Abstract
An integral part of auxin-regulated gene expression involves the interplay of two types of transcription factors, the DNA binding auxin response factor (ARF) activators and the interacting auxin/indole acetic acid (Aux/IAA) repressors. Insight into the mechanism of how these transcription factors interact with one another has recently been revealed from crystallographic information on ARF5 and ARF7 C-terminal domains (i.e., a protein-protein interaction domain referred to as domain III/IV that is related to domain III/IV in Aux/IAA proteins). Three-dimensional structures showed that this domain in ARF5 and ARF7 conforms to a well-known PB1 (Phox and Bem1) domain that confers protein-protein interactions with other PB1 domain proteins through electrostatic contacts. Experiments verifying the importance of charged amino acids in conferring ARF and Aux/IAA interactions have confirmed the PB1 domain structure. Some in planta experiments designed to test the validity of PB1 interactions in the auxin response have led to updated models for auxin-regulated gene expression and raised many questions that will require further investigation. In addition to the PB1 domain, a second protein interaction module that functions in ARF-ARF dimerization and facilitates DNA binding has recently been revealed from crystallography studies on the ARF1 and ARF5 DNA binding domains.
INTRODUCTION
Two transcription factor families, the auxin response factor (ARF) family and the auxin/indole acetic acid (Aux/IAA) family, play key roles in regulating the expression of auxin response genes. ARFs are DNA binding proteins that are targeted to TGTCTC or related auxin response elements (AuxREs) in promoters of auxin response genes and function as transcriptional activators or repressors through an activation domain (AD) that is enriched in glutamine or a repression domain (RD) that lacks a glutamine-rich region (reviewed in Guilfoyle and Hagen, 2007). Aux/IAA proteins are short-lived nuclear-localized proteins without a DNA binding domain (DBD) and function as transcriptional repressors through a conserved D/E-L-X-L-X-L or related EAR-like (ethylene-responsive element binding factor-associated amphiphilic repression motif) RD (Tiwari et al., 2004; Li et al., 2011; reviewed in Chapman and Estelle, 2009). ARF and Aux/IAA proteins contain a similar PB1 (Phox and Bem1) protein-protein interaction domain (previously referred to as domain III/IV or domain III and domain IV) in their C termini that facilitates the formation of ARF-ARF, ARF-Aux/IAA, and Aux/IAA-Aux/IAA homo- and hetero-oligomers (Han et al., 2014; Korasick et al., 2014; Nanao et al., 2014; Phyre2 fold library entry id c2m1mA [http://www.sbg.bio.ic.ac.uk/~phyre2/]; reviewed in Guilfoyle and Hagen, 2012). Interactions of ARF activators with Aux/IAA repressors through their PB1 domains on AuxREs facilitate the repression of auxin response genes when auxin concentrations in a cell are low (Tiwari et al., 2003; reviewed in Guilfoyle and Hagen, 2007, 2012). Degradation of the Aux/IAA repressors by the ubiquitin-proteasome pathway at elevated auxin concentrations relieves the repression and leads to activation of auxin response genes (reviewed in Guilfoyle and Hagen, 2007; Chapman and Estelle, 2009; Salehin et al., 2015).
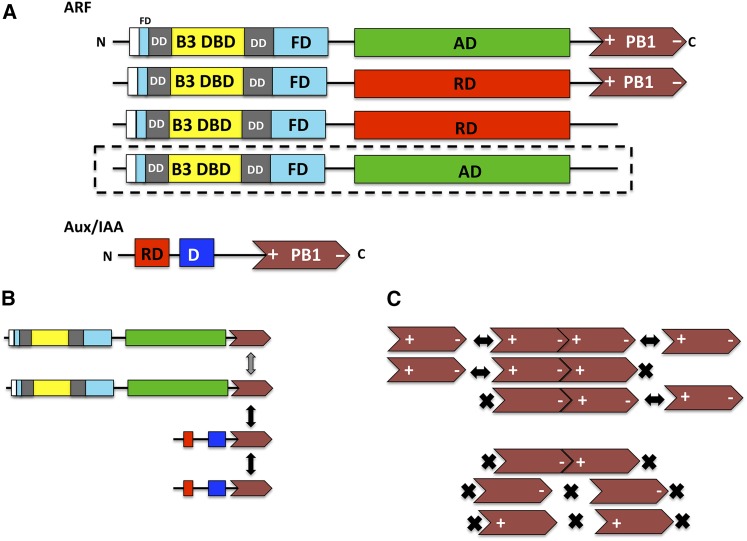
ARF proteins were originally reported to contain three domains, consisting of a B3-type DBD (the domain that contacts AuxREs and is related in amino acid sequence to DNA binding domains in RAV, LAV, and REM transcription factors), followed by an AD or RD, and terminating in a protein-protein interaction domain related to domain III/IV in Aux/IAA proteins (reviewed in Guilfoyle and Hagen, 2007, 2012). A recent crystallographic study by Boer et al. (2014) revealed two additional domains associated with ARF5 and ARF1 DBDs, and these are a dimerization domain (DD) and a Tudor-like ancillary domain found within the C-terminal region of the flanking domain (FD) (Figure 1A). The DD facilitates cooperative binding of the B3 DBD to selected AuxREs, but the function of Tudor-like ancillary domain has not been determined. Not all ARFs contain the five domains described above. Some ARFs that contain an RD lack the C-terminal domain (a PB1 domain previously referred to as domain III/IV), but whether these truncated ARFs function constitutively in repressing auxin response genes is unclear (Figure 1A). Beyond the natural ARFs, it has been reported that when domain III/IV is artificially removed from ARFs containing an AD, these modified ARFs constitutively activate auxin response genes in protoplast transfection assays and confer “high auxin” phenotypes when overexpressed in transformed plants (Wang et al., 2005; Lau et al., 2011; Krogan et al., 2012). The results with ARF activators lacking a domain III/IV support the importance of this protein-protein interaction domain in the auxin response.
Figure 1.
Diagrams of ARF and Aux/IAA Domains and the Interactions of ARF and Aux/IAA Proteins through Their PB1 Domains.
(A) ARF proteins contain a nonconserved AD (green) or RD (red) flanked by an N-terminal DBD (composed of a B3 domain in yellow, a DD in gray, and an FD in blue) and a C-terminal PB1 domain (previously referred to as domain III/IV) in brown. Parts of the DD and FD are found both N-terminal and C-terminal to the B3 domain. The Tudor-like ancillary domain is found within the C-terminal region of the FD. Both parts of the FD are related in amino sequence to a fragment of BRWD proteins, which includes the Auxin_resp domain. It is proposed that the DD may be derived from the internal portion of the FD that flanks the DD in ARFs. Some natural ARFs with RDs lack the PB1 domain. Artificial ARFs with ADs (outlined with dashes) that are created by removing the PB1 domain function in an auxin-independent manner in activating auxin response genes. Aux/IAA proteins contain an N-terminal repression domain (RD in red; domain I), followed by domain II (D in blue) with a conserved degron, and a C-terminal PB1 domain (in brown) related to that in ARF proteins. PB1 domains are diagrammed as arrows with an N-terminal basic amino acid region (+) and a C-terminal acidic region (−) required for forming dimers and oligomers in a front-to-back manner.
(B) Most common PB1 domain interactions among members of the ARF and Aux/IAA protein families in Arabidopsis as reported by Vernoux et al. (2011) are shown as Aux/IAA-Aux/IAA and Aux/IAA-ARF activator with black double-headed arrows. The gray double-headed arrow indicates interactions through PB1 domains for ARF5-ARF5 and ARF7-ARF7 reported in the crystallographic studies of Nanao et al. (2014) and Korasick et al. (2014). Front-to-back interactions are not indicated in this diagram.
