SPA proteins interact with phyA and phyB within nuclear bodies; light-induced binding of phyA and phyB to SPA proteins likely disrupts direct interaction of COP1 and SPAs, resulting in initiation of photomorphogenic development.
Abstract
Phytochromes function as red/far-red photoreceptors in plants and are essential for light-regulated growth and development. Photomorphogenesis, the developmental program in light, is the default program in seed plants. In dark-grown seedlings, photomorphogenic growth is suppressed by the action of the CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1)/SUPPRESSOR OF phyA-105 (SPA) complex, which targets positive regulators of photomorphogenic growth for degradation by the proteasome. Phytochromes inhibit the COP1/SPA complex, leading to the accumulation of transcription factors promoting photomorphogenesis; yet, the mechanism by which they inactivate COP1/SPA is still unknown. Here, we show that light-activated phytochrome A (phyA) and phytochrome B (phyB) interact with SPA1 and other SPA proteins. Fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy analyses show that SPAs and phytochromes colocalize and interact in nuclear bodies. Furthermore, light-activated phyA and phyB disrupt the interaction between COP1 and SPAs, resulting in reorganization of the COP1/SPA complex in planta. The light-induced stabilization of HFR1, a photomorphogenic factor targeted for degradation by COP1/SPA, correlates temporally with the accumulation of phyA in the nucleus and localization of phyA to nuclear bodies. Overall, these data provide a molecular mechanism for the inactivation of the COP1/SPA complex by phyA- and phyB-mediated light perception.
INTRODUCTION
Plants use light not only for photosynthesis but also as a source of information, which is important to adapt growth and development to ever-changing and often hostile environments. For light perception, plants possess several classes of photoreceptors. The cryptochromes (CRY1 and CRY2), phototropins (PHOT1 and PHOT2), and ZEITLUPE family proteins are receptors for blue, UVR8 for UV-B, and phytochromes for red (R) and far-red (FR) light (Kami et al., 2010; Rizzini et al., 2011). Phytochromes exist in two states, the inactive Pr and the biologically active Pfr form that maximally absorb in R and FR, respectively (Mancinelli, 1994). By absorption of light, phytochromes reversibly convert between the two forms, resulting in wavelength-specific Pfr:Ptot (Ptot = Pr + Pfr) ratios. The phytochrome gene family in the model plant Arabidopsis thaliana consists of five members, of which phyA and phyB play a dominant role (Franklin and Quail, 2010). PhyB, the major phytochrome species in light-grown and adult plants, is important for responses to R light and for measuring the R:FR ratio (Kami et al., 2010; Li et al., 2011). By contrast, phyA is highly abundant in dark-grown plants but rapidly degraded in light (Li et al., 2011). Responses induced by low Pfr:Ptot ratios, which are typically established by continuous irradiation with FR or light pulses of any wavelength, depend on phyA (Kami et al., 2010). As such, phyA is required for seedling establishment in light environments dominated by FR light, for instance, the undergrowth of forests (Yanovsky et al., 1995).
Depending on the light conditions, plants follow different developmental programs after germination: skotomorphogenesis in the dark and photomorphogenesis in light. The default developmental program in seed plants is photomorphogenesis, which is repressed in the absence of light. PHYTOCHROME INTERACTING FACTORs (PIFs), CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1), and members of the SUPPRESSOR OF phyA-105 (SPA) family, are crucial to inhibit photomorphogenic growth in the dark (Deng et al., 1991; Laubinger et al., 2004; Leivar et al., 2008; Shin et al., 2009). The E3 ubiquitin ligase COP1 and SPA proteins form oligomeric complexes, which target positive regulators of photomorphogenesis for degradation by the proteasome (Seo et al., 2003; Jang et al., 2005; Zhu et al., 2008). The SPA proteins (SPA1-SPA4 in Arabidopsis) are required for the E3 ubiquitin ligase function of the COP1/SPA complex and may play a role in recognition of substrates, including LONG HYPOCOTYL IN FAR-RED1 (HFR1) and LONG AFTER FAR-RED1 (LAF1) (Yang and Wang, 2006). Moreover, PIFs also form complexes with COP1 and SPA1 and enhance the substrate recruitment, autoubiquitination, and transubiquitination activity of COP1 (Xu et al., 2014). Upon activation by light, phytochromes translocate from the cytosol into the nucleus, where they trigger photomorphogenic responses by both inhibiting binding of PIFs to their target promoters and targeting them for degradation and by stabilizing the targets of the COP1/SPA complex (Kami et al., 2010; Park et al., 2012). Inactivation of COP1/SPA by phytochromes has been proposed, allowing accumulation of transcription factors, such as ELONGATED HYPOCOTYL5 (HY5), HFR1, and LAF1, which trigger photomorphogenic development (Kami et al., 2010; Li et al., 2011). Although it is established that phytochromes must inactivate the COP1/SPA complex in light-grown seedlings, the molecular mechanism is still unknown. Here, we show that both light-activated phyA and phyB compete with COP1 for binding to SPA1 and other SPA proteins, suggesting that phytochromes promote photomorphogenesis by inhibiting the direct interaction of COP1 and SPAs, leading to the inactivation of the COP1/SPA complex. A similar mechanism has been proposed for the CRY1-mediated inactivation of COP1/SPA, while CRY2 appears to employ a different method of inhibition that does not disrupt the direct interaction of COP1 and SPA1 (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011).
Many components involved in light signaling, including phytochromes, COP1, SPAs, and PIFs, form nuclear bodies (NBs) (Van Buskirk et al., 2012). The ability to form NBs often correlates with the physiological activity or degradation of these components, suggesting that NBs are critical for signal transduction and protein turnover. In this article, we show that SPA1 localization to NBs is not dependent on light, in contrast to phyA, and that the phyA G727E and phyA E777K mutants, which are impaired in NB localization, do not interact with SPA1. Yet, SPA proteins appear not to be essential for recruiting phyA into NBs
Light-induced exclusion of COP1 from the nucleus has been proposed as a mechanism to explain inactivation of COP1 in light (Osterlund and Deng, 1998). However, we show that FR-induced accumulation of HFR1, a target of COP1/SPA E3 ubiquitin ligase activity (Jang et al., 2005), precedes nuclear export of COP1 and that HFR1 is stabilized by FR light treatments that temporally correlate with phyA nuclear accumulation and NB localization. This is consistent with a mechanistic model in which disruption of the direct interaction of COP1 and SPAs by light-activated phyA is sufficient to prevent COP1/SPA-induced degradation of HFR1 independent of the dissociation of the COP1/SPA complex and COP1 relocation to the cytosol.
RESULTS
Light-Activated Phytochromes Interact with SPAs in NBs
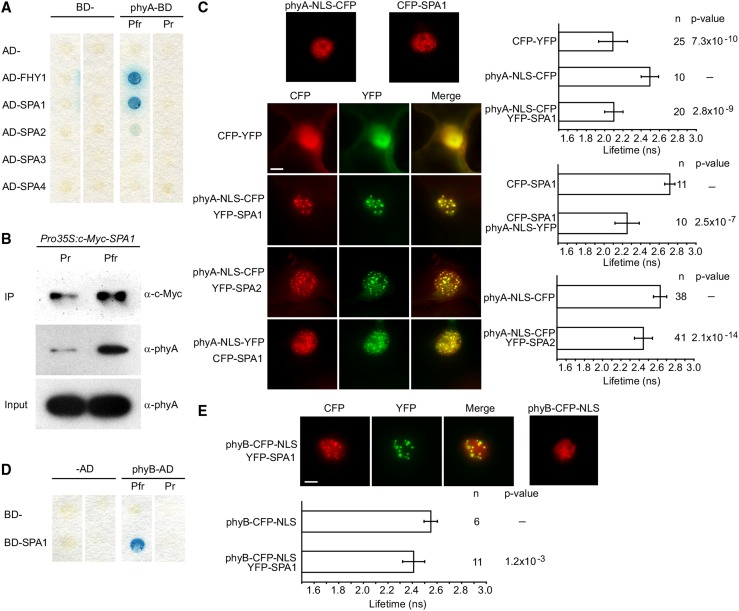
We used yeast two-hybrid screening to identify proteins that directly interact with phyA. In contrast to previous screens involving phytochromes (Ni et al., 1998), we incorporated chromophore to produce photoactive phyA. Yeast is unable to synthesize the naturally occurring chromophore of seed plant phytochromes, phytochromobilin (PΦB). However, previous reports indicate that phycocyanobilin (PCB) extracted from cyanobacteria can substitute in vivo (Kami et al., 2004). Screening on media supplemented with PCB, we identified SPA1 as a phyA-interacting protein (Figure 1A). In a similar fashion to the known phyA interactor FHY1 (Hiltbrunner et al., 2005), SPA1 preferentially bound to the active Pfr form of phyA, with no detectable interaction with the inactive phyA Pr. Supporting this observation, coimmunoprecipitation using Arabidopsis expressing c-MYC-SPA1 also demonstrated a light-dependent pull-down of phyA with SPA1 (Figure 1B). phyA has previously been shown to coimmunoprecipitate with SPA1 from FR-grown Arabidopsis seedlings; however, a direct interaction was never shown (Saijo et al., 2008), and copurification of phyA and SPA1 has been attributed to shared interaction with COP1.
Figure 1.
Light-Activated phyA and phyB Interact with SPAs in Nuclear Bodies.
