at visit j. X2ij–Xpij were the covariates including confounders and predictors for HRV, for participant i at visit j.
To describe the effect modification by potential effect modifiers, we estimated from the mixed‐effect models the HRV‐PM2.5 associations at the values corresponding to the quartiles of each effect modifier and reported them separately. We reported effect estimates, 95% CIs, and P values for the HRV‐PM2.5 associations from the linear mixed‐effect models (ie, percent change in HRV per 10 μg/m3 increase in PM2.5) for the midpoint of each quartile. Analyses were performed using SAS 9.4 (SAS Institute, Cary, NC).
Results
Cohort Characteristics and Exposure Levels
The Normative Aging Study is a cohort of aging individuals, between 55 and 100 years old at the first visit of the present study (Figure 1). We excluded N=91 participants with heart arrhythmias at baseline. An additional N=63 participants were excluded as they had missing outcome (N=13 with missing HRV) or missing exposure (N=50 with missing PM2.5 exposure), giving a total of N=573. The baseline characteristics of the participants included in the final analysis are given in Table 1. All participants were male, and 96% were white. Seventy‐nine percent of the participants were overweight, 19% were diabetics, and 71% were hypertensive. The study population included 5% current smokers and 19% heavy alcohol drinkers (≥2 drinks/d). During the study period (November 2000–December 2011), the 48‐hour PM2.5 moving average varied between 2.14 and 42.8 μg/m3 with an average exposure level of 10.5 μg/m3, which is lower than the U.S. Environmental Protection Agency's daily health National Ambient Air Quality Standard for PM2.5 (35.0 μg/m3). The baseline mean methylation level across 5 CpG positions (Figure 2) in the TLR2 promoter region ranged between 0.3%5mC and 7.1%5mC. There was no apparent difference in exposure level or methylation level across different characteristic subgroups (Table 1).
Table 1.
Baseline Characteristics of Study Participants (N=573), 48‐Hour Moving Average of PM2.5, and Averaged Methylation Across CpG Positions in the Promoter Region of TLR2 Gene
| Characteristics | N (%) | Mean (SD) | |
|---|---|---|---|
| PM2.5 (μg/m3) | TLR2 Methylation (%5mC) | ||
| Age, y | |||
| 55 to 69 | 176 (30.7) | 11.4 (5.2) | 2.9 (1.3) |
| 70 to 79 | 293 (51.1) | 11.7 (6.4) | 3.1 (1.3) |
| 80 to 89 | 97 (16.9) | 10.6 (6.1) | 2.9 (1.1) |
| >90 | 7 (1.2) | 13.0 (5.0) | 3.4 (1.0) |
| Physical activity, MET‐h/wk | |||
| <12 | 371 (64.8) | 11.4 (6.3) | 2.93 (1.2) |
| 12 to <30 | 114 (19.9) | 11.3 (5.3) | 3.14 (1.4) |
| ≥30 | 88 (15.4) | 11.7 (5.5) | 3.08 (1.2) |
| Alcohol (drinks/d) | |||
| ≥2 | 106 (18.5) | 11.6 (6.7) | 2.9 (1.1) |
| <2 | 467 (81.5) | 11.4 (5.8) | 3.0 (1.2) |
| Diabetes | |||
| Yes | 111 (19.4) | 11.0 (6.0) | 2.9 (1.0) |
| No | 462 (80.6) | 11.5 (6.0) | 3.0 (1.3) |
| Race | |||
| White | 549 (95.8) | 11.4 (5.9) | 3.0 (1.3) |
| Nonwhite | 24 (4.2) | 11.6 (7.7) | 2.8 (0.9) |
| Hypertension | |||
| Yes | 405 (70.7) | 11.3 (6.0) | 3.0 (1.3) |
| No | 168 (29.3) | 11.8 (6.0) | 2.9 (1.2) |
| BMI, kg/m2 | |||
| <25 | 123 (21.5) | 12.4 (6.8) | 3.0 (1.2) |
| ≥25 | 450 (78.5) | 11.2 (5.7) | 3.0 (1.3) |
| Smoking | |||
| Never | 166 (29.0) | 11.7 (6.0) | 3.1 (1.4) |
| Current | 26 (4.5) | 10.9 (3.8) | 3.3 (1.1) |
| Former | 381 (66.5) | 11.4 (6.1) | 2.9 (1.2) |
| Annual income >$60 000 | |||
| Yes | 272 (47.5) | 11.0 (5.4) | 3.1 (1.3) |
| No | 301 (52.5) | 11.8 (6.5) | 2.9 (1.2) |
BMI indicates body mass index; MET, metabolic equivalent of task; PM2.5, particulate matter with aerodynamic diameter <2.5 μm; TLR2, Toll‐like receptor 2.
Main Effect of Air Pollutants on HRV
Average PM2.5 exposure over the 48 hours before the day of each visit was associated with significantly lowered SDNN and LF, marginally lowered rMSSD and HF, and nonsignificantly increased HR. Every 10 μg/m3 increase in the 48‐hour PM2.5 moving average was associated with 7.74% (95% CI, −1.21% to 15.90%; P=0.09), 7.46% (95% CI, 0.99% to 13.50%; P=0.02), 14.18% (95% CI, 1.14% to 25.49%; P=0.03), and 12.94% (95% CI, −2.36% to 25.96%; P=0.09) reductions in rMSSD, SDNN, LF, and HF, respectively (Table 2). Exposure to black carbon, carbon monoxide, nitrogen dioxide, and ozone over the 48 hours before the day of each visit was not significantly associated with HR, rMSSD, SDNN, LF, and HF (Table 3).
Table 2.
Effect of 48‐Hour Moving Average of PM2.5 on HRV, Normative Aging Study, 2000–2011 (N=500)
| HRV Measure | Change in HRV Per 10 μg/m3 Increase in PM2.5 Concentration* | ||
|---|---|---|---|
| % Change | 95% CI | P Value | |
| HR | 0.42 | −0.94 to 1.80 | 0.55 |
| rMSSD | −7.74 | −15.90 to 1.21 | 0.09 |
| SDNN | −7.46 | −13.50 to −0.99 | 0.02 |
| LF | −14.18 | −25.49 to −1.14 | 0.03 |
| HF | −12.94 | −25.96 to 2.36 | 0.09 |
HF indicates high‐frequency power (0.15 to 0.4 Hz); HR, heart rate; HRV, heart rate variability; LF, low‐frequency power (0.04 to 0.15 Hz); PM2.5, particulate matter with aerodynamic diameter <2.5 μm; rMSSD, root mean square of successive differences; SDNN, standard deviation of normal‐to‐normal intervals.
Results were adjusted for age; body mass index; fasting glucose level; hypertension; smoking status; alcohol consumption; physical exercise; household income; the use of calcium channel blocker, β‐blocker, and angiotensin‐converting enzyme inhibitor; room temperature; outdoor apparent temperature; season; weekday; and visit date.
Table 3.
Effect of 48‐Hour Moving Average of Black Carbon, Carbon Monoxide, Nitrogen Dioxide, and Ozone on HRV, Normative Aging Study, 2000–2011 (N=573)
| HRV | Black Carbon | Carbon Monoxide | Nitrogen Dioxide | Ozone | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Change* | 95% CI | P Value | % Change* | 95% CI | P Value | % Change* | 95% CI | P Value | % Change* | 95% CI | P Value | |
| HR | 0.04 | −0.82 to 0.92 | 0.92 | −0.97 | −3.16 to 1.28 | 0.40 | 0.25 | −0.76 to 1.27 | 0.63 | −0.92 | −2.19 to 0.37 | 0.16 |
| rMSSD | −2.02 | −7.59 to 3.88 | 0.49 | −1.86 | −15.63 to 14.15 | 0.81 | −3.71 | −10.08 to 3.11 | 0.28 | −5.31 | −13.2 to 3.3 | 0.22 |
| SDNN | −2.28 | −6.36 to 1.98 | 0.29 | −2.19 | −12.42 to 9.23 | 0.69 | −1.93 | −6.72 to 3.1 | 0.44 | −4.13 | −10.04 to 2.17 | 0.19 |
| LF | −2.46 | −10.82 to 6.68 | 0.59 | −1.61 | −21.91 to 23.96 | 0.89 | −1.55 | −11.31 to 9.3 | 0.77 | −5.92 | −17.66 to 7.5 | 0.37 |
| HF | −1.91 | −11.45 to 8.67 | 0.71 | −2.55 | −25.13 to 26.86 | 0.85 | −7.29 | −17.7 to 4.44 | 0.21 | −5.19 | −18.57 to 10.39 | 0.49 |
HF indicates high‐frequency power (0.15 to 0.4 Hz); HR, heart rate; HRV, heart rate variability; LF, low‐frequency power (0.04 to 0.15 Hz); rMSSD, root mean square of successive differences; SDNN, standard deviation of normal‐to‐normal intervals.