(C) Front to back interactions occur through the + and – face of PB1 domains. Type I/II PB1 domains facilitate the formation of homo- and heterodimers. Oligomerization is also possible (double-headed black arrows). Type I and type II PB1 domains can interact with type I/II domains to form dimers. Oligomerization can only occur through the type I/II domain (double-headed arrow). An X indicates the loss of either the + face or the – face, which is required for interaction to occur. Type I PB1 domains can interact with type II PB1 domains to form dimers. Oligomerization cannot occur. Type I PB1 domains cannot interact with one another, and type II PB1 domains cannot interact with one another whether homotypic or heterotypic.
Aux/IAA proteins generally contain one (domain I) or two EAR-type RDs in their N-terminal region, followed by a region containing a degron (domain II), and terminating in a PB1-type protein-protein interaction domain (domain III/IV) related to that in ARFs (Figure 1A; Li et al., 2011; reviewed in Chapman and Estelle, 2009; Guilfoyle and Hagen, 2012; Salehin et al., 2015). Mutations in the conserved degron motif (GWPP) result in the stabilization of Aux/IAA proteins and generally result in plants having constitutive repression of auxin response genes and “low auxin” phenotypes. Some Aux/IAA proteins lack an apparent domain II and have extended lifetimes compared with typical Aux/IAA proteins (Dreher et al., 2006), while others lack both domains I and II and might not function as repressors (Tiwari et al., 2004). Transgenic plants expressing stabilized Aux/IAA proteins with defective RDs display “high auxin” phenotypes, and it has been suggested that these phenotypes result from interactions of the stabilized defective repressors with activator ARFs, thus preventing association of wild-type Aux/IAA repressors and resulting in constitutive expression of auxin response genes (Li et al., 2011).
This review focuses on the PB1 domain found in ARF and Aux/IAA transcription factors and also discusses the DD in the ARF DBD. In this review, the ARF DBD is defined as including the B3 domain as well as the flanking DD and FD regions (Figure 1A) that were originally shown to be required for ARF1 binding to TGTCTC AuxREs in vitro (Ulmasov et al., 1997a). The review includes (1) a brief assessment of the specificity of interactions among ARF and Aux/IAA family members, (2) a summary of the secondary and tertiary structures for the C-terminal domains in ARF5 and ARF7, (3) highlights of some experiments that verify residues in domain III/IV required for protein-protein interactions and an auxin response, (4) a brief description and reassessment of the dimerization and Tudor-like ancillary domains associated with ARF DBDs, and (5) a slate of unanswered questions related to interactions among ARF and Aux/IAA proteins and the roles of these interactions in regulating auxin response genes.
THE SPECIFICITY OF ARF AND AUX/IAA INTERACTIONS MEDIATED BY THEIR C-TERMINAL DOMAINS
In 1997, results based upon yeast two-hybrid (Y2H) assays and screens suggested that the C-terminal domains of ARFs and Aux/IAA proteins were protein-protein interaction domains with amino acid sequence similarity (Kim et al., 1997; Ulmasov et al., 1997a, 1997b; reviewed in Guilfoyle and Hagen, 2012). Since that time, numerous studies have confirmed and expanded this original work using Y2H and a variety of other protein-protein interaction assays; however, the specificity of reported ARF and Aux/IAA interactions has not been entirely consistent and remains controversial (reviewed in Guilfoyle and Hagen, 2012). The most comprehensive analysis of ARF and Aux/IAA interactions to date was performed by Vernoux et al. (2011) for Arabidopsis ARF and Aux/IAA proteins using a Y2H system and bimolecular fluorescence complementation in Nicotiana benthamiana leaves. This study concluded that Aux/IAA-Aux/IAA and Aux/IAA-ARF activator interactions are common and show some, but limited, specificity (Figure 1B). On the other hand, few interactions were found for ARF-ARF and Aux/IAA-ARF repressors, suggesting that if these interactions do occur, they are weaker and/or more specific than Aux/IAA-Aux/IAA and Aux/IAA-ARF activator interactions. While the conclusions of Vernoux et al. (2011) are largely consistent with the bulk of less detailed studies on ARF and Aux/IAA interactions, they are at variance with some studies that have reported ARF activator-ARF activator, ARF repressor-ARF repressor, ARF repressor-Aux/IAA, and ARF repressor-ARF activator interactions (reviewed in Guilfoyle and Hagen, 2012).
A major shortfall inherent in making conclusions about ARF and Aux/IAA protein interactions is that the interaction studies invariably rely on assays where the proteins may have increased stabilities and/or are expressed at much higher levels than occur under natural conditions within a plant cell (reviewed in Guilfoyle and Hagen, 2012). This shortfall can lead to false positives in Y2H and a variety of other protein-protein interaction assays (i.e., interactions observed in the assays that rarely if ever occur under natural conditions). False positives might also arise in some assays (e.g., Y2H) because identified protein partners may not be expressed under natural conditions in the same cells, same subcellular compartment, or at the same time. False negatives can occur due to poor expression levels, highly transient interactions, improper folding, or lack of posttranslational modifications of the proteins tested in different types of assays.
CRYSTAL STRUCTURES REVEAL A PB1 PROTEIN-PROTEIN INTERACTION DOMAIN IN THE C TERMINI OF ARF AND AUX/IAA PROTEINS
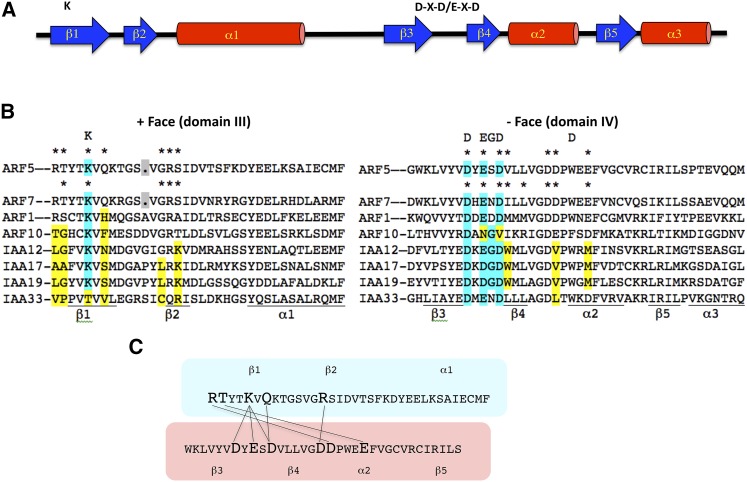
A bioinformatic analysis of domains III and IV in Aux/IAA proteins and the related domains in ARF proteins led to the proposal that these domains were, in fact, a single protein-protein interaction domain (referred to here as domain III/IV as opposed to domain III and domain IV) related in secondary structure to the PB1 protein-protein interaction domain that would form a tertiary structure related to the ubiquitin-like β-grasp fold (Guilfoyle and Hagen, 2012). Figure 2A shows that the secondary structure for domain III/IV consists of a five-stranded β-sheet (β1-β5) and three α-helices (α1-α3). The third α-helix (α3) is not part of typical PB1 domains (reviewed in Sumimoto et al., 2007) and was not originally proposed to be part of PB1 domains in ARF and Aux/IAA proteins (Guilfoyle and Hagen, 2012). Domain III/IV also resembled a PB1 domain in consisting of 80 to 100 amino acids in length, but showed little amino acid sequence identity or similarity to known PB1 domains. Interestingly, however, domain III/IV of both ARF and Aux/IAA proteins contained an invariant lysine at the same position in β1 and an acidic motif (D-X-D/E-X-D) linking β3 and β4, and these are signature residues of PB1 domains that are crucial for conferring interactions (reviewed in Sumimoto et al., 2007). The conserved lysine and other residues in the N-terminal end and the conserved acidic motif and other residues in the C-terminal end of PB1 proteins promote electrostatic and hydrogen-bonding interactions in a front-to-back orientation in the formation of homodimers, heterodimers, or larger oligomers. While some PB1 domains contain both the conserved lysine and acidic motif residues (referred to as type I/II or AB), others contain only the conserved lysine (referred to as type II or B for basic) or conserved acidic motif (referred to as type I or A for acidic). Figure 1C provides diagrams of how the different types of PB1 domains can interact in forming dimers and in some cases oligomers. However, there is specificity in PB1 domain interactions, in that not all type I PB1 domains interact with all type II PB1 domains (Lamark et al., 2003). For the most part, ARF and Aux/IAA domain III/IV resembles a type I/II or AB PB1 domain. While type I and type II PB1 domains can only form dimers, some type I/II PB1 domains not only dimerize but can also form homomultimers and heteromultimers (reviewed in Sumimoto et al., 2007).