(A) Yeast two-hybrid protein-protein interaction assay. The phyA-GAL4-DNA binding domain (phyA-BD) fusion was coexpressed with GAL4-activation domain (AD-) fusions of FHY1 and SPA1-4. Yeast cells were lifted from chromophore-supplemented plates that had been incubated for 48 h under either constant R (Pfr) or FR light (Pr). Interaction was detected by an X-Gal filter lift assay.
(B) Coimmunoprecipitation of phyA with SPA1. SPA1 was immunoprecipitated from stable transformed Arabidopsis plants expressing c-Myc-tagged SPA1 using an α-c-Myc antibody. Plants were grown in darkness (Pr) or darkness followed by a 5-min R light pulse (Pfr) prior to immunoprecipitation. α-phyA antibodies were used to detect phyA copurifying with c-Myc-SPA1.
(C) FRET-FLIM analysis of NB-localized phyA and SPA1/SPA2 CFP and YFP fusions transiently expressed under the control of the 35S promoter in N. benthamiana. Left, epifluorescent microscope visualization of subcellular localization of phyA, SPA1, and SPA2 upon transfer from darkness to light; right, fluorescence lifetime of the donor (CFP). Error bars show one sd. n = number of measurements. P values indicate t test analysis for statistically significant differences. Bar = 10 μm.
(D) Yeast two-hybrid protein-protein interaction assay as for (A) except BD-SPA1 and phyB-AD fusions were used to avoid phyB autoactivation.
(E) FRET-FLIM analysis as for (C) of NB-localized phyB-CFP and YFP-SPA1.
To establish if phyA and SPA1 interact in planta, we coexpressed cyan and yellow fluorescent protein (CFP and YFP) fusions of either SPA1 or phyA in Nicotiana benthamiana (wild tobacco) under the control of the strong 35S mosaic virus promoter. As phyA nuclear import was rather inefficient in N. benthamiana leaves, a nuclear localization signal (NLS) was included for phyA fusions to ensure sufficient phyA in leaf epidermal cell nuclei (Genoud et al., 2008; Supplemental Figure 1). phyA was subsequently activated through exposure to epifluorescent light. In N. benthamiana leaves transiently coexpressing YFP-SPA1 and phyA-NLS-CFP (or CFP-SPA1 and phyA-NLS-YFP), the two proteins colocalized to NBs (Figure 1C). Furthermore, in fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy (FRET-FLIM) analyses of NB-localized phyA and SPA1 (phyA-NLS-CFP/YFP-SPA1 or CFP-SPA1/phyA-NLS-YFP), the fluorescence lifetime of the donor (CFP) was strongly reduced as compared with negative controls, supporting an interaction of SPA1 and phyA within NBs (Figure 1C).
We also observed colocalization of phyA-NLS-CFP with YFP-SPA2, -SPA3, and -SPA4 in NBs of Agrobacterium tumefaciens-infiltrated N. benthamiana leaves, though only for SPA2 was a significant reduction in fluorescence lifetime detected (Figure 1C; Supplemental Figure 1). Yeast two-hybrid assays also detected binding of phyA to SPA2, but not SPA3 or SPA4, though weak interactions cannot be excluded (Figure 1A; Supplemental Figure 2). Thus, a positive signal in FRET-FLIM analyses was specifically observed for SPA proteins that physically interacted with phyA but not for those that only colocalized with phyA in NBs.
The major Arabidopsis phytochrome in adult plants, phyB, has been reported to interact with SPA1 in a light-independent manner (Zheng et al., 2013). Using altered conditions to avoid autoactivation, and to include full-length photoactive phytochrome, a light-dependent interaction between SPA1 and phyB was observed in yeast two-hybrid assays, suggesting that both phyA and phyB may share a similar light-dependent interaction with SPA1 (Figure 1D; Supplemental Figure 2). phyB was also observed to colocalize as a CFP-NLS fusion with YFP-SPA1 in NBs (Figure 1E). Furthermore, a significant decrease in donor fluorescence life time was measured in FRET-FLIM analyses, suggesting that phyB and SPA1 also interact in NBs.
SPA1 NBs Do Not Depend on Light
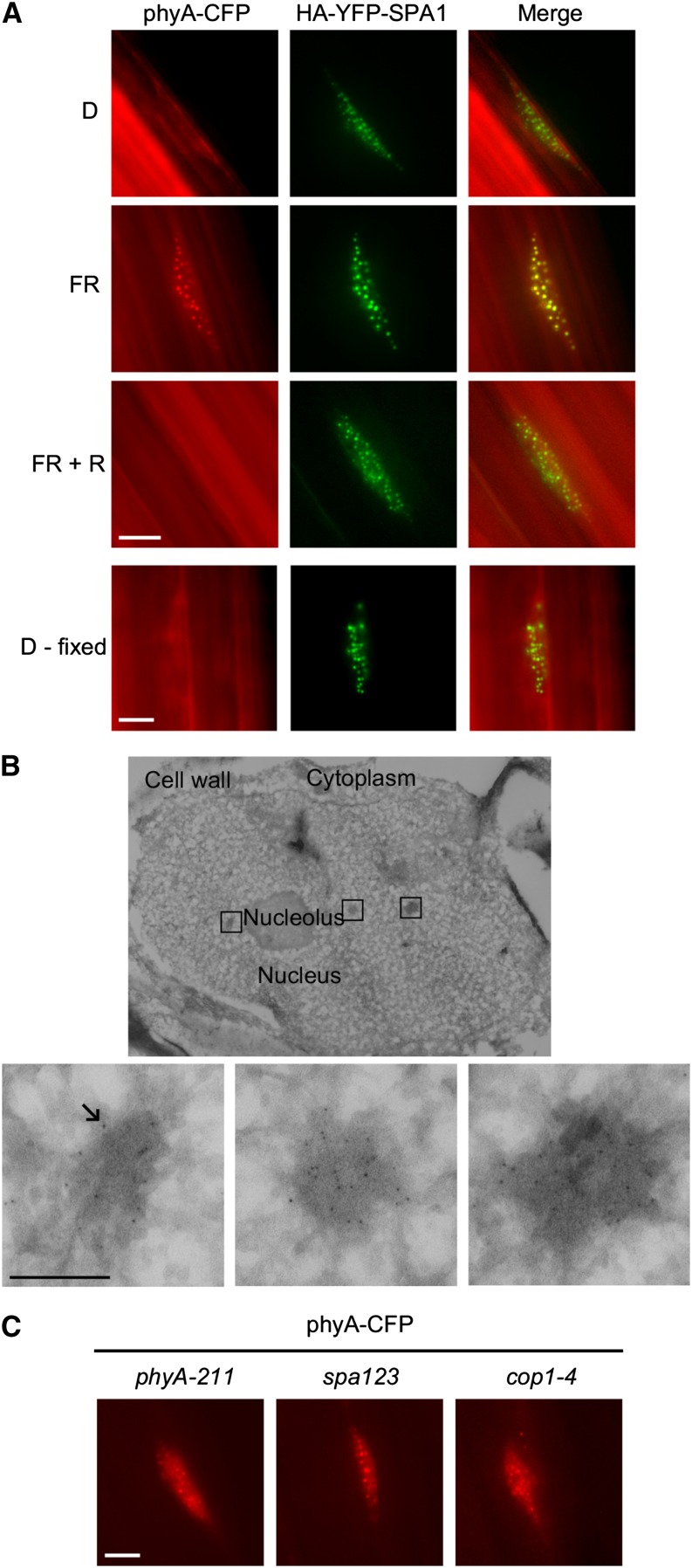
Using stable transgenic ProPHYA:PHYA-CFP and Pro35S:HA-YFP-SPA1 Arabidopsis lines, we confirmed the colocalization of phyA and SPA1 in NBs of seedlings exposed to FR (Figure 2A; Supplemental Figure 3A). Furthermore, we performed immunofluorescence and immunoelectron microscopy to show that native Arabidopsis phyA is localized to NBs after irradiation with light and that phyA-containing NBs correspond to electron-dense structures of 100 to 200 nm in diameter (Figure 2B; Supplemental Figure 4). NBs are therefore not an artifact of transgene expression, consistent with similar experiments performed on pea (Pisum sativum) phyA (Hisada et al., 2000, 2001). As SPA1 is normally a light-induced gene, we used constitutive 35S promoter-driven expression to investigate the dependence of light on protein localization (Hoecker et al., 1999; Fittinghoff et al., 2006). In contrast to phyA, SPA1 was also present in NBs of dark-grown, etiolated seedlings (Figure 2A; Supplemental Figure 3B). To exclude the possibility that brief light exposure during sample preparation or image acquisition induced the formation of SPA1-containing NBs, we fixed the seedlings with formaldehyde under green light (525 nm) prior to microscopy. Even in dark-grown seedlings expressing HA-YFP-SPA1 fixed with formaldehyde, we observed SPA1 in NBs (Figure 2A). Thus, SPA1 NB-localization does not depend on light, which is in contrast to other components involved in light signaling (phytochromes, cryptochromes, and PIFs), suggesting that SPA proteins could function in recruitment of phyA into NBs (Van Buskirk et al., 2012). To test the requirement of SPA proteins for phyA NB formation, we transformed ProPHYA:PHYA-CFP into a spa123 triple mutant background (Figure 2C). Interestingly, phyA-CFP localization was unaffected. Together with the previous observation that SPA4 only plays a minor role in dark-grown seedlings (Laubinger et al., 2004), this leads to the conclusion that another as yet uncharacterized factor is likely sufficient for NB formation. COP1 is also localized to NBs in darkness, so we also investigated phyA-CFP localization in a cop1-4 mutant background. Similar to the spa123 background, phyA-CFP still localized to NBs after activating light exposure in a cop1-4 background (Figure 2C). However, cop1-4 is a weak COP1 mutation that expresses a truncated COP1 protein (complete loss-of-function alleles are lethal), and it cannot be concluded for certain that COP1 is not involved in phyA NB localization (McNellis et al., 1994).