Results were adjusted for age; body mass index; fasting glucose level; hypertension; smoking status; alcohol consumption; physical exercise; household income; the use of calcium channel blocker, β‐blocker, and angiotensin‐converting enzyme inhibitor; room temperature; outdoor apparent temperature; season; weekday; and visit date.
Modification of PM2.5 Effect on HRV by TLR2 Methylation
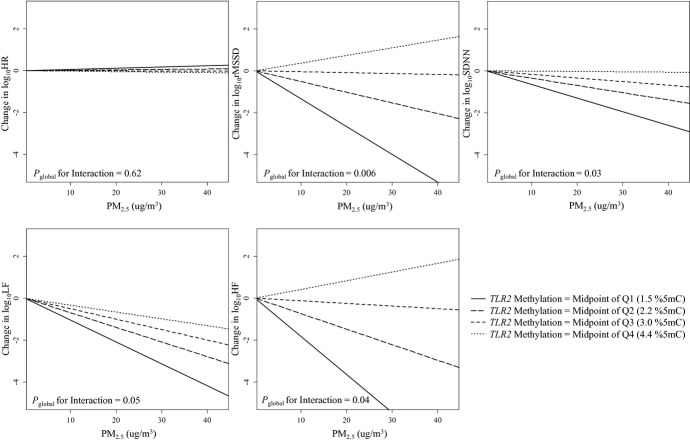
The effect of 48‐hour PM2.5 exposure at participants' homes on rMSSD, SDNN, LF, and HF was significantly modified by the level of methylation within the TLR2 promoter region (P=0.006 for rMSSD; P=0.03 for SDNN; P=0.05 for LF; and P=0.04 for HF; global test for interaction across 5 positions) (Table 4). PM2.5 exposure had no significant effect on HRV in individuals with low TLR2 methylation, but it showed increasingly negative effects on HRV as TLR2 methylation increased (Figure 3). For instance, in individuals with TLR2 methylation within the first quartile, every 10 μg/m3 increase in 48‐hour PM2.5 exposure was associated with 8.77% (95% CI, −7.55% to 27.95%; P=0.31), −0.39% (95% CI, −11.76% to 12.45%; P=0.95), −7.25% (95% CI, −27.74% to 19.06%; P=0.56), and 10.06% (95% CI, −16.25% to 44.64%; P=0.49) nonsignificant estimated changes in rMSSD, SDNN, LF, and HF, respectively (effects estimated at the midpoint of the first quartile, ie, 1.5%5mC to represent the within‐quartile effect; Table 4); on the other hand, in the fourth TLR2 methylation quartile, every 10 μg/m3 increase in 48‐hour PM2.5 exposure was associated with 26.38% (95% CI, 13.69% to 37.20%; P=0.0002), 13.94% (95% CI, 3.12% to 23.55%; P=0.01), 21.40% (95% CI, −0.39% to 38.47%; P=0.05), and 34.28% (95% CI, 13.84% to 49.87%; P=0.003) reductions in rMSSD, SDNN, LF, and HF, respectively (effects estimated at the midpoint of the fourth quartile, ie, 4.4%5mC to represent the within‐quartile effect). Analysis of individual position‐specific methylation showed notable effect heterogeneity across the 5 CpG positions, with stronger effect modifications by positions 1, 3, and 5 (Table 5).
Table 4.
Effect of PM2.5 Exposure on HRV at Different Mean TLR2 Methylation Levels, Normative Aging Study, 2000–2011 (N=500)
| Mean TLR2 Methylation | % Change* | 95% CI | P Value |
|---|---|---|---|
| HR | |||
| Midpoint of Q1 (1.5 5mc%) | −0.56 | −2.88 to 1.82 | 0.64 |
| Midpoint of Q2 (2.2 5mc%) | −0.10 | −1.99 to 1.81 | 0.92 |
| Midpoint of Q3 (3.0 5mc%) | 0.42 | −1.25 to 2.11 | 0.62 |
| Midpoint of Q4 (4.4 5mc%) | 1.34 | −0.92 to 3.66 | 0.25 |
| Pglobal for interaction* | 0.62 | ||
| rMSSD | |||
| Midpoint of Q1 (1.5 5mc%) | 8.77 | −7.55 to 27.95 | 0.31 |
| Midpoint of Q2 (2.2 5mc%) | −1.01 | −13.14 to 12.81 | 0.88 |
| Midpoint of Q3 (3.0 5mc%) | −11.11 | −20.87 to −0.16 | 0.05 |
| Midpoint of Q4 (4.4 5mc%) | −26.38 | −37.20 to −13.69 | 0.0002 |
| Pglobal for interaction* | 0.006 | ||
| SDNN | |||
| Midpoint of Q1 (1.5 5mc%) | −0.39 | −11.76 to 12.45 | 0.95 |
| Midpoint of Q2 (2.2 5mc%) | −3.85 | −12.78 to 6.00 | 0.43 |
| Midpoint of Q3 (3.0 5mc%) | −7.65 | −15.31 to 0.71 | 0.07 |
| Midpoint of Q4 (4.4 5mc%) | −13.94 | −23.55 to −3.12 | 0.01 |
| Pglobal for interaction* | 0.03 | ||
| LF | |||
| Midpoint of Q1 (1.5 5mc%) | −7.25 | −27.74 to 19.06 | 0.56 |
| Midpoint of Q2 (2.2 5mc%) | −10.88 | −27.11 to 8.96 | 0.26 |
| Midpoint of Q3 (3.0 5mc%) | −14.86 | −28.80 to 1.81 | 0.08 |
| Midpoint of Q4 (4.4 5mc%) | −21.40 | −38.47 to 0.39 | 0.05 |
| Pglobal for interaction* | 0.05 | ||
| HF | |||
| Midpoint of Q1 (1.5 5mc%) | 10.06 | −16.25 to 44.64 | 0.49 |
| Midpoint of Q2 (2.2 5mc%) | −2.82 | −22.02 to 21.10 | 0.80 |
| Midpoint of Q3 (3.0 5mc%) | −15.71 | −30.75 to 2.60 | 0.09 |
| Midpoint of Q4 (4.4 5mc%) | −34.28 | −49.87 to −13.84 | 0.003 |
| Pglobal for interaction* | 0.04 | ||
Results were adjusted for age; body mass index; fasting glucose level; hypertension; smoking status; alcohol consumption; physical exercise; household income; the use of calcium channel blocker, β‐blocker, and angiotensin‐converting enzyme inhibitor; room temperature; outdoor apparent temperature; season; weekday; and visit date. Q1, Q2, Q3, and Q4 indicate the first, second, third, and fourth quartile. HF indicates high‐frequency power (0.15 to 0.4 Hz); HR, heart rate; HRV, heart rate variability; LF, low‐frequency power (0.04 to 0.15 Hz); PM2.5, particulate matter with aerodynamic diameter <2.5 μm; rMSSD, root mean square of successive differences; SDNN, standard deviation of normal‐to‐normal intervals; TLR2, Toll‐like receptor 2.
The percent change in HRV per 10 μg/m3 increase in PM2.5 concentration.
Based on a global test for effect modification by position‐specific methylation. Interaction terms between each of the 5 TLR2 positions and PM2.5 were included in the model, and these 5 interaction terms were tested jointly for effect modification using a Wald test (global test).
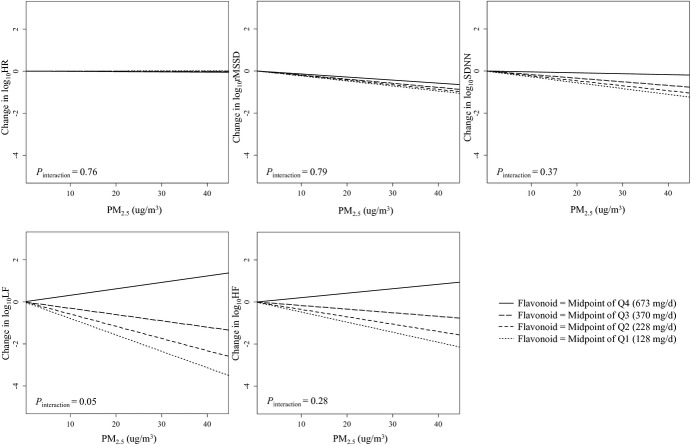
Figure 3.
The effect of particulate matter with aerodynamic diameter <2.5 μm (PM2.5) exposure on heart rate variability (HRV) at different mean Toll‐like receptor 2 (TLR2) methylation levels, Normative Aging Study, 2000–2011 (N=500). log10HR indicates log10‐transformed heart rate; log10rMSSD, log10‐transformed root mean square of the successive differences; log10SDNN, log10‐transformed standard deviation of normal‐to‐normal intervals; log10LF, log10‐transformed low‐frequency power (0.04 to 0.15 Hz); log10HF, log10‐transformed high‐frequency power (0.15 to 0.4 Hz); Q1, Q2, Q3, and Q4 indicate the first, second, third, and fourth quartiles. The association of PM2.5 with rMSSD, SDNN, LF, and HF is modified by mean TLR2 methylation levels, as indicated by the different slopes. The 4 lines in each figure represent the relationship between PM2.5 and HRV when the mean TLR2 methylation level is at the midpoints of each quartile. If there was no effect modification, the 4 lines would be the same. The Pglobal for interaction was based on a global test for effect modification by position‐specific methylation. Interaction terms between each of the 5 TLR2 positions and PM2.5 were included in the model, and these 5 interaction terms were tested jointly for effect modification using a Wald test (global test). Results were adjusted for age; body mass index; fasting glucose level; hypertension; smoking status; alcohol consumption; physical exercise; household income; the use of calcium channel blocker, β‐blocker, and angiotensin‐converting enzyme inhibitor; room temperature; outdoor apparent temperature; season; weekday; and visit date.
