Abstract
Purpose
To determine the temporal and spatial expression of Pitx2, a bicoid-like homeobox transcription factor, during postnatal development of mouse extraocular muscle and to evaluate its role in the growth and phenotypic maintenance of postnatal extraocular muscle.
Methods
Mouse extraocular muscles of different ages were examined for the expression of Pitx2 by RT-PCR, q-PCR, and immunostaining. A conditional mutant mouse strain, in which Pitx2 function is inactivated at postnatal day (P)0, was generated with a Cre-loxP strategy. Histology, immunostaining, realtime PCR, in vitro muscle contractility, and in vivo ocular motility were used to study the effect of Pitx2 depletion on extraocular muscle.
Results
All three Pitx2 isoforms were expressed by extraocular muscle and at higher levels than in other striated muscles. Immunostaining demonstrated the presence of Pitx2 mainly in extraocular muscle myonuclei. However, no obvious expression patterns were observed in terms of anatomic region (orbital versus global layer), innervation zone, or muscle fiber types. The mutant extraocular muscle had no obvious pathology but had altered muscle fiber sizes. Expression levels of myosin isoforms Myh1, Myh6, Myh7, and Myh13 were reduced, whereas Myh2, Myh3, Myh4, and Myh8 were not affected by postnatal loss of Pitx2. In vitro, Pitx2 loss made the extraocular muscles stronger, faster, and more fatigable. Eye movement recordings found saccades to have a lower peak velocity.
Conclusions
Pitx2 is important in maintaining the mature extraocular muscle phenotype and regulating the expression of critical contractile proteins. Modulation of Pitx2 expression can influence extraocular muscle function with long-term therapeutic implications.
Extraocular muscle is fundamentally distinct from other skeletal muscle and demonstrates specific anatomic divisions, unique muscle fiber types, diverse myosin isoform expression patterns, and a distinct genomic profile, as well as a differential involvement by neuromuscular disorders.1 Its unique properties are driven by the requirements of the visual system for rapid, coordinated eye movements as well as fatigue resistance.1,2 The maintenance of the mature extraocular muscle phenotype relies on cell-autonomous and non–cell-autonomous regulatory mechanisms. Dissection of these regulatory mechanisms may make it possible to exploit them to modify extraocular muscle contractile properties and influence eye movements. Achievement of such a goal may lead to treatments for strabismus and other disorders of ocular motility. Appreciation of phenotypic regulation may also lead to an understanding of disease characteristics, such as the sparing of extraocular muscle by most muscular dystrophies3 and their preferential involvement in Graves' ophthalmopathy4 and neuromuscular transmission disorders, such as myasthenia gravis.5
Signaling cascades controlled by transcription factors direct extraocular muscle expression of a diverse array of traits that adapt to the wide dynamic physiologic range required by eye movement control systems. The paired-like homeodomain transcription factor 2 (Pitx2) plays a critical developmental role in formation of extraocular muscle.6–10 The Pitx2 protein was detected as early as E13.5 in mouse extraocular muscle and Pitx2 expression by extraocular muscle precursors is essential for early development, as deletion by homologous recombination leads to complete agenesis of mouse extraocular muscle.6 Haploinsufficiency of PITX2 has also been identified as the cause of the human disorder Axenfeld-Rieger syndrome, in which patients exhibit abnormal development of the anterior segment and are at high risk of glaucoma. The degree of ocular motility or extraocular muscle abnormalities in this syndrome have not been well-characterized,11 and their exact relationship to the Pitx2 deficit is uncertain, since visual loss itself could alter extraocular muscle development.12–14
Genomic profiling demonstrated that Pitx2 is expressed at high levels in adult rodent extraocular muscle.15 Thus, we hypothesized that Pitx2 plays a role in maintaining the mature extraocular muscle phenotype. To lay the groundwork for testing the hypothesis, we examine temporal expression pattern of Pitx2 mRNA and Pitx2 protein distribution in mature mouse extraocular muscle. We then tested our hypothesis directly by characterizing extraocular muscle in a mouse strain with a conditional knockout of Pitx2, which allows for the normal expression of Pitx2 to occur until approximately P0, but then is absent in the adult.
Methods
Animal Husbandry
Two mouse strains were crossed to generate the Pitx2 conditional knockout mice: The muscle creatine kinase (Mck)-Cre mouse strain was obtained from Ronald Kahn (Harvard University).16 The Mck-Cre mice, which are backcrossed to the C57BL/6 for more than three generations, express CRE recombinase under the direction of the Mck promoter.16 The Pitx2flox/flox mouse strain, in which the DNA binding homeodomain exon 4 of the Pitx2 gene is flanked with the LoxP sequence, was obtained from Philip Gage (University of Michigan).17 MCK-CRE is expressed predominantly in skeletal and cardiac muscle at the time of normal expression of the Mck gene, which in mice is around the time of birth.18,19 In rodents, Mck expression in extraocular muscle is lower than in cardiac and limb skeletal muscle20,21 but is still sufficient to trigger loxP excision of Pitx2 within the extraocular muscle. Genotyping was performed by PCR using genomic DNA isolated from tail tips. Identical PCR were also used to confirm the excision of genomic DNA isolated from extraocular muscle of mice that were both Pitx2flox/flox and Cre positive (referred to as the conditional mutant, Pitx2Δflox/Δflox mice; their littermates Pitx2flox/flox mice were used as control animals). Animals were housed and handled in accordance with National Institutes of Health (NIH) guidelines for animal care. All procedures involving mice were approved by Institutional Animal Use and Care Committees at Case Western Reserve University, University of Kentucky, and Saint Louis University. All experiments were conducted in accordance with the principles and procedures established by the NIH and the Association for Assessment of Laboratory Animal Care and in the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Tissue Preparation and General Histology
Extraocular muscle and other muscles were dissected from C57BL/6 mice after euthanatization by CO2 asphyxiation at postnatal day (P)0, 3 weeks, and 6 weeks and from Pitx2Δflox/Δflox mice and control littermates Pitx2flox/flox at 3 weeks, 6 weeks, and 3 months of age. Pitx2Δflox/Δflox mice were not studied at P0 because such mice would not have complete Cre-mediated recombination. After dissection, the muscles were immediately frozen in liquid N2-cooled 2-methylbutane and stored at −80°C until use. Ten-micrometer sections were prepared and analyzed with hematoxylin and eosin or Gomori's trichrome.22,23
Antibodies
The P2Y4 antibody (provided by TH) or P2R10 (provided by Philip Gage) was used to identify Pitx2 protein expression. The antibody detects an epitope of Pitx2 encoded by a region in exon 4 that precedes the homeodomain and common to all three isoforms. P2R10 has been used in previous studies of several tissues, including muscle.7,24-28 The sources and working dilutions for antibodies against myosin heavy chain isoforms were as follows: mouse anti-extraocular muscle-specific myosin29 (1:100, provided by Neal Rubinstein, University of Pennsylvania), mouse anti-developmental myosin (1:50; Vector Laboratories, Burlingame, CA), mouse anti-fast myosin (1:500, clone 32; Sigma-Aldrich, St. Louis, MO), mouse anti-slow myosin (1:400; Chemicon, Temecula, CA). Mouse anti-myosin heavy chain 2A (SC-71) and mouse anti-myosin heavy chain 2B (BF-F3) were obtained as hybridomas from ATCC (Manassas, VA) and the supernatants of cell cultures were used. Mouse anti-dystrophin (1:50; Novocastra Laboratories, Newcastle-upon-Tyne, UK) was used to identify myotube boundaries. 4′,6-Diamidino-2-phenylindole (DAPI; Invitrogen, Carlsbad, CA) was used to identify the cell nuclei. Sulforhodamine 101-α-bungarotoxin (red, 1:500; Biotium, Inc., Hayward, CA) or Alexa Fluor 488-α-bungarotoxin (green, 1:500, Invitrogen-Molecular Probes, Eugene, OR) was used to identify neuromuscular junctions. All secondary antibodies were obtained from Molecular Probes and used at 1:500 dilutions: Alexa Fluor 594 goat anti-rabbit detecting Pitx2 antibody binding, Alexa Flour 488 goat anti-mouse IgG identifying the dystrophin antibody, and all the MHC antibodies, with the exception of the extraocular muscle specific myosin antibody and BF-F3, which were identified by Alexa Fluor 488 goat anti-mouse IgM.