Figure 2.
phyA Colocalizes with SPA1 in Arabidopsis Nuclear Bodies.
(A) Epifluorescence microscopy visualization of phyA-CFP and HA-YFP-SPA1 in hypocotyl cells of stable cotransformed ProPHYA:PHYA-CFP and Pro35S:HA-YFP-SPA1 Arabidopsis plants. Seedlings were grown for 4 d in darkness and treated with either no light (D), 6 h FR light, or 6 h FR followed by 6 h R light (FR + R). Additionally, 4-d-old etiolated seedlings were fixed with formaldehyde prior to microscopy (D - fixed).
(B) Immunoelectron microscopy localization of phyA in wild-type Arabidopsis Col-0 hypocotyl nuclei. Seedlings were grown in darkness for 4 d and treated with 6 h FR light followed by 5 min R light prior to fixation. Endogenous phyA was probed with α-phyA antibodies and detected with protein A-labeled 6-nm gold particles (indicated by arrow). Upper panel: overview (nucleus). Lower panels: enlarged areas (nuclear bodies). Bar = 200 nm.
(C) Epifluorescence microscopy visualization of phyA-CFP expressed from ProPHYA:PHYA-CFP in phyA-211, spa1-7 spa2-1 spa3-1 (spa123), and cop1-4 Arabidopsis backgrounds. Seedlings were grown in darkness for 4 d, followed by 6 h FR. Bars in (A) and (C) = 4 μm.
The Photosensory Domain Is Not Sufficient for the Pr/Pfr Specificity of the phyA-SPA1 Interaction
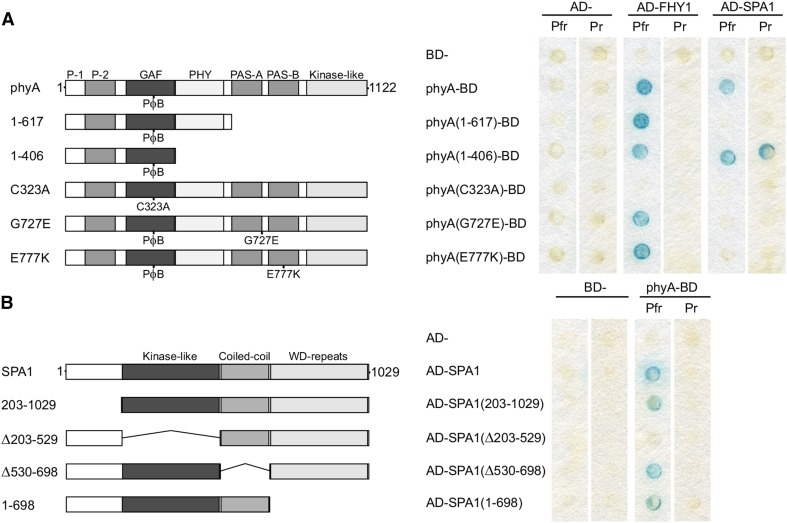
Consistent with the Pfr-dependent interaction of phyA and SPA1, we found that the phyA C323A mutant, which cannot covalently bind chromophore (Rockwell et al., 2006), does not interact with SPA1 (Figure 3A). Unexpectedly, expression of an N-terminal fragment of phyA (1 to 406) resulted in a light-independent interaction with SPA1, yet a larger fragment including the PHY domain (1 to 617) resulted in a loss of binding, despite retaining a light-dependent interaction with FHY1 (Figure 3A; Supplemental Figure 5). These results indicate a binding site located in the N-terminal 406 residues of phyA and that access is regulated through residues 407 to 1122, including the PHY domain, which is expected to form contacts with the chromophore and GAF domain (Essen et al., 2008; Yang et al., 2009).
Figure 3.
The N Terminus of phyA Interacts with the Kinase-Like Domain of SPA1.
(A) Domain and mutant analysis of phyA. phyA truncations and amino acid substitutions fused to the GAL4-DNA binding domain (BD) were coexpressed with GAL4-activation domain (AD)-SPA1. Yeast cells were lifted from chromophore-containing plates that had been incubated for 48 h under either constant R (Pfr) or FR (Pr) light. Interaction was detected by an X-Gal filter lift assay. Left, schematic of the phyA truncations and substitutions; right, X-Gal filter lift assay.
(B) Domain analysis of SPA1. SPA1 truncations or deletions fused to the GAL4 AD were coexpressed with phyA-BD. The yeast two hybrid assay was performed as described in (A). Left, schematic of SPA1 truncations and deletions; right, X-Gal filter lift assay.
Immunoblot analysis of phyA and SPA1 protein levels and quantitative assays are shown in Supplemental Figure 5.
The phyA-103 and phyA-302 mutants are insensitive to FR light due to missense mutations in the PAS-A (G727E) or PAS-B (E777K) domains of phyA, respectively (Dehesh et al., 1993; Yanovsky et al., 2002). As neither phyA mutant is recruited into NBs, where we observed the phyA-SPA1 interaction in planta, we investigated if phyA G727E or phyA E777K are altered in their interaction with SPA1. While these mutations did not affect the interaction with FHY1 in yeast, they both abolished detectable binding of SPA1 (Figure 3A). As both mutations are located in the C terminus of phyA, they are likely involved in the aforementioned C terminus-mediated light dependency of the SPA1-phyA interaction, rather than residues that directly bind SPA1. As phyA still forms NBs in the spa123 mutant background, it seems likely that other uncharacterized phyA interactions also are affected in phyA G727E and phyA E777K, leading to loss of NB localization.
The Kinase-Like Domain of SPA1 Is Essential for the Interaction with phyA
SPA proteins consist of a variable N terminus and three conserved domains: a kinase-like domain, a coiled-coil domain, and a C-terminal WD-repeat domain. Using yeast two-hybrid assays, we found that the kinase-like domain of SPA1 is essential for binding to phyA (Figure 3B; Supplemental Figure 5). By contrast, the N-terminal extension, the coiled-coil, and WD-repeat domains are not essential for the interaction with phyA, although they may contribute to the strength of the interaction.
Phytochromes Inhibit the Interaction of SPAs and COP1
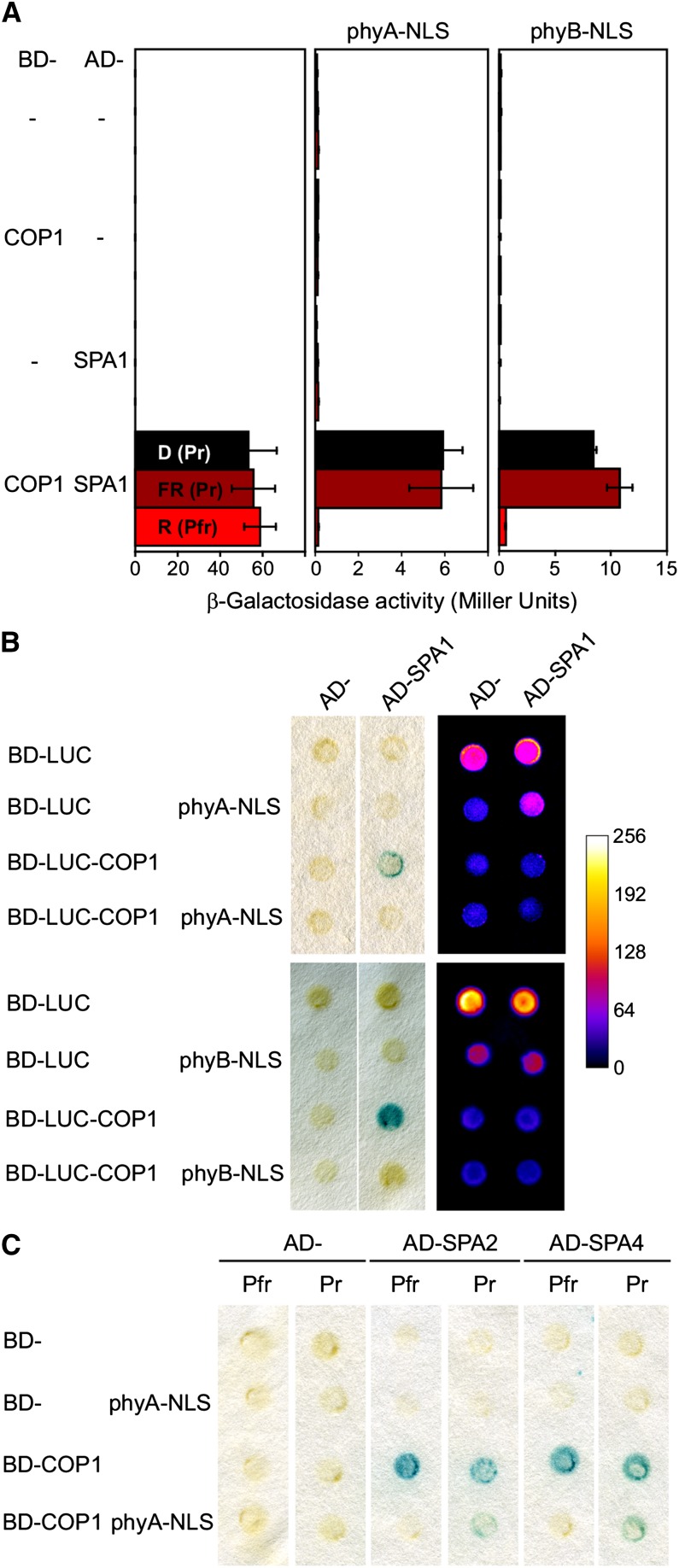
It is well established that phyA inhibits the COP1/SPA-dependent turnover of transcription factors, such as HY5, HFR1, LAF1, and CO, but the molecular mechanism has not been described (Ang et al., 1998; Seo et al., 2003; Duek et al., 2004; Jang et al., 2005; Yang et al., 2005). Using yeast three-hybrid assays, we investigated if light-activated phyA is able to regulate the interaction of COP1 and SPA proteins. Under activating light conditions, coexpression of phyA inhibited the interaction of COP1 with SPA1, whereas only weak inhibition was observed in the dark, even though the phyA protein levels were lower in light than in dark-grown yeast cells (Figures 4A and 4B; Supplemental Figure 6). Neither COP1 nor SPA1 protein abundance was affected by the presence of phyA in yeast, eliminating regulation of protein stability as a possible explanation. In addition, similar photoactivated phyA-dependent inhibition of the interaction between COP1 and both SPA2 and SPA4 was observed, indicating that phyA can bind and inhibit SPA1, SPA2, and SPA4 protein in a light-dependent manner (Figure 4C). Light-activated phyB was also observed to inhibit the interaction of COP1 and SPA1 in yeast three-hybrid assays (Figures 4A and 4B).