Figure 2.
Features of PB1 Domains in ARF and Aux/IAA Proteins.
(A) A general diagram of the secondary structure predicted for PB1 domains in ARF and Aux/IAA proteins is shown. The structure has been confirmed using crystallography for ARF5 and ARF7 domain III/IV (Korasick et al., 2014; Nanao et al., 2014). Blue arrows represent β-sheets (β1 through β5), red cylinders represent α-helices (α1 through α3), and the black line indicates unstructured loops or extensions. The K above β1 is the approximate position of the invariant Lys found in the + face, and D-X-D/E-X-D above and between β3 and β4 is the approximate position of an acidic motif found in the – face of PB1 domains.
(B) Amino acid sequence alignment for PB1 domains in selected ARF and Aux/IAA proteins are from Arabidopsis. ARF5 and ARF7 are activators in protoplasts, ARF1 is a repressor in protoplasts, ARF10 is a predicted repressor within a more divergent clade, IAA12, IAA17, and IAA19 are repressors from different clades, and IAA33 is a more divergent Aux/IAA protein that lacks both domains I and II and has an unknown function. The invariant K in the + face (originally referred to as domain III) of a type II PB1 domain is indicated above and shaded in cyan within the alignment. A conserved acidic motif in the – face (originally referred to as domain IV) of a type I PB1 domain is indicated above and shaded in cyan within the alignment (the E shown could also be an Asp). A signature Gly residue characteristic of PB1 domains (Sumimoto et al., 2007) at the position indicated above the alignment is absent from most ARFs, but present in most Aux/IAA PB1 domains. The D shown above α2 is part of the OPCA motif in typical PB1 domains but is not found at that position in most ARF and Aux/IAA proteins. Most ARFs are type I/II PB1 domains because they contain the signature residues in both faces, but ARF10 is missing part of the acidic motif in the – face and is predicted to be a type II PB1 domain, and IAA33 is missing the invariant K in the + face and is predicted to be a type I PB1 domain. Asterisks show the amino acids known to make contacts between the – and + faces of the PB1 domains for ARF5-ARF5 and AR7-ARF7 (Korasick et al., 2014; Nanao et al., 2014). Residues shaded in yellow differ in similarity or identity from those marked with asterisks in ARF5 and ARF7. The secondary structure (β-sheet and α-helix) is underlined below the alignment.
(C) Main interactions for ARF5-ARF5 PB1 domain dimers reported by Nanao et al. (2014). In the three-dimensional structure, residues in the + face (outlined in light blue) that make contact with residues in the – face (outlined in light red) are shown with lines connecting the amino acids. Relative positions of the β-sheets and α-helices are shown above the + face and below the – face.
In 2014, crystal structures for domain III/IV in ARF5 (Nanao et al., 2014) and ARF7 (Korasick et al., 2014) were published that confirmed these were type I/II PB1 domains with ubiquitin-like β-grasp folds. Similar to typical PB1 domains, structures for ARF5 and ARF7 showed that the invariant lysine is exposed on the surface of β1 in region III of domain III/IV, and the OPCA motif (octicosapeptide or OPR motif, the Phox and Cdc or PC motif, and the atypical protein kinase C interaction domain or AID motif) in region IV of domain III/IV is found in exposed loops flanking β4. The invariant lysine and OPCA motifs are found on opposite faces of the ARF5 and ARF7 domain III/IV structures as in all PB1 domains. The crystal structures confirmed that these PB1 domains interact in a front-to-back manner to dimerize and oligomerize and revealed other charged and uncharged amino acids that contribute to the interactions (Figure 2B). Most of the charged residues that interact with one another are conserved in ARFs, but there are a few differences in Aux/IAA proteins, and some uncharged residues that form contacts in ARF5 and ARF7 are not well conserved in ARF or Aux/IAA proteins. The main contacts for residues in the + face and – face of an ARF5 dimer are diagrammed in Figure 2C. Similar contacts were observed for ARF7.
The 3D structures also revealed some characteristics of domain III/IV that do not conform to typical PB1 domains. Domain III/IV contains an additional α helix (α3) at the C terminus (Korasick et al., 2014; Nanao et al., 2014). However, it has not been shown that the α3 helix, which protrudes from the main body of the ubiquitin-like β-grasp fold, is a functional part of the PB1 domain. The five-residue acidic motif that links β3 and β4 in PB1 domains generally contains a glycine at position 4 that is considered important for positioning the three acidic residues to orient correctly in producing an acidic surface (reviewed in Sumimoto et al., 2007), but this glycine is substituted with serine in ARF5 and asparagine in ARF7 (Figure 2B). This glycine is also absent in domain III/IV of other ARF activators but is found in most Aux/IAA proteins and some ARF repressors. In the typical OPCA motif (i.e., D-X-D/E-G-D-X8-D/E), the C-terminal acidic residue is separated from the cluster of acidic residues by eight amino acids (X8), but domain III/IV in ARF5 and ARF7 as well as that in most other ARF and Aux/IAA proteins do not show this same spacing. Regions III and IV of domain III/IV in ARF and Aux/IAA proteins are joined through a loop of variable length between α1 and β3, and the size of the loop is generally larger than that found in typical PB1 domains. These differences from typical PB1 domains may have arisen from domain III/IV being derived from an ancient PB1 family as suggested by Burroughs et al. (2007).
EXPERIMENTAL CONFIRMATIONS OF THE PB1 DOMAIN IN ARF AND AUX/IAA PROTEINS
Using size-exclusion chromatography, in vitro pull-down assays, and Y2H assays, Korasick et al. (2014) and Nanao et al. (2014) showed that mutations in the invariant lysine residue or the OPCA motif disrupted interactions for ARF7 and ARF5, respectively. Recombinant wild-type ARF7 and ARF5 domain III/IV formed high molecular weight aggregates when tested by size-exclusion chromatography (+ and – arrows at the top of Figure 1C), while recombinant ARF7 domain III/IV with lysine or OPCA motif mutations formed monomers (+ or – arrows at the bottom of Figure 1C) as did recombinant ARF7 and ARF5 domain III/IV with both lysine and OPCA motif mutations. On the other hand, a mixture of recombinant ARF7 or ARF5 domain III/IV with either a lysine or OPCA mutation resulted in formation of dimers (one arrow with a + and the other arrow with a − in Figure 1C), suggesting that the two electrostatic faces are the driving forces for the interactions.