Immunohistochemistry
Cryosections of extraocular muscle from P0, 3-week-old, and 6-week-old C57BL/6 mice were air dried, washed with PBS and blocked with 10% normal goat serum. To assess normal expression of Pitx2 and a correlation with known differentially expressed proteins, the sections were immunostained with the P2Y4 antibody, to detect Pitx2 expression, or doubly labeled with mouse anti-dystrophin, Alexa Fluor 488 labeled α-bungarotoxin, or MHC isoform antibodies. Cryosections from Pitx2Δflox/Δflox and Pitx2flox/flox mice were immunostained with antibodies against MHC isoforms. Sections were examined with a fluorescence microscope (Diaphot Nikon Instruments Inc., Melville, NY) and images captured with a digital camera (Spot; Diagnostic Instruments, Sterling Heights, MI) and software (SpotAdvanced; Diagnostic Instruments) before processing with Photoshop (San Jose, CA).
Extraocular Muscle Fiber Distribution
Cryosections of the rectus muscle were immunostained with mouse anti-dystrophin antibody, which marked the myofiber boundary. Twelve-bit images were collected (Retiga EXI; Q-Imaging, Vancouver, BC, Canada) with an automated microscope (DMI 6000B; Leica, Deerfield, IL) equipped with a motorized stage. Commercial image-analysis software (Metamorph; Molecular Devices, Downingtown, PA) was then used to threshold the dystrophin-immunoreactive boundaries of the muscle fibers, whereas non-muscle boundaries were manually eliminated. The number of muscle fibers within the inferior rectus muscle and the cross-sectional area of each individual fiber were automatically measured by the software, and a distribution plot produced.
RNA Extraction and RT-PCR
Total RNA was extracted from the tissues (TRIzol reagent; Invitrogen) and reverse transcription was performed (Superscript First-Strand Synthesis System; Invitrogen) according to the manufacturer's instructions. Amplification of cDNA was performed (TaqMan DNA polymerase; Invitrogen). Two sets of PCR reactions were designed to detect evidence of recombination with a common reverse primer (5′ GCC AGG CTC GAG TTA CAT TGT 3′) from the C-terminal region of Pitx2. Set 1 with the forward primer (5′ GGC AGC CGT TGA ATG TCT CTT 3′) from the N-terminal region of the Pitx2c isoform amplified a truncated product of 590 bp from extraocular muscle tissue of Pitx2Δflox/Δflox mouse compared to the 798-bp wild-type product. Set 2 with the forward primer (5′ AAG ATA AGG GCC AGC AAG GAA AGA 3′) from within the homeodomain region produced no product from extraocular muscle tissue of Pitx2Δflox/Δflox mice, while amplification from Pitx2flox/flox control mice produced a 658-bp product. Aliquots of PCR products were typically resolved by using 1% agarose gel electrophoresis with ethidium bromide (Sigma-Aldrich) for visualization and confirmation of product size.
Quantitative Real-Time PCR (qPCR)
Transcript-specific primers (Supplementary Table S1, http://www.iovs.org/cgi/content/full/50/10/4531/DC1) were designed (Primer Express 2.0 software; Applied Biosystems, Inc. [ABI], Foster City, CA) and specificity confirmed by BLAST (www.ncbi.nlm.nih.gov/blast/ provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD). qPCR was performed with SYBR green PCR core reagent (ABI) in a 24-μL reaction volume, with a sequence detection system (Prism model 7000; ABI). GAPDH was used as an internal positive loading control. Relative transcript abundance was normalized to the amount of GAPDH and calculated as the ratio to a specific baseline level or in the case of Pitx2Δflox/Δflox mice to Pitx2flox/flox and reported as the mean change (x-fold) ± SE. Change values represent averages from triplicate measurements, determined by the 2–ΔΔCT method.30
Contractility of Extraocular Muscle
The contractile function of control and conditional mutant extraocular muscles was studied as described previously.14 Whole extraocular muscles from Pitx2Δflox/Δflox and age-matched Pitx2flox/flox mice were isolated and firmly attached to a force transducer (AE801; SensoNor, Horten, Norway) and the movable arm of a servomotor (Aurora Scientific, Aurora, ON, Canada) and positioned between platinum electrodes inside a muscle chamber. The chamber was superfused with a physiological salt solution (in mM): 137 NaCl, 5 KCl, 2.0 CaCl2, 1.0 MgSO4, 1.0 Na2HPO4, 24 NaHCO3, 11 glucose, and 0.026 D-tubocurarine, bubbled with a 95% O2 and 5% CO2 gas mixture to maintain pH at 7.4 at 25°C. The muscles were stretched to the length that gave maximum tetanic force (optimal length, L0). Force measurements (P0, in Newtons) were normalized to muscle cross-sectional area (in square centimeters). The unloaded shortening velocity (V0, in L0 seconds−1) was determined with slack tests. Fatigue was induced by stimulating muscles at a frequency giving approximately one half of maximum tetanic force (50-70 Hz) for 500 ms, followed by a 1.5-second interval between contractions until force declined to 50% of initial force or 10 minutes. All results are presented as the mean ± SD of eight muscles per group. Statistical significance was determined at the 95% confidence level by using Student's t-test for paired samples as indicated; the treatment effect in the fatigue runs was determined by analysis of variance.
Ocular Motility Recording
Eye movement recordings were performed as described previously14,31,32 with the modification that since recordings were performed in the light only, animals were not treated with the ophthalmic physostigmine as ordinarily administered in our studies to limit pupil dilation in darkness, and this limits any concerns that the physostigmine may have subtle effects at the neuromuscular junction. Data were collected to generate plots of saccade peak eye velocity versus saccade amplitude, the saccade main sequence. Two different stimuli to generate the saccades were used. In the standard main-sequence stimulus, a turntable was rotated manually to produce saccades largely initiated from near-stationary eye positions. In the fatiguing stimulus, saccades were generated by rotating the animal sinusoidally at 0.2 Hz, ±30° amplitude, in which case the saccades occurred as interruptions in the vigorous slow phase movements in the opposite direction. In each session, a prefatigue, standard main sequence was collected, and then the fatiguing stimulus was run continuously for 7 minutes, collecting samples of the saccades made in response to the stimulus just before, and just after the timed 7-minute period. A postfatigue, standard main sequence was then collected.
Statistical Analysis
Data were analyzed and tested for statistical significance (P < 0.05) by ANOVA and paired two-tailed Student's t-tests. For comparison of fiber size cross-sectional areas, the pair-wise nonparametric Kolmogorov-Smirnov two-sample test was performed.
Results
Expression of Pitx2 in Extraocular Muscle and Other Muscle
The Pitx2 gene utilizes an upstream promoter to express the Pitx2a and Pitx2b isoforms that differ from each other as a result of alternative splicing, and Pitx2c is expressed by a downstream promoter located in intron 3.33-35 Pitx2d exists in humans and appears to act as a negative regulator of the other isoforms.26 Alternative translation initiation (Pitx2Cbeta) and alternative mRNA splicing (Pitx2b2) variants were identified36 but not evaluated in the present study. Transcripts of all three murine isoforms were expressed in mature extraocular muscle (Fig. 1A) and show a distinct temporal expression pattern. Postnatal Pitx2c expression declines in extraocular muscle, in marked contrast to Pitx2a and Pitx2b, which remain relatively constant, with the Pitx2a isoform being the most abundantly expressed isoform (Fig. 1B). We compared Pitx2 isoform expression across several adult skeletal muscles and heart (Fig. 1C). We arbitrarily chose heart to compare the differences. Extraocular muscle expressed all three isoforms at much higher levels than did any of the other muscles.
Figure 1.
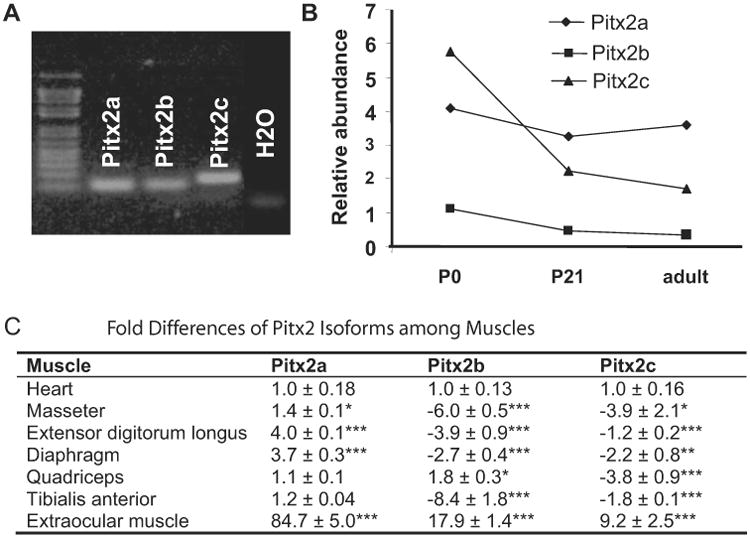
Expression of all three Pitx2 isoforms in postnatal extraocular muscle. (A) RT-PCR with Pitx2a, Pitx2b, and Pitx2c isoform-specific primers on mRNA isolated from adult extraocular muscle. H2O represents cDNA blank control with all three pairs of primers mixed. (B) Relative expression levels of the three Pitx2 isoforms compared to GAPDH in P0, P21, and adult extraocular muscle with qPCR analysis. Arbitrary units were used. Pitx2c expression declined over time, whereas Pitx2a and Pitx2b remained about the same level. Pitx2a was the most abundantly expressed isoform in adult extraocular muscle. (C) Table represents x-fold differences of Pitx2 isoform expression among striated muscles by qPCR. Data are expressed as the mean ± SD of the difference compared with expression in heart; n = 3. *P < 0.05; **P < 0.01; ***P < 0.001.