Table 5.
Effect Modification by Position‐Specific TLR2 Methylation on the Association Between PM2.5 Exposure and HRV (N=500)
| Position | Methylation | % Change* | 95% CI | P Value | % Change* | 95% CI | P Value | % Change* | 95% CI | P Value |
|---|---|---|---|---|---|---|---|---|---|---|
| HR | rMSSD | SDNN | ||||||||
| 1 | Midpoint of Q1 | 0.36 | −1.96 to 2.75 | 0.76 | 7.26 | −8.65 to 25.94 | 0.39 | 1.66 | −9.79 to 14.56 | 0.79 |
| Midpoint of Q2 | 0.42 | −1.38 to 2.24 | 0.65 | −4.56 | −15.66 to 8.01 | 0.46 | −4.30 | −12.71 to 4.93 | 0.35 | |
| Midpoint of Q3 | 0.46 | −1.22 to 2.17 | 0.60 | −13.25 | −22.84 to −2.47 | 0.02 | −8.91 | −16.51 to −0.63 | 0.04 | |
| Midpoint of Q4 | 0.53 | −1.72 to 2.83 | 0.65 | −26.01 | −36.84 to −13.31 | 0.0002 | −16.11 | −25.42 to −5.64 | 0.004 | |
| P for interaction* | 0.92 | 0.0009 | 0.02 | |||||||
| 2 | Midpoint of Q1 | 0.08 | −2.12 to 2.34 | 0.94 | −0.21 | −14.47 to 16.43 | 0.98 | −7.00 | −17.08 to 4.3 | 0.22 |
| Midpoint of Q2 | 0.28 | −1.49 to 2.09 | 0.76 | −6.61 | −17.49 to 5.72 | 0.28 | −7.36 | −15.51 to 1.58 | 0.10 | |
| Midpoint of Q3 | 0.44 | −1.23 to 2.13 | 0.61 | −11.33 | −21.1 to −0.35 | 0.04 | −7.64 | −15.31 to 0.73 | 0.07 | |
| Midpoint of Q4 | 0.75 | −1.4 to 2.95 | 0.50 | −20.06 | −31.31 to −6.98 | 0.004 | −8.20 | −17.98 to 2.74 | 0.14 | |
| P for interaction* | 0.65 | 0.03 | 0.86 | |||||||
| 3 | Midpoint of Q1 | −0.82 | −3.19 to 1.61 | 0.51 | 5.96 | −10.3 to 25.17 | 0.50 | 1.15 | −10.63 to 14.49 | 0.86 |
| Midpoint of Q2 | −0.16 | −1.99 to 1.71 | 0.86 | −3.69 | −15.24 to 9.44 | 0.56 | −3.72 | −12.44 to 5.87 | 0.43 | |
| Midpoint of Q3 | 0.37 | −1.3 to 2.07 | 0.67 | −10.77 | −20.6 to 0.28 | 0.06 | −7.44 | −15.13 to 0.94 | 0.08 | |
| Midpoint of Q4 | 1.27 | −0.85 to 3.44 | 0.24 | −21.64 | −32.48 to −9.05 | 0.00 | −13.45 | −22.51 to −3.33 | 0.01 | |
| P for interaction* | 0.18 | 0.005 | 0.05 | |||||||
| 4 | Midpoint of Q1 | −0.28 | −2.62 to 2.12 | 0.82 | 2.48 | −12.89 to 20.56 | 0.77 | −4.43 | −15.3 to 7.83 | 0.46 |
| Midpoint of Q2 | 0.11 | −1.73 to 1.99 | 0.90 | −4.88 | −16.33 to 8.13 | 0.44 | −6.13 | −14.66 to 3.25 | 0.19 | |
| Midpoint of Q3 | 0.45 | −1.22 to 2.15 | 0.60 | −10.81 | −20.64 to 0.23 | 0.06 | −7.57 | −15.25 to 0.8 | 0.08 | |
| Midpoint of Q4 | 1.10 | −1.08 to 3.32 | 0.33 | −21.06 | −32.16 to −8.15 | 0.002 | −10.25 | −19.8 to 0.45 | 0.06 | |
| P for interaction* | 0.38 | 0.01 | 0.43 | |||||||
| 5 | Midpoint of Q1 | −1.24 | −3.63 to 1.2 | 0.32 | 6.37 | −9.99 to 25.69 | 0.47 | 1.72 | −10.17 to 15.19 | 0.79 |
| Midpoint of Q2 | −0.31 | −2.15 to 1.57 | 0.75 | −3.71 | −15.29 to 9.45 | 0.56 | −3.62 | −12.39 to 6.02 | 0.45 | |
| Midpoint of Q3 | 0.48 | −1.2 to 2.18 | 0.58 | −11.31 | −21.05 to −0.36 | 0.04 | −7.82 | −15.46 to 0.52 | 0.07 | |
| Midpoint of Q4 | 1.81 | −0.34 to 4 | 0.10 | −22.79 | −33.47 to −10.39 | 0.0007 | −14.49 | −23.45 to −4.47 | 0.006 | |
| P for interaction* | 0.05 | 0.003 | 0.03 | |||||||
| LF | HF | |||||||||
| 1 | Midpoint of Q1 | −11.35 | −30.67 to 13.36 | 0.34 | 1.00 | −22.87 to 32.27 | 0.94 | |||
| Midpoint of Q2 | −13.74 | −28.64 to 4.28 | 0.13 | −9.74 | −26.73 to 11.19 | 0.34 | ||||
| Midpoint of Q3 | −15.64 | −29.53 to 0.98 | 0.06 | −17.68 | −32.53 to 0.45 | 0.06 | ||||
| Midpoint of Q4 | −18.73 | −36.26 to 3.64 | 0.10 | −29.38 | −46.1 to −7.48 | 0.01 | ||||
| P for interaction* | 0.61 | 0.06 | ||||||||
| 2 | Midpoint of Q1 | −17.70 | −34.99 to 4.19 | 0.11 | −5.94 | −27.43 to 21.91 | 0.64 | |||
| Midpoint of Q2 | −16.04 | −30.54 to 1.48 | 0.07 | −11.67 | −28.3 to 8.83 | 0.24 | ||||
| Midpoint of Q3 | −14.72 | −28.69 to 1.99 | 0.08 | −15.90 | −30.99 to 2.48 | 0.09 | ||||
| Midpoint of Q4 | −12.02 | −30.33 to 11.11 | 0.28 | −23.77 | −41.18 to −1.22 | 0.04 | ||||
| P for interaction* | 0.67 | 0.22 | ||||||||
| 3 | Midpoint of Q1 | 4.93 | −18.73 to 35.46 | 0.71 | 10.57 | −16.54 to 46.49 | 0.48 | |||
| Midpoint of Q2 | −6.44 | −23.08 to 13.79 | 0.51 | −4.44 | −22.97 to 18.55 | 0.68 | ||||
| Midpoint of Q3 | −14.64 | −28.62 to 2.06 | 0.08 | −14.96 | −30.18 to 3.58 | 0.11 | ||||
| Midpoint of Q4 | −26.97 | −41.87 to −8.26 | 0.007 | −30.26 | −45.83 to −10.2 | 0.005 | ||||
| P for interaction* | 0.03 | 0.01 | ||||||||
| 4 | Midpoint of Q1 | −12.69 | −31.92 to 11.98 | 0.29 | −0.15 | −24.04 to 31.25 | 0.99 | |||
| Midpoint of Q2 | −13.86 | −29.23 to 4.84 | 0.14 | −8.49 | −26.31 to 13.62 | 0.42 | ||||
| Midpoint of Q3 | −14.86 | −28.79 to 1.8 | 0.08 | −15.14 | −30.32 to 3.36 | 0.10 | ||||
| Midpoint of Q4 | −16.72 | −33.93 to 4.96 | 0.12 | −26.43 | −42.95 to −5.13 | 0.02 | ||||
| P for interaction* | 0.77 | 0.09 | ||||||||
| 5 | Midpoint of Q1 | −1.26 | −23.57 to 27.57 | 0.92 | 15.29 | −12.68 to 52.22 | 0.32 | |||
| Midpoint of Q2 | −9.27 | −25.47 to 10.46 | 0.33 | −3.32 | −21.96 to 19.78 | 0.76 | ||||
| Midpoint of Q3 | −15.39 | −29.24 to 1.17 | 0.07 | −16.41 | −31.27 to 1.68 | 0.07 | ||||
| Midpoint of Q4 | −24.79 | −40.16 to −5.46 | 0.01 | −34.57 | −49.1 to −15.88 | 0.001 | ||||
| P for interaction* | 0.10 | 0.002 | ||||||||
HF indicates high‐frequency power (0.15 to 0.4 Hz); HR, heart rate; HRV, heart rate variability; LF, low‐frequency power (0.04 to 0.15 Hz); PM2.5, particulate matter with aerodynamic diameter <2.5 μm; Q1, Q2, Q3, and Q4 indicate the first, second, third, and fourth quartile; rMSSD, root mean square of successive differences; SDNN, standard deviation of normal‐to‐normal intervals; TLR2, Toll‐like receptor 2.