Korasick et al. (2014) used Y2H assays to confirm the in vitro chromatography results and further showed that wild-type ARF7 domain III/IV interacted with wild-type IAA17, an IAA17 lysine mutant, an IAA17 OPCA motif mutant, but not an IAA17 with mutations in both the lysine and OPCA motif. The IAA17 results provided support that IAA17 has a functional PB1 domain (domain III/IV) that facilitates interactions with the ARF7 PB1 domain. Nanao et al. (2014) used size-exclusion chromatography to show that recombinant wild-type IAA12 formed trimers when tested in a buffer that reduced aggregation, but recombinant IAA12 with mutations in lysine, the OPCA motif, or both the lysine and OPCA motifs were reduced to monomers. Likewise, recombinant wild-type domain III/IV of ARF5 could interact with recombinant wild-type IAA12 in a buffer that reduced aggregation, but mutation of the lysine and OPCA motif of ARF5 abolished the interaction. Y2H and in vitro pull-down assays showed that there was no directionality in domain III/IV of ARF5 and IAA12 (i.e., IAA12 could interact with wild-type domain III/IV of ARF5 through either its positive or negative face). Conclusions that can be drawn from these experiments and those described in the preceding paragraph provide support for domain III/IV in ARF and Aux/IAA proteins being a type I/II PB1 domain (Figure 1C).
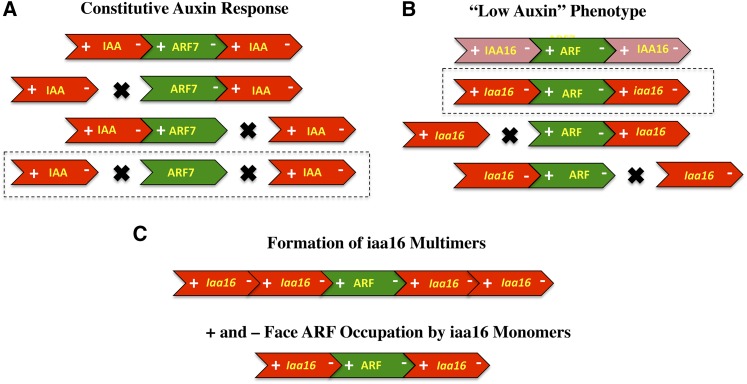
Nanao et al. (2014) used Arabidopsis thaliana protoplast transfection assays to confirm the in vitro and Y2H results in an in planta system. In one type of assay, stabilized versions (domain II mutants) of IAA17 and IAA19 with wild-type domain III/IV, a lysine mutation, an OPCA motif mutation, or lysine plus OPCA motif mutations were tested in wild-type protoplasts containing an integrated DR5:GUS (β-glucuronidase) reporter gene. In this case, transfection of stabilized wild-type IAA17 and IAA19 repressed auxin-responsive reporter gene expression as did lysine or OPCA motif mutants of IAA17 and IAA19 (although repression was reduced somewhat). Most of the repression was lost, however, when both the lysine and OPCA motifs were mutated. In a second type of assay, wild-type ARF7 or ARF7 with mutations in the lysine, OPCA motif, or both the lysine and OPCA motifs were transfected into nph4 mutant (an ARF7 knockout) Arabidopsis protoplasts with a DR5:GUS transgene. Transfection of all ARF7 effector genes resulted in enhanced expression of the reporter gene in auxin-treated protoplasts, and lysine, OPCA, or lysine plus OPCA mutations further enhanced reporter gene expression compared with wild-type ARF7. However, only ARF7 with lysine plus OPCA mutations showed enhanced reporter gene expression in the absence of auxin. This latter result suggested auxin-independent expression was dependent on mutating both the positive and negative faces of the PB1 domain of ARF7 to prevent interaction with endogenous IAA repressors in the protoplasts (Wang et al., 2005). Taken together, the protoplast transfection assays suggest that either the positive or negative face of ARF7 can promote an interaction with Aux/IAA proteins and confer an enhanced auxin response, but when both faces are compromised, ARF7 confers high constitutive expression of DR5:GUS (Figure 3A). These results also suggest that ARF7 does not require the formation of oligomers mediated by the PB1 domain to bring about auxin-independent expression of the reporter gene.
Figure 3.
Diagrams Showing Results and Interpretation of Studies Performed in Transformed Arabidopsis Plants with iaa16 and Arabidopsis Protoplasts with ARF7 PB1 Domain Mutations.
(A) Results of Nanao et al. (2014) showed that overexpression of ARF7 in arf7 mutant Arabidopsis mesophyll protoplasts conferred auxin-independent (constitutive) expression of an integrated DR5:GUS auxin-responsive reporter gene only when both the + and – faces of the ARF7 PB1 domain were mutated (outlined in dashes). Green arrows show the wild type or PB1 mutant transfected ARF7 proteins. Red arrows depict endogenous Aux/IAA repressors.
(B) Results of Korasick et al. (2014) showed that overexpression of stabilized iaa16-1 in transformed Arabidopsis plants was only effective in conferring a “low auxin” phenotype if there were no mutations in either the + face or – face of the iaa16-1 PB1 domain (outlined in dashes). Overexpression of wild-type IAA16 or stabilized iaa16-1 with mutations in either face of the PB1 domain results in plants with wild-type phenotype. Light-red arrows show wild-type IAA16 and dark-red arrows show iaa16-1 with wild-type PB1domains or iaa16-1 with a mutation in either the + or – face of the PB1 domain. Green arrows depict endogenous ARF activators.
(C) Korasick et al. (2014) proposed that the results in (B) revealed that IAA16 proteins might have to form multimers, not dimers, with ARFs to function in repressing auxin response genes (top arrows). Alternatively, it may not be multimerization of IAA16 that is required for repression, but simply that IAA16 must dimerize with both the + and – face of an ARF activator to bring about repression (i.e., IAA16-IAA16 oligomerization may be not required; bottom arrows).
It might be predicted that in planta expression of a lysine or OPCA mutation in a stabilized Aux/IAA protein would lead to “low auxin” phenotypes just like a stabilized Aux/IAA protein without these mutations (i.e., either face of a stabilized Aux/IAA protein might dimerize with a wild-type ARF activator to confer repression). Korasick et al. (2014) tested this prediction using a stabilized iaa16-1 protein (a domain II mutant) with no lysine or OPCA motif mutation, with a lysine mutation, or with an OPCA motif mutation using transformed Arabidopsis plants. Surprisingly, expression of the domain III/IV mutant proteins resulted in wild-type-like plants or plants overexpressing an unstable wild-type IAA16 protein, unlike the stabilized iaa16-1 mutant, which showed a stunted phenotype with a smaller rosette diameter, decreased fertility, and loss of apical dominance. These results led Korasick et al. (2014) to conclude that Aux/IAA proteins must multimerize to repress auxin response genes and confer an auxin response (Figures 3B and 3C). While the above results are consistent with the conclusions drawn, they do not appear to be in complete agreement with protoplast transfection assays as described by Nanao et al. (2014) with stabilized IAA17 and IAA19. These latter results showed that loss of repression of a DR5:GUS reporter gene was greater with a lysine plus OPCA mutant compared with a lysine or OPCA mutant (i.e., although the single mutants did appear to lose some repression compared with the stabilized Aux/IAA proteins without domain III/IV mutations).