Temporal and Spatial Distribution of Pitx2 within Extraocular Muscle
We evaluated the temporal and spatial distribution Pitx2 protein in postnatal mouse extraocular muscle. Extraocular muscle, unlike other skeletal muscle, has a great deal of regional and fiber-type and neuromuscular synaptic diversity, and therefore, we evaluated whether a correlation of Pitx2 expression could be made with extraocular muscle anatomic divisions (global versus orbital, midportion versus distal ends), myosin isoform expression, or innervation. Our hypothesis was that Pitx2 would be associated with anatomic region, known differentially regulated muscle proteins, or innervation patterns. Pitx2 was expressed at P0 and appeared predominantly in the orbital layer (Fig. 2A), and by 3 weeks of age, expression spread to the global layer (Fig. 2B) with the majority of Pitx2-positive nuclei still in the orbital layer. In the adult extraocular muscle, Pitx2 immunoreactivity was identified in the global and orbital regions with no clear predominance (Fig. 2C). Furthermore, there was no predominance of Pitx2 immunoreactivity in the midsection or distal ends of the muscle.
Figure 2.
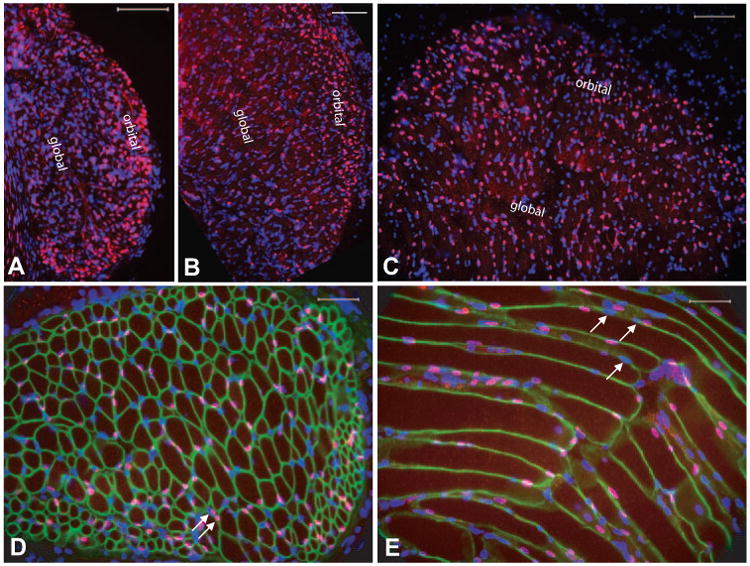
Differential expression of Pitx2 by extraocular muscle. Immunostaining with antibody against Pitx2 showed that Pitx2 was mostly expressed in the orbital layer at P0 (A), starting to be more expressed in the global layer at 3 weeks (B) and evenly distributed in global and orbital layer at adult (C). Cross-section of extraocular muscle (D) was immunolabeled with antibody to dystrophin (green) to reveal the myofiber boundary and with antibody to Pitx2 (red). Counterstain with DAPI (blue) identifies all nuclei. Pitx2 expression is localized within the boundary of dystrophin immunoreactivity, indicating that the majority of Pitx2 immunoreactive nuclei (showing purple) are myonuclei. Occasional Pitx2 immuno-reactive nuclei were outside the sarcolemma, but closely adjacent, and are likely to be satellite cells (arrows). Longitudinal section of adult extraocular muscle (E) was immunolabeled the same way as in (D). As appreciated in (D), not all myonuclei are Pitx2 immunoreactive as evidenced by DAPI-only positive nuclei (arrows). Scale bar: (A-C) 100 μm; (D, E) 50 μm.
Pitx2 immunoreactivity was localized exclusively to the nucleus (Fig. 2D), which is consistent with its role as a transcriptional factor.24,25 Most Pitx2-positive nuclei appeared to be myonuclei, as evidenced by their localization within the ring of dystrophin immunoreactivity, which marks the periphery of the muscle fibers. We found a few immunoreactive nuclei immediately adjacent but outside the myofibers (Fig. 2D, arrows), and those were likely to be nuclei of satellite cells. Pitx2 immunoreactive nuclei were identified along the length of fibers (Fig. 2E); however, all myonuclei of a given fiber were not uniformly Pitx2 immunoreactive (Fig. 2E, arrows).
Because the Myh13 gene (extraocular muscle-specific MHC or EOM-MHC) does contain a potential Pitx2 binding domain in its cis-regulatory domain,37 we expected that Pitx2 expression would be associated with EOM-MHC expression. However, no such association was found. Pitx2-positive (Fig. 3A, arrow) and -negative (Fig. 3A, yellow dashed arrow) nuclei were found in EOM-MHC-expressing fibers.
Figure 3.
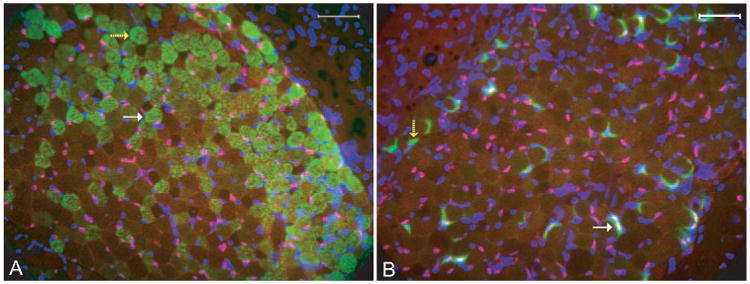
No obvious pattern was observed between Pitx2 expression and neuromuscular junction or myosin-expressing muscle fibers. Cross-sections of normal adult extraocular muscle were doubly stained with rabbit anti-Pitx2 (red) and mouse anti-extraocular muscle MHC (A, green) or Alexa Fluor 488-α-bungarotoxin (B, green). Nuclei were revealed by counterstain with DAPI (blue). Pitx2 was colocalized in nuclei and thus showed purple. White arrows: a muscle fiber positive for both EOM-MHC and Pitx2 in (A) and to a neuromuscular junction that is also positive for Pitx2 in (B). Yellow dashed arrows: a muscle fiber that is positive only for EOM-MHC in (A) and a neuromuscular junction that is not stained with Pitx2 in (B). Scale bar, 50 μm.
The developmental (d)MHC (Mhy3) is exclusively expressed in the orbital layer of rat extraocular muscle38 and we found the same to be true in mouse. Again, both Pitx2-positive and -negative dMHC fibers were found in normal extraocular muscle serial sections (data not shown). Fibers expressing the type I MHC and type II MHC also contained a mixture of Pitx2 immunoreactive and negative nuclei (data not shown). Therefore, in mouse extraocular muscle, there is no correlation between Pitx2 expression and the MHC isoforms examined.
We used labeled α-bungarotoxin to mark the location of neuromuscular junctions and evaluated whether Pitx2 expression was related to innervation. Again, both Pitx2-positive (Fig. 3B, arrow) and -negative (Fig. 3B, yellow dashed arrow) end-plates were found, indicating no relation existed between the neuromuscular junctions and Pitx2 immunoreactivity.
In summary, Pitx2 was present in the extraocular muscle myonuclei, but not in a fiber-type or region-specific pattern. Furthermore, there was no specificity with respect to MHC expression, point of innervations, or whether a fiber was singly or multiply innervated.