The percent change in HRV per 10 μg/m3 increase in PM2.5 concentration. Results were adjusted for age; body mass index; fasting glucose level; hypertension; smoking status; alcohol consumption; physical exercise; household income; the use of calcium channel blocker, β‐blocker, and angiotensin‐converting enzyme inhibitor; room temperature; outdoor apparent temperature; season; weekday; and visit date.
To examine effect modification by position‐specific TLR2 methylation, an interaction term between position‐specific TLR2 methylation and PM2.5 was included in the model.
Association of Flavonoid and Methyl Nutrients Intakes With TLR2 methylation
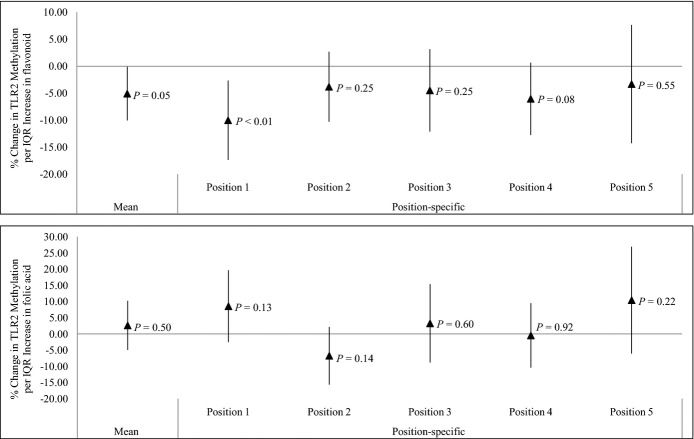
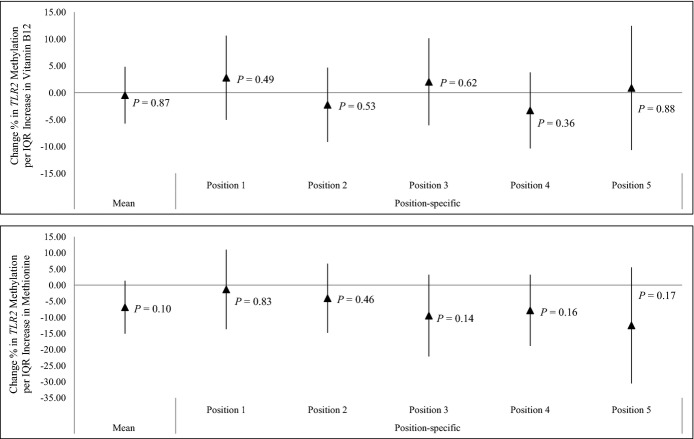
The average daily flavonoid intake over the preceding year was negatively associated with mean TLR2 methylation, considered as the average methylation level across 5 CpG positions (5.09% reduction; 95% CI, 0.12% to 10.06%; per an interquartile‐range increase in flavonoid intake; P=0.05) (Figure 4). In position‐specific analyses (Figure 4 and Table 6), methylation at TLR2 promoter positions 1 and 4 showed the strongest associations with flavonoid intake. We did not observe significant association between methyl nutrients (folic acid, vitamin B12, and methionine) intakes in the previous year and either mean or position‐specific TLR2 methylation (Figures 4 and 5).
Figure 4.
Effect of dietary flavonoid (N=497) and folic acid (N=482) intake on mean and position‐specific Toll‐like receptor 2 (TLR2) methylation. IQR indicates interquartile range. Results were adjusted for age, body mass index, total caloric intake, total vitamin C intake, total fiber intake, smoking status, household income, and physical activity.
Table 6.
Effect of Dietary Flavonoid Intake on Mean and Position‐Specific TLR2 Methylation, Adjusted for Total Fruit and Vegetable Intake
| Original Model (N=497)* | Adjusted for Total Fruit and Vegetable Intake (N=482)* | |||||
|---|---|---|---|---|---|---|
| % Change | 95% CI | P Value | % Change | 95% CI | P Value | |
| Position 1 | −10.02 | −17.37 to −2.66 | <0.01 | −8.47 | −15.94 to −1.01 | 0.03 |
| Position 2 | −3.81 | −10.31 to 2.69 | 0.25 | −2.51 | −9.33 to 4.30 | 0.48 |
| Position 3 | −4.49 | −12.11 to 3.14 | 0.25 | −4.28 | −12.11 to 3.55 | 0.28 |
| Position 4 | −6.05 | −12.74 to 0.65 | 0.08 | −6.61 | −13.51 to 0.29 | 0.06 |
| Position 5 | −3.31 | −14.26 to 7.64 | 0.55 | −4.82 | −16.12 to 6.48 | 0.41 |
| Mean | −5.09 | −10.06 to −0.12 | 0.05 | −4.99 | −10.16 to 0.17 | 0.06 |
TLR2 indicates Toll‐like receptor 2.
Results were adjusted for age, year of visit, body mass index, total caloric intake, total vitamin C intake, total fiber intake, smoking status, household income, and physical activity.
In addition to all the covariates in the original model except total vitamin C and fiber intake, results were adjusted for total fruit and vegetable intake.
Figure 5.
Effect of dietary vitamin B12 (N=497) and methionine (N=482) intake on mean and position‐specific Toll‐like receptor 2 (TLR2) methylation. IQR indicates interquartile range. Results were adjusted for age, body mass index, total caloric intake, total vitamin C intake, total fiber intake, smoking status, household income, and physical activity.
Effect Modification by Flavonoid and Methyl Nutrients Intakes
Higher average daily flavonoid intake over the preceding year significantly weakened the association between PM2.5 exposure and LF reduction (P=0.05), and produced a nonsignificant attenuation of the effect of PM2.5 exposure on rMSSD, SDNN, and HF (P=0.79 for rMSSD; P=0.37 for SDNN; P=0.28 for HF) (Table 7 and Figure 6). PM2.5 exposure had no significant effect on LF in individuals with high flavonoid intake, but it was associated with increasingly negative effects on LF as flavonoid intakes decreased (Figure 6). In particular, in individuals at the first quartile of flavonoid intake, every 10 μg/m3 increase in 48‐hour PM2.5 exposure was associated with a 16.48% (95% CI, −1.61% to 31.35%; P=0.07) reduction in LF (effects estimated at the midpoint of the first quartile, ie, 128 mg/d to represent the within‐quartile effect), while no effect was seen for the second, third, and fourth flavonoid quartile (Table 7). The effect modification by flavonoid intake was abrogated when mean TLR2 methylation and the interaction between mean TLR2 methylation and PM2.5 exposure were fitted in the model (Table 8).
Table 7.