A SECOND PROTEIN INTERACTION MODULE IS FOUND IN DBD OF ARF PROTEINS
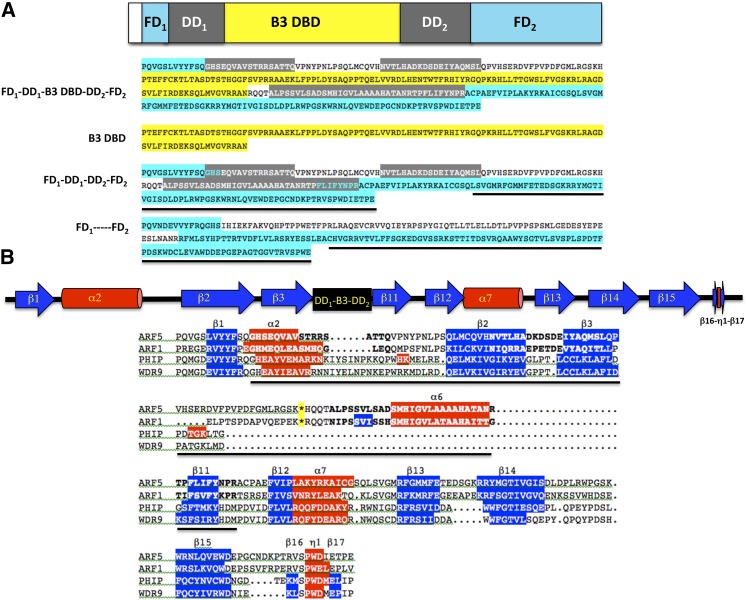
Crystallographic studies revealed that ARFs contain a second protein interaction module (i.e., a homodimerization domain referred to as DD; Boer et al., 2014) located in their DBDs. The DD is divided into components that are adjacent to the N- and C-terminal ends of the B3 domain (DD in Figure 1 and DD1 and DD2 in Figure 4A). These components fold into a “taco”-like tertiary structure that facilitates the formation of ARF dimers and cooperative DNA binding on appropriately spaced AuxREs organized as inverted repeats (Boer et al., 2014). Homodimerization of ARF1 and ARF5 through their DDs facilitates ARF binding to adjacent inverted repeat AuxREs with spacing of five to nine bases and explains specificity for ARF1 and ARF5 binding to these types of DNA target sites. Residues important for dimerization and conserved in ARFs were also revealed in these studies, and mutations in some of these residues in ARF5 impaired the ability of ARF5 to function in vivo. The in vivo studies support the conclusion by Boer et al. (2014) that DBD dimerization is required for ARF5 to function properly in auxin response gene expression.
Figure 4.
The ARF B3 DBD Is Flanked by a Dimerization Domain and a Fragment Found in BRWD Proteins.
(A) The diagram at the top shows the extended DBD of ARFs with the B3 domain flanked by two fragments of the dimerization domain (DD1 and DD2) and two fragments of a segment conserved in BRWD proteins (FD1 and FD2). Below the diagram is the amino acid sequence of the ARF5 extended DBD (FD1-DD1-B3-DD2-FD2) color coded as shown in the diagram. Sequences boxed in gray correspond to the DD motifs reported by Boer et al. (2014). The B3 domain sequence removed from the extended DBD is boxed in yellow. The FD1-DD1-DD2-FD2 minus the B3 domain sequence is shown below the B3 domain sequence, and N- and C-terminal regions of the DD (boxed in gray) related to the BRWD protein fragment are highlighted with cyan lettering. FD1—–FD2 shows the fragment of an Acanthamoeba castellani BRWD protein related to FD1-FD2 and part of the DD in ARFs (boxed in cyan). The sequence with a secondary structure related to the Tudor domain is underlined in ARF5 and the A. castellani BRWD fragment.
(B) The diagram at the top shows the secondary structure of ARF5 (as determined in Boer et al., 2014 and Phyre2 analysis) that corresponds to conserved structures within a fragment of BRWD proteins (as determined by Phyre2 analysis). The black box shows a portion of the DD and B3 domain in ARFs that is not conserved in BRWD proteins. Below the diagram is an amino acid sequence alignment for ARF5, ARF1, mouse pleckstrin homology domain interacting protein (PHIP) BRWD fragment, and human WDR9 BRWD fragment. The predicted secondary structures from Phyre2 analysis are indicated in terms of β-sheet (sequences boxed in blue) or α-helix (sequences boxed in red) using the nomenclature of Boer et al. (2014). The experimentally determined structure for ARF1 and ARF5 (Boer et al., 2014) closely resembles the predicted structures except that the α6 is more extensive. The sequence corresponding to the DD in ARFs is underlined and residues in regions involved in dimerization are in bold letters for ARF1 and ARF5 (Boer et al., 2014). The asterisk (boxed in yellow) is the position of the excised B3 domain in ARF1 and ARF5.
While the PB1 domain is found in all eukaryotes as a protein-protein interaction domain, the ARF DD is not. It was proposed that the B3 domain must have inserted into the DD in the generation of ARF proteins, but the source of the DD is unknown and it has no apparent structural or amino acid sequence similarity to known protein-protein interaction domains or other types of protein domains. What then was the origin of the DD in ARFs? An alternative interpretation for the DD presence in ARF proteins is that the ARF B3 domain inserted into a part of a bromodomain and WD40 repeat (BRWD) protein that included the DD or something that evolved into the DD. The ARF DD (DD1 and DD2 in Figure 4A) is bordered on its N- and C-terminal ends by motives that show strong amino acid sequence and predicted secondary structure similarity to a fragment in BRWD proteins (i.e., based on protein BLAST and Phyre2 FASTA alignments and secondary structure predictions; Figure 4). BRWD proteins typically contain seven or eight WD40 repeats (Pfam 00400) and one or two bromodomains (Pfam00439), but the portion of BRWD proteins that bears similarity to the sequence found in ARFs called the ARF domain or Auxin_resp domain (Pfam 06507) lies outside of both the WD40 repeats and bromodomains in BRWD proteins and has no known function. At the same time, the BRWD fragment that resembles a part of the ARF DBD is a highly conserved region found in BRWD proteins in eukaryotic organisms. Here, the BRWD-like domains in ARFs are referred to as the flanking domains (FD1 and FD2) as opposed to the ancillary domain as designated by Boer et al. (2014) to avoid confusion with the activation domain. A part of FD2 contains the Auxin_resp domain sequence.
Phyre2 results suggest that the BRWD motif is more similar in secondary structure to the related motifs (FD1 and FD2) in ARF1 and ARF5 crystal structures than to the next closest structure, a five-stranded β-barrel-like structure in the Tudor domain (Pfam 00567) of the human PHD-finger protein 20 in FD2 (referred to as the ancillary domain or AD in Boer et al., 2014; Figure 4B). The C-terminal end of the BRWD motif also resembles the Tudor domain found in ARF DBDs. While the C-terminal region FD2 in ARF1, ARF5, and BRWD proteins (∼50 amino acid residues between β13 and β17) resembles a Tudor domain in structure (Boer et al., 2014), there is little amino acid sequence similarity in this region of FD2 to Tudor domains (Supplemental Figure 1). On the other hand, there is substantial similarity in secondary structure and amino acid sequence of the Auxin_resp domain of ARFs to the Auxin_resp domain fragment in BRWD proteins (Figures 4A and 4B; Supplemental Figure 1). When the BRWD fragment encompassing FD1 and FD2 is subjected to BLAST and Phyre2 searches, the FD1 and FD2 of ARFs are retrieved as highly related sequences and secondary structures, encompassing ∼100 amino acids in FD2 (consisting of β11-β17 and α7) as well as 50 amino acids in FD1 (consisting of β1 and the N-terminal part of α2) (Figures 4A and 4B; Supplemental Figure 1). FD1 has not been classified as part of the Auxin_resp domain previously, but the related sequence and predicted secondary structure (β1 and α2) is found amino terminal to FD2 in ARF and BRWD proteins. While the ARF1 and ARF5 DDs show limited structural and amino acid similarity to BRWD regions between FD1 and FD2, there is secondary structural similarity found in these BRWD regions and ARFs (α2, β2, β3, and β11, but not in α6). Some amino acid sequence similarity is found in the BRWD region between FD1 and FD2 within α2, β2, β3, and β11 (Supplemental Figure 1). Furthermore, the N terminus of DD1 overlaps with FD1 in α2, and the C terminus of DD2 overlaps with FD2 in β11 (Figures 4A and 4B). In the scenario proposed here, the ARF DD is not an isolated domain but is part of a larger BRWD-related domain that functions as a dimerization domain in ARF proteins. However, it remains unclear how the B3 domain might have inserted into a fragment of a BRWD protein to generate the extended DBD in ARFs (Ulmasov et al., 1997a; Boer et al., 2014) or if the BRWD-like fragment in ARFs plays any additional roles besides ARF dimerization (e.g., in recruitment of chromatin modifying complexes by recognizing methylated histones). The region between FD1 and FD2 in BRWD proteins is not well conserved in terms of amino acid sequence or length across species, and this region is proposed here to be the origin of DDs in the ARF DBD of land plants.