Generation of Pitx2 Conditional Knockout Mouse
We exploited Cre-loxP-mediated gene recombination to inactivate Pitx2 in postnatal extraocular muscle. The goal was to allow normal prenatal extraocular muscle development and then turn off Pitx2 expression at birth. Figure 4A depicts a typical result of PCR amplification of DNA from a Pitx2flox/flox mouse showing a single floxed band of 1092 bp. The same PCR amplification reaction of DNA extracted from extraocular muscle of Pitx2Δflox/Δflox mice, which also express MCK-CRE demonstrated that the majority of DNA was recombined, and amplification produced a 495-bp mutant band (Fig. 4B, lane 1). As expected, RT-PCR using a forward primer binding within the homeodomain region and a reverse primer to the C-terminal common region did not amplify any product (Fig. 4C, lane 2) from extraocular muscle tissue of Pitx2Δflox/Δflox mice. RT-PCR with the forward primer from the N-terminal region of Pitx2c isoform and the reverse primer from the C-terminal common region produced a truncated product of 590 bp (Fig. 4D, lane 2) from the extraocular muscle of Pitx2Δflox/Δflox mice.
Figure 4.
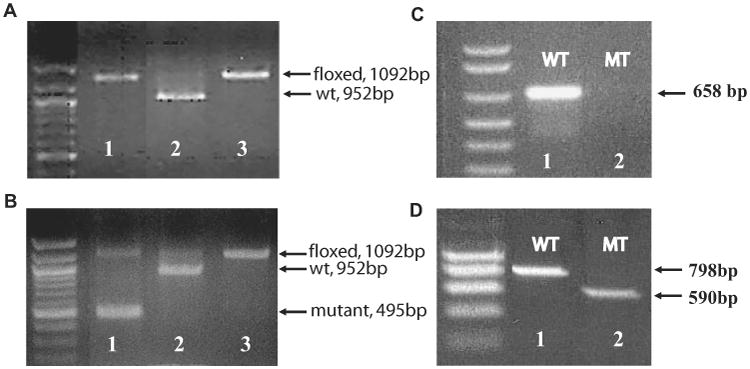
Generation of Pitx2 conditional mutants. (A) Genotyping PCR results of tail DNA. Lane 1: one mouse that was homozygous for Pitx2flox; lane 2: a control for Pitx2flox/+; lane 3: positive control for Pitx2flox/flox bands. (B) the same genotyping PCR result of extraocular muscle tissue from the same mouse in (A), showing that the majority of DNA was recombined and had the 495-bp mutant band (lane 1). Lanes 2 and 3: the same as in (A). (C) RT-PCR result with the forward primer from within the homeodomain region and the reverse primer from the C-terminal common region showing no product from extraocular muscle tissue of mutant mouse (lane 2). (D) is RT-PCR results with the forward primer from the N-terminal region of Pitx2c isoform and the reverse primer from the C-terminal common region, showing a truncated product of 590 bp from the mutant mouse extraocular muscle (lane 2).
We examined the expression of Pitx2 protein in extraocular muscle of Pitx2Δflox/Δflox mice at p0, p21, and 3 months of age with the antibody P2R10. At P0 there was no difference in the number and pattern of Pitx2-positive nuclei between Pitx2Δflox/Δflox and Pitx2flox/flox mice. By p21, a dramatic reduction in the number of positive nuclei was observed, and at 3 months only rare nuclei were immunoreactive in the Pitx2Δflox/Δflox mice (data not shown). Thus, we successfully generated a Pitx2 conditional mutant mouse strain with no functional Pitx2 RNA or protein.
General Phenotypic Evaluation
The Pitx2Δflox/Δflox mice demonstrated no gross phenotypic differences from control littermates. Biweekly observation of their activity levels showed no differences among littermates. Body weights were not different between Pitx2Δflox/Δflox and Pitx2flox/flox mice from 3 weeks (males, 10.37 g ± 0.49 g vs. 10.1 g ± 0.8 g, P = 0.78; females 10.9 g ± 2.0 g vs. 9.8 g ± 2.0 g, P = 0.32) to 6 months (males, 34.4 g ± 1.0 g vs. 34.5 g ± 1.1 g, P = 0.97; females 26.4 g ± 3.9 g vs. 25.6 g ± 1.7 g, P = 0.61) of age. Extensive necropsies by an Animal Research Center veterinarian of three Pitx2Δflox/Δflox mice and three Pitx2flox/flox littermates at 3 months of age found no gross abnormalities. We did identify cardiac abnormalities through specialized assessment, which will be described in a separate publication.
Histology of Pitx2Δflox/Δflox Extraocular Muscle
We performed standard histologic evaluation of extraocular muscle at 3, 8, and 12 weeks of age. Extraocular muscle had no signs of disease; distinct global and orbital regions were identified (Fig. 5A, 5B). The number of fibers was no different in extraocular muscle from Pitx2Δflox/Δflox and Pitx2flox/flox mice in either the global layer (control, 337 ± 19; mutant, 354 ± 27; P = 0.43) or the orbital layer (control, 525 ± 53; mutant, 513 ± 62; P = 0.81). Central myonuclei, a sensitive marker of muscle disease, were not observed.
Figure 5.
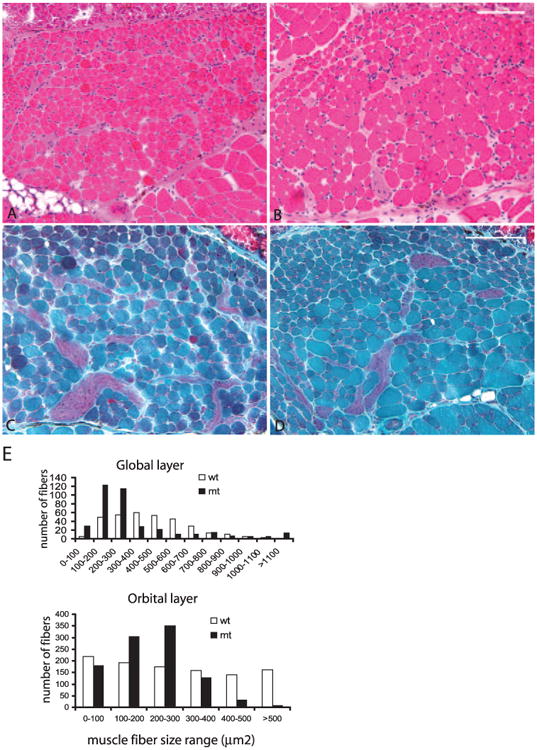
Altered extraocular muscle fiber sizes in Pitx2Δflox/Δflox. H & E (A, B) and Gomori's trichrome staining (C, D) of extraocular muscle sections from 3-month-old Pitx2Δflox/Δflox (B, D) and littermate control Pitx2flox/flox (A, C) showed no centralized nuclei. However, in the Pitx2Δflox/Δflox mice, the muscle fiber size was altered. Fiber size measurement (E) confirmed that myofiber sizes for both orbital and global layers are significantly smaller than in the control extraocular muscle (P < 0.001 for the nonparametric Kolmogorov-Smirnov two-sample test) despite the fact that a small number of fibers in the global layer in the mutant are extremely large. Scale bar, 100 μm.
An obvious alteration of extraocular muscle fiber size was present in 8-week- and 3-month-old Pitx2Δflox/Δflox mice (Figs. 5B, 5D). Some muscle fibers appeared to be larger while others appeared smaller than those in control mice (Fig. 5A, 5C). An assessment of fiber size distribution found a shift toward smaller fibers in both the global and orbital layers of Pitx2Δflox/Δflox extraocular muscle (Fig. 5E). Within the global layer, however, a small percentage of fibers shifted toward larger sizes. Extraocular muscles of 3-week-old mice were identical between Pitx2Δflox/Δflox mice and the Pitx2flox/flox control.