Figure 4.
Phytochromes Inhibit the Interaction of COP1 with SPA Proteins.
(A) Yeast three-hybrid analysis of the effects of phyA and phyB on the COP1-SPA1 interaction. COP1 and SPA1 were expressed as a standard yeast two-hybrid protein-protein interaction pair (BD-COP1 and AD-SPA1). Phytochromes were coexpressed as additional proteins, with C-terminal nuclear localization signals (phyA/B-NLS). Yeast cells were grown on chromophore-supplemented plates for 72 h under either constant darkness (D), R, or FR light, and the interaction of COP1 and SPA1 detected using ONPG. Values are the average of nine assays; error bars display 1 sd.
(B) Yeast three-hybrid analysis, including a LUC fusion of COP1 (BD-LUC-COP1) and AD-SPA1, performed as in (A) under constant R light. Left, X-Gal filter lift assays; right, in vivo luciferase activity. Plates were sprayed with 5 mM luciferin and imaged using a CCD camera. Arbitrary light signal intensity is indicated in the adjacent scale.
(C) Yeast three-hybrid analysis of the effects of phyA on the COP1-SPA2/4 interaction. Performed as for (A) in either darkness (Pr) or constant R light (Pfr), and interaction was detected by X-Gal filter lift assay.
Immunoblot analysis of BD-LUC-COP1, AD-SPA1, and phyA-NLS protein levels is shown in Supplemental Figure 6.
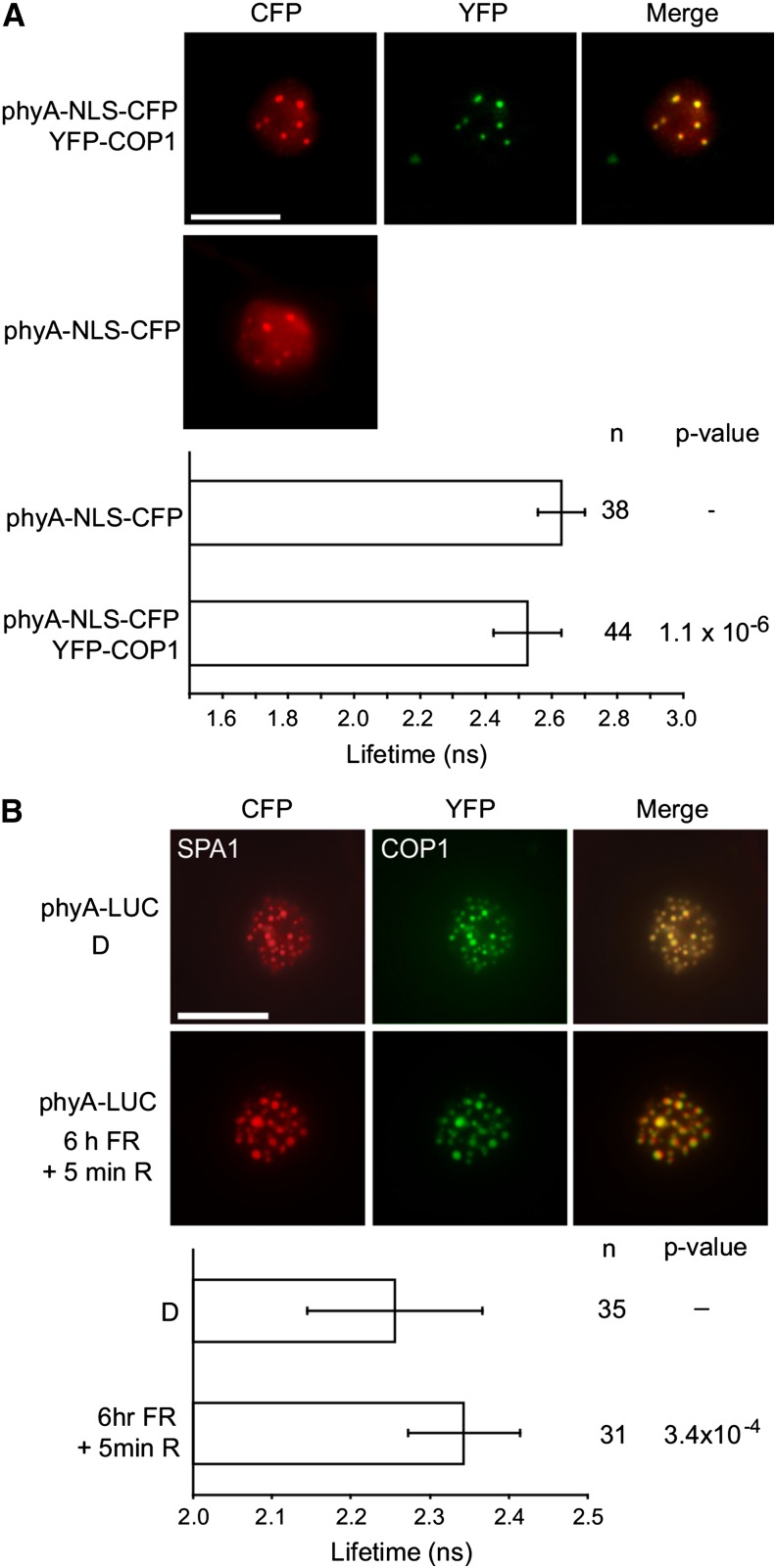
Based on yeast three-hybrid data (Figure 4), COP1 would be expected to be excluded from NBs under FR light conditions where phyA is recruited into NBs. However, phyA has also been indicated to interact with COP1 (Seo et al., 2004; Viczián et al., 2012), suggesting that phyA-induced inactivation of the COP1/SPA complex does not require dissociation of COP1 from NBs in FR. Using FRET-FLIM measurements of phyA and COP1 expressed in N. benthamiana, we confirmed that light-activated phyA was associated with COP1 in planta within NBs (Figure 5A). To investigate if the interaction of COP1 and SPA1 is altered by phyA in planta, we coexpressed CFP-COP1 and YFP-SPA1 as a FRET pair in N. benthamiana, with the addition of a firefly luciferase (LUC) fusion of PHYA (PHYA-LUC). Following activating light conditions (6 h FR followed by 5 min R light) the fluorescence lifetime of the CFP donor was increased in FRET-FLIM measurements, consistent with disruption of the direct interaction of the COP1 and SPA1 molecules within the complex (Figure 5B; Supplemental Figure 7). To ensure that irradiation during FRET-FLIM measurement did not affect the complex, samples were also fixed following light treatments, obtaining similar results (Supplemental Figure 7).
Figure 5.
Reorganization of the COP1/SPA1 Complex by phyA upon Irradiation with Light.
(A) FRET-FLIM analysis of NB-localized phyA and COP1 CFP and YFP fusions in transiently transformed N. benthamiana plants. Upper panels show epifluorescent microscope visualization of subcellular localization. The lower panel displays the fluorescence lifetime of the donor (CFP).
(B) FRET-FLIM analysis of the disruption of the interaction between COP1 and SPA1. CFP-SPA1, YFP-COP1, and phyA-LUC were cotransformed into N. benthamiana. Plants were grown in darkness (D) or darkness followed by 6 h FR and a 5-min R pulse to activate phyA nuclear transport and NB formation. Upper panels show epifluorescent microscope visualization of subcellular localization. The lower panel displays the fluorescence lifetime of the donor (CFP).
Error bars show 1 sd. n = number of measurements. P values indicate t test analysis for statistically significant differences. Expression of constructs in (A) and (B) was driven by the 35S promoter. Bars = 10 μm.