It seems unlikely that the Auxin_resp domain and additional amino acid residues (including FD1 and sequences flanking FD1 and FD2) originated in ARFs and inserted into BRWD proteins because this domain or sequence motif is found in BRWD proteins of protists, fungi, algae, plants, and animals as revealed from BLAST searches. Furthermore, the Auxin_resp domain is not found in ARFs before plants emerged from an aquatic to land environment. Freshwater algae ancestors of land plants, the charophytes, are reported to contain precursor ARF-like proteins with a B3 domain and domain III/IV, but lack the Auxin_resp domain (De Smet et al., 2011; Finet et al., 2013). The charophyte Klebsormidium flaccidum has an ARF-like protein with an AP2/ERF-related DBD, a B3 domain, and a C-terminal domain III/IV containing the conserved lysine and OPCA motifs found in PB1 domains, but no sequence related to the DD or BRWD found in typical ARF proteins (annotated gene sequence kf100094_0070 in 40528 Klebsormidium flaccidum genome V1.0; Hori et al., 2014). Interestingly, sequences related to the Auxin_resp domain and FD1 with secondary structure predictions resembling segments of ARF FD1 and FD2 are also found in the parasitic protozoan Toxoplasma gondii cloroquine resistance marker gene (NCBI Conserved Domain Architecture Retrieval Tool), although these conserved sequence motifs are separated by lengthy stretches of intervening sequences (Supplemental Figure 1).
Based upon the recent crystallographic studies (Boer et al., 2014; Korasick et al., 2014; Nanao et al., 2014), it might be expected that ARFs, in general, can homodimerize or homo-oligomerize through two protein interaction modules, the DD and PB1 domains (Figure 1A). Both modules might also participate in the formation of ARF heterodimers or hetero-oligomers. Boer et al. (2014) suggested that the PB1 domain in ARF5 might stabilize dimers of the DD and that the PB1 domains in Aux/IAAs might act as competitive inhibitors in preventing ARF oligomerization through the PB1 domain (Boer et al., 2014). How these two protein interaction modules might function together in regulating the expression of auxin response genes will require additional investigation. The ARF DD-FD as well as the PB1 domain might also function in recruiting other transcription factors (Ulmasov et al., 1995; Walcher and Nemhauser, 2012; reviewed in Guilfoyle and Hagen, 2012) that function in a cooperative manner to regulate auxin response genes.
UNANSWERED QUESTIONS AND CONTROVERSIES
The crystal structures of the PB1 domain in ARF5 and ARF7 and dimerization domains in ARF1 and ARF5 DBDs along with other experiments described above provided new insight into how ARF and Aux/IAA proteins interact to regulate auxin response gene expression. On the other hand, these studies have exposed many issues that need to be addressed, and some of these are listed below.
(1) The apparent selective interactions between ARF activators and Aux/IAA proteins remain to be assessed. ARF repressor-Aux/IAA interactions have been documented, but reports on these interactions are less common than ARF activator-Aux/IAA interactions, and interactions may be weak (Vernoux et al., 2011; reviewed in Guilfoyle and Hagen, 2012). Why do ARFs that lack a Q-rich middle region and are predicted to be repressors, but contain the invariant lysine and OPCA motifs, appear to interact weakly, if at all, with Aux/IAA proteins? Is there specificity for individual ARF activator and Aux/IAA repressor interactions, as has been suggested (Knox et al., 2003; Muto et al., 2007; Weijers et al., 2005), and, if so, what residues and contacts in the PB1 domain confer this specificity? There is an interesting amino acid difference located between β1 and β2 in ARF activators when compared with ARF repressors and Aux/IAA proteins (Figure 2B). ARF activators have one less residue than ARF repressors or Aux/IAA proteins in this region. Could this missing residue facilitate ARF activator-Aux/IAA interactions and prevent ARF repressor-Aux/IAA interactions? A second interesting difference between ARF and AuxIAA proteins is found between β4 and α2. The G-D-V-P motif is found in almost all Aux/IAA proteins, but a G-D-D-P motif is found in almost all ARF proteins. This extra aspartic acid and an additional glutamic acid in α2 (Figure 2B) of ARF proteins might be expected to influence electrostatic protein interactions (see asterisks in Figure 2B). Amino acid compositions within the nonconserved loops (especially that between α1 and β3) and within the α3 helix that protrudes from the ubiquitin-like β-grasp fold (Figure 2A) are also candidates for conferring specificity in ARF and Aux/IAA interactions.
There are some PB1 domain swap experiments that might shed initial light on the specificity of ARF and Aux/IAA protein interactions. If the PB1 domain from an ARF activator is swapped for the PB1 domain in an ARF repressor or an Aux/IAA protein or if the PB1 domain in an Aux/IAA protein is swapped for that in an ARF activator or an ARF repressor, what are the phenotypes when the chimeric ARF or chimeric stabilized Aux/IAA protein is overexpressed in plants? Do the chimeric ARF activators fail to be auxin responsive or do they confer constitutive activation or perhaps repression? Do stabilized chimeric Aux/IAA proteins fail to bring about constitutive repression?
(2) While some ARFs (e.g., ARF1 and ARF2) that have been shown or are predicted (e.g., ARF10 and ARF11) to be repressors contain a C-terminal PB1 domain, others do not (e.g., ARF3 and ARF17). What role does the PB1 domain play in ARF repressors? It might be predicted that this domain would facilitate the formation of homo-oligomers similar to those observed with ARF5 and ARF7 (Korasick et al., 2014; Nanao et al., 2014) and possibly hetero-oligomers with other ARF repressors. Formation of dimers or oligomers might increase the capacity of these ARFs to repress transcription of auxin response genes by interacting with the corepressor TOPLESS or a related corepressor (Causier et al., 2012) through a conserved R/K-L-F-G-V repression motif found in ARF repressors (Ikeda and Ohme-Takagi, 2009). If ARF repressor-Aux/IAA interactions do occur on auxin response genes, these could conceivably increase repression beyond that of the ARF repressors themselves. On the other hand, how many repressors are needed to curtail expression of an auxin response gene, and what would be the outcome of Aux/IAA destruction at elevated auxin levels on an inactive gene associated with an ARF repressor?