Effect of PM2.5 Exposure on HRV at Different Daily Flavonoid Intake Levels, Normative Aging Study, 2000–2011 (N=513)
| Flavonoid Intake | % Change* | 95% CI | P Value |
|---|---|---|---|
| HR | |||
| Midpoint of Q1 (128 mg/d) | 0.06 | −1.65 to 1.80 | 0.95 |
| Midpoint of Q2 (228 mg/d) | 0.00 | −1.51 to 1.53 | 1.00 |
| Midpoint of Q3 (370 mg/d) | −0.09 | −1.45 to 1.30 | 0.90 |
| Midpoint of Q4 (673 mg/d) | −0.26 | −1.98 to 1.48 | 0.76 |
| P for interaction | 0.76 | ||
| rMSSD | |||
| Midpoint of Q1 (128 mg/d) | −5.32 | −16.91 to 7.88 | 0.41 |
| Midpoint of Q2 (228 mg/d) | −4.94 | −15.25 to 6.62 | 0.39 |
| Midpoint of Q3 (370 mg/d) | −4.41 | −13.82 to 6.03 | 0.39 |
| Midpoint of Q4 (673 mg/d) | −3.25 | −15.28 to 10.49 | 0.63 |
| P for interaction | 0.79 | ||
| SDNN | |||
| Midpoint of Q1 (128 mg/d) | −6.14 | −14.57 to 3.11 | 0.19 |
| Midpoint of Q2 (228 mg/d) | −5.22 | −12.74 to 2.95 | 0.20 |
| Midpoint of Q3 (370 mg/d) | −3.89 | −10.81 to 3.56 | 0.30 |
| Midpoint of Q4 (673 mg/d) | −1.00 | −10.03 to 8.93 | 0.84 |
| P for interaction | 0.37 | ||
| LF | |||
| Midpoint of Q1 (128 mg/d) | −16.48 | −31.35 to 1.61 | 0.07 |
| Midpoint of Q2 (228 mg/d) | −12.55 | −26.37 to 3.87 | 0.13 |
| Midpoint of Q3 (370 mg/d) | −6.65 | −20.07 to 9.03 | 0.39 |
| Midpoint of Q4 (673 mg/d) | 7.31 | −12.16 to 31.11 | 0.49 |
| P for interaction | 0.05 | ||
| HF | |||
| Midpoint of Q1 (128 mg/d) | −10.42 | −28.69 to 12.54 | 0.34 |
| Midpoint of Q2 (228 mg/d) | −7.78 | −24.49 to 12.63 | 0.43 |
| Midpoint of Q3 (370 mg/d) | −3.89 | −19.74 to 15.09 | 0.67 |
| Midpoint of Q4 (673 mg/d) | 4.96 | −16.93 to 32.63 | 0.68 |
| P for interaction | 0.28 |
Results were adjusted for age; body mass index; total caloric intake; total vitamin C intake; total fiber intake; fasting glucose level; hypertension; smoking status; alcohol consumption; physical exercise; household income; the use of calcium channel blocker, β‐blocker, and angiotensin‐converting enzyme inhibitor; room temperature; outdoor apparent temperature; season; weekday; and visit date. HF indicates high‐frequency power (0.15 to 0.4 Hz); HR, heart rate; HRV, heart rate variability; LF, low‐frequency power (0.04 to 0.15 Hz); PM2.5, particulate matter with aerodynamic diameter <2.5 μm; Q1, Q2, Q3, and Q4 indicate the first, second, third, and fourth quartile; rMSSD, root mean square of successive differences; SDNN, standard deviation of normal‐to‐normal intervals.
The percent change in HRV per 10 μg/m3 increase in PM2.5 concentration.
Figure 6.
The effect of particulate matter with aerodynamic diameter <2.5 μm (PM2.5) exposure on heart rate variability (HRV) at different flavonoid intake levels, Normative Aging Study, 2000–2011 (N=513). log10HR indicates log10‐transformed heart rate; log10rMSSD, log10‐transformed root mean square of the successive differences; log10SDNN, log10‐transformed standard deviation of normal‐to‐normal intervals; log10LF, log10‐transformed low‐frequency power (0.04 to 0.15 Hz); log10HF, log10‐transformed high‐frequency power (0.15 to 0.4 Hz); Q1, Q2, Q3, and Q4 indicate the first, second, third, and fourth quartiles. The association of PM2.5 on rMSSD, SDNN, LF, and HF is modified by flavonoid intake, as indicated by the different slopes. The 4 lines in each figure represent the relationship between PM2.5 and HRV when the flavonoid intake is at the midpoints of each quartile. If there was no effect modification, the 4 lines would be the same. Results were adjusted for age; body mass index; total caloric intake; total vitamin C intake; total fiber intake; fasting glucose level; hypertension; smoking status; alcohol consumption; physical exercise; household income; the use of calcium channel blocker, β‐blocker, and angiotensin‐converting enzyme inhibitor; room temperature; outdoor apparent temperature; season; weekday; and visit date.
Table 8.
Effect Modification by Flavonoid Intake on the Association Between PM2.5 Exposure and HRV, Adjusted for Total Fruit and Vegetable Intake, and Adjusted for Mean TLR2 Methylation
| HRV Measure | Original Model (N=513)* | Adjusted for Total Fruit and Vegetable Intake (N=482)* | Adjusted for Mean TLR2 Methylation (N=423)* | ||||||
|---|---|---|---|---|---|---|---|---|---|
| % Change* | 95% CI | P for Interaction | % Change* | 95% CI | P for Interaction | % Change* | 95% CI | P for Interaction | |
| HR | −0.18 | −1.32 to 0.98 | 0.76 | −0.30 | −1.48 to 0.90 | 0.62 | 0.89 | −0.72 to 2.52 | 0.30 |
| rMSSD | 1.17 | −7.38 to 10.51 | 0.79 | 0.67 | −8.45 to 10.7 | 0.89 | 0.07 | −11.28 to 12.87 | 0.99 |
| SDNN | 2.98 | −3.43 to 9.81 | 0.37 | 2.77 | −3.88 to 9.88 | 0.42 | 2.28 | −6.49 to 11.87 | 0.62 |
| LF | 14.63 | 0.27 to 31.03 | 0.05 | 15.81 | 0.50 to 33.45 | 0.04 | 10.48 | −8.39 to 33.24 | 0.30 |
| HF | 8.98 | −6.93 to 27.62 | 0.28 | 7.65 | −9.17 to 27.59 | 0.40 | 7.65 | −12.28 to 32.10 | 0.48 |
HF indicates high‐frequency power (0.15 to 0.4 Hz); HR, heart rate; HRV, heart rate variability; LF, low‐frequency power (0.04 to 0.15 Hz); PM2.5, particulate matter with aerodynamic diameter <2.5 μm; rMSSD, root mean square of successive differences; SDNN, standard deviation of normal‐to‐normal intervals; TLR2, Toll‐like receptor 2.
Results were adjusted for age; body mass index; total caloric intake; total vitamin C intake; total fiber intake; fasting glucose level; hypertension; smoking status; alcohol consumption; physical exercise; household income; the use of calcium channel blocker, β‐blocker, and angiotensin‐converting enzyme inhibitor; room temperature; outdoor apparent temperature; season; weekday, and visit date.
In addition to all the covariates in the original model except total vitamin C and fiber intake, results were adjusted for total fruit and vegetable intake.
In addition to all the covariates in the original model, results were adjusted for mean TLR2 methylation, and the interaction between mean TLR2 methylation and PM2.5 concentration.
The additional percent change in HRV associated with 10 μg/m3 PM2.5 concentration, compared to participants with 1 interquartile range lower flavonoid intake level.
Correlation of Plasma Inflammatory Markers With TLR2 Methylation
Increased TLR2 methylation level was consistently correlated with increased plasma intercellular adhesion molecule 1 level, across 5 CpG positions (P<0.0001, P<0.0001, P=0.0002, P=0.0004, and P=0.0003, respectively) (Table 9). IL‐8 and vascular endothelial growth factor were also positively correlated with TLR2 methylation level at position 3 (P=0.01, P=0.02, respectively) (Table 9).
Table 9.
Correlation Between Mean Blood TLR2 Methylation and Plasma Inflammatory Markers
| TLR2 Methylation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Position 1 | Position 2 | Position 3 | Position 4 | Position 5 | ||||||
| Correlation Coefficient | P Value* | Correlation Coefficient | P Value* | Correlation Coefficient | P Value* | Correlation Coefficient | P Value* | Correlation Coefficient | P Value* | |
| IL‐6 | −0.067 | 0.08 | −0.025 | 0.52 | 0.067 | 0.08 | 0.040 | 0.30 | 0.020 | 0.60 |
| IL‐8 | −0.004 | 0.93 | 0.006 | 0.88 | 0.095 | 0.01 | 0.053 | 0.17 | 0.049 | 0.20 |
| IL‐1β | −0.031 | 0.42 | 0.040 | 0.31 | 0.046 | 0.23 | 0.019 | 0.63 | 0.015 | 0.70 |
| TNF‐α | −0.043 | 0.27 | −0.056 | 0.15 | 0.006 | 0.87 | −0.009 | 0.81 | −0.039 | 0.32 |
| TNFγ | −0.030 | 0.44 | −0.007 | 0.86 | −0.017 | 0.66 | −0.029 | 0.45 | −0.022 | 0.58 |
| CRP | 0.032 | 0.35 | −0.007 | 0.85 | 0.011 | 0.76 | 0.024 | 0.49 | 0.047 | 0.17 |
| ICAM‐1 | 0.138 | <0.0001 | 0.159 | <0.0001 | 0.127 | 0.0002 | 0.123 | 0.0004 | 0.124 | 0.0003 |
| VEGF | −0.022 | 0.57 | −0.016 | 0.68 | 0.091 | 0.02 | 0.050 | 0.20 | 0.046 | 0.24 |
CRP indicates C‐reactive protein; ICAM‐1, intercellular adhesion molecule 1; IL, interleukin; TLR2, Toll‐like receptor 2; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
P value represents Prob>|r| under H0: ρ (Spearman's rank correlation coefficient)=0.
Sensitivity Analyses
The major sources of dietary flavonoids and folic acid are fruits and vegetables, which contain other nutrients that are likely to affect DNA methylation. Therefore, in sensitivity analysis of models including flavonoids, we adjusted for daily total fruit and vegetable intake—instead of total vitamin C and fiber intake—to examine the robustness of our findings. This adjustment only resulted in minor changes in the effect estimates for the association between flavonoid and TLR2 methylation, as well as of the effect modification by flavonoid of the effect of PM2.5 exposure on HRV (Tables 6 and 8).