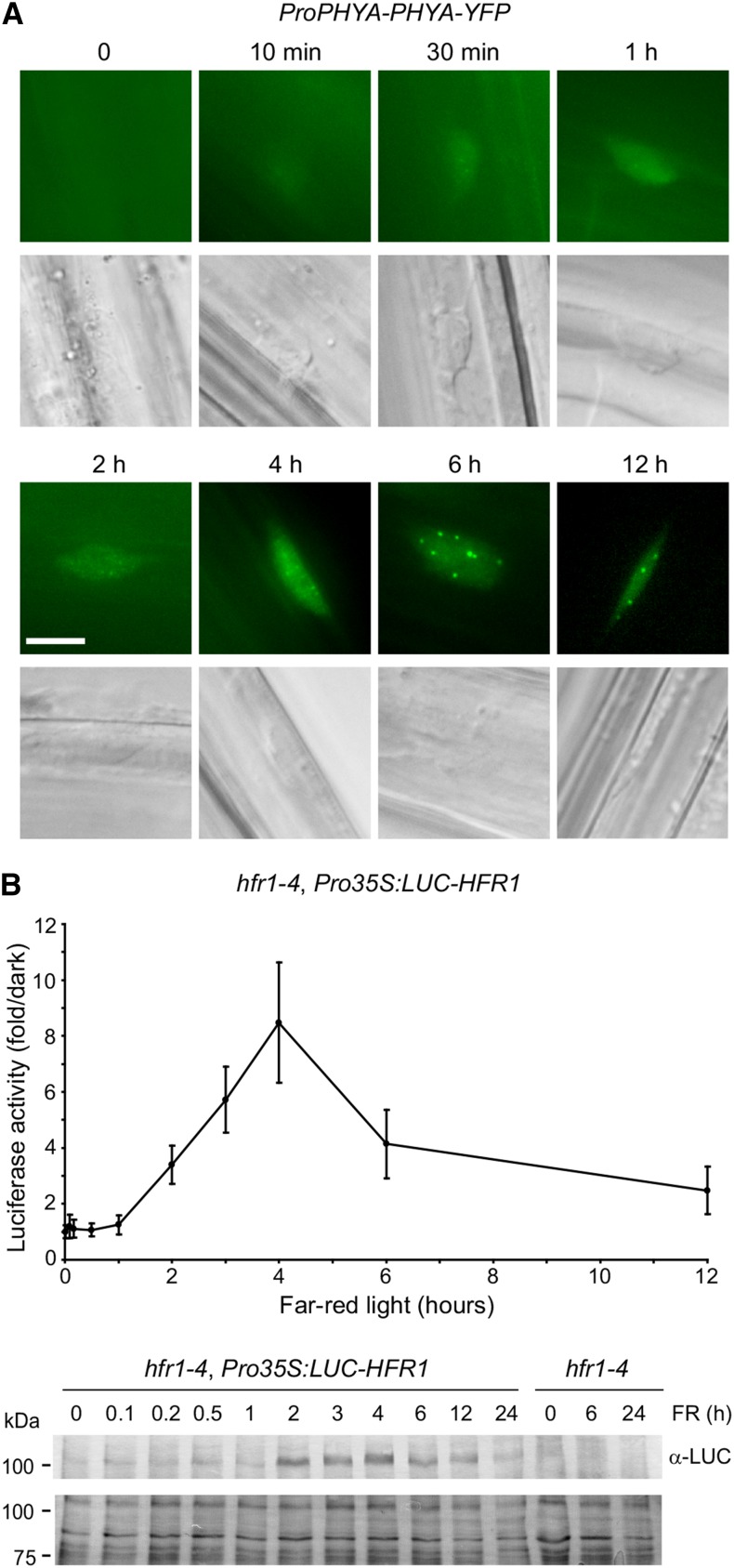
To further characterize phyA-dependent inactivation of COP1/SPA activity in plants, we measured the timing of stabilization by light of the positive photomorphogenic factor HFR1, which is a target of the COP1/SPA complex (Jang et al., 2005). Localization of phyA-YFP to the nucleus began within minutes of FR exposure and formation of late NBs began after 2 to 4 h FR light exposure, with localization at a maximum after 6 h (Figure 6A). Using stably transformed hfr1-4, Pro35S:LUC-HFR1 lines, we quantified the protein abundance over the length of FR exposure by measuring luciferase activity and by immunoblots (Figure 6B; Supplemental Figure 8). Stabilization began after 1 h, reaching a peak at 4 to 6 h, similar to phyA nuclear accumulation and localization to NBs, consistent with these events being linked. Longer exposures to FR resulted in destabilization of HFR1, potentially by activation of an unknown feed-back mechanism.
Figure 6.
Temporal Correlation of HFR1 Accumulation and phyA Nuclear Localization.
(A) Time course of phyA nuclear accumulation and localization to nuclear bodies. phyA-YFP localization was observed in dark-grown phyA-211 ProPHYA:PHYA-YFP seedlings exposed to various lengths of FR light (10 μmol m−2 s−1). Bar = 5 μm.
(B) HFR1 accumulates under FR light. The luciferase activity of stable transformed hfr1-4, Pro35S:LUC-HFR1 Arabidopsis plants was measured in dark-grown seedlings exposed to various lengths of FR light (10 μmol m−2 s−1). Below, immunoblot detection of LUC-HFR1 in 5 μg plant extracts. Lower panel shows amido black-stained membrane as a loading control.
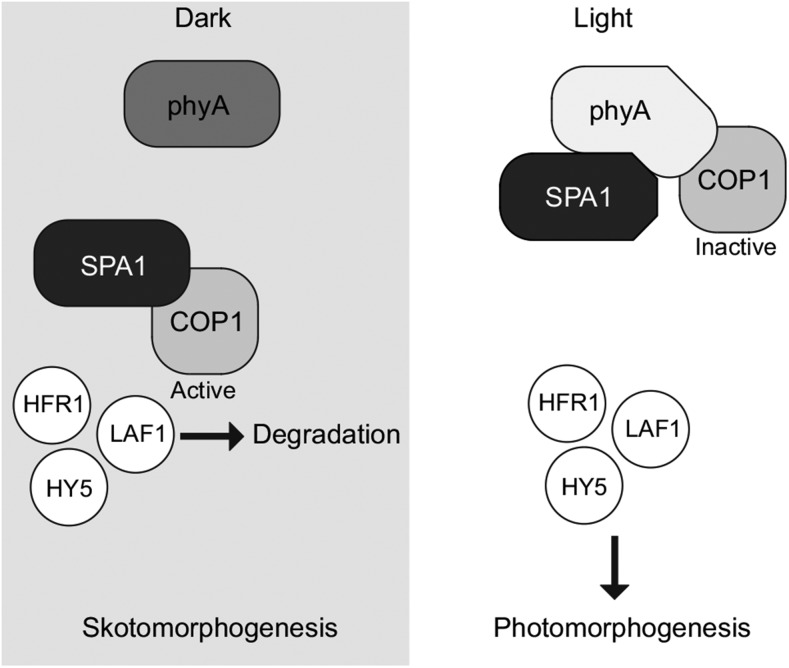
Thus, it appears probable that in planta light-induced binding of phyA to SPA proteins can disrupt the direct interaction of COP1 and SPAs and thus inactivate the COP1/SPA complex, resulting in rapid accumulation of transcription factors initiating photomorphogenic development. COP1 and SPAs may be retained in complex through independent interactions with phyA, yet the direct interaction of COP1 and SPAs would remain disrupted in plants exposed to light (Figure 7). Interaction of phyA and SPAs may induce additional events, such as modification of COP1 and SPAs or regulation of SPA protein stability, leading to sustained downregulation of COP1/SPA activity in light-grown plants.
Figure 7.
A Model for Light-Dependent Induction of Photomorphogenesis.
In darkness, SPA1 and other SPAs bind and activate the E3 ubiquitin ligase COP1. Activated COP1 is able to target positive photomorphogenic factors, including HFR1, LAF1, and HY5, for degradation by the proteasome. Thus, the skotomorphogenic program is established, resulting in hypocotyl elongation and repression of cotyledon development. Light-activated phyA (and phyB) can bind SPA1 and other SPAs and disrupt the direct COP1-SPA interaction. COP1, lacking a direct activating interaction with SPA, can no longer target photomorphogenic factors for degradation. These factors accumulate and promote the photomorphogenic program, inhibiting hypocotyl elongation and promoting cotyledon and leaf development. Through the direct interaction with phyA, COP1 can remain in complex even under conditions that promote photomorphogenesis.
DISCUSSION
SPA proteins, which are represented in Arabidopsis by SPA1-4, have important functions in regulating photomorphogenesis (Hoecker et al., 1999; Laubinger et al., 2004). SPA1 and SPA2 have been shown to be the primary SPAs involved in repression of photomorphogenesis at the seedling stage, while SPA3 and SPA4 have roles during adult plant development (Laubinger and Hoecker, 2003; Laubinger et al., 2004). Here, we have shown that phyA preferentially binds to SPA1 and SPA2 and these interactions correlate well with the primary function of phyA during seedling development and lesser effects on adult plant growth (Laubinger et al., 2004). Though SPA1 is dispensable for most of adult plant development, it is required for proper control of flowering (Laubinger et al., 2006). Therefore, interaction of SPA1 and phyA is also consistent with the role of phyA in regulation of flowering in short-day conditions with FR extension (Laubinger et al., 2006). In addition, we also observed Pfr-dependent binding of SPA1 to phyB, which also plays a role in adult plants.