Altered Gene Expression in Pitx2Δflox/Δflox Mice
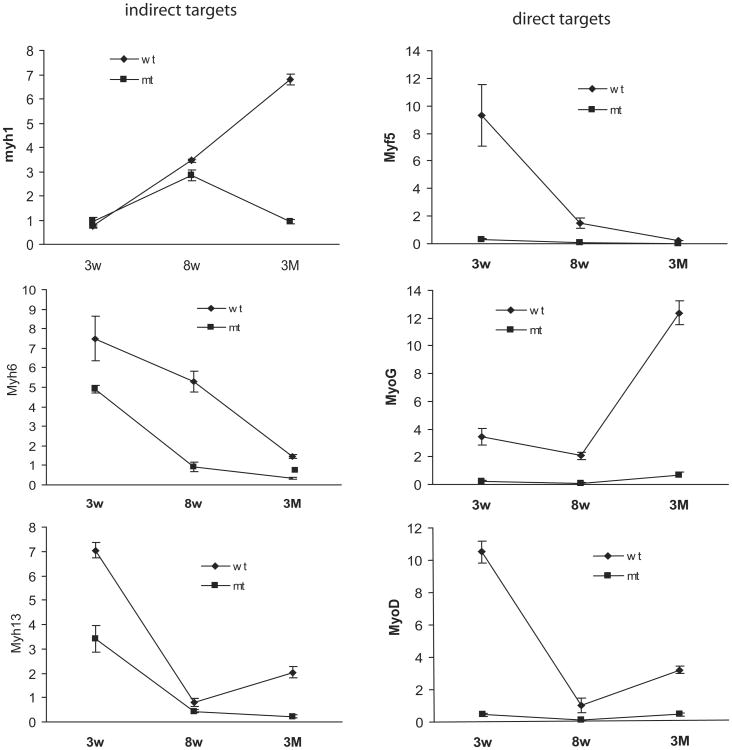
Myogenic regulatory factors are thought to act downstream of Pitx2 during extraocular muscle development.6,34 We measured the expression of selected myogenic regulatory factors (Myf5, Myog, and Myod) to evaluate the impact of Pitx2 loss at 3 weeks, 8 weeks, and 3 months (Fig. 6E). Transcript levels of Myf5, Myog, and Myod were decreased dramatically in the Pitx2Δflox/Δflox mice compared with Pitx2flox/flox littermates. We then assessed transcript levels of major MHC isoforms. Myh 1 (type IIX), Myh 6 (α-cardiac), Myh 7 (MHC 1 or slow MHC), and Myh13 (EOM-MHC) transcripts were reduced, most dramatically at 3 months. In contrast, Myh2 (2A), Myh3 (developmental), Myh4 (2B), and Myh8 (neonatal) expression were not significantly affected.
Figure 6.
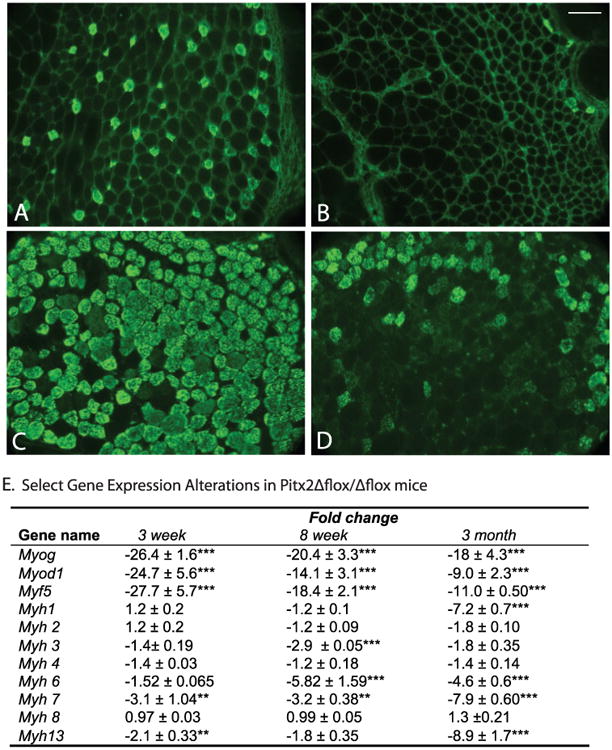
Alteration of gene expression in Pitx2Δflox/Δflox mice. Cross-sections of 3-month-old extraocular muscle from Pitx2Δflox/Δflox mice (B, D) and Pitx2flox/flox control mice (A, C) were immunolabeled with antibodies specific to MHC 1 (A, B) and to EOM-MHC (C, D). The immunoreactivity was dramatically decreased for MHC 1, and moderately decreased for EOM-MHC. Scale bars, 50 μm. (E) Selected gene expression alterations in Pitx2Δflox/Δflox mice determined by qPCR. Data represent mean ± SD of the differences between Pitx2Δflox/Δflox mice versus age-matched Pitx2flox/flox mice, n = 3. Changes ≤ 2-fold were not considered to be of biological significance. *P < 0.05; **P < 0.01; ***P < 0.001.
Comparison of relative expression levels of myogenic regulatory factors and Myh isoforms in extraocular muscle from Pitx2Δflox/Δflox and Pitx2flox/flox mice at 3-week, 8-week and 3-month time points revealed that genes with expression levels affected by Pitx2 fell into two categories (Fig. 7). The first category, which we defined as the direct effect, included myogenic transcription factors Myf5, Myog, and Myod, whose expression was suppressed rapidly after Pitx2 loss. In the second, which we defined as the indirect effect, Myh isoforms were expressed at similar levels in both mouse strains at 3 weeks of age and then they decreased over time in the Pitx2Δflox/Δflox mice.
Figure 7.
Comparison of gene abundance between Pitx2Δflox/Δflox and Pitx2flox/flox mice. Relative expression levels were normalized to GAPDH by qPCR analysis. Arbitrary units were used for the purpose of illustration. Levels of Myh isoform expression (shown are Myh1, Myh6, and Myh13) in the Pitx2Δflox/Δflox are at similar or slightly lower level to their wild-type counterparts at early time points and decreased over time, suggesting that these genes may be indirect targets of Pitx2. Levels of myogenic transcription factors (Myf5, MyoG, and MyoD) were low from early time points and stayed low throughout; therefore, they may be direct targets of Pitx2 action.
Myosin Heavy Chain Content
We assessed MHC distribution by immunohistochemistry. MHC I immunoreactivity was greatly reduced in extraocular muscle from Pitx2Δflox/Δflox mice at 3 weeks, 8 weeks (data not shown), and 3 months (Fig. 6B compared with 6A), consistent with decreased transcript abundance at this age (Fig. 6E). The distribution of type 2 MHC was not altered (data not shown) as assessed by pan-type 2 MHC antibody, although Myh 1 (for MHC 2X fiber) had sevenfold reduction in its transcript level at 3 months. Furthermore, immunostaining with MHC 2A- and 2B-specific antibodies found no significant differences in the content of these isoforms at 3 weeks to 3 months of age between Pitx2Δflox/Δflox mice and Pitx2flox/flox littermates (data not shown). Consistent with the almost nine-fold reduction in Myh13 transcript level at 3 months, EOM-MHC immunoreactivity was greatly reduced in Pitx2Δflox/Δflox extraocular muscle, although some positive fibers were identified (Figs. 6C, 6D). By qPCR, the transcript levels of both Myh7 and Myh13 genes were less than 1% of total myosin mRNA in wild-type mice (0.55% for Myh7 and 0.078% for Myh13 at 3 months of age). In the mutant mice, the percentage decreased to 0.014% for Myh7 and 0.004% for Myh13. Myh1, Myh2, and Myh4 are the predominant isoforms expressed making up more than 90% of all myosin gene transcripts.
Evaluation for Cre Toxicity
We examined whether the MCK-Cre transgene itself has an effect on the phenotypes we observed. We compared the histology, gene expression patterns, and MHC1 (Myh 7) immunostaining of extraocular muscle from MCKCre/Pitx2+/+, Pitx2Δflox/+, Pitx2Δflox/Δflox, and Pitx2flox/flox mice. We did not observe fiber size alterations in MCK-Cre/Pitx2+/+ mice; nor did we find differences in gene expression patterns by qPCR (data not shown). Expression of MHC1 (assessed by immunostaining) was identical in the extraocular muscle of MCK-Cre/Pitx2+/+, Pitx2flox/flox, or Pitx2+/+ mice. Therefore, the mere presence of the MCK-Cre transgene did not appear to affect extraocular muscle. The Pitx2Δflox/+ mice, on the other hand, showed a phenotype intermediate between the Pitx2Δflox/Δflox mice and their control littermates Pitx2flox/flox, suggesting dose-dependent effects of Pitx2, as had been observed in developmental studies.6
Contractile Properties of Extraocular Muscle from Pitx2Δflox/Δflox Mice
We tested Pitx2Δflox/Δflox extraocular muscle function in vitro to determine the influence of the histologic and gene expression changes on contractility. The Pitx2flox/flox demonstrated contractile properties similar to those reported in wild-type mice.14 As shown in Table 1, the mutant extraocular muscle had a 20% higher peak tetanic force (P0) and 9% greater maximum velocity of shortening (V0). In contrast, the endurance time (time to a 50% reduction in force from the start of the fatigue protocol) was 555 ± 50.1 seconds for the Pitx2Δflox/Δflox extraocular muscle with four of eight specimens unable to complete the fatigue protocol compared with 600 ± 0 seconds for Pitx2flox/flox littermates, all of which completed the protocol. The force of contraction at the end of the fatigue protocol was reduced to 48.0% of initial force among the mutants, compared with 59.1% among controls.