Due to the high tissue and cell‐type specificity of DNA methylation, DNA methylation in whole blood might be affected by differences in the proportions of leukocyte cell types.36 We therefore examined the correlation between the percentage of specific leukocyte types (ie, eosinophils, lymphocytes, neutrophils, monocytes, and basophils) and TLR2 methylation level. Among these 5 leukocyte types, the percentage of lymphocytes and neutrophils in total blood cells were correlated with TLR2 methylation (Table 10). Therefore, we performed sensitivity analysis adjusted for lymphocyte and neutrophil percentages. Our findings were not affected by this adjustment (Table 11).
Table 10.
Correlation Between Mean Blood TLR2 Methylation Level and Proportions of Different Leukocyte Cell Types
| Cell Type (%) | Spearman Correlation With Mean TLR2 Methylation* | |
|---|---|---|
| Correlation Coefficient | P Value* | |
| Eosinophils | 0.051 | 0.145 |
| Lymphocytes | 0.121 | 0.001 |
| Neutrophils | −0.118 | 0.001 |
| Monocytes | 0.018 | 0.604 |
| Basophils | 0.012 | 0.730 |
TLR2 indicates Toll‐like receptor 2.
Mean TLR2 methylation level across 5 CpG sites.
P value represents Prob>|r| under H0: ρ (Spearman's rank correlation coefficient)=0.
Table 11.
Effect Modification by Position‐Specific TLR2 Methylation Level on the Association Between PM2.5 Exposure and HRV, Adjusted for Cell Types
| Position | % Change*, Original Model, N=500* | % Change*, Adjusted for Cell Types, N=498* |
|---|---|---|
| HR | ||
| 1 | 0.05 | 0.03 |
| 2 | 0.17 | 0.26 |
| 3 | 0.53 | 0.60 |
| 4 | 0.36 | 0.44 |
| 5 | 0.83* | 0.85* |
| rMSSD | ||
| 1 | −10.07* | −9.64* |
| 2 | −5.60* | −6.10* |
| 3 | −7.35* | −7.45* |
| 4 | −6.55* | −6.48* |
| 5 | −8.29* | −7.88* |
| SDNN | ||
| 1 | −5.34* | −5.01* |
| 2 | −0.34 | −0.72 |
| 3 | −3.87* | −3.97* |
| 4 | −1.62 | −1.56 |
| 5 | −4.58* | −4.21* |
| LF | ||
| 1 | −2.45 | −1.65 |
| 2 | 1.75 | 1.39 |
| 3 | −8.77* | −8.90* |
| 4 | −1.22 | −1.23 |
| 5 | −7.09 | −6.46 |
| HF | ||
| 1 | −9.72 | −8.72 |
| 2 | −5.31 | −6.35 |
| 3 | −11.01* | −11.31* |
| 4 | −7.63 | −7.74 |
| 5 | −14.20* | −13.33* |
HF indicates high‐frequency power (0.15 to 0.4 Hz); HR, heart rate; HRV, heart rate variability; LF, low‐frequency power (0.04 to 0.15 Hz); PM2.5, particulate matter with aerodynamic diameter <2.5 μm; rMSSD, root mean square of successive differences; SDNN, standard deviation of normal‐to‐normal intervals; TLR2, Toll‐like receptor 2.
Results were adjusted for age, body mass index, fasting glucose level, hypertension, smoking status, alcohol consumption, physical exercise, household income, medication use (β‐blocker, calcium channel blocker, angiotensin‐converting enzyme inhibitor), room temperature, outdoor apparent temperature, season, weekday, and visit date.
Results were adjusted for all covariates in the original model and the percentage of neutrophil and lymphocyte.
P≤0.05, statistically significant.
The additional percent change in HRV associated with 10 μg/m3 increase in PM2.5 concentration, compared to participants with one %5mC lower TLR2 methylation level.
Discussion
This study on a cohort of aging male Boston‐area residents demonstrated the adverse impact of short‐term PM2.5 exposure estimated at the home address on HRV. In addition, novel findings showed that individuals with higher methylation in the TLR2 gene promoter are more susceptible to reduced HRV after PM2.5 exposure, compared to those with lower methylation levels. Furthermore, higher flavonoid intake in the year preceding the examination significantly lowered TLR2 methylation, and attenuated the association between PM2.5 exposure and the LF measure of HRV.
PM exposure is a modifiable factor that contributes to cardiovascular morbidity and mortality, particularly in the acute exposure period, as emphasized by the recent American Heart Association statement on air pollution.2 Consistent with our previous study,30 we observed the strongest effect on HRV from PM2.5 among major types of air pollutants. The negative association between short‐term PM2.5 exposure and HRV has been repeatedly observed in general population samples,37 cardiac and hypertensive patients,38 asthmatic adults,39 young adults,40 and older adults,4,30 as well as in a recent meta‐analysis of 29 different studies (N=18 667).41 The present study showed effects of PM2.5 exposure across 4 different indices of HRV, but not on HR. This finding suggests that short‐term PM2.5 exposure may affect the cardiac interbeat intervals before any appreciable influence is detected on HR. SDNN, a marker of the cyclic components, represents the total variability over the 7 minutes of ECG monitoring. LF variability is linked to the activity of both the sympathetic and parasympathetic nervous system, whereas HF and rMSSD are sensitive to high‐frequency heart period fluctuations and reflect the parasympathetic nervous activity. The HRV reductions associated with PM2.5 in these 4 indices may reflect pathophysiological changes in cardiac autonomic balance following exposure, which could be a potential mechanism linking air pollution and cardiac morbidity and mortality in the aging population.5
Many epidemiological studies4,30 have presented a wide interindividual variability in responses to short‐term air pollution, possibly due to biological characteristics.7 For example, preexisting cardiac conditions, diabetes, and polymorphism of genes involved in endothelial function may confer higher susceptibility to autonomic dysfunction induced by PM2.5 exposure.4,30 However, these characteristics are either nonmodifiable or already well‐recognized conditions with specific treatment guidelines. This study revealed that increased TLR2 methylation is linked with intensified inflammatory responses, as reflected in elevated plasma IL‐8, intercellular adhesion molecule 1, and vascular endothelial growth factor level. In addition, we demonstrated that low TLR2 methylation, an epigenetic mechanism ensuring immunoregulation, reduces the adverse effects of PM2.5 exposure on HRV. TLR2 provides a critical line of protection by inducing regulatory T cells to limit immune‐mediated damage.13–14 Netea et al reported a 50% decrease in the number of CD4+CD25+ regulatory T cells accompanied by an impaired release of immunosuppressive cytokines in TLR2 null mice compared with wild‐type mice, demonstrating the role of TLR2 in immunoregulation.14 These findings support our hypothesis that high TLR2 methylation, an epigenetic process associated with suppressed TLR2 expression, conveys susceptibility to PM exposure‐induced HRV reductions by decreasing immunoregulation.
Our data presented notable effect heterogeneity across the 5 CpG positions within the TLR2 promoter region. The distance from position 1 to position 5 is <20 base pairs; thus, these sites are likely to share most of the same functional complexes and traits (ie, CpG islands, DNaseI hypersensitivity, nuclease accessibility, and histone modifications [UCSC genome browser]) (Figure 2). However, differential methylation patterns across 5 positions have been observed in the ENCODE project, as identified by the Illumina Infinium Human Methylation 450 Bead Array platform. This suggests that variable methylation patterns exist at these sites even though the CpGs share common functional complexes.
TLR2 methylation is sensitive to dietary and other behavioral factors.42 In particular, flavonoids, a major subtype of polyphenols, are established inhibitors for methylation‐induced gene silencing.17–18 Flavonoids inhibit the activity of DNA methyltransferase in a dose–response manner, reverse DNA methylation, and can reactivate silenced genes.17–18,43 Our study showed that dietary flavonoid intake over the previous year is negatively associated with TLR2 methylation, indicating that adequate flavonoid intake may be essential to maintain a low, protective TLR2 methylation level. In addition, we showed that increased flavonoid intake alleviates the adverse effect of PM2.5 exposure on LF, and the effect modification is diminished when the model is adjusted for TLR2 methylation. This finding suggests that, at least in part, the average daily flavonoid intake over the preceding year affects individual susceptibility to postexposure HRV reduction by modulating TLR2 methylation level. However, formal mediation analysis was not performed in the present study due to the lack of appropriate statistical methodology to identify the potential pathway between 2 effect modifiers in repeated‐measure data.
We also investigated a group of methyl nutrients including folic acid, vitamin B12, and methionine, which are essential components in the 1‐carbon metabolism pathway that is expected to favor higher blood DNA methylation.19 However, no evidence to date demonstrates whether methyl nutrients can specifically modulate immune genes.44 Our analysis did not show significant correlation between methyl nutrients intakes and TLR2 methylation status. In our previously published study, PM2.5 was negatively associated with HRV in subjects with lower vitamin B12 and methionine intakes, but not in the higher‐intake groups. Our findings indicated that the effect modification by vitamin B12 and methionine intake reported in our previous study is likely to act through a different mechanism, independent of TLR2 methylation.