SPAs form a light-independent complex with COP1 (Hoecker and Quail, 2001). This complex has been shown to both enhance the E3 ubiquitin ligase activity of COP1 and to bind and target for degradation many transcription factors that promote photomorphogenesis, including HFR1, LAF1, and HY5 (Seo et al., 2003; Jang et al., 2005; Yang et al., 2005). The stability of these positive factors is increased upon exposure to light, indicating a light-dependent inactivation of COP1 (Kami et al., 2010; Li et al., 2011). Here, we demonstrated that the phyA- and phyB-SPA interactions disturb the COP1/SPA complex under activating light conditions. Indeed, loss of SPA1 has been shown to enhance light responses, and it has been suggested that phyA might inactivate SPA1 in response to FR light (Hoecker et al., 1998). Moreover, multiple spa-null seedlings are hypersensitive to FR light, particularly in the absence of a functional SPA1, consistent with less activated phyA being required for inactivating the lower levels of SPA proteins in these lines (Balcerowicz et al., 2011). We propose that under light conditions where phyA and phyB accumulate in the nucleus, activated phyA and phyB bind to SPAs, disrupting the COP1-SPA interaction and thereby preventing the degradation of positive photomorphogenic factors (Figure 7). Even after disruption of the direct COP1-SPA interaction, COP1 can be retained in complex due to a direct interaction with phyA. Although the coiled coil domain of SPA1, which mediates the interaction with COP1 (Hoecker and Quail, 2001), is not blocked directly through SPA1-phyA binding, it is possible that phyA introduces a steric hindrance that interferes with binding of SPA1 to COP1.
Phytochromes binding to COP1/SPA may have additional effects, such as regulation of SPA protein turnover (Balcerowicz et al., 2011) or, hypothetically, modification of COP1 and SPAs, which may be important to ensure sustained downregulation of COP1/SPA activity in light-grown plants. It is interesting that the kinase-like domain of SPA1 was found to be essential for the interaction with phyA, as it has been shown that the coiled coil and WD-repeats produce a functional SPA1 but that the kinase-like domain is required for the destabilization of SPA1 in FR (Yang and Wang, 2006). SPA1 and SPA2 are destabilized in seedlings exposed to FR (Balcerowicz et al., 2011), and it is possible this is a consequence of the light-regulated interaction with phyA. Interestingly, phyA degradation in R is reduced in spa123 triple mutant background (Debrieux et al., 2013), suggesting that binding of SPAs to activated phyA could contribute to light-enhanced phyA protein turnover, which may be important to avoid overactivation of the phyA downstream signaling pathway in R. phyA is also stabilized in the cop1-4 mutant in presence of sucrose, which is in contrast to spa123, where the effect occurs on standard Murashige and Skoog growth medium (Debrieux et al., 2013). Given that phyA degradation is only reduced but not inhibited in spa123 and cop1-4 and stabilization partially depends on sucrose, it seems unlikely that COP1/SPA are key components of the as yet unknown general phyA degradation mechanism.
A light-independent phyB-SPA1 interaction has been reported and proposed to enhance the activity of COP1/SPA1 in FR light, stabilizing HY5 and thereby counteracting phyA signaling (Zheng et al., 2013). Using altered conditions, including photoactive full-length phyB, we have shown the interaction between phyB and SPA1 to be dependent on light. Moreover, in yeast three-hybrid competition assays, light-activated phyB disrupted the direct interaction of COP1 and SPA1 that would presumably downregulate the activity of COP1/SPA1 in planta. Consistent with a negative effect of phyB on COP1/SPA1 activity in R, HY5 protein levels are increased under these conditions in a phyB-dependent manner (Osterlund et al., 2000). However, HY5 transcript levels also are elevated upon exposure to light, making it difficult to establish to what extent stabilization of the protein and increased mRNA levels contribute to HY5 protein accumulation in R (Osterlund et al., 2000).
A direct COP1-phyA interaction has been reported (Seo et al., 2004; Viczián et al., 2012). However, these experiments are not consistent, showing either an interaction between the N-terminal PAS-GAF region of phyA and COP1 (Viczián et al., 2012) or between the PAS-A/PAS-B region of phyA and the COP1 WD-repeat domain (Seo et al., 2004). As neither interaction has been shown to be light dependent, they do not explain how phyA mediates a light-specific repression of COP1 activity, though there is potential for other regulation such as light-dependent phosphorylation to be involved. We observed that COP1 and phyA interact within NBs in planta; therefore, exclusion of COP1 from the complex is unlikely in FR light. Indeed, COP1 has been reported to reaccumulate in the nucleus in shade conditions that are rich in FR light (Pacín et al., 2013). It is fortunate that a COP1-phyA interaction cannot be detected in yeast using full-length phyA, as this allowed the detection of the light-induced phyA-mediated disruption of the COP1/SPA interaction. Interestingly, expression of the N terminus of phyA (1 to 406), which binds to COP1 independently of light, results in constitutive signaling in plants (Viczián et al., 2012). However, this fragment can bind both COP1 and SPA1 independent of light, making it difficult to establish which interaction of this fragment represses COP1 function in planta, i.e., the effect observed by Viczián et al. (2012) is not necessarily due to interaction of phyA 1-406 with COP1 but might be due to its binding to SPA proteins or both.
COP1, SPA1, and phyA are present in the same complex in NBs of FR-grown plants. Yet, in vivo experiments revealed that light-activated phyA binding to SPA1 reorganizes the complex, likely increasing the spatial distance and therefore disrupting the direct interaction between COP1 and SPA1. Given that phyA NB formation is observed only upon irradiation with light, even in lines expressing constitutively nuclear-localized phyA (Genoud et al., 2008), it seems likely that in planta binding of phyA to COP1 is Pfr dependent. Thus, binding of activated phyA to COP1 also may contribute to disruption of the direct interaction between COP1 and SPA1.
Both light-activated CRY1 and CRY2 have been shown to interact with SPA proteins, resulting in inactivation of the COP1/SPA complex and initiation of downstream signaling in response to blue light (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011). However, the mechanism of inactivation differs. CRY2 stabilizes the interaction of COP1 and SPAs, but nevertheless reduces the activity of the COP1/SPA complex (Zuo et al., 2011). By contrast, CRY1 inactivates the COP1/SPA complex by binding to SPA proteins and inhibiting their association with COP1 (Lian et al., 2011; Liu et al., 2011). Interestingly, it appears likely that phyA and CRY1 use a similar mechanism to downregulate the activity of the COP1/SPA complex; yet, phyA shares an interaction with the SPA1 kinase-like domain similar to CRY2 (Zuo et al., 2011), whereas CRY1 binds to the SPA1 WD-repeat domain (Liu et al., 2011). We have shown HFR1 protein accumulation in seedlings within 2 h of irradiation with FR, similar to previous studies where it was found that HFR1 is rapidly stabilized by blue light, peaking 2 h after light exposure (Duek et al., 2004). It seems likely that different photoreceptors employ a similar mechanism to trigger light-induced accumulation of HFR1, which is degraded in dark-grown seedlings in a COP1-dependent manner (Duek et al., 2004).
Here, we have shown that SPAs interact with phyA and phyB within NBs in planta, though we cannot exclude that they also interact in the nucleoplasm, which could not be determined by FRET-FLIM due to the lower abundance in this fraction. Many other components of light signaling have been found to form nuclear bodies as well; however, the function of these electron-dense structures is still unclear (Van Buskirk et al., 2012). Other photoreceptors, including the blue-light-absorbing cryptochromes, and the UV-B receptor UVR8, localize to nuclear bodies in light and either inactivate or alter the function of the COP1/SPA complex, potentially forming a converging point for light signaling pathways (Van Buskirk et al., 2012).
METHODS
Plant Material
Arabidopsis thaliana Columbia-0 (Col-0) ProPHYA:PHYA-CFP and Pro35S:HA-YFP-SPA1 were created by Agrobacterium tumefaciens-mediated cotransformation of Col-0 with pPHYA40-PHYA and pPPO70v1HA-SPA1 (Davis et al., 2009); plasmids are described in the Supplemental Methods. spa123 ProPHYA:PHYA-CFP and cop1-4 ProPHYA:PHYA-CFP were obtained by transforming pPHYA40-PHYA into spa1-7 spa2-1 spa3-1 (Balcerowicz et al., 2011) and cop1-4 (McNellis et al., 1994) backgrounds, respectively. The lines hfr1-4 Pro35S:LUC-HFR1, phyA-211 ProPHYA:PHYA-YFP, spa1-7 ProSPA1:HA-YFP-SPA1, and spa1-7 Pro35S:HA-YFP-SPA1 were created by Agrobacterium-mediated transformation of hfr1-4 (Sessa et al., 2005), phyA-211 (Reed et al., 1994), and spa1-7 (Fittinghoff et al., 2006) with pCHF91-HFR1, pPPO30A-PHYA, pSPA1HAYFP-SPA1, and pPPO70v1HA-SPA1. The c-MYC-SPA1 line has been described previously (= Pro35S:TAP-SPA1 = Pro35S:2xIgG-BD-9xc-MYC-SPA1; Saijo et al., 2003).