Table 1. Contractile Characteristics of Pitx2Δflox/Δflox Extraocular Muscle.
| Pitx2Δflox/Δflox | Pitx2flox/flox | |
|---|---|---|
| V0 (L0/sc) | 11.1 ± 0.8* | 10.2 ± 0.6 |
| P0 (N/cm2) | 6.9 ± 0.7** | 5.7 ± 0.5 |
| Fatigue index (% of force at time = 0) | 48.0 ± 4*** | 591 ± 6 |
| Endurance (fatigue duration, sec) | 555 ± 50.1* | 600 ± 0 |
Data represent the mean ± SD; n = 8 animals for each group.
P < 0.05;
P < 0.01;
P < 0.005, compared with control littermates.
Eye Movement Recordings
The oculomotor consequences of the Pitx2Δflox/Δflox mutation were evaluated in vivo. Saccades require the highest levels of extraocular muscle force and are thus particularly sensitive to alterations of extraocular muscle function. Therefore, saccade performance was used to assess ocular motility and quantified by linear regression on the main sequences (i.e., plots of saccade peak velocity versus saccade amplitude). Pitx2Δflox/Δflox mice exhibited the main sequence of the usual mouse configuration (i.e., the relationship of peak velocity to saccade amplitude was fairly linear, with abducting saccades being slower than adducting). Each mouse's main sequence was further parameterized by predicting from the regression slope and offset, the peak velocity for saccades of 20° amplitude in abducting or adducting directions. Table 2 shows the mean values of this velocity for the Pitx2Δflox/Δflox and Pitx2flox/flox mice before and after fatiguing and for both the standard and fatiguing stimulus. It also tabulates the effect of fatiguing on the predicted peak velocities, as measured by the contrast functions (post − pre)/(post + pre). Peak velocities of mutants were lower than in control animals for both the standard and fatigue-stimulus data sets. Some of the P-values failed a 0.05 threshold, but the consistency of the trend argues that the difference is real, and a conclusion that the Pitx2Δflox/Δflox and Pitx2flox/flox mice are not different could lead to a type II error. The contrast data indicate that peak velocities were reduced marginally after the 7-minute fatiguing period, but roughly equally for the Pitx2Δflox/Δflox and Pitx2flox/flox littermates.
Table 2. Saccade Velocity Measurement.
| Stimulus Type | Pre/Post Fatigue | Direction | Pitx2Δflox/Δflox | Pitx2flox/flox | P* |
|---|---|---|---|---|---|
| Standard | Pre | Abduct | 738 ± 48 deg/s | 828 ± 138 deg/s | 0.16 |
| Post | Abduct | 710 ± 71 deg/s | 805 ± 80 deg/s | 0.045 | |
| Pre | Adduct | 985 ± 64 deg/s | 1090 ± 73 deg/s | 0.02 | |
| Post | Adduct | 956 ± 82 deg/s | 1030 ± 73 deg/s | 0.12 | |
| Contrast | Abduct | −0.02 ± 0.04 | −0.01 ± 0.04 | 0.66 | |
| Contrast | Adduct | −0.02 ± 0.017 | −0.03 ± 0.01 | 0.13 | |
| Fatiguing | Pre | Abduct | 763 ± 67 deg/s | 856 ± 94 deg/s | 0.07 |
| Post | Abduct | 726 ± 65 deg/s | 839 ± 79 deg/s | 0.018 | |
| Pre | Adduct | 972 ± 72 deg/s | 1094 ± 72 deg/s | 0.011 | |
| Post | Adduct | 934 ±55 deg/s | 1060 ± 79 deg/s | 0.008 | |
| Contrast | Abduct | −0.02 ± 0.02 | −0.009 ± 0.03 | 0.27 | |
| Contrast | Adduct | − 0.02 ± 0.02 | −0.02 ± 0.01 | 0.72 |
Data represent the mean ± SD; n = 6 animals.
Pitx2Δflox/Δflox vs. Pitx2flox/flox mice by Student's t-test.
Discussion
Our results demonstrate that Pitx2, in addition to its critical role in development, modulates the mature extraocular muscle phenotype. The conditional knockout of Pitx2 produced alterations in key myogenic regulators and myosin isoforms as well as altered fiber size distribution. In vitro contractility was modified, producing a faster and stronger muscle that was more susceptible to fatigue. Eye movement recordings did not identify fatigue, but saccadic peak velocities were lower (see the Discussion section). Despite the high expression level of Pitx2 in wild-type mice, the conditional removal of Pitx2 expression postnatally did not lead to degeneration of extraocular muscle as might have been expected. In fact, the extraocular muscle of the Pitx2Δflox/Δflox mice did not demonstrate any signs of muscle disease (i.e., fiber degeneration or centrally displaced myonuclei), but rather developed a “new” extraocular muscle with distinct morphology, gene expression, and functional characteristics. The findings demonstrate that Pitx2 in the mature extraocular muscle does not fulfill a critical maintenance role, but rather modulates the extraocular muscle phenotype.
Understanding of the transcriptional control of the mature extraocular muscle phenotype is extremely limited and is based entirely on analogy with other striated muscle and developmental studies of extraocular muscle.1 Despite Pitx2's critical importance in development and its high expression in mature extraocular muscle, it is clear from our study that other transcription factors are essential for the maintenance of the adult extraocular muscle phenotype, given that a functional extraocular muscle is produced despite the absence of Pitx2. It is possible that Pitx3 compensates for the loss of Pitx2, as appears to be the case in skeletal muscle.39 Despite our extensive evaluation of the normal expression pattern of Pitx2, a correlation of expression could not be made to global and orbital regions, fiber type specificity, points of innervations, tendon insertions, all of which are sites of regional specialization in skeletal and extraocular muscle.1,40,41 Therefore, the expression pattern provided limited insight into Pitx2's functional role.
The rapid reduction of myogenic regulatory factors in the Pitx2Δflox/Δflox mice would suggest that Pitx2 acts upstream of them, as has been hypothesized during development (Fig. 6).17,34 This notion is speculative, however, as there is very little information on target genes for Pitx2.25,42,43 The Pitx2Δflox/Δflox mice had dramatically lower levels of myogenic regulators detected at 3 weeks compared with the controls, which we defined as supporting a direct effect of Pitx2. Myogenic regulatory factors (MyoD, Myf5, myogenin) are not up-regulated in the Pitx2Δflox/Δflox indicating that they have no role in compensating for the absence of Pitx2.
Pitx2 was found in the majority of myonuclei, but without a specific pattern of expression related to region, fiber-type, MHC or innervation. In skeletal muscle, the region-specific expression of certain transcripts is well appreciated—for example, subsynaptic and myotendinous nuclei have enhanced mRNA expression for proteins that are preferentially found in those regions.44,45 Pitx2 is involved in myotendinous development in the forelimb,46 and again differential expression of Pitx2 to the insertion points of the extraocular muscle could have been expected. However, the link between transcription at individual myonuclei and ultimate protein expression is complex.47 For example, despite the requirement of troponin across the length of the myofiber, its gene is not expressed at all myonuclei.48 If Pitx2 expression were stochastic or pulsatile, our immunohistochemical studies would reflect the snapshot of expression of the transcriptional factor, which is likely to have a short half life.
Extraocular muscle fibers are generally smaller and show a broader range of fiber size than do other skeletal muscles. In the Pitx2-deficient extraocular muscle, the global region had a distinct prominence of large fibers, but quantitative assessment found that these large fibers made up only a small percentage of the total fibers and that the fiber size distribution was skewed toward smaller fibers in both the global and orbital layers. In skeletal muscle, fiber size is controlled by intrinsic factors and factors extrinsic to the muscle (i.e., mechanical load, hormones, diffusible factors, and motoneuron activity),49 and the same appears true in extraocular muscle, although the specific mechanisms are likely to differ.1 The alteration in fiber size distribution in the Pitx2Δflox/Δflox indicates that Pitx2 plays a role in fiber size.