This study has several strengths, including its prospective study design and relatively large sample size that enabled us to determine effect modifications by TLR2 methylation and flavonoid intakes. To demonstrate the power of the present study, we performed simulation‐based power calculation using linear mixed‐effect models with compound symmetry covariance structure, accounting for covariates adjustments. Specifically, we generated 1000 simulated data sets, each with N=500 independent subjects, in which each subject had 2 (correlated) observations. The exposure, effect modifier, and outcome were assumed to be normally distributed. A random intercept linear mixed‐effect model (with compound symmetry correlation structure) was then fitted to each of the simulated data sets. Using effect sizes for both the main effect of PM2.5 exposure and the effect modification after adjustment for covariates estimated from our data, we expected to have >90% power to detect the observed main effect of PM on HRV indices, and >80% power to detect the observed effect modification by TLR2 methylation and flavonoid intakes at α=0.05 level. Exposure misclassification, which is inherent in epidemiological studies on air pollution, is a limitation of the present study. Most previous studies relied on a few monitoring sites in the study area as surrogates for recent PM2.5 exposure history; that approach introduced measurement errors leading to underestimation of the effect.45 We reduced misclassification by using a novel, validated, hybrid prediction model to assess temporally and spatially resolved PM2.5 exposure level,24 and the observed effect size of PM2.5 exposure on HRV is larger compared to a previous study from the same cohort using monitoring site data.4 This prediction model accounted for the spatial variability of PM2.5 concentration within the study area and produced more accurate estimates of recent PM2.5 exposure at a 10×10‐km resolution. The Boston metropolitan area comprises 15 corresponding model grid cells, and the use of the cell‐specific predictions for each participant enabled us to assign different exposure levels to participants on a relatively small grid based on their residential areas.26 Although measurement error cannot be completely avoided, it was unlikely to be associated with participants' HRV status. Therefore, misclassification is likely nondifferential and is expected to bias our result toward the null. Acknowledging that confounding is a critical concern in any observational study, we included in regression models an extensive list of covariates. We conducted further analysis to evaluate the sensitivity of our results to covariate specification, and our results were stable and robust. We also examined the correlation between 48‐hour and 1‐year moving average of PM2.5 to rule out the possibility that the observed effect was partially due to correlation of the 48‐hour moving average with long‐term exposure to PM2.5. These 2 exposure metrics were not correlated (R2=0.07); therefore, the association between the 48‐hour moving average of PM2.5 and HRV was unlikely to be confounded by long‐term PM2.5 exposure. In addition, all HRV measures were conducted at the same time of the day to eliminate confounding due to diurnal variation. While residual confounding due to unmeasured variables is not unlikely, chances that the observed association and effect modification reflected bias resulting from residual confounding are minimized.
We acknowledge several other limitations in the present study. During the follow‐up of the present study—when data on PM exposure are available—there were 636 eligible Normative Aging Study participants (Figure 1). However, our analysis only included a study population of 573 participants due to missing PM exposure or HRV data. To examine whether the final analysis sample is different from the eligible population, we compared key baseline characteristics (age, race, physical activity, alcohol consumption, smoking status, hypertension, diabetes, and household annual income) between participants excluded in the final analysis due to missing exposure or outcome (N=63) and the participants for final analysis (N=573), and no significant difference was observed (data not shown). In addition, in the analysis investigating effect modification by TLR2 methylation, 73 of 573 participants were excluded due to lack of DNA samples for methylation analysis. In the analysis of flavonoid intake and TLR2 methylation, 60 of 573 participants were excluded due to lack of FFQ data. However, based on the study operations, it is reasonable to assume that the missingness was independent of participants' exposure level, HRV status, and methylation states. We compared the HRV measures between those with complete data (N=500/N=513) and those without (N=73/N=60), and no apparent difference was observed (data not shown). Therefore, selection bias due to informative missingness in this study was unlikely. Both the within‐person and between‐person effects contributed to the effect modification by TLR2 methylation. However, during the study period, the effect modification by TLR2 methylation was mostly due to between‐person effects, as when the models were adjusted for subject identification, the significance of effect modification was diminished (data not shown). Due to the limited number of within‐subject repeated visits, we were unable to identify the exact magnitude of effect modification by TLR2 methylation within subject. In addition, our findings are limited to male older individuals who were residing in a lightly polluted urban area. Therefore, the conclusion might not be generalizable to young adults, females, or people living in other areas, due to differential environmental and physiological factors.
Finally, alveolar inflammation and the subsequent systemic inflammation are essential, but not the only mechanism linking PM2.5 exposure and cardiac dysfunction. For example, particles may perturb the autonomic nervous system balance via interaction with lung receptors; a small fraction of PM (ultrafine particles, and some PM constituents such as organic compounds and metals) may translocate into the systemic circulation.2 Due to the less‐than‐comprehensive analysis on plasma inflammatory marker profile and limited sample size, we were unable to draw a definite conclusion on the mechanistic pathway underlying the protective effect of low TLR2 methylation status and high flavonoid intake. Future studies are warranted to identify the precise pathophysiological processes of PM‐induced cardiovascular responses, as well as the mechanistic underpinnings of the effect of TLR2 methylation and dietary flavonoid intake.
The United States is an aging society, and older people are especially susceptible to air pollution–triggered cardiovascular disease. As air pollution remains an important ubiquitous public health threat, PM2.5‐associated HRV reduction is an alarming signal of influences on the autonomic regulation of heart rate in older people. These alterations may represent a primary sentinel effect to identify the influence of PM2.5 among older individuals or even reflect intermediate mechanisms that may favor acute cardiovascular events.2–3 Providing preventive strategies at the individual level is especially important in cardiovascular research, which was recognized as an important new emphasis in the American Heart Association statement on air pollution.2 Our study provides a novel perspective on individual susceptibility to adverse cardiovascular effects following short‐term air pollution; our findings suggest high TLR2 methylation as a modifiable “epigenetic predisposition” for cardiovascular responses that are associated with short‐term PM2.5 exposure in older people. In addition, based on our data, increasing flavonoid intake—which could be achieved by increasing intake of 1 or more of the major sources of dietary flavonoids (tea, beans, wine, berries, and citrus fruits)—may help maintain a low TLR2 methylation level and mitigate PM2.5 exposure‐induced cardiovascular system impairment.
In summary, our findings suggest that the epigenetic regulation of TLR2‐related immunity may determine vulnerability of older individuals when confronted with air pollution peaks, and that adequate dietary flavonoid intake may ensure a protective epigenetic status against the adverse effects of PM2.5 exposure.
Sources of Funding
This study was supported by NIH grants R01ES015172, R21ES021895, R01ES021733, R01ES021357, and P30ES000002; and U.S. Environmental Protection Agency funding (RD‐83479801). The VA Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center, Boston, Massachusetts. David Sparrow was supported by a VA Research Career Scientist award.
Disclosures
None.