Growth Conditions
Wild tobacco (Nicotiana benthamiana) plants were greenhouse cultivated (temperature, 26°C day/19°C night; humidity, 62%; photoperiod, 14 h). Germination of Arabidopsis seeds was induced by stratification on half-strength Murashige and Skoog agar plates for 4 d at 4°C, followed by 4 h white or R light induction. Subsequently, plates were transferred to either complete darkness, continuous FR (720 nm, 12 μmol m−2 s−1, or 1 μmol m−2 s−1 where indicated for spa1-7), or continuous R light (670 nm, 30 μmol m−2 s−1), each at 22°C.
Transient Transformation of N. benthamiana Leaves
The leaves of 4- to 6-week-old N. benthamiana plants were infiltrated with Agrobacterium C58 as previously described (Grefen et al., 2008). The p19 protein from tomato bushy stunt virus was used for suppression of transgene silencing (Voinnet et al., 2003). Sixteen hours after infiltration, N. benthamiana plants were transferred to complete darkness (26°C) for 2 d to allow the accumulation of phyA. Transient expression and localization of the fusion proteins in plant epidermal leaf cells were detected using epifluorescence microscopy or CCD camera visualization after spraying with 1 mM d-luciferin (Pro35S:PHYA-LUC). The constructs used for transient expression in N. benthamiana leaves are described in the Supplemental Methods; expression of constructs in N. benthamiana was driven by the 35S promoter.
FRET-FLIM Analysis
All FLIM measurements were performed as previously described (Wanke et al., 2011), with the following modifications for CFP-YFP FRET. A pulsed 440-nm diode laser (Picoquant LDH-D-C-440), operating at a repetition rate of 20 MHz, was used for excitation, in conjunction with LD01-439/8-12.5 (Semrock) cleanup interference filters. A dichroic beam splitter plate (λcut-on = 455 nm at 45° incident angle) was used with a long-pass interference filter (LP02-458RU-25) and a band-pass interference filter (BrightLine Basic FF01-469/35-25; Semrock) to exclusively detect the donor (CFP) signal. To survey the cell nuclei, FLIM images were obtained by raster scanning the samples using a feedback controlled piezo-driven sample stage (P-517.3CD; PI Physik Instrumente). Only photons originating from nuclear bodies were selected for analysis. Time-correlated single-photon-counting histograms were deconvolved from the instrument response function and fitted to exponential decays to provide the average lifetime.
Yeast Interaction Assays
All yeast two- and three-hybrid plasmids (described in the Supplemental Methods) were cotransformed into Saccharomyces cerevisiae strain Y190 (Harper et al., 1993) using a Frozen-EZ yeast transformation kit (ZymoResearch), followed by growth selection on synthetic media lacking leucine and tryptophan. Transformed yeast were suspended in sterile double-distilled water and, under green light (525 nm) conditions, 5 μL plated onto selective media lacking leucine and tryptophan (and methionine for yeast three-hybrid assays) supplemented with 20 μM phycocyanobilin (purified from Spirulina as previously described; Kunkel et al., 1993). Plates were incubated for 48 h (yeast two-hybrid assay) or 72 h (yeast three-hybrid assay) at 26°C in either darkness, constant R light (670 nm, 1.7 μmol m−2 s−1), or FR light (720 nm, 13 μmol m 2 s−1). X-Gal filter lift assays were performed as previously described (Breeden and Nasmyth, 1985) except yeast were lifted from plates and freeze/thawed five times with liquid N2 under green light. For quantitative ortho-nitrophenyl-β-galactoside (ONPG) assays, yeast was grown as above, harvested in green light, and used for liquid ONPG assays as previously described in the Clontech yeast two-hybrid manual.
Immunolocalization
Arabidopsis seedlings were fixed in microtubule stabilizing buffer with a formaldehyde concentration of 4% (w/v) for 45 min, followed by 8% for a further 120 min. Hypocotyls were embedded in 10% (w/v) gelatin and then infiltrated with a solution of 2.1 M sucrose and 1.8% (w/v) polyvinylpyrrolidone and mounted on stubs. Samples were frozen in liquid nitrogen, and ultrathin (70 nm) sections were cut at −115°C using a cryo-ultramicrotome (Leica). Sections were transferred to Pioloform and carbon-coated grids and blocked for 30 min in PBS, 0.5% (w/v) BSA, and 0.5% (w/v) milk powder. Grids were probed with rabbit α-phyA serum (Agrisera; 1:300 in blocking buffer) for 60 min and washed six times with blocking buffer. Bound antibodies were detected with 6-nm gold particle labeled protein-A (1:50 in blocking buffer; Aurion). Grids were stained with 1% (v/v) uranyl acetate and embedded in a thin layer of methyl cellulose containing 0.3% (v/v) uranyl acetate and imaged with a transmission electron microscope (Leo 906).
Luciferase Quantification
Four-day-old seedlings were harvested and frozen in liquid N2 under green light. Tissue was disrupted using glass beads and a Silamat while frozen and proteins extracted in LUCI buffer (100 mM K2PO4, pH 7.8, 0.05% [v/v] Tween 20, protease inhibitor cocktail [Sigma-Aldrich P2714], 20 μM MG132, and 1 mM DTT) under green light. Lysate (100 μL) was then assayed in triplicate in a luminometer, injecting 50 μL LUCII buffer (80 mM glycyl-glycine, 60 mM ATP, and 40 mM MgSO4, pH 7.8) and 100 μL 10 mM d-luciferin. Protein concentration was determined by amido-black as previously described (Popov et al., 1975).
Luciferase Assay
Yeast cultured on selective media supplemented with 20 μM PCB and grown for 2 d in constant R light (670 nm, 1.7 μmol m−2 s−1) were sprayed with 5 mM d-luciferin and visualized by 5-min exposure with a CCD camera.
Immunoblotting
Seedlings were treated as for luciferase quantification, except proteins were extracted in 100 mM Tris-HCl, pH 8.0, 250 mM NaCl, 1 mM EDTA, 10 mM NaF, 15 mM glycero-phosphate, 50 μM MG132, protease inhibitor cocktail (Sigma-Aldrich P2714), 0.1% SDS, and 1 mM DTT. Protein concentration was determined as above, and 5 μg each sample separated by 8% SDS-PAGE electrophoresis, blotted to polyvinylidene fluoride membrane, blocked, and probed. Primary antibody (LUC-1 Sigma-Aldrich L2164) was used at 1:2000 overnight at 4°C, secondary antibody (α-mouse AP Vector Laboratories) was used at 1:10,000 for 1 h, and detection performed with 5-bromo-4-chloro-3-indolyl phosphate and Nitro blue tetrazolium.
In Vivo Coimmunoprecipitation Assays
For the in vivo coimmunoprecipitation assay, 4-d-old dark-grown seedlings expressing SPA1 tagged with an alternative TAP tag containing 9xc-MYC (Saijo et al., 2003) were pretreated with 50 μM MG132 for 5 h and either kept in darkness or given a pulse of 3000 μmol m−2 R light followed by 10-min dark incubation. Total proteins were extracted with 0.8 mL native extraction buffer (100 mM phosphate buffer, pH 7.8, 150 mM NaCl, 0.1% Nonidet P-40, 1× protease inhibitor [Sigma-Aldrich P9599], 1 mM PMSF, 50 μM MG132, 5 μM β-mercaptoethanol, 25 mM β-glycerophosphate, 10 mM sodium fluoride, and 2 mM sodium orthovanadate) and cleared by centrifugation at 16,000g for 15 min at 4°C. For each sample, 20 μL Dynabeads (Life Technologies 10002D) were preincubated with 1 μg α-MYC antibody (Cell Signaling Technology; 2276S) at 4°C for an hour and washed twice with the native extraction buffer. Total protein extracts (500 μg) along with antibody bound beads in 1 mL total volume were incubated at 4°C in the dark for 1 h. The beads were washed three times with the binding buffer (with 0.2% Nonidet P-40), dissolved in 1× SDS loading buffer, and incubated at 65°C for 10 min. The immunoprecipitated proteins were separated on 6.5% SDS-PAGE gel and transferred on to polyvinylidene fluoride membrane. α-phyA (073D) (1:500) and α-MYC (1:5000) (Sigma-Aldrich SAB4700447) antibodies were used to detect phyA and TAP-SPA1 proteins.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: At2g32950 (COP1), At2g37678 (FHY1), At1g02340 (HFR1), At1g09570 (PHYA), At2g18790 (PHYB), At2g46340 (SPA1), At4g11110 (SPA2), At3g15354 (SPA3), and At1g53090 (SPA4).
Supplemental Data
Supplemental Figure 1. phyA Colocalizes with SPA3 and SPA4 in Nicotiana benthamiana Nuclear Bodies.
Supplemental Figure 2. Light-Activated phyA Interacts with SPA1 and SPA2 in Yeast.
Supplemental Figure 3. Pro35S:HA-YFP-SPA1 Rescues spa1-7.
Supplemental Figure 4. Immunohistochemistry of phyA and phyA-YFP Nuclear Bodies.
Supplemental Figure 5. Truncations and Mutants of phyA and SPA1 Are Stable in Yeast.
Supplemental Figure 6. Immunoblot Detection of Yeast Three-Hybrid Proteins from Figure 4.
Supplemental Figure 7. Reorganization of the COP1/SPA1 Complex by phyA upon Irradiation with Light.
Supplemental Figure 8. HFR1 Accumulates in FR Light.
Supplemental Methods. Yeast Interaction Assays, Immunoblotting, Immunohistochemistry, and Cloning of Constructs.