MHC expression forms the basis of fiber-type classification of skeletal muscle and is a major determinant of a muscle's contractile properties. Extraocular muscles are unique in their expression of nearly all known MHC and the coexpression of certain MHC in a single fiber.38 Pitx2Δflox/Δflox mice had significant reductions in gene and protein expression of MHC-1 and extraocular muscle-specific MHC, the Myh13 gene product, and no alteration in MHC-2. Figure 6 illustrates the striking dissociation in the pattern of expression of Myh1 and Myh13. Myh1 in wild-type continued to demonstrate increased expression at 3 months compared with a decrease in mutant mice, in contrast to the uniformly lower and falling level of Myh13. The Myh13 gene has Pitx2 binding sites in the promoter region, and the absence of Pitx2 would be expected to reduce Myh13 mRNA levels, as observed. Consistent with the reduction in transcript level, the protein level of extraocular muscle-specific MHC is also downregulated, although we consistently observe a few positive fibers in the orbital layer. The pattern for the extraocular muscle-specific MHC to be relatively retained is consistent with the dissociation of protein expression and regulation of MHC genes based on promoter analysis.37,50 We did not attempt to classify extraocular muscle fibers of the Pitx2Δflox/Δflox mice because of the diversity in fiber size and MHC content, which are major components of the generally accepted classification.1
In vitro functional studies of extraocular muscle demonstrated Pitx2Δflox/Δflox extraocular muscle had higher velocity of shortening and greater peak tetanic force but fatigued more rapidly. The increased shortening velocity is consistent with the observed decrease in the slow MHC isoform. Force of contraction is dependent on number of contractile units per muscle cross-sectional area. We found no evidence of an increase in the number of the Pitx2Δflox/Δflox extraocular muscle fibers, but the presence of the population of larger fibers could have led to higher overall force generation. We found no reduction in the connective tissue matrix leading to an improved “packing” of contractile units. Extraocular muscle is highly resistant to fatigue.1,20,51 The susceptibility to fatigue of the Pitx2Δflox/Δflox extraocular muscle was significantly increased: half the Pitx2Δflox/Δflox extraocular muscles did not complete our standard fatigue protocol, whereas all the control muscles did.
In contrast to the increases in maximum tension and greater fatigability observed in vitro, the eye movements recorded in vivo were slightly slower (suggesting lessened peak tensions) and no more fatigable than in control animals. Although apparently contradictory, the in vivo results could arise if, in the in vivo condition, the extraocular muscles are equilibrated at what is the fatigued state of the in vitro experiment. In other words, since the extraocular muscles are continuously activated in the in vivo preparation, they may never achieve the rested, prefatigue state found of the in vitro experiment, in which the extraocular muscles are activated only intermittently. Alternatively, the fatigability of extraocular muscles of Pitx2Δflox/Δflox revealed by the in vitro assay may not occur in vivo. These results suggest the hazards of relying purely on in vitro studies to assess extraocular muscle function in studies of therapeutic manipulation.
The Pitx2Δflox/Δflox mouse ablates Pitx2 expression limited to tissues that express Mck, and not other tissues, such as midbrain motor neurons, leading to the retention of normal innervations, and therefore the Pitx2 effect is only related to its loss from the extraocular muscle.27,52 This effect is an important advantage of the conditional knockout over complete knockouts of Pitx2 during development that are compromised by potential loss of innervation as a contributor to muscle agenesis. organotypic nerve-muscle cocultures have demonstrated that extraocular muscle explants will only survive long-term culture with oculomotor motoneurons and not spinal motoneurons.53 An in vivo parallel, the Wnt1−/− mouse, shows loss of the midbrain, including oculomotor and trochlear motoneurons, aberrant extraocular muscle innervation, and impaired myogenesis.54 The motoneuron pool-specific requirement for extraocular muscle survival makes it clear that neurotrophic mechanisms direct key features of the extraocular muscle phenotype.
The results demonstrate that Pitx2 functions to sustain a specific extraocular muscle phenotype. Its loss does not lead to degeneration of extraocular muscle—as might have been expected—but rather to altered expression of key myogenic regulatory factors and muscle specific genes and proteins. Extraocular muscle anatomy is largely maintained, but contractile function is altered consistent with alterations in myosin isoform expression. Our results demonstrate that alterations of transcriptional expression can engineer modification of extraocular muscle contraction with the ultimate expectation that appropriate targeted gene expression intervention could lead to a therapeutic intervention.
Supplementary Material
Acknowledgments
Supported by the National Institutes of Health Grants R01 EY015306 (HJK) and EY012998 (FHA) and the Swedish Research Council (TAH).
Footnotes
Disclosure: Y. Zhou, None; G. Cheng, None; L. Dieter, None; T.A. Hjalt, None; F.H. Andrade, None; J.S. Stahl, None; H.J. Kaminski, None
References
- 1.Spencer RF, Porter JD. Biological organization of the extraocular muscles. Prog Brain Res. 2005;151:43–80. doi: 10.1016/S0079-6123(05)51002-1. [DOI] [PubMed] [Google Scholar]
- 2.Leigh RJ, Zee DS. The Neurology of Eye Movements. 4th. New York: Oxford University Press; 2006. p. 776. [Google Scholar]
- 3.Andrade FH, Porter JD, Kaminski HJ. Eye muscle sparing by the muscular dystrophies: lessons to be learned? Microsc Res Tech. 2000;48:192–203. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<192::AID-JEMT7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 4.Prabhakar BS, Bahn RS, Smith TJ. Current perspective on the pathogenesis of Graves' disease and ophthalmopathy. Endocr Rev. 2003;24:802–835. doi: 10.1210/er.2002-0020. [DOI] [PubMed] [Google Scholar]
- 5.Conti-Fine BM, Milani M, Kaminski HJ. Myasthenia gravis: past, present, and future. J Clin Invest. 2006;116:2843–2854. doi: 10.1172/JCI29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diehl AG, Zareparsi S, Qian M, Khanna R, Angeles R, Gage PJ. Extraocular muscle morphogenesis and gene expression are regulated by Pitx2 gene dose. Invest Ophthalmol Vis Sci. 2006;47:1785–1793. doi: 10.1167/iovs.05-1424. [DOI] [PubMed] [Google Scholar]
- 7.Gage PJ, Rhoades W, Prucka SK, Hjalt T. Fate maps of neural crest and mesoderm in the mammalian eye. Invest Ophthalmol Vis Sci. 2005;46:4200–4208. doi: 10.1167/iovs.05-0691. [DOI] [PubMed] [Google Scholar]
- 8.Lin CR, Kioussi C, O'Connell S, et al. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 9.Piedra ME, Icardo JM, Albajar M, Rodriguez-Rey JC, Ros MA. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell. 1998;94:319–324. doi: 10.1016/s0092-8674(00)81475-0. [DOI] [PubMed] [Google Scholar]
- 10.Ryan AK, Blumberg B, Rodriguez-Esteban C, et al. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- 11.Hjalt TA, Semina EV. Current molecular understanding of Axenfeld-Rieger syndrome. Expert Rev Mol Med. 2005;7:1–17. doi: 10.1017/S1462399405010082. [DOI] [PubMed] [Google Scholar]
- 12.Brueckner JK, Porter JD. Visual system maldevelopment disrupts extraocular muscle-specific myosin expression. J Appl Physiol. 1998;85:584–592. doi: 10.1152/jappl.1998.85.2.584. [DOI] [PubMed] [Google Scholar]
- 13.Cheng G, Mustari MJ, Khanna S, Porter JD. Comprehensive evaluation of the extraocular muscle critical period by expression profiling in the dark-reared rat and monocularly deprived monkey. Invest Ophthalmol Vis Sci. 2003;44:3842–3855. doi: 10.1167/iovs.03-0170. [DOI] [PubMed] [Google Scholar]