(3) The composition/stoichiometry of ARF and Aux/IAA proteins on auxin response genes remains to be investigated. Why might Aux/IAA repressors have to multimerize to function in the auxin response as suggested by Korasick et al. (2014), or is it possible that instead of forming multimers, they function as dimers with ARF activators, but have to interact with both the negative and positive face of an ARF activator and thus prevent ARF-ARF interactions on the face unoccupied by the Aux/IAA repressor (Figure 3C)? In experiments described by Korasick et al. (2014), stabilized iaa16 with an invariant lysine or OPCA motif mutation could occupy only one face of an ARF PB1 domain, and this might be insufficient for repression. On the hand, multimerization proposed for ARF and Aux/IAA proteins could result in homomeric and heteromeric complexes (e.g., ARF7-ARF7-ARF5, ARF5-ARF5-ARF12-ARF12, ARF7-ARF7-IAA14-IAA17-IAA29, etc.), which could substantially increase the repertoire of the auxin response. While dimerization is common for PB1 domains, there are examples of multimerization of type I/II PB1 domains in forming homotypic front-to-back arrays (Wilson et al., 2003; reviewed in Seibenhener et al., 2007). Whether Aux/IAA repressors typically function only by forming short or long homo- or hetero-oligomers is still an open question.
PB1 domains can be highly specific or promiscuous in conferring interactions (Lamark et al., 2003). Some types of I/II PB1 domains (e.g., p62) not only interact with themselves, but also with a number of other PB1 domains (reviewed in Seibenhener et al., 2007), and this raises questions about whether interactions of the PB1 domains in ARF and Aux/IAA proteins are limited to interactions among these two families of proteins. There are a variety of other PB1 domain proteins in plants, including other transcription factors (Chardin et al., 2014), protein kinases, and scaffold proteins detected in BLAST searches that could potentially interact with ARF or Aux/IAA proteins.
(4) Not all ARF and Aux/IAA PB1 domains fit the criteria for type I/II domains, and a few would appear to fall into the class of type I or type II. The IAA33 PB1 domain lacks the invariant lysine in α1, but contains the OPCA motif, and might be classified as having a type I domain (Figure 2B). Conversely, the PB1 domains in IAA29, IAA31, and IAA32 as well as ARF10 (Figure 2B), ARF14, ARF15, ARF16, ARF20, and ARF21 lack one or more conserved acidic residues in the OPCA motif but contain the invariant lysine in α1, and these might be classified as type II domains. However, a single substitution for an aspartic acid residue in the acidic cluster of the OPCA motif in IAA29, IAA31, and IAA32 may not be sufficient to inactivate the type I PB1 domain, and these might still function as type I/II PB1 domains. Interestingly, ARF14, ARF15, ARF20, and ARF21 belong to a clade of ARF genes in Arabidopsis, and this clade is missing in most other plant species (reviewed in Guilfoyle and Hagen, 2007). On the other hand, ARF10 and ARF16 have homologs in other plants, and these also lack an apparent OPCA motif. Do these putative type I and type II PB1 domain proteins serve functions different from most ARF and Aux/IAA proteins with type I/II PB1 domains? If multimerization of Aux/IAA proteins or occupation of both the + face and – face of a ARF activator by an Aux/IAA protein is required to bring about repression of auxin response genes, can Aux/IAA proteins with type I or type II PB1 domains function in isolation or do they require Aux/IAA partners with PB1 domains that can interact with the opposite face of an ARF activator?
(5) A recent article showed that rice (Oryza sativa) OsIAA23-R5, a suppressor of the domain II mutant OsIAA23-3, contained a tryptophan-to-serine mutation within the OPCA motif in domain III/IV that changed the “low auxin” phenotype of OsIAA23-3 to a wild-type phenotype (Ni et al., 2014). OsIAA23 interacted with five different putative OsARF activators in Y2H assays, but OsIAA23-R5 failed to interact with these ARFs. In addition, the putative OsARF activators with a tryptophan-to-serine substitution at the same position in domain III/IV failed to interact with wild-type OsIAA23 in Y2H assays. These results appear to be inconsistent with amino acid substitutions required to disrupt type I/II PB1 interaction. The big question is, how does this single tryptophan-to-serine substitution in the acidic face of the OsIAA23 or the OsARF proteins prevent protein-protein interactions in the Y2H system and revert plants expressing OsIAA23 from having a “low auxin” phenotype to wild-type phenotype?
(6) Several studies have shown that domain III/IV in selected ARFs or Aux/IAA proteins can interact with other transcription factors that lack a PB1 domain (e.g., Arabidopsis MYB77 interacting with ARF7, BREVIS RADIX interacting with ARF5, the bHLH protein BIGPETAL interacting with ARF8, and the sunflower [Helianthus annuus] HaIAA27 protein interacting with the heat shock transcription factor HaHSFA9; reviewed in Guilfoyle and Hagen, 2012). It is unclear how these interactions occur and which amino acids in the PB1 domains of ARF or Aux/IAA proteins are required for these interactions? It is worth noting that there is precedence for PB1 domains in other systems forming scaffolds or functioning as adaptors with other proteins that lack a PB1 domain (Nakamura et al., 2006; reviewed in Sumimoto et al., 2007); therefore, it may not be too surprising that PB1 domains in ARF and Aux/IAA proteins may show some promiscuous behavior in interacting with proteins that lack PB1 domains.
(7) There are examples of PB1 domains being regulated (reviewed in Sumimoto et al., 2007). An interesting recent study showed that p62, a type I/II PB1 domain protein found in animals, is phosphorylated at a serine residue in its + face by a cyclic AMP protein kinase (PKA) and associates with cyclic AMP degrading phosphodiesteraser-4 (PDE4) through its – face (Christian et al., 2014). The phosphorylation by PKA confers a negative charge to the + face and disrupts binding of the basic face with partner PB1 domain proteins as well as homopolymerization of p62. The loss of p62 homopolymerization impairs autophagosome formation and subsequent degradation of cellular components via the lysosome, and the loss of PB1 partner binding to p62 impacts signaling pathways related to cancer development, nutrient homeostasis, and immunological responses.
A recent investigation in plants suggests that PB1 domain-dependent interactions of ARF7 and ARF19 with Aux/IAA proteins may be regulated by phosphorylation (Cho et al., 2014). The GSK3-like kinase BIN2 was shown to phosphorylate two serine residues in the glutamine-rich AD of ARF7 (i.e., residues Ser-698 and Ser-707) during lateral root formation, and this leads to disruption of interactions of ARF7 with Aux/IAA repressors. ARF19 was also shown to be phosphorylated by BIN2, and this similarly suppressed interactions with Aux/IAA proteins, but the sites of phosphorylation were not identified. This phosphorylation is reported to increase DNA binding of ARF7 and ARF19 to target promoters and enhance transcription, presumably by disrupting the interactions with Aux/IAA repressors off of DNA. Unlike the studies on p62, however, the results of Cho et al. (2014) do not reveal a mechanism for the disruption of ARF-Aux/IAA interactions and raise some questions. By what mechanism does phosphorylation of ARF7 (or ARF19) in the AD influence the disruption of the ARF and Aux/IAA protein PB1 interaction in C termini of the proteins? Do ARF7 and ARF19 interact with Aux/IAA repressors off of ARF DNA targets, and are these Aux/IAA-ARF complexes inhibited from binding AuxREs? Why does auxin increase DNA binding for ARF7 but decrease DNA binding for ARF2 (Walcher and Nemhauser, 2012), and is this directly related to phosphorylation of the ARFs and ARF PB1 interactions with Aux/IAA proteins? How general are observations made with BIN2 phosphorylation of ARF7, given that one or both of the serine phosphorylation sites are poorly conserved in ARF7 homologs found in other species as well as different in ARFs found in the same species?
(8) It remains unclear whether all ARFs are capable of dimerization through the DD in the extended ARF DBD. Some Arabidopsis ARFs (e.g., ARF3, ARF4, ARF10, ARF16, and ARF17) and ARFs in other species (e.g., Physcomitrella patens) have DDs that are less conserved in amino acid sequence than most ARFs and appear to lack residues important for dimerization that were observed with ARF1 and ARF5 (Boer et al., 2014). However, it was shown that ARF3 (which lacks a PB1 domain), like ARF1 and ARF5, was capable of dimerizing in protoplasts using FRET-FLIM analysis. It remains to be shown whether ARFs can heterodimerize as well as homodimerize through their DD. Whether the BRWD-like fragments in ARFs play any functional role or simply flank the DD domain remains to be assessed.