Supplementary Material
Acknowledgments
We thank Ute Hoecker (University of Cologne, Germany) for providing SPA2, SPA3, and SPA4 cDNA clones as well as spa1-7 and spa1-7 spa2-1 spa3-1 seeds. We also thank Claudia König, Rebecca Kühn (University of Tübingen, Germany), and Martina Krenz (University of Freiburg, Germany) for excellent technical work, the greenhouse facility of the Center for Plant Molecular Biology (University of Tübingen; Gert Huber, Johanna Schröter, and Sofia Riegger) for taking care of Arabidopsis and N. benthamiana plants, as well as Klaus Harter and Alfred J. Meixner for hosting C.M. and S.z.O.-K., respectively. We also thank the Riken Bioresource Center, Japan, for supplying the SPA1 cDNA clone. This study was supported by the Excellence Initiative of the German Federal and State Governments (EXC 294) to A.H., by grants from the German Research Foundation (Deutsche Forschungsgemeinschaft) to A.H. (HI 1369/3-1 and HI 1369/4-1) and F.S. (SCHL 1940/2-1 and SFB1101 project D02), by the National Science Foundation to E.H. (IOS-1120946), and by the Human Frontier Science Program to E.H. and A.H. (RGP0025/2013).
AUTHOR CONTRIBUTIONS
D.J.S., C.M., S.z.O.-K., F.S., Y.-D.S., L.Z., E.H., and A.H. designed research. D.J.S., C.M., S.z.O.-K., B.E., L.Z., P.J., and Y.-D.S. performed research and analyzed data. D.J.S., C.M., S.z.O.-K., Y.-D.S., L.Z., E.H., and A.H. wrote the article.
Glossary
- R
red
- FR
far-red
- NB
nuclear body
- PCB
phycocyanobilin
- FRET-FLIM
fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy
- Col-0
Columbia-0
- ONPG
ortho-nitrophenyl-β-galactoside
References
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222. [DOI] [PubMed] [Google Scholar]
- Balcerowicz M., Fittinghoff K., Wirthmueller L., Maier A., Fackendahl P., Fiene G., Koncz C., Hoecker U. (2011). Light exposure of Arabidopsis seedlings causes rapid de-stabilization as well as selective post-translational inactivation of the repressor of photomorphogenesis SPA2. Plant J. 65: 712–723. [DOI] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. (1985). Regulation of the yeast HO gene. Cold Spring Harb. Symp. Quant. Biol. 50: 643–650. [DOI] [PubMed] [Google Scholar]
- Davis A.M., Hall A., Millar A.J., Darrah C., Davis S.J. (2009). Protocol: Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrieux D., Trevisan M., Fankhauser C. (2013). Conditional involvement of constitutive photomorphogenic1 in the degradation of phytochrome A. Plant Physiol. 161: 2136–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh K., Franci C., Parks B.M., Seeley K.A., Short T.W., Tepperman J.M., Quail P.H. (1993). Arabidopsis HY8 locus encodes phytochrome A. Plant Cell 5: 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.W., Caspar T., Quail P.H. (1991). cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5: 1172–1182. [DOI] [PubMed] [Google Scholar]
- Duek P.D., Elmer M.V., van Oosten V.R., Fankhauser C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14: 2296–2301. [DOI] [PubMed] [Google Scholar]
- Essen L.O., Mailliet J., Hughes J. (2008). The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. USA 105: 14709–14714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittinghoff K., Laubinger S., Nixdorf M., Fackendahl P., Baumgardt R.L., Batschauer A., Hoecker U. (2006). Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 47: 577–590. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Städele K., Růzicka K., Obrdlik P., Harter K., Horák J. (2008). Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol. Plant 1: 308–320. [DOI] [PubMed] [Google Scholar]
- Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15: 2125–2130. [DOI] [PubMed] [Google Scholar]
- Hisada A., Hanzawa H., Weller J.L., Nagatani A., Reid J.B., Furuya M. (2000). Light-induced nuclear translocation of endogenous pea phytochrome A visualized by immunocytochemical procedures. Plant Cell 12: 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisada A., Yoshida T., Kubota S., Nishizawa N.K., Furuya M. (2001). Technical advance: an automated device for cryofixation of specimens of electron microscopy using liquid helium. Plant Cell Physiol. 42: 885–893. [DOI] [PubMed] [Google Scholar]
- Hoecker U., Quail P.H. (2001). The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276: 38173–38178. [DOI] [PubMed] [Google Scholar]
- Hoecker U., Tepperman J.M., Quail P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496–499. [DOI] [PubMed] [Google Scholar]
- Hoecker U., Xu Y., Quail P.H. (1998). SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10: 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang J.Y., Seo H.S., Chua N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C., Lorrain S., Hornitschek P., Fankhauser C. (2010). Light-regulated plant growth and development. In Plant Development, Timmermans M.C.P., ed (Academic Press), pp. 29–66. [DOI] [PubMed] [Google Scholar]
- Kami C., Mukougawa K., Muramoto T., Yokota A., Shinomura T., Lagarias J.C., Kohchi T. (2004). Complementation of phytochrome chromophore-deficient Arabidopsis by expression of phycocyanobilin:ferredoxin oxidoreductase. Proc. Natl. Acad. Sci. USA 101: 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T., Tomizawa K., Kern R., Furuya M., Chua N.H., Schäfer E. (1993). In vitro formation of a photoreversible adduct of phycocyanobilin and tobacco apophytochrome B. Eur. J. Biochem. 215: 587–594. [DOI] [PubMed] [Google Scholar]
- Laubinger S., Hoecker U. (2003). The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J. 35: 373–385. [DOI] [PubMed] [Google Scholar]
- Laubinger S., Fittinghoff K., Hoecker U. (2004). The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Marchal V., Le Gourrierec J., Wenkel S., Adrian J., Jang S., Kulajta C., Braun H., Coupland G., Hoecker U. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222. [DOI] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Li, G., Wang, H., and Wang Deng, X. (2011). Phytochrome signaling mechanisms. The Arabidopsis Book 9: e0148, doi/10.1199/tab.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., Sun S.X., Li L., Yang H.Q. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25: 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25: 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli, A.L. (1994). The physiology of phytochrome action. In Photomorphogenesis in Plants, R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 211–270. [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667. [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Deng X.W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16: 201–208. [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Pacín M., Legris M., Casal J.J. (2013). COP1 re-accumulates in the nucleus under shade. Plant J. 75: 631–641. [DOI] [PubMed] [Google Scholar]
- Park E., Park J., Kim J., Nagatani A., Lagarias J.C., Choi G. (2012). Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N., Schmitt M., Schulzeck S., Matthies H. (1975). Eine störungsfreie Mikromethode zur Bestimmung des Proteingehaltes in Gewebehomogenaten. Acta Biol. Med. Ger. 34: 1441–1446. [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L., Favory J.J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G.I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106. [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Su Y.S., Lagarias J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57: 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Sullivan J.A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W. (2003). The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17: 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Zhu D., Li J., Rubio V., Zhou Z., Shen Y., Hoecker U., Wang H., Deng X.W. (2008). Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol. Cell 31: 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Watanabe E., Tokutomi S., Nagatani A., Chua N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18: 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999. [DOI] [PubMed] [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Possenti M., Mittempergher F., Becker J., Morelli G., Ruberti I. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19: 2811–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk E.K., Decker P.V., Chen M. (2012). Photobodies in light signaling. Plant Physiol. 158: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viczián A., Ádám É., Wolf I., Bindics J., Kircher S., Heijde M., Ulm R., Schäfer E., Nagy F. (2012). A short amino-terminal part of Arabidopsis phytochrome A induces constitutive photomorphogenic response. Mol. Plant 5: 629–641. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wanke D., Hohenstatt M.L., Dynowski M., Bloss U., Hecker A., Elgass K., Hummel S., Hahn A., Caesar K., Schleifenbaum F., Harter K., Berendzen K.W. (2011). Alanine zipper-like coiled-coil domains are necessary for homotypic dimerization of plant GAGA-factors in the nucleus and nucleolus. PLoS ONE 6: e16070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L., Bu Q., Huang X., Deng X.W., Huq E. (2014). PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell 26: 1992–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang H. (2006). The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J. 47: 564–576. [DOI] [PubMed] [Google Scholar]
- Yang J., Lin R., Hoecker U., Liu B., Xu L., Wang H. (2005). Repression of light signaling by Arabidopsis SPA1 involves post-translational regulation of HFR1 protein accumulation. Plant J. 43: 131–141. [DOI] [PubMed] [Google Scholar]
- Yang X., Kuk J., Moffat K. (2009). Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc. Natl. Acad. Sci. USA 106: 15639–15644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M.J., Casal J.J., Whitelam G.C. (1995). Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: weak de-etiolation of the phyA mutant under dense canopies. Plant Cell Environ. 18: 788–794. [Google Scholar]
- Yanovsky M.J., Luppi J.P., Kirchbauer D., Ogorodnikova O.B., Sineshchekov V.A., Adam E., Kircher S., Staneloni R.J., Schäfer E., Nagy F., Casal J.J. (2002). Missense mutation in the PAS2 domain of phytochrome A impairs subnuclear localization and a subset of responses. Plant Cell 14: 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., et al. (2013). Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25: 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Maier A., Lee J.H., Laubinger S., Saijo Y., Wang H., Qu L.J., Hoecker U., Deng X.W. (2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.