- 14.McMullen CA, Andrade FH, Stahl JS. Functional and genomic changes in the mouse ocular motor system in response to light deprivation from birth. J Neurosci. 2004;24:161–169. doi: 10.1523/JNEUROSCI.3234-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porter JD, Khanna S, Kaminski HJ, et al. Extraocular muscle is defined by a fundamentally distinct gene expression profile. Proc Natl Acad Sci U S A. 2001;98:12062–12067. doi: 10.1073/pnas.211257298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruning JC, Michael MD, Winnay JN, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 17.Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- 18.Trask RV, Billadello JJ. Tissue-specific distribution and developmental regulation of M and B creatine kinase mRNAs. Biochim Biophys Acta. 1990;1049:182–188. doi: 10.1016/0167-4781(90)90039-5. [DOI] [PubMed] [Google Scholar]
- 19.Lyons GE, Muhlebach S, Moser A, et al. Developmental regulation of creatine kinase gene expression by myogenic factors in embryonic mouse and chick skeletal muscle. Development. 1991;113:1017–1029. doi: 10.1242/dev.113.3.1017. [DOI] [PubMed] [Google Scholar]
- 20.McMullen CA, Hayess K, Andrade FH. Fatigue resistance of rat extraocular muscles does not depend on creatine kinase activity. BMC Physiol. 2005;5:12. doi: 10.1186/1472-6793-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porter JD, Merriam AP, Gong B, et al. Postnatal suppression of myomesin, muscle creatine kinase and the M-line in rat extraocular muscle. J Exp Biol. 2003;206:3101–3112. doi: 10.1242/jeb.00511. [DOI] [PubMed] [Google Scholar]
- 22.Thompson SW. Selected histochemical and histopathological methods. Springfield, IL: Charles C Thomas; 1966. [Google Scholar]
- 23.Sheehan DC, Hrapchak BB. Theory and Practice of Histotechnology. Columbus, OH: Battelle Memorial Institute; 1987. [Google Scholar]
- 24.Green PD, Hjalt TA, Kirk DE, et al. Antagonistic regulation of Dlx2 expression by PITX2 and Msx2: implications for tooth development. Gene Expr. 2001;9:265–281. doi: 10.3727/000000001783992515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hjalt TA, Semina EV, Amendt BA, Murray JC. The Pitx2 protein in mouse development. Dev Dyn. 2000;218:195–200. doi: 10.1002/(SICI)1097-0177(200005)218:1<195::AID-DVDY17>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 26.Cox CJ, Espinoza HM, McWilliams B, et al. Differential regulation of gene expression by PITX2 isoforms. J Biol Chem. 2002;277:25001–25010. doi: 10.1074/jbc.M201737200. [DOI] [PubMed] [Google Scholar]
- 27.Martin DM, Skidmore JM, Philips ST, et al. PITX2 is required for normal development of neurons in the mouse subthalamic nucleus and midbrain. Dev Biol. 2004;267:93–108. doi: 10.1016/j.ydbio.2003.10.035. [DOI] [PubMed] [Google Scholar]
- 28.Martin DM, Skidmore JM, Fox SE, Gage PJ, Camper SA. Pitx2 distinguishes subtypes of terminally differentiated neurons in the developing mouse neuroepithelium. Dev Biol. 2002;252:84–99. doi: 10.1006/dbio.2002.0835. [DOI] [PubMed] [Google Scholar]
- 29.Rubinstein NA, Porter JD, Hoh JF. The development of longitudinal variation of myosin isoforms in the orbital fibers of extraocular muscles of rats. Invest Ophthalmol Vis Sci. 2004;45:3067–3072. doi: 10.1167/iovs.04-0106. [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahl JS. Calcium channelopathy mutants and their role in ocular motor research. Ann N Y Acad Sci. 2002;956:64–74. doi: 10.1111/j.1749-6632.2002.tb02809.x. [DOI] [PubMed] [Google Scholar]
- 32.Stahl JS, van Alphen AM, De Zeeuw CI. A comparison of video and magnetic search coil recordings of mouse eye movements. J Neurosci Methods. 2000;99:101–110. doi: 10.1016/s0165-0270(00)00218-1. [DOI] [PubMed] [Google Scholar]
- 33.Gage PJ, Suh H, Camper SA. The bicoid-related Pitx gene family in development. Mamm Genome. 1999;10:197–200. doi: 10.1007/s003359900970. [DOI] [PubMed] [Google Scholar]
- 34.Kitamura K, Miura H, Miyagawa-Tomita S, et al. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra-and periocular mesoderm and right pulmonary isomerism. Development. 1999;126:5749–5758. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- 35.Kitamura K, Miura H, Yanazawa M, Miyashita T, Kato K. Expression patterns of Brx1 (Rieg gene), Sonic hedgehog, Nkx2.2, Dlx1 and Arx during zona limitans intrathalamica and embryonic ventral lateral geniculate nuclear formation. Mech Dev. 1997;67:83–96. doi: 10.1016/s0925-4773(97)00110-x. [DOI] [PubMed] [Google Scholar]
- 36.Lamba P, Hjalt TA, Bernard DJ. Novel forms of Paired-like homeodomain transcription factor 2 (PITX2): generation by alternative translation initiation and mRNA splicing. BMC Mol Biol. 2008;9:31. doi: 10.1186/1471-2199-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schachat F, Briggs MM. Phylogenetic implications of the superfast myosin in extraocular muscles. J Exp Biol. 2002;205:2189–2201. doi: 10.1242/jeb.205.15.2189. [DOI] [PubMed] [Google Scholar]
- 38.Brueckner JK, Itkis O, Porter JD. Spatial and temporal patterns of myosin heavy chain expression in developing rat extraocular muscle. J Muscle Res Cell Motil. 1996;17:297–312. doi: 10.1007/BF00240928. [DOI] [PubMed] [Google Scholar]
- 39.L'Honore A, Coulon V, Marcil A, et al. Sequential expression and redundancy of Pitx2 and Pitx3 genes during muscle development. Dev Biol. 2007;307:421–433. doi: 10.1016/j.ydbio.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 40.Hughes BW, Kusner LL, Kaminski HJ. Molecular architecture of the neuromuscular junction. Muscle Nerve. 2006;33:445–461. doi: 10.1002/mus.20440. [DOI] [PubMed] [Google Scholar]
- 41.Heinemeier KM, Olesen JL, Haddad F, et al. Expression of collagen and related growth factors in rat tendon and skeletal muscle in response to specific contraction types. J Physiol. 2007;582:1303–1316. doi: 10.1113/jphysiol.2007.127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kioussi C, Briata P, Baek SH, et al. Identification of a Wnt/Dvl/beta-catenin→Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 43.Wei Q, Adelstein RS. Pitx2a expression alters actin-myosin cytoskeleton and migration of HeLa cells through Rho GTPase signaling. Mol Biol Cell. 2002;13:683–697. doi: 10.1091/mbc.01-07-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon AM, Hoppe P, Burden SJ. Spatial restriction of AChR gene expression to subsynaptic nuclei. Development. 1992;114:545–553. doi: 10.1242/dev.114.3.545. [DOI] [PubMed] [Google Scholar]
- 45.Jarvinen TA, Jozsa L, Kannus P, et al. Mechanical loading regulates the expression of tenascin-C in the myotendinous junction and tendon but does not induce de novo synthesis in the skeletal muscle. J Cell Sci. 2003;116:857–866. doi: 10.1242/jcs.00303. [DOI] [PubMed] [Google Scholar]
- 46.Holmberg J, Ingner G, Johansson C, Leander P, Hjalt TA. PITX2 gain-of-function induced defects in mouse forelimb development. BMC Dev Biol. 2008;8:25. doi: 10.1186/1471-213X-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allen DL, Roy RR, Edgerton VR. Myonuclear domains in muscle adaptation and disease. Muscle Nerve. 1999;22:1350–1360. doi: 10.1002/(sici)1097-4598(199910)22:10<1350::aid-mus3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 48.Newlands S, Levitt LK, Robinson CS, et al. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harridge SD. Plasticity of human skeletal muscle: gene expression to in vivo function. Exp Physiol. 2007;92:783–797. doi: 10.1113/expphysiol.2006.036525. [DOI] [PubMed] [Google Scholar]
- 50.Briggs MM, Schachat F. The superfast extraocular myosin (MYH13) is localized to the innervation zone in both the global and orbital layers of rabbit extraocular muscle. J Exp Biol. 2002;205:3133–3142. doi: 10.1242/jeb.205.20.3133. [DOI] [PubMed] [Google Scholar]
- 51.Andrade FH, McMullen CA. Lactate is a metabolic substrate that sustains extraocular muscle function. Pflugers Arch. 2006;452:102–108. doi: 10.1007/s00424-005-0010-0. [DOI] [PubMed] [Google Scholar]
- 52.Sclafani AM, Skidmore JM, Ramaprakash H, Trumpp A, Gage PJ, Martin DM. Nestin-Cre mediated deletion of Pitx2 in the mouse. Genesis. 2006;44:336–344. doi: 10.1002/dvg.20220. [DOI] [PubMed] [Google Scholar]
- 53.Porter JD, Hauser KF. Survival of extraocular muscle in long-term organotypic culture: differential influence of appropriate and inappropriate motoneurons. Dev Biol. 1993;160:39–50. doi: 10.1006/dbio.1993.1284. [DOI] [PubMed] [Google Scholar]
- 54.Porter JD, Baker RS. Absence of oculomotor and trochlear motoneurons leads to altered extraocular muscle development in the Wnt-1 null mutant mouse. Brain Res Dev. 1997;100:121–126. doi: 10.1016/s0165-3806(97)00020-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.