Of several charophytes investigated, De Smet et al. (2011) found they had ARF-like proteins with B3 and domain III/IV, but lacked the ARF domain related to the BRWD fragment (Auxin_resp domain), and this has led to the suggestion that the Auxin_resp domain was introduced only after plants moved from a water to a land environment (Finet et al., 2013). It will be interesting to determine if ARF-like proteins in charophytes are incapable of dimerizing through their DBDs. It will also be important to determine how general cooperative binding of ARFs on inverted repeat AuxREs of specific spacing is for conferring auxin responsiveness to the large family of genes that are regulated by auxin.
Important questions are: How many auxin response genes contain inverted repeat AuxREs of appropriate spacing to facilitate cooperative binding of ARFs? Also, is dimerization conferred by DDs important for auxin-responsive gene expression in genes that lack these types of AuxREs?
CONCLUSIONS
The availability of three-dimensional structural information for ARF DBDs, including the dimerization domain, and ARF and Aux/IAA PB1 oligomerization domains will be valuable for designing experiments (1) to test the intrinsic specificity of ARF and Aux/IAA interactions and ARF interactions with AuxREs in conferring auxin responses, (2) to create novel members of ARF and Aux/IAA proteins that might be used to modify plant growth and development, (3) to determine how other transcription factors and signaling proteins might interact with ARF and Aux/IAA proteins, and (4) to investigate the higher order structures (e.g., homo- and heterodimers and homo- and hetero-oligomers) of ARF and Aux/IAA proteins on and off of DNA target sites of various AuxRE compositions (e.g., single binding sites, inverted repeats, and tandem direct repeats). Along with structural studies on the TIR1/AFB auxin receptors and the Aux/IAA degron domain, the structural studies summarized in this review have provided substantial new insight into the auxin signaling pathway and the interplay of ARF and Aux/IAA proteins in regulating auxin-responsive gene expression.
Supplemental Data
Supplemental Figure 1. Amino Acid Alignments for the BRWD-Like Fragment in Selected Arabidopsis ARFs and BRWD Proteins.
Supplementary Material
Acknowledgments
Support was provided by Food for the 21st Century Program at the University of Missouri. I thank Gretchen Hagen for comments on the article.
AUTHOR CONTRIBUTIONS
The author is solely responsible for the content of this article.
Footnotes
Articles can be viewed online without a subscription.
References
- Boer D.R., Freire-Rios A., van den Berg W.A., Saaki T., Manfield I.W., Kepinski S., López-Vidrieo I., Franco-Zorrilla J.M., de Vries S.C., Solano R., Weijers D., Coll M. (2014). Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589. [DOI] [PubMed] [Google Scholar]
- Burroughs A.M., Balaji S., Iyer L.M., Aravind L. (2007). Small but versatile: the extraordinary functional and structural diversity of the β-grasp fold. Biol. Direct 2: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Estelle M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43: 265–285. [DOI] [PubMed] [Google Scholar]
- Chardin C., Girin T., Roudier F., Meyer C., Krapp A. (2014). The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 65: 5577–5587. [DOI] [PubMed] [Google Scholar]
- Cho H., et al. (2014). A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 16: 66–76. [DOI] [PubMed] [Google Scholar]
- Christian F., Krause E., Houslay M.D., Baillie G.S. (2014). PKA phosphorylation of p62/SQSTM1 regulates PB1 domain interaction partner binding. Biochim. Biophys. Acta 1843: 2765–2774. [DOI] [PubMed] [Google Scholar]
- De Smet I., et al. (2011). Unraveling the evolution of auxin signaling. Plant Physiol. 155: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K.A., Brown J., Saw R.E., Callis J. (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finet C., Berne-Dedieu A., Scutt C.P., Marlétaz F. (2013). Evolution of the ARF gene family in land plants: old domains, new tricks. Mol. Biol. Evol. 30: 45–56. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2012). Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. 190: 82–88. [DOI] [PubMed] [Google Scholar]
- Han M., Park Y., Kim I., Kim E-H., Yu T-K., Rhee S., Suh J-Y (2014). Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc. Natl. Acad. Sci. USA 111: 18613–18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., et al. (2014). Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Ohme-Takagi M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50: 970–975. [DOI] [PubMed] [Google Scholar]
- Kim J., Harter K., Theologis A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94: 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick D.A., Westfall C.S., Lee S.G., Nanao M.H., Dumas R., Hagen G., Guilfoyle T.J., Jez J.M., Strader L.C. (2014). Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. USA 111: 5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K., Grierson C.S., Leyser O. (2003). AXR3 and SHY2 interact to regulate root hair development. Development 130: 5769–5777. [DOI] [PubMed] [Google Scholar]
- Krogan N.T., Ckurshumova W., Marcos D., Caragea A.E., Berleth T. (2012). Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 194: 391–401. [DOI] [PubMed] [Google Scholar]
- Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., Johansen T. (2003). Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278: 34568–34581. [DOI] [PubMed] [Google Scholar]
- Lau S., De Smet I., Kolb M., Meinhardt H., Jürgens G. (2011). Auxin triggers a genetic switch. Nat. Cell Biol. 13: 611–615. [DOI] [PubMed] [Google Scholar]
- Li H., Tiwari S.B., Hagen G., Guilfoyle T.J. (2011). Identical amino acid substitutions in the repression domain of AUX/IAA proteins have contrasting effects on auxin signaling. Plant Physiol. 155: 1252–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H., Watahiki M.K., Nakamoto D., Kinjo M., Yamamoto K.T. (2007). Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol. 144: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Uhlik M.T., Johnson N.L., Hahn K.M., Johnson G.L. (2006). PB1 domain-dependent signaling complex is required for extracellular signal-regulated kinase 5 activation. Mol. Cell. Biol. 26: 2065–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanao M.H., et al. (2014). Structural basis for oligomerization of auxin transcriptional regulators. Nat. Commun. 5: 3617. [DOI] [PubMed] [Google Scholar]
- Ni J., Zhu Z., Wang G., Shen Y., Zhang Y., Wu P. (2014). Intragenic suppressor of Osiaa23 revealed a conserved tryptophan residue crucial for protein-protein interactions. PLoS ONE 9: e85358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M., Bagchi R., Estelle M. (2015). SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener M.L., Geetha T., Wooten M.W. (2007). Sequestosome 1/p62—more than just a scaffold. FEBS Lett. 581: 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumimoto H., Kamakura S., Ito T. (2007). Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci. STKE 2007: re6. [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen G., Guilfoyle T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen G., Guilfoyle T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1997a). ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Liu Z.-B., Hagen G., Guilfoyle T.J. (1995). Composite structure of auxin response elements. Plant Cell 7: 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcher C.L., Nemhauser J.L. (2012). Bipartite promoter element required for auxin response. Plant Physiol. 158: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Hagen G., Guilfoyle T.J. (2013). ARF-Aux/IAA interactions through domain III/IV are not strictly required for auxin-responsive gene expression. Plant Signal. Behav. 8: e24526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tiwari S.B., Hagen G., Guilfoyle T.J. (2005). AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17: 1979–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Benkova E., Jäger K.E., Schlereth A., Hamann T., Kientz M., Wilmoth J.C., Reed J.W., Jürgens G. (2005). Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 24: 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.I., Gill D.J., Perisic O., Quinn M.T., Williams R.L. (2003). PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell 12: 39–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.