References
- World Health Organization. Burden of Disease From Ambient Air Pollution for 2012—Summary of Results. 2014Geneva, Switzerland: WHO; 2014 [Google Scholar]
- Brook RD, Rajagopalan S, Pope CA, III, Brook JR, Bhatnagar A, Diez‐Roux AV, Holguin F, Hong Y, Luepker RV, Mittleman MA, Peters A, Siscovick D, Smith SC, Jr, Whitsel L, Kaufman JDAmerican Heart Association Council on Epidemiology and Prevention, Council on the Kidney in Cardiovascular Disease, Council on Nutrition, Physical Activity & Metabolism. Particulate matter air pollution and cardiovascular disease: an update to the scientific statement from the American Heart Association. Circulation. 2010; 121:2331-2378. [DOI] [PubMed] [Google Scholar]
- Baccarelli A, Cassano PA, Litonjua A, Park SK, Suh H, Sparrow D, Vokonas P, Schwartz J. Cardiac autonomic dysfunction: effects from particulate air pollution and protection by dietary methyl nutrients and metabolic polymorphisms. Circulation. 2008; 117:1802-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Baccarelli A, Wilker E, Suh H, Sparrow D, Vokonas P, Wright R, Schwartz J. Lipid and endothelium‐related genes, ambient particulate matter, and heart rate variability—the VA Normative Aging Study. J Epidemiol Community Health. 2010; 64:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA, III, Verrier RL, Lovett EG, Larson AC, Raizenne ME, Kanner RE, Schwartz J, Villegas GM, Gold DR, Dockery DW. Heart rate variability associated with particulate air pollution. Am Heart J. 1999; 138:890-899. [DOI] [PubMed] [Google Scholar]
- Stein PK, Barzilay JI, Chaves PH, Mistretta SQ, Domitrovich PP, Gottdiener JS, Rich MW, Kleiger RE. Novel measures of heart rate variability predict cardiovascular mortality in older adults independent of traditional cardiovascular risk factors: the Cardiovascular Health Study (CHS). J Cardiovasc Electrophysiol. 2008; 19:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunekreef B, Holgate ST. Air pollution and health. Lancet. 2002; 360:1233-1242. [DOI] [PubMed] [Google Scholar]
- Stone PH, Godleski JJ. First steps toward understanding the pathophysiologic link between air pollution and cardiac mortality. Am Heart J. 1999; 138:804-807. [DOI] [PubMed] [Google Scholar]
- Schins RP, Lightbody JH, Borm PJ, Shi T, Donaldson K, Stone V. Inflammatory effects of coarse and fine particulate matter in relation to chemical and biological constituents. Toxicol Appl Pharmacol. 2004; 195:1-11. [DOI] [PubMed] [Google Scholar]
- Zeka A, Sullivan JR, Vokonas PS, Sparrow D, Schwartz J. Inflammatory markers and particulate air pollution: characterizing the pathway to disease. Int J Epidemiol. 2006; 35:1347-1354. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006; 124:783-801. [DOI] [PubMed] [Google Scholar]
- Becker S, Fenton MJ, Soukup JM. Involvement of microbial components and toll‐like receptors 2 and 4 in cytokine responses to air pollution particles. Am J Respir Cell Mol Biol. 2002; 27:611-618. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhou S, Chi Y, Wen X, Hoellwarth J, He L, Liu F, Wu C, Dhesi S, Zhao J, Hu W, Su C. CD4+CD25+ Treg induction by an HSP60‐derived peptide SJMHE1 from Schistosoma japonicum is TLR2 dependent. Eur J Immunol. 2009; 39:3052-3065. [DOI] [PubMed] [Google Scholar]
- Netea MG, Sutmuller R, Hermann C, Van der Graaf CA, Van der Meer JW, van Krieken JH, Hartung T, Adema G, Kullberg BJ. Toll‐like receptor 2 suppresses immunity against Candida albicans through induction of IL‐10 and regulatory T cells. J Immunol. 2004; 172:3712-3718. [DOI] [PubMed] [Google Scholar]
- Furuta T, Shuto T, Shimasaki S, Ohira Y, Suico MA, Gruenert DC, Kai H. DNA demethylation‐dependent enhancement of toll‐like receptor‐2 gene expression in cystic fibrosis epithelial cells involves SP1‐activated transcription. BMC Mol Biol. 2008; 9:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol. 2012; 863:359-376. [DOI] [PubMed] [Google Scholar]
- Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (−)‐epigallocatechin‐3‐gallate inhibits DNA methyltransferase and reactivates methylation‐silenced genes in cancer cell lines. Cancer Res. 2003; 63:7563-7570. [PubMed] [Google Scholar]
- Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007; 137:223S-228S. [DOI] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005; 135:1382-1386. [DOI] [PubMed] [Google Scholar]
- Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, Schwartz J. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene‐environment interactions in an elderly cohort. Epidemiology. 2012; 23:332-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camm AJ, Malik M, Bigger JT, Breithardt G, Cerutti S, Cohen RJ, Coumel P, Fallen EL, Kennedy HL, Kleiger RE, Lombardi F, Malliani A, Moss AJ, Rottman JN, Schmidt G, Schwartz PJ, Singer DH. Heart rate variability: standards of measurement, physiological interpretation and clinical use. Task force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation. 1996; 93:1043-1065. [PubMed] [Google Scholar]
- Baccarelli A, Tarantini L, Wright RO, Bollati V, Litonjua AA, Zanobetti A, Sparrow D, Vokonas P, Schwartz J. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA Normative Aging Study. Epigenetics. 2010; 5:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang SC, Mehta AJ, Alexeeff SE, Gryparis A, Coull B, Vokonas P, Christiani D, Schwartz J. Residential black carbon exposure and circulating markers of systemic inflammation in elderly males: the Normative Aging Study. Environ Health Perspect. 2012; 120:674-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloog I, Chudnovsky A, Koutrakis P, Schwartz J. Temporal and spatial assessments of minimum air temperature using satellite surface temperature measurements in Massachusetts, USA. Sci Total Environ. 2012; 432:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigano J, Kloog I, Goldberg R, Coull BA, Mittleman MA, Schwartz J. Long‐term exposure to PM2.5 and incidence of acute myocardial infarction. Environ Health Perspect. 2013; 121:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloog I, Ridgway B, Koutrakis P, Coull BA, Schwartz JD. Long‐ and short‐term exposure to PM2.5 and mortality: using novel exposure models. Epidemiology. 2013; 24:555-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanosky JD, Paciorek CJ, Suh HH. Predicting chronic fine and coarse particulate exposures using spatiotemporal models for the northeastern and midwestern United States. Environ Health Perspect. 2009; 117:522-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloog I, Nordio F, Coull BA, Schwartz J. Incorporating local land use regression and satellite aerosol optical depth in a hybrid model of spatiotemporal PM2.5 exposures in the mid‐Atlantic states. Environ Sci Technol. 2012; 46:11913-11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloog I, Koutrakis P, Coull BA, Lee HJ, Schwartz J. Assessing temporally and spatially resolved PM2.5 exposures for epidemiological studies using satellite aerosol optical depth measurements. Atmos Environ. 2011; 45:6267-6275. [Google Scholar]
- Park SK, O'Neill MS, Vokonas PS, Sparrow D, Schwartz J. Effects of air pollution on heart rate variability: the VA Normative Aging Study. Environ Health Perspect. 2005; 113:304-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett WC, Sampson L, Stampfer MJ, Rosner B, Bain C, Witschi J, Hennekens CH, Speizer FE. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol. 1985; 122:51-65. [DOI] [PubMed] [Google Scholar]
- Rimm EB, Giovannucci EL, Stampfer MJ, Colditz GA, Litin LB, Willett WC. Reproducibility and validity of an expanded self‐administered semiquantitative food frequency questionnaire among male health professionals. Am J Epidemiol. 1992; 135:1114-1126. [DOI] [PubMed] [Google Scholar]
- Cassidy A, O'Reilly EJ, Kay C, Sampson L, Franz M, Forman JP, Curhan G, Rimm EB. Habitual intake of flavonoid subclasses and incident hypertension in adults. Am J Clin Nutr. 2011; 93:338-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat SA, Haytowitz DB, Prior RL, Gu L, Hammerstone J, Gebhardt SE. USDA Database for Proanthocyanidin Content of Selected Foods. 2004. http://www.ars.usda.gov/SP2UserFiles/Place/80400525/Data/PA/PA.pdf
- Bhagwat SA, Gebhardt SE, Haytowitz DB, Holden JM, Harnly JM. USDA Database for the Flavonoid Content of Selected Foods Release 2.1. 2007. http://www.ars.usda.gov/SP2UserFiles/Place/80400525/Data/Flav/Flav02-1.pdf
- Cowley AW, Jr, Nadeau JH, Baccarelli A, Berecek K, Fornage M, Gibbons GH, Harrison DG, Liang M, Nathanielsz PW, O'Connor DT, Ordovas J, Peng W, Soares MB, Szyf M, Tolunay HE, Wood KC, Zhao K, Galis ZS. Report of the National Heart, Lung, and Blood Institute working group on epigenetics and hypertension. Hypertension. 2012; 59:899-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min KB, Min JY, Cho SI, Paek D. The relationship between air pollutants and heart‐rate variability among community residents in Korea. Inhal Toxicol. 2008; 20:435-444. [DOI] [PubMed] [Google Scholar]
- Chuang KJ, Chan CC, Chen NT, Su TC, Lin LY. Effects of particle size fractions on reducing heart rate variability in cardiac and hypertensive patients. Environ Health Perspect. 2005; 113:1693-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power KL, Balmes J, Solomon C. Controlled exposure to combined particles and ozone decreases heart rate variability. J Occup Environ Med. 2008; 50:1253-1260. [DOI] [PubMed] [Google Scholar]
- Chuang KJ, Chan CC, Su TC, Lee CT, Tang CS. The effect of urban air pollution on inflammation, oxidative stress, coagulation, and autonomic dysfunction in young adults. Am J Respir Crit Care Med. 2007; 176:370-376. [DOI] [PubMed] [Google Scholar]
- Pieters N, Plusquin M, Cox B, Kicinski M, Vangronsveld J, Nawrot TS. An epidemiological appraisal of the association between heart rate variability and particulate air pollution: a meta‐analysis. Heart. 2012; 98:1127-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegria‐Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. 2011; 3:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005; 68:1018-1030. [DOI] [PubMed] [Google Scholar]
- Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate's role. Adv Nutr. 2012; 3:21-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeger SL, Thomas D, Dominici F, Samet JM, Schwartz J, Dockery D, Cohen A. Exposure measurement error in time‐series studies of air pollution: concepts and consequences. Environ Health Perspect. 2000; 108:419-426. [DOI] [PMC free article] [PubMed] [Google Scholar]